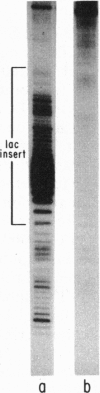
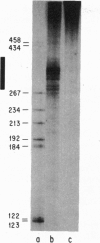
Abstract
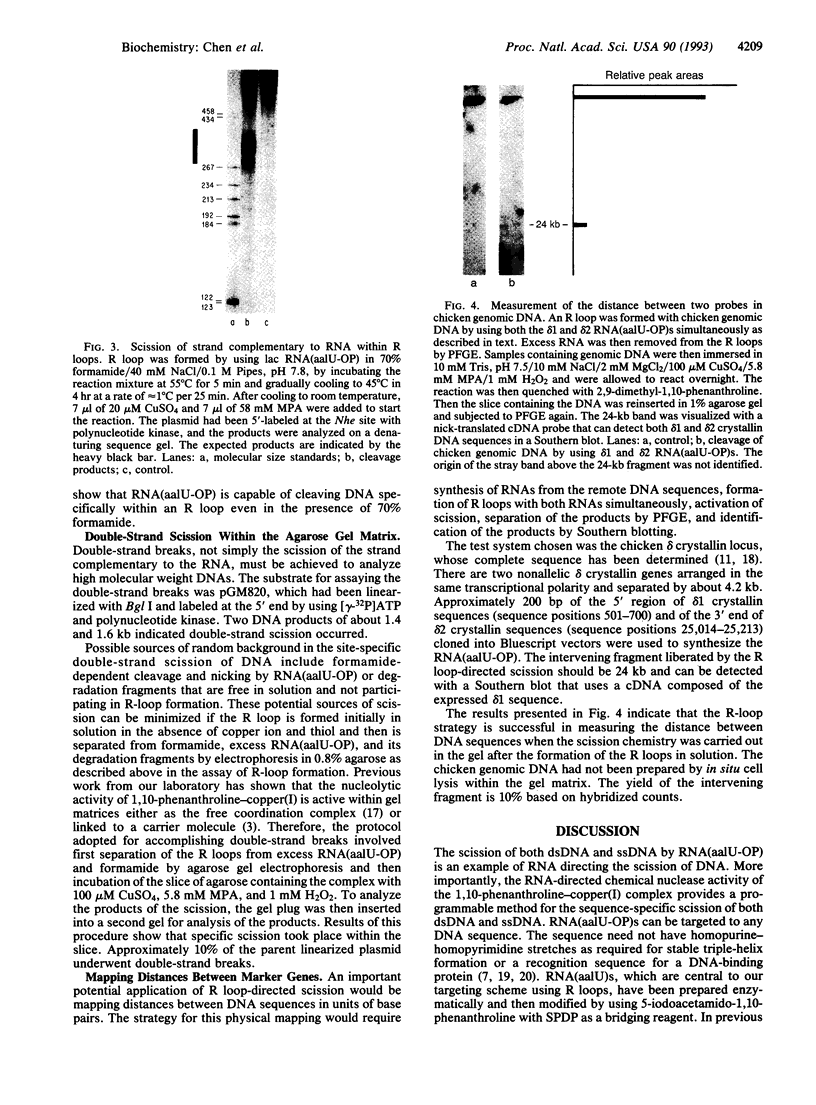
RNAs modified with the chemical nuclease 1,10-phenanthroline-copper(I) can achieve the sequence-specific scission of single- and double-stranded DNA targets. The RNAs are prepared in vitro by using 5-(3-aminoallyl)-UTP as the sole source of UTP and can be readily modified with 1,10-phenanthroline by using N-succinimidyl 3-(2-pyridyl-dithio)propionate (SPDP) to cross-link the ligand to the aminoallyl moiety. Single-stranded DNAs are efficiently cleaved at multiple sites because 1,10-phenanthroline is incorporated at several uridines in the sequence. Sequence-specific double-stranded scission of duplex DNA can also be accomplished with 1,10-phenanthroline-derivatized RNA within R loops. These triple-stranded structures form in 70% formamide and involve the displacement of one strand of DNA by the RNA of identical sequence. R loop-directed scission is the first method for DNA scission applicable to any sequence. A unique application of R loop-targeted nucleolytic scission, which relies on its ability to cut DNA at any sequence, is the determination of the distance between two marker DNA sequences within a target. In this case, 1,10-phenanthroline-linked RNAs are prepared from the two distinct sequences and used to cut the DNA fragment after R-loop formation. The size of the fragment liberated by these methods is a direct measure in base pairs of the distance between the two DNA sequences. For example, the distance separating two chicken delta crystallin (delta 1 and delta 2) genes has been confirmed as 24 kilobases by this method.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carlsson J., Drevin H., Axén R. Protein thiolation and reversible protein-protein conjugation. N-Succinimidyl 3-(2-pyridyldithio)propionate, a new heterobifunctional reagent. Biochem J. 1978 Sep 1;173(3):723–737. doi: 10.1042/bj1730723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H., Sigman D. S. Chemical conversion of a DNA-binding protein into a site-specific nuclease. Science. 1987 Sep 4;237(4819):1197–1201. doi: 10.1126/science.2820056. [DOI] [PubMed] [Google Scholar]
- Chen C. H., Sigman D. S. Nuclease activity of 1,10-phenanthroline-copper: sequence-specific targeting. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7147–7151. doi: 10.1073/pnas.83.19.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B. C., Orgel L. E. Nonenzymatic sequence-specific cleavage of single-stranded DNA. Proc Natl Acad Sci U S A. 1985 Feb;82(4):963–967. doi: 10.1073/pnas.82.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer G. B., Dervan P. B. Sequence-specific cleavage of single-stranded DNA: oligodeoxynucleotide-EDTA X Fe(II). Proc Natl Acad Sci U S A. 1985 Feb;82(4):968–972. doi: 10.1073/pnas.82.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Ebright Y. W., Pendergrast P. S., Gunasekera A. Conversion of a helix-turn-helix motif sequence-specific DNA binding protein into a site-specific DNA cleavage agent. Proc Natl Acad Sci U S A. 1990 Apr;87(8):2882–2886. doi: 10.1073/pnas.87.8.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker D. J., Vickers T. A., Bruice T. W., Freier S. M., Jenison R. D., Manoharan M., Zounes M. Pseudo--half-knot formation with RNA. Science. 1992 Aug 14;257(5072):958–961. doi: 10.1126/science.1502560. [DOI] [PubMed] [Google Scholar]
- François J. C., Saison-Behmoaras T., Barbier C., Chassignol M., Thuong N. T., Hélène C. Sequence-specific recognition and cleavage of duplex DNA via triple-helix formation by oligonucleotides covalently linked to a phenanthroline-copper chelate. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9702–9706. doi: 10.1073/pnas.86.24.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François J. C., Saison-Behmoaras T., Chassignol M., Thuong N. T., Helene C. Sequence-targeted cleavage of single- and double-stranded DNA by oligothymidylates covalently linked to 1,10-phenanthroline. J Biol Chem. 1989 Apr 5;264(10):5891–5898. [PubMed] [Google Scholar]
- Herfort M. R., Garber A. T. Simple and efficient subtractive hybridization screening. Biotechniques. 1991 Nov;11(5):598, 600, 602-4. [PubMed] [Google Scholar]
- Koob M., Szybalski W. Cleaving yeast and Escherichia coli genomes at a single site. Science. 1990 Oct 12;250(4978):271–273. doi: 10.1126/science.2218529. [DOI] [PubMed] [Google Scholar]
- Kuwabara M. D., Sigman D. S. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry. 1987 Nov 17;26(23):7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- Langer P. R., Waldrop A. A., Ward D. C. Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6633–6637. doi: 10.1073/pnas.78.11.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Murakawa G. J., Chen C. H., Kuwabara M. D., Nierlich D. P., Sigman D. S. Scission of RNA by the chemical nuclease of 1,10-phenanthroline-copper ion: preference for single-stranded loops. Nucleic Acids Res. 1989 Jul 11;17(13):5361–5375. doi: 10.1093/nar/17.13.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson J. M., Wawrousek E. F., Borras T., Hawkins J. W., Norman B. L., Filpula D. R., Nagle J. W., Ally A. H., Piatigorsky J. Sequence of the chicken delta 2 crystallin gene and its intergenic spacer. Extreme homology with the delta 1 crystallin gene. J Biol Chem. 1986 Jan 15;261(2):552–557. [PubMed] [Google Scholar]
- Nickerson J. M., Wawrousek E. F., Hawkins J. W., Wakil A. S., Wistow G. J., Thomas G., Norman B. L., Piatigorsky J. The complete sequence of the chicken delta 1 crystallin gene and its 5' flanking region. J Biol Chem. 1985 Aug 5;260(16):9100–9105. [PubMed] [Google Scholar]
- Rosbash M., Blank D., Fahrner K., Hereford L., Ricciardi R., Roberts B., Ruby S., Woolford J. R-looping and structural gene indentification of recombinant DNA. Methods Enzymol. 1979;68:454–469. doi: 10.1016/0076-6879(79)68035-7. [DOI] [PubMed] [Google Scholar]
- Sargent T. D. Isolation of differentially expressed genes. Methods Enzymol. 1987;152:423–432. doi: 10.1016/0076-6879(87)52049-3. [DOI] [PubMed] [Google Scholar]
- Strobel S. A., Dervan P. B. Site-specific cleavage of a yeast chromosome by oligonucleotide-directed triple-helix formation. Science. 1990 Jul 6;249(4964):73–75. doi: 10.1126/science.2195655. [DOI] [PubMed] [Google Scholar]
- Thomas M., White R. L., Davis R. W. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford J. L., Jr, Rosbash M. The use of R-looping for structural gene identification and mRNA purification. Nucleic Acids Res. 1979 Jun 11;6(7):2483–2497. doi: 10.1093/nar/6.7.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]