Abstract
N6-threonylcarbamoyl-adenosine (t6A) is one of the few RNA modifications that is universally present in life. This modification occurs at high frequency at position 37 of most tRNAs that decode ANN codons, and stabilizes cognate anticodon–codon interactions. Nearly all genetic studies of the t6A pathway have focused on single-celled organisms. In this study, we report the isolation of an extensive allelic series in the Drosophila ortholog of the core t6A biosynthesis factor Kae1. kae1 hemizygous larvae exhibit decreases in t6A that correlate with allele strength; however, we still detect substantial t6A-modified tRNAs even during the extended larval phase of null alleles. Nevertheless, complementation of Drosophila Kae1 and other t6A factors in corresponding yeast null mutants demonstrates that these metazoan genes execute t6A synthesis. Turning to the biological consequences of t6A loss, we characterize prominent kae1 melanotic masses and show that they are associated with lymph gland overgrowth and ectopic generation of lamellocytes. On the other hand, kae1 mutants exhibit other phenotypes that reflect insufficient tissue growth. Interestingly, whole-tissue and clonal analyses show that strongly mitotic tissues such as imaginal discs are exquisitely sensitive to loss of kae1, whereas nonproliferating tissues are less affected. Indeed, despite overt requirements of t6A for growth of many tissues, certain strong kae1 alleles achieve and sustain enlarged body size during their extended larval phase. Our studies highlight tissue-specific requirements of the t6A pathway in a metazoan context and provide insights into the diverse biological roles of this fundamental RNA modification during animal development and disease.
Keywords: Drosophila, Kae1, N6-threonylcarbamoyladenosine, t6A
INTRODUCTION
Beyond the four canonical ribonucleosides present in RNA, nearly all life forms bear an array of modified bases in various transcript populations. Over 100 chemical modifications have been documented, with dozens often present in individual species; the majority of distinct modified ribonucleosides occur on tRNA (Machnicka et al. 2013). Altogether, RNA modifications collectively play diverse roles in RNA quality control, secondary structure, transcript stability, translational efficiency, translational fidelity, and regulation of RNA–protein interactions (Gu et al. 2014; Hopper and Huang 2015) and can influence disease and cancer progression (Grewal 2015; Kirchner and Ignatova 2015).
While many post-transcriptional modifications of RNA are broadly conserved, only a few are universal across known life forms. One prominent example is N6-threonylcarbamoyladenosine (t6A), which is found at high frequency at position 37 of tRNAs that decode ANN codons (Grosjean et al. 1995). This modification was recognized decades ago, and it stabilizes cognate codon–anticodon interactions through a stacking interaction that stabilizes the weak interaction with the first codon base, which comprises a weak A–U pair (Weissenbach and Grosjean 1981; Yarian et al. 2002; Agris 2008). However, the enzymes that generate this modification remained elusive until relatively recently.
Based on the inference that all organisms harbor t6A-modified tRNAs (Sprinzl and Vassilenko 2005), bioinformatic isolation of universally conserved genes of unknown function (Galperin and Koonin 2004) yielded a short list of candidate genes that first led to the identification of YrdC/Sua5 as a t6A pathway component (El Yacoubi et al. 2009). Subsequently, members of the universal gene family Kae1/Qri7/YgjD were recognized to be essential for t6A biosynthesis, along with its partner proteins (in yeast) Pcc1 and Bud32 (El Yacoubi et al. 2011; Srinivasan et al. 2011). Of note, mutations in components of the Kae1, Pcc1, Bud32, and Cgi121 complex (collectively termed KEOPS) had been isolated some years earlier, via divergent approaches intended at identifying factors in telomere maintenance (Downey et al. 2006) and splicing/transcription (Kisseleva-Romanova et al. 2006). At the time, the molecular role of the KEOPS complex was obscure, but its deep conservation justified solving the atomic structures of the archaeal Kae1/Bud32 natural fusion protein (Hecker et al. 2008) and the entire eukaryotic KEOPS complex (Mao et al. 2008). These studies assigned Kae1 as an ASKHA-type ATPase, Bud32 as a kinase, and Pcc1 as a dimerization subunit, with Kae1 physically interacting with both Bud32 and Pcc1.
Only later, with the recognition that these genes are involved in t6A accumulation, did efforts become possible to reconstitute the biosynthesis of t6A-modified tRNA in vitro, using purified bacterial YrdC (Sua5) and YgjD (Kae1) and two bacterial-specific genes that cluster with YgjD, namely YjeE and YeaZ (Deutsch et al. 2012). Similarly, yeast Sua5 and KEOPS complex sufficed to generate t6A-modified tRNA in vitro (Perrochia et al. 2013). The proposed mechanism is that Sua5 converts bicarbonate and threonine via a series of intermediates into threonylcarbamoyladenylate, which is transferred to the active site of Kae1 within the KEOPS complex to modify tRNA position A37.
Notably, Kae1 is the only truly universally conserved component of KEOPS (Galperin and Koonin 2004), and eukaryotes contain a paralog of Kae1 termed Qri7 that is mitochondrially localized (Huh et al. 2003). The fact that no other KEOPS components localize to mitochondria led to a suspicion that Qri7 represents a primal core factor that does not require additional presumably accessory KEOPS components. Indeed, subcellular redirection of Qri7 to the cytoplasm rescued the slow growth phenotypes of all KEOPS members that are defective for t6A accumulation (Wan et al. 2013). Moreover, in vitro synthesis of t6A-modified tRNA was reconstituted solely with purified Sua5, Qri7, and substrate tRNA, in the presence of threonine, bicarbonate, and ATP (Wan et al. 2013). Thus, Kae1/Qri7 is the central downstream enzyme in the generation of t6A-modified tRNA.
The assignment of core genes in the t6A pathway permits organismal phenotypes to be correlated with the absence of these modified tRNAs. Budding yeast t6A pathway mutants (El Yacoubi et al. 2009, 2011; Srinivasan et al. 2011) are very slow-growing, while the bacterial pathway (e.g., in Bacillus subtilis and Escherichia coli) is essential for viability and cell morphology (Arigoni et al. 1998; Handford et al. 2009). These seemingly general defects befit loss of a fundamental RNA modification pathway, but do not lend insight into particular processes. Otherwise, mutants of KEOPS complex members in single-celled organisms have impact on telomere maintenance (Downey et al. 2006), transcription (Kisseleva-Romanova et al. 2006), and genome stability (Oberto et al. 2009), although it remains unclear how directly the loss of t6A relates to these phenotypes.
However, certain phenotypes now seem to be directly related to the biochemical function of t6A in improving translational fidelity. Of note, yeast sua5 was originally isolated as a suppressor of cyc1 alleles harboring an aberrant upstream start codon (uORF) (Hampsey et al. 1991; Na et al. 1992), implicating Sua5 in start site selection; AUG is indeed recognized by a t6A-modified tRNA. Correspondingly, Sua5 affects various aspects of normal translation, as mutants exhibit increased leaky scanning through AUG, frameshifting, and nonsense suppression (Lin et al. 2010). One prominent locus controlled via endogenous uORFs is the amino acid starvation-responsive transcription factor GCN4 (Hinnebusch 2005). Indeed, not only does sua5 derepress GCN4, KEOPS mutants (kae1 and pcc1) induce a gene expression signature that is substantially enriched for Gcn4 targets (Daugeron et al. 2011). These precedents suggest that it may be possible to make linkages between altered translation and organismal phenotypes, even for this “housekeeping” cellular activity.
To date, genetic analysis of t6A pathway components has mostly been limited to single cell organisms, such as microbes and yeast. One study on the Drosophila melanogaster Bud32/PRPK homolog utilized RNAi-mediated knockdowns to define growth related defects upon bud32 depletion, although biochemical evidence was not presented for an impact on t6A biogenesis in this condition (Ibar et al. 2013). Interestingly, the growth defects were related to PI3K/TOR signaling, and could be partially restored by expression of activated S6-kinase (Ibar et al. 2013).
In this study, we describe the isolation of a large allelic series of D. melanogaster kae1 mutants, ranging from weak hypomorphs, strong mutants, to null alleles. These present various growth-related defects, developmental delay, and melanotic tumors. We present extensive biochemical and genetic evidence that Drosophila kae1 and its partners are core factors in t6A biogenesis. Notably, our phenotypic analysis reveals substantial differences in the genetic requirements for t6A in proliferating versus nonproliferating tissues. In particular, analysis of mitotic tissues in full knockout and in clonal settings indicates an essential requirement for kae1, whereas various nonproliferating tissues can be maintained in the null state for weeks. Moreover, kae1 appears to act as a tumor suppressor in the larval hematopoietic tissue. These data highlight biological complexity in the requirements of a general regulatory factor, and the utility of the Drosophila model to further understand the roles of t6A modification during animal development and disease.
RESULTS
Isolation of the “Black Spot” complementation group
During efforts to isolate mutants in the 72D-F region, we conducted an F2 EMS mutagenesis screen against Df(3L)ED220, which deletes 324,210 bp of chromosome 3L:16,080,584–16,404,777 (see Materials and Methods). We screened ∼7000 crosses of animals bearing mutagenized chromosomes to this deficiency, and recovered 64 lethals. Within this collection, we recognized numerous alleles for which hemizygous larvae bore characteristic melanotic masses, which we colloquially termed “Black Spot” mutants (Fig. 1A). These mutants failed to complement each other, and they typically exhibited an extended larval period prior to lethality (Fig. 1B–D; Supplemental Fig. 1). Notably, while Drosophila larvae complete their larval stages around day 5 and commence pupation, some Black Spot mutants survive as larvae for up to 18 d. Systematic tests against all of the lethal alleles we obtained in this genomic interval revealed additional noncomplementing mutations. Many of these mutants lacked black spots or generated them with low frequency (Fig. 1B–D). Several of these alleles were able to develop to pupal stages or even into pharate adults as hemizygotes, suggesting they might represent hypomorphs. Overall, these eight alleles appear to represent an extensive allelic series.
FIGURE 1.
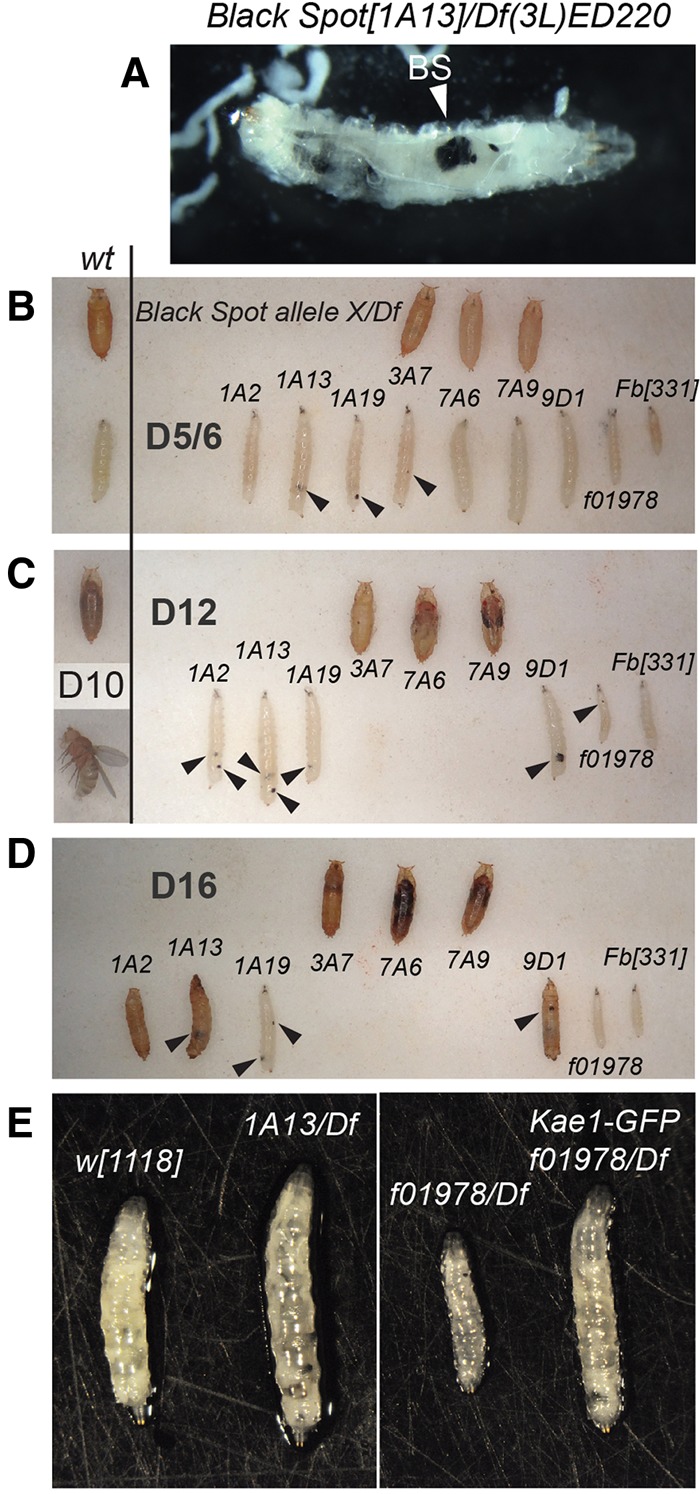
Isolation and characterization of Black Spot mutants. (A) Exemplar of the Black Spot phenotype. [1A13]/Df(3L)ED220 larvae exhibit melanotic nodules of various sizes; a particularly large one is labeled with an arrowhead (“BS”). (B–D) Complementation tests identified a collection of Black Spot mutants, only some of which actually generate melanotic masses with high penetrance (examples labeled with arrowheads). Normal (wt) animals complete their third instar larval stage by day ∼5, and proceed to ∼5 d of pupal development. Pupae containing pharate animals are visible by day ∼10, and they eclose shortly thereafter as adult flies. Wt animals are shown as a reference to the left of B and C. In contrast, many hemizygous Black Spot alleles are substantially developmentally delayed and maintain their larval phase until days 12 (C) to 16 (D), with some living mutants detected at day 18 (not shown). All of the larvae collected in this summary were mobile and thus alive. Hypomorphic mutants could pupate and even advance to the pharate stage; however, it is likely that some of the pupae shown at the latest days are deceased. See Supplemental Figure 1 for additional notes regarding the larval extended phase, lethal phase, and black spot penetrance at different ages in the different hemizygous mutants. (E) Several of the Black Spot mutants attain larger larval size than normal; shown are [1A13] hemizygotes as an example. Other alleles such as [f10978] are growth deficient. All alleles tested could be rescued by kae1 genomic transgenes; shown is rescue of [f10978]/Df larva.
Black Spot mutants affect the Drosophila ortholog of Kae1
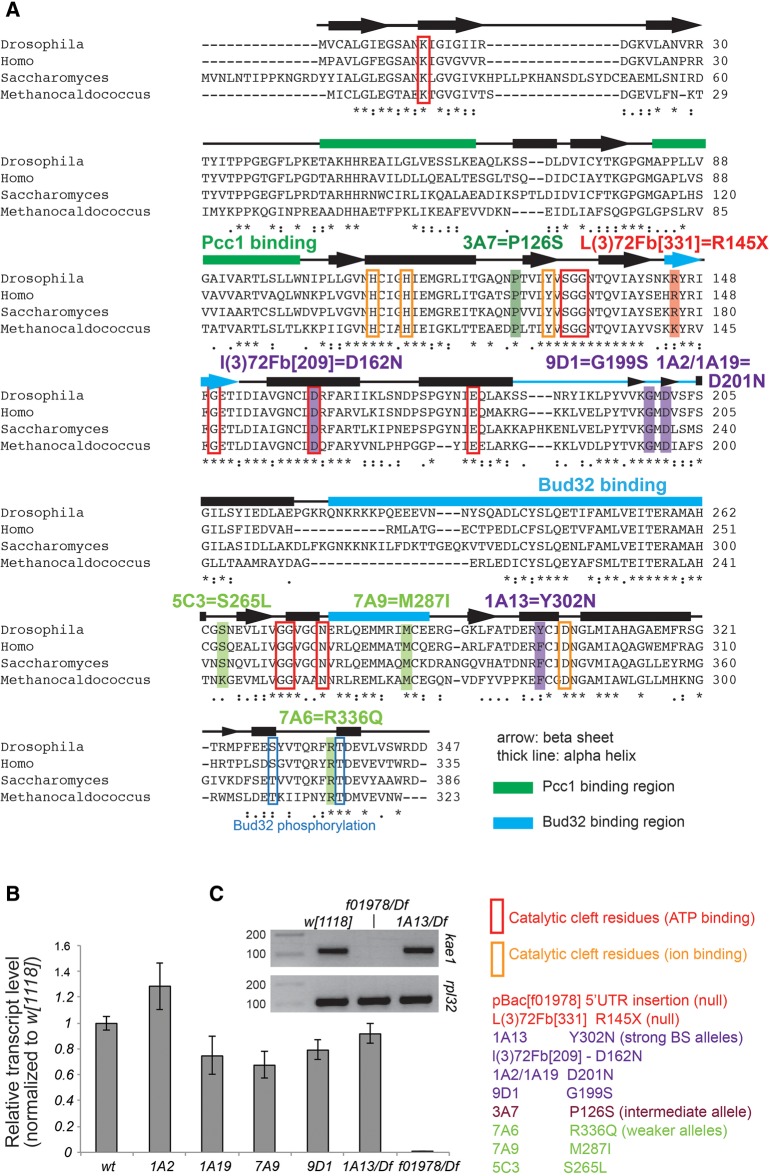
To identify the gene mutated in Black Spot mutants, we used P-element mapping (Zhai et al. 2003) to narrow the region of interest within the deficiency (Supplemental Fig. 2). Coding exons from genes in the region were amplified by PCR from DNA extracted from hemizygous mutant larvae and sequenced. We identified one locus (CG4933), the fly ortholog of Kae1, that harbored nonconservative changes in all eight alleles (Fig. 2A). Subsequently, we identified two uncharacterized mutants in 72F-73A (l(3)72Fb[209] and l(3)72Fb[331], V Bedian, pers. comm. to FlyBase) that failed to complement Black Spot mutants. Sequencing showed that the l(3)72Fb alleles exhibit missense and nonsense mutations in CG4933, respectively. Notably, while l(3)72Fb[209] mutates a residue in the ATP-binding pocket, none of the other point mutations directly affect any functionally characterized regions (e.g., other residues that bind ATP or metal ion, or Kae1 faces that bind its partner KEOPS proteins Pcc1 or Bud32, Fig. 2A). Finally, we found that PBac{WH}CG4933[f01978], which carries a piggyBac insertion into the CG4933 5′ UTR (dos Santos et al. 2015), also failed to complement Black Spot mutants. These observations strongly support that CG4933 corresponds to Black Spot. Consistent with this, qRT-PCR analysis of several independent point mutants showed moderate decreases in CG4933 transcripts, and these were undetectable in f01978 hemizygotes (Fig. 2B,C).
FIGURE 2.
Summary of Drosophila kae1 (CG4933) mutant alleles. (A) An alignment of fruit fly, human, yeast, and archaeal orthologs of Kae1. Functionally/structurally defined regions are highlighted, including the binding regions for Pcc1 and Bud32, and the ATP- and ion-binding regions. All Black Spot alleles recovered from our screen revealed that point mutations affected well-conserved residues. Notably, none of the mutations directly affects a known functional site, although a few mutations are in the vicinity of such regions. The noncomplementing allele l(3)72Fb[331], identified from a public stock collection, bears a nonsense mutation in kae1. Note that 1A2 and 1A19 induce the same coding change, even though these were isolated from different chromosomes in the screen. (B) qRT-PCR analysis of kae1 from various homozygous or hemizygous mutants. All of our point alleles maintain substantial levels of kae1 transcripts, although several of them are reduced by 20%–30%. The pBac insertion into the 5′ UTR of kae1, f[10978], appears to be a null allele as no transcript was detected. (C) Agarose gel analysis of qRT-PCR amplicons confirms the absence of kae1 transcripts in f[10978]/Df larvae.
To confirm that Black Spot phenotypes were functionally due to loss of CG4933, we generated untagged and GFP-tagged genomic transgenes using the P[acman] backbone. Both CG4933 constructs rescued independent lethal hemizygotes (1A13 and f01978) to adult viability, and no melanotic masses were observed in rescued animals (e.g., Figs. 1E, 3 and other data not shown). In summary, we have isolated metazoan mutants of kae1, encoding a universal tRNA modification factor, representing the first extensive allelic series of a t6A pathway factor in any organism.
FIGURE 3.
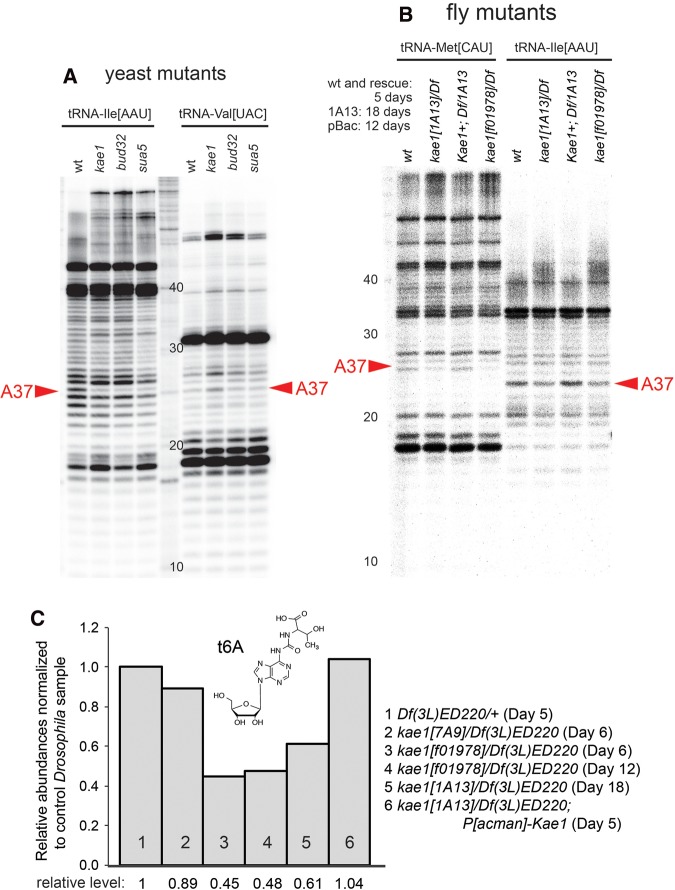
Analysis of t6A-modified tRNAs in yeast KEOPS and Drosophila kae1 mutants. (A) Analysis of yeast t6A pathway mutants is shown as a control. Primer extension assays yield a ladder of bands indicating the presence of stalled products. These include a band at position A37 of tRNA-Ile[AAU] of wild-type yeast that is strongly depleted in yeast t6A pathway mutants, thus indicating a t6A-modified nucleoside. tRNA-Val[UAC] is shown as a control for a non-t6A-bearing substrate. The ladder positions are inaccurate due to incomplete digestion; therefore, sizes are labeled according to the input primer. (B) Analysis of Drosophila kae1 mutants. Modifications of the A37 positions of tRNA-Met[CAU] and tRNA-Ile[AAU] are decreased although not eliminated in two strong hemizygous conditions of kae1. The reduction observed in kae1[1A13]/Df was rescued by a P[acman]-kae1 genomic transgene (“kae1+”). Note that mutant larvae were collected at an advanced age in their extended larval period to separate them from potential maternal contributions; the normal larval period ends at ∼5 d. (C) LC-MS/MS analysis of total tRNAs from Drosophila kae1 hemizygous mutant larvae. We observe a minor decrease in total t6A-modified tRNAs in the hypomorphic allele [7A9] and stronger decreases in the strong/null alleles [f01978] and [1A13]. The levels of t6A-modified tRNAs did not decrease further in exceptionally long-lived kae1 mutant larvae, and the loss of t6A was rescued by a kae1 genomic transgene.
Decrease, but not elimination, of t6A-modified tRNAs in kae1 mutants
Although Sua5/KEOPS genes are well-conserved across different domains of life, their involvement in t6A synthesis has not formally been demonstrated outside of unicellular models. We initially utilized primer extension assays to assess potential defects in the accumulation of t6A-modified tRNAs. As positive controls, we assayed RNAs isolated from wild-type yeast and sua5, kae1, and bud32 knockouts (Srinivasan et al. 2011). Extensions using an end-labeled primer specific for tRNA-Ile[AAU] showed that t6A pathway mutants were specifically depleted of a stalled product at the normally modified position A37, whereas primer extension patterns for control tRNA-Val[UAC] were not substantially altered (Fig. 3A). Similar assays of third instar larvae bearing various strong Drosophila kae1 alleles in trans to a deficiency also revealed alteration at A37 (Fig. 3B). However, the loss of modified tRNAs in the total population were notably modest compared to the yeast mutants, since yeast kae1 mutants retain only a few percent of total t6A (El Yacoubi et al. 2011; Srinivasan et al. 2011); see also Figure 4.
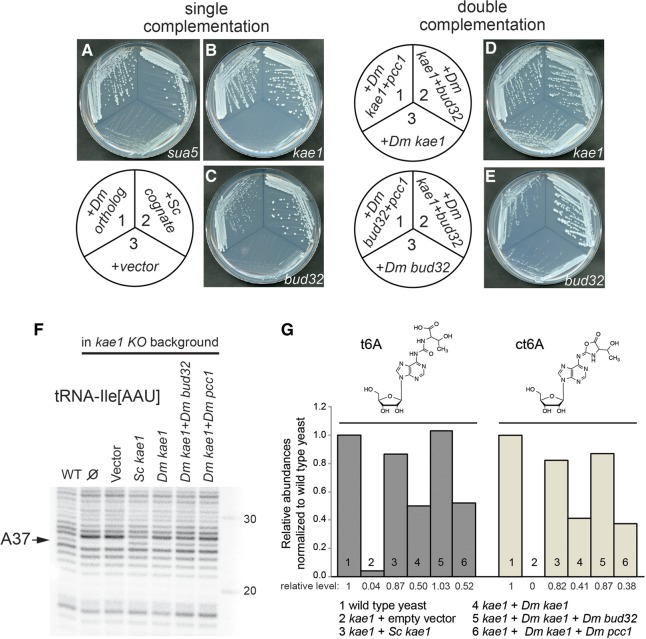
FIGURE 4.
Genetic and molecular complementation of yeast t6A mutants with Drosophila orthologs. (A–C) Single complementation tests. The schematic depicts the transformations of cognate yeast genes and their fly orthologs, assayed for complementation of sua5 (A), kae1 (B), and bud32 (C) null mutants. Comparison of single colony sizes indicates substantial growth rescue by fly proteins, although not as robust as the yeast counterparts. (D,E) Double complementation tests. The schematics depict combinations of fly orthologs assayed for complementation of kae1 (D) and bud32 (E) null mutants. Improved growth rescue by supplying combinations of fly factors was noticeable, particularly for Drosophila kae1 + bud32. (F) Primer extension analysis demonstrating molecular rescue of t6A-modified tRNA-Ile by yeast and fly factors. (G) Mass spectrometry analysis of total yeast tRNAs. kae1 mutant yeast has borderline detectable levels of t6A-modified tRNAs, which are restored by complementation with yeast KAE1 and by Drosophila KEOPS genes. Consistent with the behavior in yeast growth (D) and in primer extension assays (F), the combination of Drosophila kae1 + bud32 conferred the best rescue of t6A-modified tRNAs.
Inspection of modENCODE data (Brown et al. 2014) shows that Drosophila kae1 is ubiquitously expressed but present at highest levels in ovaries and the early blastoderm embryo, indicating its strong maternal deposition. Some maternal gene products can support organismal processes for days (Lecourtois and Schweisguth 1995; Lu et al. 2000), and tRNAs themselves are typically expected to be relatively stable (Hanoune and Agarwal 1970), although loss of some tRNA modifications is documented to render tRNAs unstable (Alexandrov et al. 2006). Because of these considerations, we sought to extend the analysis of kae1 zygotic mutants as late as possible. Bearing in mind that normal Drosophila complete their larval stages by ∼5 d, we collected RNA from 12-d and 18-d kae1 mutant larvae. However, we did not observe further decreases in the level of t6A-modified tRNAs even at these extended timepoints (Fig. 3).
To provide a global assessment on the amount of t6A-modified tRNAs in kae1 mutants, we purified total tRNA, enzymatically hydrolyzed the tRNA to ribonucleosides, and analyzed these using liquid chromatography-coupled triple quadrupole mass spectrometry (LC-MS/MS) using a dynamic MRM method (Su et al. 2014). This analysis showed a mild (∼10%) decrease of t6A-modified tRNAs in hemizygotes of the weak allele kae1[7A9], and more substantial decreases (∼40%) in hemizygotes of the strong/null alleles kae1[1A13] and kae1[f01978] (Fig. 3C). We also observed full restoration of t6A-modified tRNAs in LC-MS/MS analysis of P[acman]-CG4933/+; kae1[1A13]/Df larvae, consistent with the viability rescue conferred by the genomic transgene.
Together, these observations provide evidence that kae1 is required for normal accumulation t6A-modified tRNAs in a metazoan setting. We cannot rule out that the remaining modified tRNAs are exceptionally stable. However, quantitative analysis did not reveal further decrease of t6A-modified tRNAs in kae1[f01978]/Df between 6- and 12-d larvae, suggesting that perdurance is not the major underlying cause of the remaining t6A. An alternate scenario is that they are contributed by the mitochondrial pool, which is presumably generated by the mitochondrial Kae1 paralog Qri7 (annotated as Drosophila CG14231). We note that Qri7 generates only a minute fraction of total t6A-modified tRNAs in yeast (Srinivasan et al. 2011), suggesting a potential difference between these eukaryotic species.
Drosophila Sua5/KEOPS genes complement corresponding yeast null mutants
Because the fly null mutants did not fully eliminate t6A, we sought supporting evidence on the role of Drosophila Kae1 in this pathway. Therefore, we conducted genetic replacement assays in a simpler heterologous host system, namely, in S. cerevisiae mutants in t6A pathway genes. As described, these are slow-growing but can be maintained in the null condition and are nearly completely deficient for t6A (El Yacoubi et al. 2011; Srinivasan et al. 2011).
To judge the overall relationship of the fly orthologs to their yeast counterparts, we tested for direct complementation of several yeast t6A mutants (Fig. 3A) by their Drosophila orthologs. We cloned the open reading frames for Drosophila kae1, bud32, and sua5 into the yeast expression plasmid, pADH416. Because yeast mutants in these three genes grow very poorly (Srinivasan et al. 2011), we transformed the Drosophila expression constructs into the corresponding yeast heterozygous diploid mutants. As controls, the same diploids were transformed with plasmids expressing the yeast protein, and with an empty vector. The transformed diploids were sporulated and then dissected to yield haploid segregants. Haploids containing plasmids expressing the Drosophila or yeast protein, or the empty vector, were identified and then streaked to individual colonies on selective medium.
The results of growth after ∼66 h at 30°C are shown in Figure 4A–C. Colony size is a sensitive measure of growth rate for yeast. Drosophila sua5, whose product generates the substrate for the KEOPS complex, yielded growth rescue that was comparable to its yeast ortholog (Fig. 4A). On the other hand, the two KEOPS components Drosophila kae1 and bud32 both conferred substantial rescue, as compared with vector control transformants, but growth was not as robust as with the cognate yeast factors (Fig. 4B,C). The relative degree of complementation was also apparent on the dissection plates on which the haploid spores germinated. Representative tetrads are shown in Supplemental Figure 3 for the kae1 and bud32 heterozygotes. In both cases, colonies identified as containing the Drosophila gene were larger than colonies containing vector or no plasmid, but not as large as wild-type colonies.
As the rescues by individual fly KEOPS proteins were less robust than with the cognate yeast factors, we tested if their function could be improved by supplying multiple Drosophila factors. This proved to be the case. Figure 4D shows a kae1 mutant expressing Drosophila Kae1 + Pcc1 (sector 1), Drosophila Kae1 + Bud32 (sector 2), or Drosophila Kae1 alone (sector 3). Coexpression with either Kae1 partner improved growth rescue. Similarly, coexpression of Drosophila Bud32 and Kae1 yielded enhanced rescue of yeast bud32 (Fig. 4E, sector 2), relative to Drosophila Bud32 alone (sector 3). Notably, expressing Drosophila Bud32 and Pcc1 together did not improve complementation of yeast bud32 relative to Bud32 alone (Fig. 4E, cf. sectors 1 and 3). This is consistent with the fact that Bud32 and Pcc1 do not interact directly in the complex, whereas both of them physically contact different faces of Kae1 in the KEOPS complex (Hecker et al. 2008; Mao et al. 2008). The growth improvements in these double complementation tests were also visible on tetrad dissection plates (Supplemental Fig. 3).
Rescue of t6A-modified tRNAs in yeast kae1 mutants by Drosophila KEOPS factors
We performed biochemical analysis of tRNA modification in mutant and rescued yeast. Primer extensions using a probe against tRNA-Ile[AAU] recapitulated strong loss in the A37 band in kae1 mutant yeast transformed with an empty vector, which was fully restored by the S. cerevisiae KAE1 expression construct (Fig. 4F). Comparison with yeast transformed with various combinations of Drosophila KEOPS factors showed strong agreement between growth properties and accumulation of t6A. That is, we observed moderate rescue by fly kae1 alone, but much stronger improvement by including other members of the Drosophila KEOPS complex (Fig. 4F).
We extended the above analysis of individual tRNAs to the total tRNA population using LC-MS/MS. These assays were strongly correlated with the relative degree of rescue observed in primer extension analysis. In particular, this quantitative method allowed us to distinguish greater restoration of t6A by Drosophila kae1 + bud32, relative to kae1 + pcc1 (Fig. 4G). In particular, the rescue by Drosophila kae1 + bud32 was equivalent to the cognate rescue by yeast kae1. Finally, we note that LC-MS/MS analysis of yeast samples also detected cyclic t6A (ct6A). It has been reported that ct6A is formed by cyclization of t6A and was observed in diverse domains of life including yeast, but not in any vertebrates tested (Miyauchi et al. 2013). Consistent with this, we did not detect ct6A above background in the analysis of fly tRNAs (data not shown).
In the yeast profilings, trends in ct6A levels closely mirrored those of t6A (Fig. 4G). ct6A levels were below the limit of detection in kae1 mutants, and rescued to similar relative levels as with t6A by the KEOPS expression constructs. However, ct6A peak areas were approximately fivefold smaller than corresponding levels of t6A. Despite efforts to minimize hydrolysis by conducting the analysis at neutral pH, it is possible that a portion of ct6A was hydrolyzed to t6A during sample preparation. In any case, these results clearly demonstrate the biochemical activity of Drosophila KEOPS genes including Kae1 in the biogenesis of N6-threonylcarbamoyladenosine-modified tRNAs. Since we did not detect ct6A in Drosophila samples, we will continue to generally refer to t6A in the remainder of this study.
kae1 mutants exhibit aberrant hematopoietic development and activity
Having established the role of Drosophila Kae1 and its partners in t6A biosynthesis, we sought to examine the nature of developmental defects caused by absence of this tRNA modification pathway. We began with the analysis of the namesake “Black Spots,” which resemble phenotypes associated with derangement in the development and/or deployment of the hematopoietic system (Minakhina and Steward 2006). Therefore, we investigated the status of the lymph gland as well as circulating blood cells in the hemolymph.
We first examined kae1[1A13]/Df(3L)ED220 larvae, which exhibit a strong molecular defect in t6A biogenesis (Fig. 3C) and generate “black spots” with nearly complete penetrance (Fig. 1A). The hemolymph of control TM6B sibling third instar larvae exhibit normal distributions of blood cells (Fig. 5A), primarily plasmatocytes (identifiable as ∼10 µM diameter cells) and more rare lamellocytes (40–50 µM diameter cells). In contrast, hemolymph smears of ∼10-d-old kae1[1A13] hemizygous larvae showed increased density of blood cells, and in particular, massive overrepresentation of lamellocytes (Fig. 5B). We also identified large circulating aggregates that resembled lymph gland fragments (Fig. 5C), as well as apparent phalloidin-rich tumors that were packed with lamellocytes (Fig. 5D). The latter could be melanized and were identifiable as “black spots,” but they were not necessarily pigmented, suggesting they had not yet initiated the melanogenesis cascade.
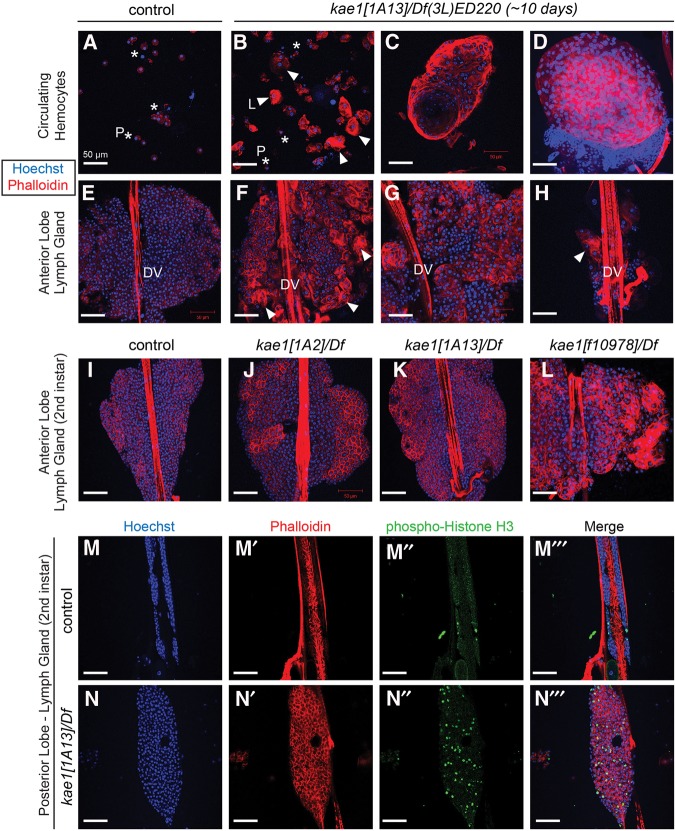
FIGURE 5.
Hematopoietic overgrowth and blood cell defects in kae1 mutants. All samples were stained with Hoechst (blue) to label DNA and rhodamine phalloidin (red) to label F-actin; all scale bars, 50 µm. Control animals were TM6B siblings of crosses used to generate kae1/Df(3L)ED220 larvae and are thus heterozygous for kae1. (A) Control hemolymph shows the typical profile in which most circulating hemocytes are plasmatocytes ([P] asterisks). (B) kae1[1A13] hemizygous hemolymph shows increased density of blood cells, as well as numerous lamellocytes, often present in clusters ([L] arrowheads). Lamellocytes (∼40–50 µm in diameter) are larger than plasmatocytes (10 µm in diameter). (C) Large aggregates bearing organized structure were observed in circulation as were overt tumors that are packed with plasmatocytes and lamellocytes (D). The latter correspond to the melanotic masses observed in live “black spot” larvae. (E–H) Anterior lobe of lymph gland from third instar in control (E) and from aged kae1/Df(3L)ED220 larvae (F–H); the dorsal vessel (DV) is labeled. The kae1 mutants exhibit strong overcommitment to lamellocyte fate in the lymph gland (F) and typically have overgrown lobes (G). Frequently, the anterior lobe was observed in the process of dispersal or was detached entirely from the dorsal vessel (H). (I–L) Analysis of anterior lymph gland lobes in second instar larvae. Compared with control (I), all kae1 mutants exhibit ectopic lamellocytes (J–L). The hypomorphs [1A2] (J) and [1A13] (K) are relatively normally sized, but the null [f01978] (L) is strongly overgrown. (M,N) Analysis of mitotic cells (labeled with anti-phospho-Histone H3, green) in posterior lobes of lymph glands from control (M) and [1A13]/Df (N). The kae1 mutant has overgrown posterior lobes that harbor more actively dividing cells.
Notably, lamellocyte overproduction is a frequent feature of other characterized mutants bearing melanotic masses (Lemaitre et al. 1995; Minakhina and Steward 2006). Therefore, we analyzed the status of the Drosophila hematopoietic tissue, the lymph gland. The compact disposition of the anterior lobe in normal third instar lymph gland is shown in Figure 5E and shows few lamellocytes. In contrast, ∼10-d-old kae1[1A13] hemizygous larval lymph glands show abundant maturation of lamellocytes and are typically enlarged (Fig. 5F,G). Often, we find that the anterior lobe was in the process of dispersing, or completely detached from the dorsal vessel (Fig. 5H). This correlates with the observation of apparent lymph tissue in the circulatory system (Fig. 5C). We observed similar phenotypes with the hypomorphic combination kae1[1A2]/Df(3L)ED220, except that its hematopoietic alterations were weaker and/or slower to develop (Supplemental Fig. 4). That is, defects were less robust at day 8 relative to day 10 larvae.
As mentioned, most kae1 mutants exhibit a vastly extended larval stage. Thus, we considered it plausible that the enlarged lymph tissues developed progressively over this aberrant larval period. To ask whether kae1 mutants exhibit defects during the normal course of larval development, we examined [1A2] (weak), [1A13] (strong), and [f10978] (null) kae1 hemizygotes at the second instar. We observed overrepresentation of lamellocytes in anterior lymph gland lobes in all genotypes, although [1A2] was not overgrown and [1A13] was only slightly enlarged relative to control (Fig. 5I–K). However, [f10978] exhibited both considerable overgrowth and ectopic commitment to lamellocyte fates (Fig. 5L). This is despite the fact that [f10978] develops modest frequencies of black spot compared with [1A13] (Fig. 1), indicating that hematopoietic tumors can exist without melanization of nodules. We also note that [f10978] exhibit lymph gland overgrowth despite the comparably small size of these larvae (Fig. 1), indicating partial uncoupling of organism size and individual organ size.
To verify their status as “tumors,” we stained second instar lymph glands for phospho-Histone H3 (pH3), a marker of mitotic cells. Figure 5M,N shows that kae1[1A13]/Df mutants also show expansion of their posterior lobes, although not as robust as with the anterior lobe. Nevertheless, these tissues show clear increases in the frequency of anti-pH3-labeled cells, indicating that the expanded hematopoietic tissues are accounted for by increased cell division, as well as alteration in cell fate toward lamellocytes.
Essential requirement for kae1 in imaginal discs but not throughout the larval CNS
A conspicuous aspect of the strong kae1 alleles were their nearly complete absence of imaginal discs. These begin as small cell populations that are set aside in the embryo, and proliferate during larval stages to increase their numbers ∼1000-fold (Milán et al. 1996), generating the tissues that will form the adult structures during metamorphosis. We mounted larval brain-imaginal disc complexes from wild-type and two strong kae1 mutants for comparison (Fig. 6A–C). Discs were completely absent in most larvae, or at best yielded tiny tissue rudiments that might potentially be discs, but cannot confidently be assigned to any particular disc identity. In contrast, the CNS was slightly smaller but remained present. Closer inspection revealed that the optic lobes were strongly reduced in size in kae1 mutants, whereas the ventral nerve cord (VNC) was less affected (Fig. 6A–C). Notably, the optic lobes are the main proliferating centers in the larval CNS, whereas the VNC is primarily composed of post-mitotic neurons.
FIGURE 6.
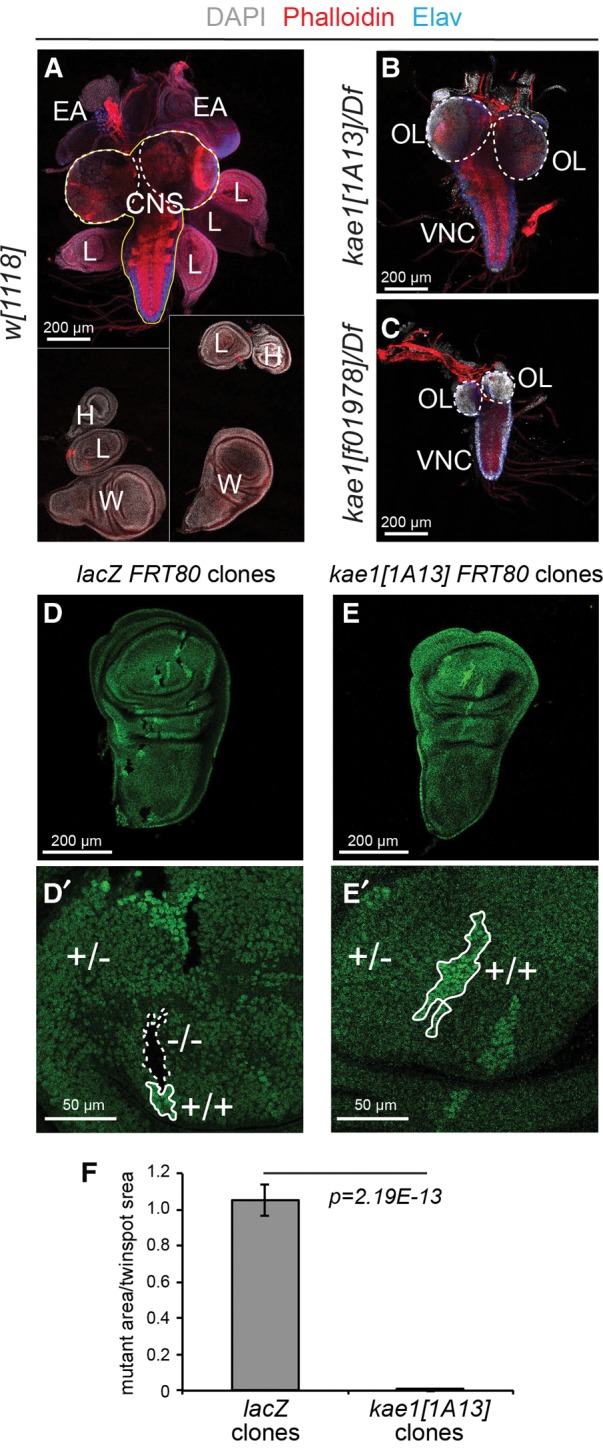
Essential requirement for kae1 in imaginal disc cells. (A–C) Preparations of third instar brain-disc complexes, stained for DAPI (gray), phalloidin (red) and the post-mitotic neuronal marker Elav (blue). Scale bars, 200 μm. (A) Wt complexes show the CNS (in outline) and an array of discs that generate the adult body structures. Because not all the discs remain attached to each other during dissection, a composite image that captures all of the discs is shown. These include pairs of wing (W), haltere (H), and eye-antennal (EA) discs, and six leg (L) discs. In kae1[1A13] (B) and kae1[f01978] (C) hemizygotes, essentially no imaginal discs are found, even though the CNS can be readily identified. The CNS can be divided into the optic lobes (OL, dotted lines) and the ventral nerve cord (VNC). The CNS is mildly smaller in kae1[1A13]/Df larvae (B) and substantially reduced in kae1[f01978]/Df larvae (C); however, note that the OL is preferentially reduced. The VNC harbors a stronger proportion of post-mitotic neurons than the OLs, as evidenced by Elav staining. (D,E) Analysis of mitotic clones generated in wing imaginal discs of control lacZ FRT80 (D) and kae1 FRT80 (E) chromosomes; enlargements are shown in D′ and E′. The clones are generated in a heterozygous (+/−) ub-GFP FRT80 background, which produces an intermediate level of GFP. Homozygous mutant clones (−/−) lack GFP whereas their sibling twin-spot clones (+/+) express elevated levels of GFP. Seventy-two hours after clone induction, all three cell populations are identified in the control experiment (D), whereas homozygous kae1 mutant cells were not recovered in wing discs (E). (F) Quantification of relative sizes of clones and their sibling twin-spots. n = 26 lacZ clones and 20 kae1 clones analyzed. Scale bars, 200 μm, except D′, E′, 50 μm.
We further examined the role of kae1 in proliferating tissues by generating negatively marked somatic clones in wing imaginal discs. In this system, both daughters of a mitotic recombination event are marked and remain distinguished as clones. The homozygous mutant clone is detected by loss of a marker (e.g., ubi-GFP) whereas its homozygous wild-type “twinspot” clone expresses the marker more intensely than the surrounding heterozygous tissue. Control FRT80 clones exhibit similar relative sizes to their twinspots (Fig. 6D,D′). In contrast, FRT80 kae1[1A13] clones were never recovered in imaginal discs, even though their twinspots were sizable (Fig. 6E,E′). We quantified the differences in clone/twinspot sizes in Figure 6F. These data imply substantial growth and/or competitive disadvantage of cells lacking the t6A modification pathway, which must have been removed from development after only a few cell divisions at most. We note that somatic clones of kae1 should still have inherited at least some Kae1 protein, mRNA, and/or t6A-modified tRNAs, yet failed to survive. From this, we infer that perdurance is not a major cause for the ability of kae1 null zygotic mutants to survive for over two and a half weeks (Fig. 1).
Differential requirements of kae1 in mitotic and nonproliferating larval tissues
The clonal analyses caused us to evaluate more carefully the extent to which different tissues were affected in kae1 mutants. It is clear that kae1-deficient cells exhibit growth defects in some contexts. Nevertheless, our original isolation of kae1 mutants as extremely long-lived larvae exhibiting hematopoietic expansion (Fig. 1) indicates that many cells and tissues can not only survive, but seemingly thrive in the kae1 null state for weeks. For example, differentiated post-mitotic blood cells such as lamellocytes (Krzemien et al. 2010) are both overrepresented and actively functional in encapsulation during the extended larval phase of kae1 mutants (Fig. 5D; Supplemental Fig. 4).
We surveyed other tissues that were maintained in kae1 mutant larvae, including the gut (Supplemental Fig. 5), salivary glands (Fig. 7A–F), fat body, and somatic musculature (Fig. 7G–I). For the most part, all of these tissues are dominantly or solely composed of nonproliferating cells during the larval stages. Therefore, these observations solidify the notion that mitotic cells are preferentially affected by loss of kae1. However, this does not mean that defects were lacking in these other larval tissues. Interestingly, quantitative analysis showed tissue-specific and allele-specific differences between their behavior. For example, even though kae1[1A13]/Df mutants maintain only marginally more t6A than kae1[f10978]/Df mutants (Fig. 3C), we observed progressively smaller salivary glands in kae1 mutants with less t6A (Fig. 7J). More strikingly, the size of salivary gland cells was mostly unaffected in [1A13] hemizygotes, whereas they were tiny by comparison in [f10978] hemizygotes (Fig. 7K). Thus, the slight amount of t6A in a strongly hypomorphic kae1 condition can maintain salivary gland cell size, even though these glands evidently have fewer cells than normal, whereas the null condition compromises both cell and organ size in the salivary gland. In contrast, the absolute size of the body wall musculature is largely unaffected in kae1[1A13]/Df, and only slightly reduced in the kae1[f10978]/Df mutants (Fig. 7L,M). Indeed, the small size of kae1 null larvae (Fig. 1E) may be sufficient to explain the small size of its muscles, which are expected to scale to body size.
FIGURE 7.
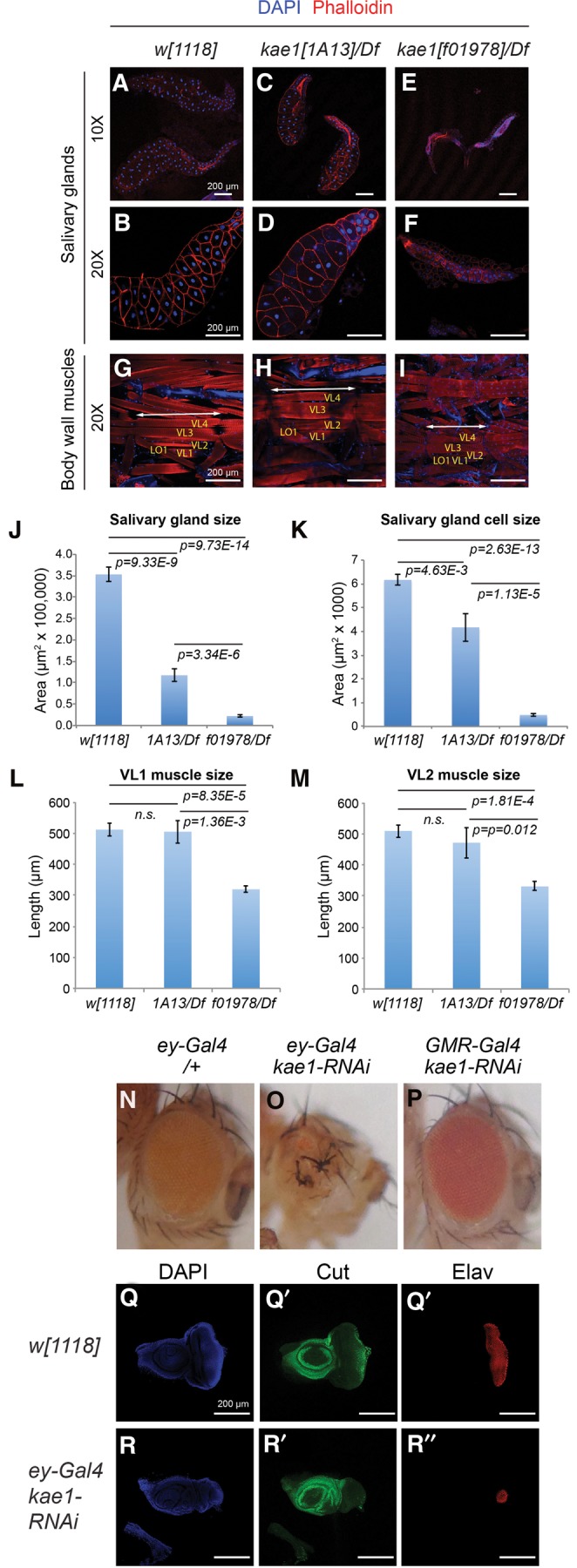
Differential requirements of kae1 in mitotic vs. nonproliferating tissues. Larval tissues were analyzed from kae1[1A13] and kae1[f01978] hemizygotes, which fully lack imaginal discs (Fig. 6). (A–I) Immunostaining of dissected tissues, using DAPI (blue) and phalloidin (red). (A–F) Salivary glands (and some attached fat bodies) imaged at 10× to visualize entire glands (A–C) and at 20× to emphasize individual cell sizes (D–F). (G–I) Body wall musculature. Selected muscles VL1-4 and LO1 are labeled. The length of a body segment is denoted with the double arrow lines. (J) Quantifications of salivary gland sizes (n = 10 all genotypes) and (K) salivary gland cell sizes (n = 15 cells from the distal tip measured in each of eight glands, all genotypes). Although kae1[1A13]/Df larvae exhibit severe defects in mitotic tissues, the overall salivary gland size and cell size are more modestly affected. The null allele kae1[f01978] is severely compromised in both aspects of the salivary gland. (L) Quantifications of VL1 muscle length and (M) VL2 muscle length. kae1[1A13]/Df larvae exhibit normal muscle sizes, while the null allele is only ∼40% reduced. Error bar shows standard error; all statistical analyses were unpaired Student's t-tests. (N–P) Adult eyes bearing Gal4 driver alone (N) or expressing kae1-RNAi under control of ey-Gal4 (O) or GMR-Gal4 (P). The former is active in the proliferating eye primordium, whereas the latter is active in post-mitotic photoreceptor neurons. (Q,R) Third instar eye-antennal imaginal discs stained for DAPI, Cut (green, to label cone cells) and Elav (red, to label photoreceptors). Compared to the control disc (Q), depletion of kae1 using ey-Gal4 induces severely defective eye development. All scale bars, 200 μm.
Distinct effects of depleting kae1 in mitotic and post-mitotic contexts of an individual tissue
Our extensive phenotypic analyses reveal strikingly distinct requirements for Kae1 in mitotic and nonproliferating larval tissues. To test for potentially distinct requirements for kae1 in mitotic and post-mitotic contexts within the same tissue, we turned to an RNAi approach. The developing eye is convenient for this purpose, as characterized drivers can be used to express transgenes in the undifferentiated, mitotic portion of the eye primordium (ey-Gal4) or in the post-mitotic photoreceptors (GMR-Gal4). Analysis of ey-Gal4 > UAS-kae1-RNAi flies showed strongly malformed eyes, which were small and rough (Fig. 7N,O). We obtained similar results with independent RNAi transgenes (data not shown). In contrast, GMR-Gal4 > UAS-kae1-RNAi produced eyes that developed normally, with all transgenes tested (Fig. 7P).
Cellular analysis demonstrated that depletion of kae1 using ey-Gal4 caused developmental defects. Compared to the normal size and differentiation status of normal third instar eye discs, stained for the photoreceptor marker Elav and cone cell marker Cut, ey-Gal4 > UAS-kae1-RNAi discs exhibit rudimentary eye discs with disturbed patterning (Fig. 7Q,R). On the other hand, knockdown with GMR-Gal4 did not affect larval eye development (data not shown). Therefore, kae1 is critically required in mitotic, but not in post-mitotic, eye cells.
DISCUSSION
Confirmation of the t6A modification pathway in a metazoan
To date, nearly all studies of the t6A biogenesis pathway have been conducted in unicellular organisms such as bacteria or yeast. The first functional study of a metazoan t6A component involved knockdown of the mitochondrial Kae1 paralog Qri7 (OSGEP-L) in Caenorhabditis elegans, which induced defective mitochondrial morphology and function (Oberto et al. 2009). Subsequently, a knockdown study of the D. melanogaster Bud32/PRPK reported growth defects relating to defective TOR signaling (Ibar et al. 2013). However, neither study assessed biochemical defects in t6A accumulation per se. Furthermore, knockdown approaches do not generally achieve genetic null states.
We demonstrate loss of t6A-modified tRNAs in Drosophila kae1 mutants, using both primer extension of individual species and mass spectrometry to examine the bulk population. Moreover, we demonstrate substantial cross-species rescue by fly Kae1 (and its partners) in yeast kae1 mutants, both in terms of growth capacity and in recovery of t6A-modified tRNAs. While this work was in preparation, similar observations using the kae1 transposon insertion alleles were reported by the Glavic group (Rojas-Benitez et al. 2015). Our findings corroborate and extend their results. Nevertheless, our detection of substantial t6A-modified tRNAs in Drosophila kae1 mutants was unexpected.
Previous characterization of yeast kae1 mutants (El Yacoubi et al. 2011; Srinivasan et al. 2011), which we confirm here, showed nearly complete absence of t6A-modified tRNAs. This raises two general possibilities. First, it may be that t6A-modified tRNAs are exceptionally stable in Drosophila. In this scenario, even though we assayed timepoints going well past the established range of maternal effects (e.g., up to 18 d, or the equivalent of a week-old adult fly), those modified tRNAs that were maternally deposited or generated from maternal kae1 mRNA and/or protein persisted all that time. Alternatively, many species, including yeast and Drosophila, express a mitochondrial Kae1 paralog named Qri7. Although mitochondrially derived t6A appears to comprise a minute portion of the total pool in single-celled organisms, the situation might be different in a metazoan. For example, the substantial populations of nonproliferating tissues that tolerate deletion of kae1 include the larval musculature, which sustain larval locomotion in mutants for weeks. Therefore, the remaining t6A-modified tRNAs in kae1 null mutant larvae may potentially be contributed by the abundant mitochondria in muscles. In the future, characterization of a Drosophila Qri7 (CG14231) knockout would provide a useful comparison, especially to see if Qri7 loss preferentially affects tissues with exceptional needs for oxidative phosphorylation.
An extensive allelic series of kae1 mutants reveal diverse biology of t6A
The first allele in a t6A biogenesis component, yeast sua5-1, was isolated in a forward genetic screen as an extragenic suppressor of a mutant cyc1 allele bearing an aberrant uORF (Hampsey et al. 1991). UV and X-rays were used as mutagens in these screens, and cloning of sua5-1 revealed a single-base substitution resulting in an S107F missense alteration (Na et al. 1992). Subsequently, other genetic efforts hit various KEOPS components of the t6A pathway, but these studies utilized screening of a deletion collection (Downey et al. 2006) or a multicopy suppressor scheme (Kisseleva-Romanova et al. 2006). All of these studies pre-dated the connection of Sua5/KEOPS factors to t6A biogenesis, and for the most part, studies have focused on generation of straight or conditional knockout alleles (Arigoni et al. 1998; El Yacoubi et al. 2009, 2011; Handford et al. 2009; Srinivasan et al. 2011).
We describe the first extensive set of point mutations in a t6A pathway component. These mutations sort along a comprehensive allelic series that includes null alleles, strong mutants, and weak hypomorphs. We document allele-specific phenotypic severities in different tissues, and also note that biochemically strong hypomorphs exhibit modest phenotypes in some settings (e.g., exhibit normal somatic musculature) but phenocopy null alleles in other settings (e.g., completely lack imaginal discs). These discrepancies highlight the utility of the allelic series in dissecting multiple requirements of essential housekeeping genes in regulating fundamental cellular processes in vivo. Future studies can continue to exploit these alleles to elucidate the biology of t6A modification. For example, it remains unclear what underlying causes provoke the dramatic hematopoietic responses in kae1 mutants, and our alleles provide unique means for querying this. Of note, substantial hematopoietic overgrowth is observed in both strong hypomorphs and the null condition, but only the former proceeds reliably toward generation of melanotic nodules.
The kae1 alleles we report should also be valuable for understanding the mechanism of t6A modification. Extensive structural studies delineate the Kae1 active site and binding surfaces with its partners Pcc1 and Bud32 (Hecker et al. 2008; Mao et al. 2008; Wan et al. 2013; Zhang et al. 2015). Interestingly, only one of our many point mutations directly affects any of these functionally characterized regions (Fig. 2A). This highlights the usage of a forward genetic screen to identify relevant mutations. This mutant collection can be utilized as a basis of structure-function studies to assess how these uncharacterized residues might affect formation of the KEOPS complex, productive t6A transfer, or potentially the specificity of substrate tRNA recognition.
Differential requirements of t6A in proliferating and nonproliferating cells
Studies of the biology of t6A modification pathway in metazoan systems are only in their infancy. The Glavic group reported foundational studies using transgenic RNAi against Drosophila Prpk/Bud32 (Ibar et al. 2013), and their recent work (Rojas-Benitez et al. 2015) and our current study extend this to genuine kae1 mutants. There are general similarities between the phenotypes of fruit fly and unicellular organism mutants, in that these genes are lethal (as in bacteria) and exhibit substantially defective growth (as in yeast). Of course, one of the major opportunities specifically available in higher eukaryotes is to examine potential cell- and tissue-specific requirements. Here, we find evidence for differential requirements for kae1 in distinct cell types.
Quite noticeably, strong kae1 mutants completely lack imaginal discs. The progenitor populations of discs are set aside during embryonic development, and they proliferate extensively during larval stages to generate the tissues that will form the adult body plan during metamorphosis. Although we cannot precisely assess the timing of effective depletion of maternal stores of t6A-modifed tRNAs (i.e., that are directly deposited into the oocyte, or that are generated via maternally deposited kae1 mRNA and/or protein), our clonal studies demonstrate a timely impact on removal of the kae1 genomic locus. That is, even though there exists substantial likelihood of Kae1 and t6A-modified tRNAs inherited from the mother cell prior to mitotic recombination, kae1 mutant disc cells are quickly disadvantaged and rapidly removed during the course of normal development.
Overall, we note these kae1 phenotypes are strikingly similar to certain other Drosophila mutants. For example, we and others previously characterized mutants of the central E1 and E2 enzymes in the SUMO pathway (the heterodimeric pair Aos1/Uba2 and Ubc9, respectively), and these develop a range of defects that are strikingly similar to what we documented for kae1 mutants. Phenotypes common to loss of Kae1 and SUMO include extended larval phases before lethality, presence of melanotic tumors, and preferential defects in mitotic versus nonproliferating cells, as evidenced by their lack of imaginal discs while retaining diverse other larval tissues (Kalamarz et al. 2012; Kanakousaki and Gibson 2012). Similarly, a number of zygotic cell cycle mutants specifically abrogate development of imaginal discs while generally sparing other endoreplicated and post-mitotic larval tissues (Gatti and Baker 1989). Altogether, these analyses highlight how mutations in seemingly “general” cellular regulators can reveal tissue-specific requirements. Moreover, they suggest that several fundamental regulatory pathways are preferentially required in highly mitotic tissues. Whether this simply indicates that diverse processes mutate to a common terminal zygotic phenotype, reflecting the distinct biology of mitotic and nonproliferating cells, or whether there are actually mechanistic links between t6A, SUMO, and the cell cycle, remains to be dissected in the future.
MATERIALS AND METHODS
Genetic screen for kae1 alleles
The region uncovered by Df(3L)ED220 was subjected to a lethal F2 screen after EMS mutagenesis. In total, we conducted nine rounds of mutagenesis using isogenized w[1118] males (rounds 1 and 2) or w ; FRT80B males (subsequent rounds). The animals were desiccated and fed 25mM EMS in 20% sucrose, then mated with w; TM3 Sb/TM6B females. Individual F1 TM6B males were crossed to w; Df(3L)ED220 w+/TM6B females (Bloomington #8077). Vials were scored for lethality by the absence of empty non-Tb pupal cases. Owing to the w+ marker integral to Df(3L)ED220, stocks of new lethal alleles were recovered by collecting white-eyed flies carrying TM6B, and re-balancing. In total, we conducted nine rounds of mutagenesis, and scored >7000 viable crosses to the deficiency, yielding 64 lethals.
The kae1 alleles [1A2], [1A13], and [1A19] were recovered in the w[1118] background, while [3A7], [5C3], [7A6], [7A9], and [9D1], FRT80B/TM6B were recovered in the w[1118]; FRT80B background. We subsequently identified other alleles of kae1 in stocks obtained from the Bloomington Stock Center, namely pBac-CG4933[f01978]/TM6B (#18497), l(3)72Fb[209]/TM6B (#4793), and l(3)72Fb[331]/TM6B (#4794).
Construction of CG4933 rescue transgenes
We amplified the genomic region encompassing CG4933 and part of the upstream flanking gene using the primers below. The fragment was cloned into pGEM-T-easy and sequence verified. The fragment was then excised with NotI and cloned into NotI digested attB-P[acman] (Venken et al. 2006), which had been treated with calf intestinal phosphatase.
4925AR TAAGCAATTATTTGATAGTCAATCTTTCG
CG4933R AAGGAACAGGCCCTTTAAGTGGG
We generated GFP-tagged CG4933 genomic transgene using recombineering, utilizing a two-step positive/negative selection (Warming et al. 2005). We performed positive/negative galK selection mediated by growth on MacConkey agar (positive selection for integration of the galK cassette) and growth on media containing 2-deoxygalactose (negative selection for replacement of galK cassette with GFP[S65T] CDS). Sequences (galK cassette and GFP[S65T] CDS) were amplified with primers described below from pgalK and pBD1010 (Barry Dickson), respectively. Purified PCR products were electroporated into SW102 bacteria (Warming et al. 2005) with the appropriate attB p[acman] CG4933 vector and selection was carried out as described previously (Venken et al. 2006).
Primers for galK recombineering of GFP into C-terminus of CG4933:
C term - galK insertion
CT4933galK-F
GTGACTCAGCGATTTCGAACGGACGAAGTGCTGGTCAGCTGGCGGGATGACCCTGTTGACAATTAATCATCGGCA
CT4933galK-R
TAGTTATTTAATAAACAAACGATGCACAGAAATAAGTTCTTGTGGTGATTATCAGCACTGTCCTGCTCCTT
C term - GFP insertion
CT4933GFP-F
GTGACTCAGCGATTTCGAACGGACGAAGTGCTGGTCAGCTGGCGGGATGACATGAGTAAAGGAGAAGAACTTTTC
CT4933GFP-R
TAGTTATTTAATAAACAAACGATGCACAGAAATAAGTTCTTGTGGTGATTATTTGTATAGTTCATCCATGCC
Immunostaining and clonal analysis
Analysis of hematopoietic system
Developmentally synchronized larvae from crosses w; kae1[IA2]/TM6B X Df(3L)ED220/TM6B and w; kae1[IA13]/TM6B X Df(3L)ED220/TM6B were collected, washed in phosphate buffered saline, pH 7.4, and either imaged without dissection using a Leica MZFLIII, or were dissected for hemolymph contents, lymph glands, and tumors, at 4, 8, and 10 d after egg laying, according to previously described methods (Small et al. 2012). To obtain larval blood cells and tumors, animals were bled gently, and hemolymph contents, including tumors, were recovered directly on slides. Larval samples were fixed in 4% paraformaldehyde in PBS, pH 7.4, stained with Rhodamine Phalloidin (Invitrogen) and Hoechst 33258 (Molecular Probes), and imaged with Zeiss laser Scanning 510 confocal microscope. Images were recorded at 8 bit. AxioVision LE 4.5 or Zeiss LSM5 AxioVision software was used to process images.
Analysis of other larval tissues
Imaginal discs, brains, and muscles were dissected and stained following previously described protocol (Lai and Rubin 2001). Antibodies used were mouse anti-Cut (DSHB, 1:50), rat anti-Elav (DSHB, 1:500), Alexa Fluor-568 phalloidin (Life Technologies, 1:1000). Appropriate secondary antibodies conjugated to Alexa 488, 568, and 647 were then used. Tissues were mounted in VECTASHIELD Antifade Mounting Medium with DAPI.
Clonal analysis
We analyzed hsFlp; [w+, arm-lacZ] FRT80B/[w+, ubi-GFP] FRT80B and hsFlp; kae1[1A13] FRT80B/[w+, ubi-GFP] FRT80B animals. Larvae were heat-shocked at 37°C for 40 min at 72 h after egg laying, and were dissected 3 d after heat shock.
RT-PCR
Total RNA from homozygous or hemizygous larvae were extracted with TRIzol (Invitrogen), followed by DNase treatment using TURBO DNA-free Kit (Ambion). cDNA were synthesized using SuperScript III First-Strand Synthesis System with random primers (Invitrogen). QPCR reaction was performed with CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Primers used were Kae1_RT_F, TTTGGGTATTGAAGGCAGTG, and Kae1_RT_R, CAGGAAACCTTCTCCTGGTG, and RPL32_RT_F, CCGCTTCAAGGGACAGTATC, and RPL32_RT_R, ACGTTGTGCACCAGGAACTT.
tRNA primer extension assays
Two micrograms of tRNA extracted with mirVana miRNA isolation kit (Ambion) was mixed with 1 pmol of [γ-32P]-labeled primer in 5 μL 1× First-strand buffer (Invitrogen), heated to 95°C and then slowly cooled down to 37°C. Annealed RNA/primer was collected by brief centrifugation, added with 5 μL retro-transcription reaction mix, including 0.5 mM dNTPs, 0.01 M DTT, 200 units SuperScript III (Invitrogen) in 1× First-strand buffer and incubated at 50°C for 50 min, followed by heat inactivation at 70°C for 15 min. Reaction samples were then electrophoresed with 15% polyacrylamide–urea sequencing gels (40 cm long).
S.c. Ile primer: gggatcgaaccgctgatc
S.c. Met primer: aggttcgaactctcgacc
S.c. Val primer: aggttcgaactcgggatc
D.m. Ile primer: gggatcgaacccgcgac
D.m. Met primer: aggtttcgatcctcggacc
D.m. Val Primer: gggatcgaaccggagacc
Yeast strains and plasmids
All yeast strains are derivatives of W303 a/α diploids homozygous for trp1 leu2 ura3 his3 ade2. The following derivatives of W303 were used: EJP51 KAE1/kae1::kanMX6, RS2586 BUD32/bud32::kanMX6, and YYF1 SUA5/sua5::kanMX6. PCR was used to amplify the Drosophila cDNAs for KAE1, BUD32, PCC1, and SUA5 open reading frames. The cDNAs were cloned into a yeast expression plasmid vector, pADH416 or pADH415, with selectable markers URA3 or LEU2, respectively. These plasmids express the Drosophila proteins from the ADH promoter on a single copy plasmid. As controls, the corresponding yeast genes were expressed under their own promoters in the single copy vector pRS316. The plasmid expressing yeast Kae1 was described previously (Srinivasan et al. 2011) and yeast Pcc1, Bud32, and Sua5 expressing plasmids are pXZ1, pXZ2, and pXZ3, respectively.
Mass spectrometric analysis of tRNA modifications
Chemicals
All chemicals and reagents used were obtained at the highest purity available and were used without further purification unless stated. Benzonase, bacterial alkaline phosphatase, butylated hydroxytoluene, acetonitrile, and buffer salts were purchased from Sigma-Aldrich. Coformycin was obtained from the National Cancer Institute. Phosphodiesterase I was purchased from Worthington. Tetrahydrouridine was purchased from Calbiochem. Water purified through a Milli-Q system (Millipore) was used throughout our studies. Sartorius Vivaspin 500-brand centrifugal filter units were used for dialysis and sample concentration.
Extraction and purification of Drosophila and yeast RNA
Total RNAs were isolated using TRIzol and then subjected to a two-step purification to obtain tRNA. First, a PureLink RNA Mini extraction kit (Life Technologies) was used to precipitate the ribosomal RNA in the sample. In the second step, size-exclusion chromatography (SEC-HPLC) was performed using an Agilent Bio SEC-3 column with 10 mM ammonium acetate at pH 7 as the mobile phase. Neutral pH was selected to minimize hydrolysis of ct6A. tRNA and rRNA were collected as separate fractions, vacuum concentrated and reconstituted in water. tRNA concentrations were determined by UV spectroscopy at 260 nm. In the yeast samples, the proportion of tRNA to ribosomal RNA was much higher so only SEC-HPLC fractionation was required to obtain pure tRNA for analysis.
Nucleoside preparation
Four micrograms of purified tRNA from each sample was digested using a mixture of Benzonase (21.5 U), bacterial alkaline phosphatase (0.5 U) and phosphodiesterase I (0.21 U) in a final reaction volume of 100 µL. Enzymes were dialyzed into 10 mM ammonium acetate at pH 7 immediately prior to use by washing twice with 10 kDa MWCO Vivaspin 500 spin filters. The reaction mixture was supplemented with MgCl2, CaCl2, and ZnCl2, each at a concentration of 0.1 mM. Nucleobase deaminase inhibitors coformycin and tetrahydrouridine were added at a concentration of 10 µg/mL and 50 µg/mL, respectively, and butylated hydroxytoluene (an antioxidant) was added at a concentration of 0.5 mM. The digestion was allowed to proceed for 2 h at 37°C and was stopped upon removal of the enzymes by microfiltration with 10 kDa spin filters.
LC-MS/MS analysis of RNA nucleosides
Ribonucleosides were separated using a Synergy Fusion RP, 2.5 µm particle size, 100 Å pore size, 100 mm length, 2 mm inner diameter from Phenomenex, on an Agilent 1290 series HPLC system equipped with a diode array detector. Mobile phase A was 5 mM ammonium acetate and mobile phase B was pure acetonitrile. Gradient elution started with 100% A for 1 min, increased to 10% B after 10 min, 40% after 14 min, 80% after 20 min and regeneration of starting conditions with 100% A for 5 additional minutes. The flow rate was 0.35 mL/min and the column temperature 35°C. The effluent from the column was directed through the DAD before entering the Agilent 6430 triple quadrupole mass spectrometer in dynamic multiple reaction monitoring (MRM) mode. The MS was operated in positive ion mode with the following parameters: electro-spray ionization (ESI-MS), fragmentor voltage (average) 80 V, cell accelerator voltage 2 V, N2-gas temperature 350°C, N2-gas flow 10 L/min, nebulizer 40 psi, capillary 3500 V.
Modified nucleosides were identified based on having both the correct retention time and mass transition. Under our chromatography conditions, t6A eluted at 8.6 min while ct6A eluted at 6.8 min. Mass transitions were m/z 413 → 281 for t6A and m/z 395 → 263 for ct6A, both corresponding to the neutral loss of ribose. 15N-2-Deoxyadenosine was added to each sample as an internal standard.
Data analysis
In each sample, the UV peak areas at 260 nm of the four canonical ribonucleosides—cytidine, uridine, guanosine, and adenosine—were integrated and summed together. Peaks corresponding to detection of MRM transitions for the modified nucleosides were first normalized to the peak area of the internal standard (15N-2-deoxyadenosine) to account for inter-sample detection fluctuations, followed by a second normalization to the summed peak areas of the canonical nucleosides to account for the amount of injected RNA. The adjusted peak areas of the five samples were calculated as a percentage of the corresponding wild-type control sample.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Fuqu Hu for Kae1 structural comparisons and the Bloomington Stock Center for stocks. Work in the Govind laboratory was supported by NSF-1121817, NASA award NNX15AB42G, and a Howard and Vicki Palefsky Fellowship to J.R. Work in the Dedon laboratory was supported by the Singapore National Research Foundation under its Singapore-MIT Alliance for Research and Technology, the National Institute of Environmental Health Science (ES017010, ES022858, ES002109), the National Science Foundation (MCB-1412379), and a fellowship from the Deutsche Forschungsgemeinschaft (S.M.K.). Work in the Sternglanz laboratory was supported by P01-GM088297. Work in the Lai laboratory was supported by the Burroughs Wellcome Foundation, the National Institutes of Health (R01-GM083300), the National Institute of Neurological Disorders and Stroke (R01-NS083833), and Memorial Sloan-Kettering Core grant P30-CA008748.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.053934.115.
REFERENCES
- Agris PF. 2008. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep 9: 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96. [DOI] [PubMed] [Google Scholar]
- Arigoni F, Talabot F, Peitsch M, Edgerton MD, Meldrum E, Allet E, Fish R, Jamotte T, Curchod ML, Loferer H. 1998. A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol 16: 851–856. [DOI] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, May G, Stoiber M, Duff MO, Booth BW, Wen J, Park S, Suzuki AM, et al. 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512: 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron MC, Lenstra TL, Frizzarin M, El Yacoubi B, Liu X, Baudin-Baillieu A, Lijnzaad P, Decourty L, Saveanu C, Jacquier A, et al. 2011. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res 39: 6148–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch C, El Yacoubi B, de Crécy-Lagard V, Iwata-Reuyl D. 2012. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J Biol Chem 287: 13666–13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM, FlyBase Consortium. 2015. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res 43: D690–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, et al. 2006. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124: 1155–1168. [DOI] [PubMed] [Google Scholar]
- El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crécy-Lagard V. 2009. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res 37: 2894–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, Iwata-Reuyl D, Murzin AG, de Crécy-Lagard V. 2011. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J 30: 882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. 2004. ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res 32: 5452–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M, Baker BS. 1989. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev 3: 438–453. [DOI] [PubMed] [Google Scholar]
- Grewal SS. 2015. Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochim Biophys Acta 1849: 898–907. [DOI] [PubMed] [Google Scholar]
- Grosjean H, Sprinzl M, Steinberg S. 1995. Posttranscriptionally modified nucleosides in transfer RNA: their locations and frequencies. Biochimie 77: 139–141. [DOI] [PubMed] [Google Scholar]
- Gu C, Begley TJ, Dedon PC. 2014. tRNA modifications regulate translation during cellular stress. FEBS Lett 588: 4287–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Na JG, Pinto I, Ware DE, Berroteran RW. 1991. Extragenic suppressors of a translation initiation defect in the cyc1 gene of Saccharomyces cerevisiae. Biochimie 73: 1445–1455. [DOI] [PubMed] [Google Scholar]
- Handford JI, Ize B, Buchanan G, Butland GP, Greenblatt J, Emili A, Palmer T. 2009. Conserved network of proteins essential for bacterial viability. J Bacteriol 191: 4732–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanoune J, Agarwal MK. 1970. Studies on the half life time of rat liver transfer RNA species. FEBS Lett 11: 78–80. [DOI] [PubMed] [Google Scholar]
- Hecker A, Lopreiato R, Graille M, Collinet B, Forterre P, Libri D, van Tilbeurgh H. 2008. Structure of the archaeal Kae1/Bud32 fusion protein MJ1130: a model for the eukaryotic EKC/KEOPS subcomplex. EMBO J 27: 2340–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Huang HY. 2015. Quality control pathways for nucleus-encoded eukaryotic tRNA biosynthesis and subcellular trafficking. Mol Cell Biol 35: 2052–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Ibar C, Cataldo VF, Vásquez-Doorman C, Olguín P, Glavic A. 2013. Drosophila p53-related protein kinase is required for PI3K/TOR pathway-dependent growth. Development 140: 1282–1291. [DOI] [PubMed] [Google Scholar]
- Kalamarz ME, Paddibhatla I, Nadar C, Govind S. 2012. Sumoylation is tumor-suppressive and confers proliferative quiescence to hematopoietic progenitors in Drosophila melanogaster larvae. Biol Open 1: 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanakousaki K, Gibson MC. 2012. A differential requirement for SUMOylation in proliferating and non-proliferating cells during Drosophila development. Development 139: 2751–2762. [DOI] [PubMed] [Google Scholar]
- Kirchner S, Ignatova Z. 2015. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet 16: 98–112. [DOI] [PubMed] [Google Scholar]
- Kisseleva-Romanova E, Lopreiato R, Baudin-Baillieu A, Rousselle JC, Ilan L, Hofmann K, Namane A, Mann C, Libri D. 2006. Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J 25: 3576–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzemien J, Oyallon J, Crozatier M, Vincent A. 2010. Hematopoietic progenitors and hemocyte lineages in the Drosophila lymph gland. Dev Biol 346: 310–319. [DOI] [PubMed] [Google Scholar]
- Lai EC, Rubin GM. 2001. neuralized functions cell-autonomously to regulate a subset of Notch-dependent processes during adult Drosophila development. Dev Biol 231: 217–233. [DOI] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. 1995. The neurogenic suppressor of sairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev 9: 2598–2608. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Meister M, Govind S, Georgel P, Steward R, Reichhart JM, Hoffmann JA. 1995. Functional analysis and regulation of nuclear import of dorsal during the immune response in Drosophila. EMBO J 14: 536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CA, Ellis SR, True HL. 2010. The Sua5 protein is essential for normal translational regulation in yeast. Mol Cell Biol 30: 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC. 2000. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics 155: 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. 2013. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res 41: D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao DY, Neculai D, Downey M, Orlicky S, Haffani YZ, Ceccarelli DF, Ho JS, Szilard RK, Zhang W, Ho CS, et al. 2008. Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol Cell 32: 259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milán M, Campuzano S, García-Bellido A. 1996. Cell cycling and patterned cell proliferation in the wing primordium of Drosophila. Proc Natl Acad Sci 93: 640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakhina S, Steward R. 2006. Melanotic mutants in Drosophila: pathways and phenotypes. Genetics 174: 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K, Kimura S, Suzuki T. 2013. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol 9: 105–111. [DOI] [PubMed] [Google Scholar]
- Na JG, Pinto I, Hampsey M. 1992. Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae. Genetics 131: 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberto J, Breuil N, Hecker A, Farina F, Brochier-Armanet C, Culetto E, Forterre P. 2009. Qri7/OSGEPL, the mitochondrial version of the universal Kae1/YgjD protein, is essential for mitochondrial genome maintenance. Nucleic Acids Res 37: 5343–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrochia L, Crozat E, Hecker A, Zhang W, Bareille J, Collinet B, van Tilbeurgh H, Forterre P, Basta T. 2013. In vitro biosynthesis of a universal t6A tRNA modification in Archaea and Eukarya. Nucleic Acids Res 41: 1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Benitez D, Thiaville PC, de Crécy-Lagard V, Glavic A. 2015. The levels of a universally conserved tRNA modification regulate cell growth. J Biol Chem 290: 18699–18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small C, Paddibhatla I, Rajwani R, Govind S. 2012. An introduction to parasitic wasps of Drosophila and the antiparasite immune response. J Vis Exp 7: e3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Vassilenko KS. 2005. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 33: D139–D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, Karzai AW, Sternglanz R. 2011. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J 30: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC. 2014. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat Protoc 9: 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751. [DOI] [PubMed] [Google Scholar]
- Wan LC, Mao DY, Neculai D, Strecker J, Chiovitti D, Kurinov I, Poda G, Thevakumaran N, Yuan F, Szilard RK, et al. 2013. Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system. Nucleic Acids Res 41: 6332–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenbach J, Grosjean H. 1981. Effect of threonylcarbamoyl modification (t6A) in yeast tRNA Arg III on codon-anticodon and anticodon-anticodon interactions. A thermodynamic and kinetic evaluation. Eur J Biochem 116: 207–213. [DOI] [PubMed] [Google Scholar]
- Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, Guenther R, Miskiewicz A, Agris PF. 2002. Accurate translation of the genetic code depends on tRNA modified nucleosides. J Biol Chem 277: 16391–16395. [DOI] [PubMed] [Google Scholar]
- Zhai RG, Hiesinger PR, Koh TW, Verstreken P, Schulze KL, Cao Y, Jafar-Nejad H, Norga KK, Pan H, Bayat V, et al. 2003. Mapping Drosophila mutations with molecularly defined P element insertions. Proc Natl Acad Sci 100: 10860–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Collinet B, Perrochia L, Durand D, van Tilbeurgh H. 2015. The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli. Nucleic Acids Res 43: 1804–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.