Abstract
Background
Sexual reproduction is an obligate phase in the life cycle of most eukaryotes. Meiosis varies among organisms, which is reflected by the variability of the gene set associated to the process. Diatoms are unicellular organisms that belong to the stramenopile clade and have unique life cycles that can include a sexual phase.
Results
The exploration of five diatom genomes and one diatom transcriptome led to the identification of 42 genes potentially involved in meiosis. While these include the majority of known meiosis-related genes, several meiosis-specific genes, including DMC1, could not be identified. Furthermore, phylogenetic analyses supported gene identification and revealed ancestral loss and recent expansion in the RAD51 family in diatoms. The two sexual species Pseudo-nitzschia multistriata and Seminavis robusta were used to explore the expression of meiosis-related genes: RAD21, SPO11-2, RAD51-A, RAD51-B and RAD51-C were upregulated during meiosis, whereas other paralogs in these families showed no differential expression patterns, suggesting that they may play a role during vegetative divisions. An almost identical toolkit is shared among Pseudo-nitzschia multiseries and Fragilariopsis cylindrus, as well as two species for which sex has not been observed, Phaeodactylum tricornutum and Thalassiosira pseudonana, suggesting that these two may retain a facultative sexual phase.
Conclusions
Our results reveal the conserved meiotic toolkit in six diatom species and indicate that Stramenopiles share major modifications of canonical meiosis processes ancestral to eukaryotes, with important divergences in each Kingdom.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1983-5) contains supplementary material, which is available to authorized users.
Keywords: Meiosis, Diatoms, Sexual reproduction, SPO11, RAD51
Background
The process of sexual reproduction is a hallmark for all the major eukaryotic groups [1–3]. It is believed that all asexual eukaryotes have evolved from sexual ancestors [1, 4], and it has been proposed that the last eukaryotic common ancestor (LECA) possessed the full set of genes known to be involved in meiosis [5–8]. Meiosis is not performed exactly in the same way in different groups: differences can be found for instance in the mechanisms of crossover formation and in the structure of the synaptonemal complex, and these differences are reflected in a variable set of meiosis-related genes [9].
Our understanding of the meiotic process, and consequently our knowledge of the gene repertoire required, is relatively strong for Opisthokonta and plants. However, despite the fact that a number of publications have recently appeared on a variety of unicellular organisms [5, 6, 9–11], information on most groups that contribute to the greater part of eukaryotic diversity are still scarce.
Among aquatic protists, diatoms are an important group of microalgae, as they are one of the major primary producers in freshwater and marine ecosystems [12, 13] and often dominate planktonic and benthic microalgal assemblages. They are a relatively recent lineage of unicellular organisms belonging to the SAR (Stramenopiles, Alveolata, Rhizaria) supergroup [14]. The Stramenopiles include both unicellular and multicellular members, as well as phototrophic, heterotrophic and parasitic members. Diatoms are the only free-living unicellular Stramenopiles for which the sexual cycle can be fully controlled in the laboratory for some species [15].
Diatoms are divided in two major groups, centrics, with radial symmetry, and pennates, with bilateral symmetry. They are unique among unicellular eukaryotes as they are encased in intricately patterned siliceous cell walls, consisting of two halves (thecae) of which one (the epitheca) is slightly larger than the other (the hypotheca). Diatoms spend the majority of their life cycle as diploid cells and multiply by mitotic divisions. Repeated cell divisions are, however, constrained by the inflexible arrangement of the silica wall. As a consequence of this rigid structure, diatom cells progressively decrease in size after cell division, which may lead to cell death and extinction of the clonal lineage unless large cell size is restored [16]. Although some species have been shown to employ alternative routes to escape the progressive cell miniaturization, the majority of diatom species restore cell size via sexual reproduction [15]. Thus, besides the fundamental goal of sexual reproduction to generate genetic diversity within a population, in diatoms the sexual phase also plays a key role in cell size restitution [15, 16].
The best studied model diatoms are Thalassiosira pseudonana and Phaeodactylum tricornutum, for which genome sequences are available [17, 18], and gene function can be studied with conventional tools for manipulation [19–21]. In contrast to most diatoms [22], sexual stages have never been observed for these two species, and laboratory strains do not reduce in size and only reproduce vegetatively. This has imposed a limitation for the study of processes related to sexual reproduction, well-documented in other diatoms [15]. The few examples of molecular studies include the identification, in Thalassiosira weissflogii, of sex-induced genes (SIG) reported to contain epidermal growth factor (EGF)-like domains, putatively encoding for components of stramenopile mastigonemes [23], and investigations of the genetic basis for sex determination, in Seminavis robusta, by linkage mapping [24].
The rapid increase in molecular data for unicellular eukaryotes has made it possible to perform comparative genomic studies to search for genes involved specifically in meiosis, allowing to assess the evolutionary history of the molecular mechanisms underlying the sexual phase. The “meiosis detection toolkit” approach provided evidence for the conservation of several of these genes in eukaryotes [5, 6, 10, 25]. The inventory of meiotic genes includes both genes that are known to play meiosis-specific roles and genes that are required for meiosis, but whose expression and functions are shared with non-meiotic processes (Table 1). Mutations in the first category of genes result in severe disruptions specific to meiosis, without documented effects on non-meiotic functions [26, 27]. Examples of genes included in the second category are genes related to DNA replication, maintenance of chromosome structure (e.g. MCM [28], SMC [29] and RAD21 [30]), as well as genes related to DNA repair (homologs of RAD51 [31], homologs of the bacterial MutS and MutL genes [32]). However, it has been shown that some genes thought to be meiosis-specific may also be conserved in parthenogenetic organisms. The expression of SPO11, the gene responsible for the creation of double strand breaks (DSBs) in homologous chromosomes, and other meiotic genes, was detected in both cyclical and obligate parthenogenetic monogonont rotifers [33] and during parthenogenesis in the microcrustacean Daphnia pulex [25]. SPO11 in Candida albicans, and SPO11, HOP1 and DMC1 in Giardia intestinalis, have been shown to function during parasexual genetic recombination [34, 35]. Likewise, in the haptophyte Emiliania huxleyi, genotypes that appear to have lost the ability to form haploid stages still retain SPO11, DMC1, and RAD51 [36]. Thus, determining how the meiotic toolkit has been conserved in different eukaryotic lineages requires comparison of representatives for which sex and meiosis can be directly observed.
Table 1.
Functional roles of meiotic genes searched in the diatom genomes
| Protein | Role in meiosis |
|---|---|
| DNA replication and chromosome maintenance | |
| Mcm2-7 | Mcm2-7 form hexamer and are involved in DNA replication [28] |
| Mcm8 | Mcm8 and Mcm9 are involved in meiotic recombination [81, 83, 84] |
| Mcm9 | Mcm8 and Mcm9 are involved in meiotic recombination [83, 84] |
| Smc1-Smc3 | Part of sister chromatids cohesin subunit, act as a heterodimer |
| Smc2-Smc4 | Heterodimer, essential for chromosome assembly and segregation, part of core condensing subunits |
| Smc5-Smc6 | Heterodimer, involved in DNA repair and checkpoint response, binds to single stranded DNA (ssDNA) |
| Pds5 | Involved in maintenance of sister chromatid cohesion in late prophase |
| Scc3 | Interacts with cohesin complex Smc1-Smc3 and Rad21/Rec8 and helps in holding cohesin ring together |
| Rec8/Syn1a | Meiotic homolog of Rad21, involved in holding sister chromatids together during meiotic recombination |
| Rad21 | Holds Smc1 and Smc3 together thus holding sister chromatids together during meiosis and mitosis |
| DNA double strand break formation | |
| Spo11-1a | Creates double strand breaks (DSBs) in homologous chromosomes in meiotic recombination |
| Spo11-2a | Creates DSBs in homologous chromosomes in meiotic recombination in plants |
| Spo11-3/Top VIA | Required for endoreduplication of DNA |
| DNA damage sensing and response | |
| Rad50 | DNA binding ATPase, holds broken DNA strands while Mre11 trims DSBs |
| Mre11 | 3′–5′ dsDNA exonuclease and ssDNA endonuclease; trims broken DNA ends after DSBs and hairpins |
| Xrs2/Nbs1 | Component of Mre11-Rad50-Xrs2, involved in homologous recombination and non-homologous end joining [125] |
| Crossover regulation | |
| Mer3a | DNA helicase that unwinds double stranded DNA during meiotic cross over formation [126] |
| Hop1a | Binds to DSBs, component of lateral and axial synaptonemal complex |
| Red1/Asy3a | Component of synaptonemal complex, interacts with Hop1 and facilitates meiosis I chromosome disjunction [48, 49] |
| Zip1/Zyp1a | Transverse filament protein involved in synaptonemal complex formation during meiosis [45, 47] |
| Zip2a,Zip3/Hei10a, Zip4a |
Components of synaptonemal complex required for morphogenesis of the synaptonemal complex [45, 64] |
| Dmc1a | Meiotic member of Rad51-RadA-RecA superfamily of proteins, binds to ssDNA end of DSBs and is involved in inter-homologous recombination |
| Hop2a | Homology search together with Mnd1, works in Dmc1 dependent homology search pathway downstream of Rad51 |
| Mnd1a | Together with Hop2 works in homology searching and is also required in stable DNA heteroduplex |
| Msh4a-Msh5a | Heterodimer, together with Mlh1/Mhl3 heterodimer directs Holliday junction resolution with crossover interference |
| Double-strand break repair (recombinational repair) | |
| Rad51, Xrcc2, Xrcc3 | Mediate homologous pairing and strand invasion, involved in DNA repair mechanisms in mitosis and meiosis. Part of Rad51-RadA-RecA superfamily, exhibit multiple paralogs |
| Rad52 | Binds to ssDNA and initiates homologous recombination, stimulates Rad51 mediated strand invasion |
| Rad1 | 5′-3′ endonuclease, required in meiotic crossing over, functions during nucleotide excision repair |
| Msh2 | Forms heterodimer with Msh3 or Msh6, works in DNA mismatch repair |
| Msh6 | Forms heterodimer with Msh2, works in DNA mismatch repair |
| Mlh1 | DNA mismatch repair protein, forms heterodimers with Mlh2, Mlh3 and Pms1, interacts with Msh2/Msh6 and Msh4/Msh5 |
| Mlh2 | DNA mismatch repair protein, forms heterodimer with Mlh1 |
| Mlh3 | Forms heterodimer with Mlh1, interacts with Msh4/Msh5 to promote meiotic crossovers |
| Pms1 | Forms heterodimer with Mlh1, involved in DNA mismatch repair |
| Mms4/Eme1 | Interacts with Mus81 and is involved in interference insensitive, class II crossovers during meiotic recombination [104] |
| Mus81 | Interacts with Mms4 and is involved in interference insensitive, class II crossovers during meiotic recombination [104] |
| Fancm | DNA helicase required for genome stability, involved in limiting meiotic crossovers [127] |
| Other accessory proteins required during meiosis | |
| Fen1 | Fen1 functions during homologous recombination mediated DNA repair by removing divergent sequences at DNA break ends [128] |
| Exo1 | A double-stranded DNA-specific 5′–3′ exonuclease [129] |
| Dna2 | A conserved DNA nuclease involved in DNA stability [130] |
| Brca1 | Regulates meiotic spindle assembly [131] |
| Brca2 | Involved in DNA damage-induced Rad51 foci formation during meiosis [132] |
The T. pseudonana genome was included in a study assessing phylogenetic distribution of core meiotic proteins [6], however, as mentioned above, this species is currently considered asexual, and further datasets have become available for other species for which sexual reproduction can be controlled in the laboratory, such as members of the Pseudo-nitzschia genus [37, 38].
With the aim to improve the definition of the meiosis toolkit for diatoms, we assembled an expanded list of meiotic genes for eukaryotes [6, 9, 33] and looked for the presence of homologs in five diatom genomes and in transcriptome sequence data. We produced an inventory of putative meiotic genes and combined this information with gene expression data for two sexually reproducing species, demonstrating that their expression is indeed increased during sexual reproduction. Phylogenetic analyses for these genes revealed the presence of multiple paralogs for the RAD51 family, the presence of two diatom homologs of SPO11 and the presence of a single RAD21 gene.
Results
Identification of meiotic genes in diatoms
Homology searches for 60 meiotic proteins [6, 9] were performed in five diatom genomes, those of Thalassiosira pseudonana, a centric species, Phaeodactylum tricornutum, Fragilariopsis cylindrus, Pseudo-nitzschia multiseries and Pseudo-nitzschia multistriata, and in the de novo transcriptome of Seminavis robusta (Table 2), all pennate species. The reference transcriptome for the latter species was produced using data from vegetatively as well as sexually reproducing samples.
Table 2.
Protein, gene model or transcript IDs for the genes involved in meiosis analyzed in this study
| Protein name | Accession numbers of proteins used as query | Thalassiosira pseudonana protein ID | Phaeodactylum tricornutum protein ID | Fragilariopsis cylindrus protein ID | Pseudo-nitzschia multiseries protein ID | Pseudo-nitzschia multistriata gene model ID | Seminavis robusta Transcript ID |
|---|---|---|---|---|---|---|---|
| DNA replication and chromosome maintenance | |||||||
| Mcm2 | NP_001185154.1 | 29936 | 18622 | 204899 | 209470 | 0061270.1 | Semro_comp78811_c0_seq1 |
| Mcm3 | Q9FL33.1 | 34975 | 51597 | 264318 | 318351 | 0004850.1 | Semro_comp50104_c0_seq1 |
| Mcm4 | NP_179236.3 | 269123 | 51412 | 146869 | 203268 | 0078600.1b | Semro_comp82592_c0_seq1 |
| Mcm5 | NP_001189521.1 | 31609 | 11490 | 224321 | 255529 | 0118810.1 | Semro_comp83065_c1_seq1 |
| Mcm6 | AED95141.1 | 26545 | 468 | 193082 | 321321 | 0022580.1 | Semro_comp61600_c0_seq1 |
| Mcm7 | P43299.2 | 262526 | 13243 | 184349 | 243980 | 0109820.1 | Semro_comp70058_c0_seq2 |
| Mcm8 | NP_187577.1 | 261512 | 52561 | 189062 | 213178 | 0068370.1 | Semro_comp79168_c0_seq3 |
| Mcm9 | NP_179021.3 | 37362 | 981 | 156569 | 183315 | 0056900.1 | Semro_comp59174_c0_seq1 |
| Smc1 | AEE79265.1 | 35499 | 25506 | 212269 | 162817 | 0116990.1 | Semro_comp83927_c0_seq1 |
| Smc2 | NP_201047.1 | 1393 | 30352 | 210755 | 191984 | 0096310.1 | Semro_comp61213_c0_seq1 |
| Smc3 | NP_180285.4 | 259020 | 52607 | 208027 | 251818 | 0079810.1 | Semro_comp78328_c0_seq1 |
| Smc4 | AED95695.1 | 42365 | 44165 | 212991 | 144962 | 0030660.1 | Semro_comp76089_c0_seq1 |
| Smc5 | AED92224.1 | 9851 | 54192 | 193562 | 286374 | 0102810.1 | Semro_comp64598_c0_seq1 |
| Smc6 | NP_196383.1 | 1743 | 36853 | 177172 | 165557 | 0090050.1 | Semro_comp65344_c0_seq1 |
| Pds5 | NP_177883.5 | 5929 | 1590 | 136077 | 203285 | 0089060.1 | Semro_comp82484_c0_seq6 |
| Scc3 | AEC10920.1 | 8747 | 51870 | 234878 | 38865 | 0079350.1 | Semro_comp20575_c0_seq1 |
| Rec8/Syn1a | NP_196168.1 | NF | NF | NF | NF | NF | NF |
| Rad21 | NP_851110.1 | 8557 | 44595 | 245879 | 324402 | 0072170.1 | Semro_comp80503_c0_seq6 |
| Double-strand break formation | |||||||
| Spo11-1a | AEE75304.1 | NF | NF | NF | NF | NF | NF |
| Spo11-2a | AEE34178.1 | 263510 | 36531 | 242364 | 156625 | 0108120.1b | Semro_comp74200_c0_seq1 |
| Spo11-3/Top VIA | NP_195902.1 | 42646 | 24838 | 239125 | 251788 | 0081370.1 | Semro_comp59497_c0_seq2 |
| DNA damage sensing and response | |||||||
| Rad50 | AEC08614.1 | 9195 | 51876 | 243939 | 320939 | 0001820.1 | Semro_comp61512_c0_seq1 |
| Mre11 | NP_200237.1 | 34332 | 54699 | 275781 | 233741 | 0086370.1 | Semro_comp82091_c0_seq3 |
| Xrs2/Nbs1 | ABA54896.1 | NF | NF | NF | NF | NF | NF |
| Crossover regulation | |||||||
| Mer3a | AAX14498.1 | 11979 | 39994 | 239915 | 285411 | 0087420.1 | Semro_comp60890_c0_seq1 |
| Hop1/Asy1a | AEE34638.1 | NF | NF | NF | NF | NF | NF |
| Red1/Asy3a | AEC10782.1 | NF | NF | NF | NF | NF | NF |
| Zip1/Zyp1a | AEE30217.1 | NF | NF | NF | NF | NF | NF |
| Zip2a | NP_011265.1 | NF | NF | NF | NF | NF | NF |
| Zip3/Hei10a | NP_175754.2 | NF | NF | NF | NF | NF | NF |
| Zip4a | ABO71664.1 | NF | NF | NF | NF | NF | NF |
| Dmc1a | AAC49617.1 | NF | NF | NF | NF | NF | NF |
| Hop2a | CAF28783.1 | NF | NF | NF | NF | NF | NF |
| Mnd1a | NP_194646.2 | 25513 | 54296 | 273989 | 295346 | 0080640.1 | Semro_comp20014_c0_seq1 |
| Msh4a | AAT70180.1 | 261368 | 51916 | 144820 | 259109 | 0116300.1b | Semro_comp57561_c0_seq2 |
| Msh5a | NP_188683.3 | 16039 | 52173 | 149505 | 183820 | 0023810.1 | Semro_comp80580_c0_seq6 |
| Double-strand break repair (recombinational repair) | |||||||
| Rad51-A | BAE99388.1 | 261303 | 51999 | 165795 (A1) 197408 (A2) | 212272 (A1) 1352 (A2) | 0056780.1 (A1) 0086180.1 (A2) | Semro_comp76648_c0_seq1 |
| Rad51-B | NP_180423.3 | 261577 | 40092 | 241710 | 325273 | 0105810.1 | Semro_comp71219_c0_seq1 |
| Rad51-C | CAC14091.1 | 257784b | 54137 | 201530 | 29459 | 0104040.1 | Semro_comp71710_c0_seq3 |
| Rad51-D | NP_001077479.1 | NF | NF | NF | NF | NF | NF |
| Xrcc2 | NP_201257.2 | NF | NF | NF | NF | NF | NF |
| Xrcc3 | NP_200554.1 | 2081c | 31781 | 242664c | 292867c | comp26486_c0_seq1c | Semro_comp70556_c0_seq2c |
| Rec-A | BAE99388.1 | 267595 | 51425 | 186275 | 166360 | 0063260.1 | Semro_comp77000_c0_seq1 |
| Rad52 | CAA86623.1 | 25447 | 49083 | 238228 | 50181 | 0088620.1 | Semro_comp79910_c0_seq1 |
| Rad1 | Q9LKI5.2 | 22869 | 30908 | 208467 | 230429 | 0074360.1b | Semro_comp82187_c0_seq2 |
| Msh2 | AEE76112.1 | 32661 | 19604 | 159571 | 153636 | 0083140.1 | Semro_comp80478_c0_seq1 |
| Msh6 | NP_001190656.1 | 261781 | 53969 | 212924 | 190397 | 0084190.1 | Semro_comp80580_c0_seq6 |
| Mlh1 | NP_567345.2 | 263509 | 54331 | 136590 | 257081 | 0125040.1 | Semro_comp75421_c0_seq1 |
| Mlh2 | NP 013135.1 | NF | NF | NF | NF | NF | NF |
| Mlh3 | NP_195277.5 | NF | NF | NF | NF | NF | NF |
| Pms1 | AAM00563.1 | 264783 | 14607 | 248102 | 242883 | 0117080.1 | Semro_comp73186_c0_seq1 |
| Mms4/Eme1 | AAF06816.1 | NF | NF | NF | NF | NF | NF |
| Mus81 | NP_194816.2 | NF | 36625 | 241086 | 63674 | 0100930.1b | Semro_comp84506_c0_seq2 |
| Fancm | NP_001185141.1 | 11922 | 47619 | 248113 | 68428 | 0010100.1 | Semro_comp74927_c0_seq1c |
| Accessory proteins required during meiosis | |||||||
| Fen1 | AED93576.1 | 269347 | 48638 | 206746 | 260195 | 0006800.1 | Semro_comp51200_c0_seq1 |
| Exo1 | Q8L6Z7.2 | 4742 | 48206 | 261553 | 110816 | 0067080.1 | Semro_comp61722_c0_seq1 |
| Dna2 | NP_001184943.1 | 10652 | 35426 | 241656 | 326992 | 0027070.1 | Semro_comp83726_c0_seq1 |
| Brca1 | AAO39850.1 | NF | NF | NF | NF | NF | NF |
| Brca2 | AEE81814.1 | 6763 | 36784 | 253990 | 284242 | 0067160.1 | Semro_comp82255_c0_seq4 |
Genes marked with a are genes that do not have known functions outside meiosis. Arabidopsis thaliana meiotic proteins were used as query sequence; whenever the specific query gene was not found/present in A. thaliana, Saccharomyces cerevisiae proteins were used (accession numbers in bold). Protein IDs are given for the diatom genomes available at the JGI portal, gene models IDs are given for the Pseudo-nitzschia multistriata genome and transcripts IDs for Seminavis robusta. Actual gene model IDs for P. multistriata include the prefix PSNMU-V1.4_AUG-EV-PASAV3
bcorrected gene model, NF not found, cgene model might need validation
Of the 60 meiosis-related genes known to play roles in DNA duplication, chromosome maintenance and stability, and DNA repair, 42 were found to be present in all diatom genomes, with the exception of MUS81 endonuclease, which could not be found in the T. pseudonana genome. Of the 15 genes known to be exclusive to meiosis (marked with an "a" in Tables 1 and 2), five were detected in all the diatom genomes and the transcriptome surveyed (Table 2). These genes include SPO11-2, a meiosis-specific gene required for the formation of double-strand breaks (DSBs) in paired chromosome homologs and highly conserved throughout eukaryotic lineages [39, 40]. The other four genes include MND1, whose protein product forms a heterodimer with Hop2 and facilitates Dmc1 dependent crossover formation [41, 42], MSH4 and MSH5, whose products form a complex and are thought to stabilize crossover intermediates [43, 44], and MER3, whose product is thought to function in the synaptonemal complex [45, 46]. The other meiosis-specific SPO11 gene, SPO11-1, and nine more meiosis-specific genes, ZIP1, ZIP2, ZIP3, ZIP4, RED1, HOP1, HOP2, DMC1 and REC8, could not be identified in any of the diatom genomes. Zip1-4, Red1 and Hop1 are known to be involved in formation of the synaptonemal complex [45, 47–49]. Seven other genes (XRS2, RAD51-D, XRCC2, MLH2, MLH3, MMS4 and BRCA1), known to function during DNA damage repair, were not detected in our search (Table 2). In a few cases the gene models retrieved were incomplete, when possible these incorrect gene models were manually corrected (marked with "b" in Table 2).
Phylogenetic analyses
To support the identification of diatom homologs of the meiotic genes analyzed, we created maximum likelihood phylogenetic trees for the 42 meiotic proteins. All the meiotic toolkit proteins of diatoms clustered together with significant bootstrap support and the branching confirmed relatedness with the respective homolog in other eukaryotes (Additional file 1).
The REC-8/RAD21, SPO11 and RAD51 gene families were analyzed in more detail to verify hypotheses on the putative roles of the different paralogs. RAD21-REC8 have interchangeable roles in different organisms, REC8 being generally required for meiosis [50, 51]. A single RAD21-REC8 homolog was identified in each diatom genome (Table 2) and the sequences clustered with the mitotic RAD21 from other eukaryotes (Fig. 1 and Additional file 2).
Fig. 1.
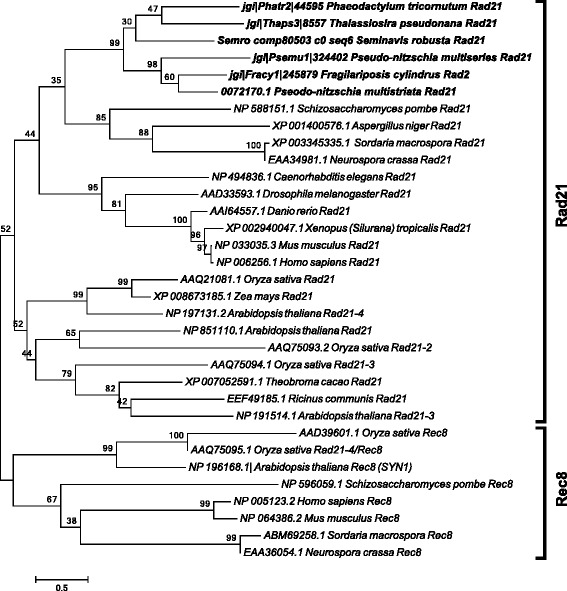
Phylogenetic tree of Rad21 proteins inferred from maximum likelihood analysis. Numbers on the branches indicate bootstrap support from 1000 replicates. Among-site substitution rate heterogeneity was corrected using two gamma-distributed substitution rate categories and WAG with frequencies (WAG + F) substitution model for amino acid substitutions. Diatom sequences are indicated in bold
The SPO11 gene family has a conserved and central role in meiotic recombination [40]. SPO11-1 is required for meiosis in animals. In plants, SPO11-1 and SPO11-2 are the meiosis-specific homologs, whereas SPO11-3/TOP VIA is involved in vegetative growth [27, 52]. Phylogenetic analysis of SPO11 paralogs in diatoms revealed that the two paralogs SPO11-2 and SPO11-3/TOP VIA clustered closely to the respective SPO11 homologs from plants (Fig. 2 and Additional file 3).
Fig. 2.
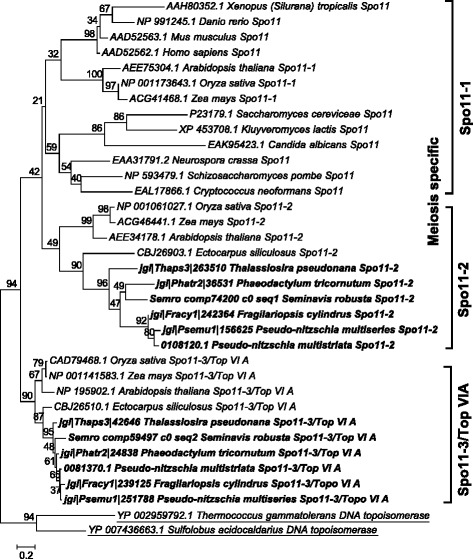
Phylogenetic tree of Spo11 proteins inferred from maximum likelihood analysis. Archaeal topoisomerase VIA protein sequences (underlined) were used as an out-group. Numbers on the branches indicate bootstrap support from 1000 replicates. Among-site substitution rate heterogeneity was corrected using two gamma-distributed substitution rate categories and LG substitution model for amino acid substitutions. Diatom sequences are indicated in bold
RAD51 is an important gene family whose members are employed in homologous recombination during both mitotic and meiotic DNA repair whereas DMC1 functions exclusively during meiosis [53, 54]. None of the diatom Rad51 homologs clustered with Dmc1 representatives from other organisms (Fig. 3). However, in some sexually reproducing organisms, such as Drosophila melanogaster and Caenorhabditis elegans, DMC1 is missing and other RAD51 homologs exert its role [55, 56]. This could thus also be the case in diatoms.
Fig. 3.
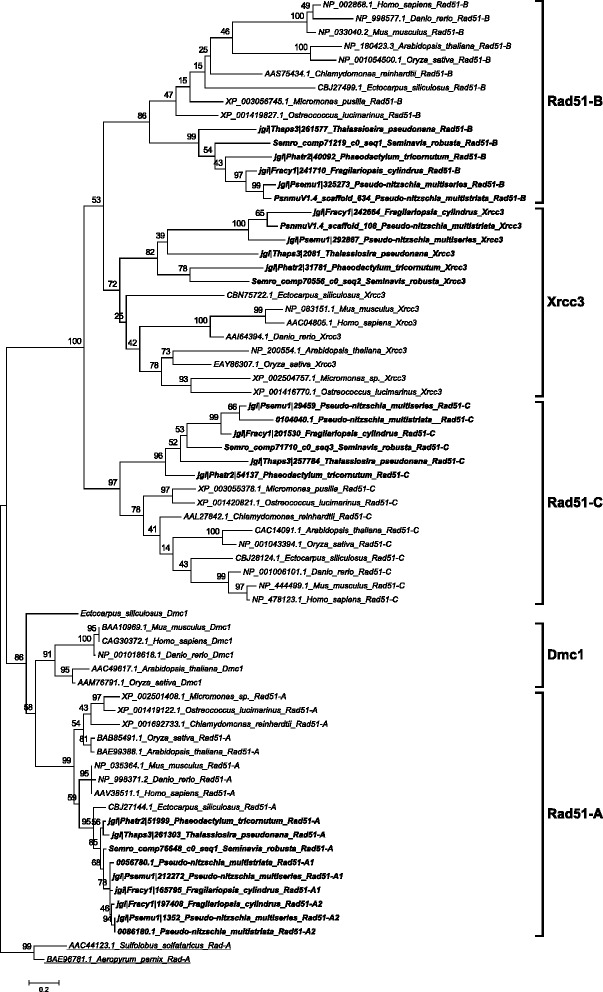
Phylogenetic tree of Rad51 proteins inferred from maximum likelihood analysis. Archaeal Rad-A protein sequences (underlined) were used as an out-group. Numbers on the branches indicate bootstrap support from 1000 replicates. Among-site substitution rate heterogeneity was corrected using two gamma-distributed substitution rate categories and LG substitution model for amino acid substitutions. Diatom sequences are indicated in bold
When searching for homologs of the other members of the RAD51 family in diatom genomes, orthologs could be found for all the sequences except for RAD51-D and XRCC2 (Table 2). The Rec-A/Rad51 domain consists of 230 amino acids and contains two conserved consensus motifs, Walker A and Walker B [57], that are found in ATPases and endow hydrolysis and ATPase activity [58]. This domain was present and complete in the diatom Rad51 proteins. For all the diatom Xrcc3 homologs, excluding the Phaeodactylum tricornutum Xrcc3 protein 31781, the Rad51 domain was predicted with low e-values. A phylogenetic analysis was performed including all diatom Rad51 homologs (Fig. 3 and Additional file 4). From this analysis, we found that the Xrcc3 proteins clustered correctly with the Xrcc3 sequences from other organisms. In addition, we found two copies of RAD51-A in the two Pseudo-nitzschia species and in F. cylindrus, which were named RAD51-A1 and RAD51-A2.
Of the members of the Rec-A/Rad51 family, Rec-A has been reported to be functioning in the chloroplast [59, 60]. An analysis using SignalP 3.0 [61] and ASAFind [62] for the P. multistriata, T. pseudonana, and P. tricornutum protein sequences revealed the presence of a signal peptide for chloroplastic transport (data not shown), supporting the hypothesis of a role for this homolog in the chloroplast rather than in the nucleus.
Gene expression analyses in S. robusta
In the diatom S. robusta, which has a described and controllable sexual phase, the two mating types (MT+ and MT-) can form mating pairs and reproduce sexually once they are below the sexual size threshold, which lies around 50 μm [63]. RNA extracts from synchronized co-cultures of two S. robusta strains of opposite mating type were collected at multiple time points during meiosis (when pairing cells can be observed) and after meiosis (auxosporulation, when the production of an elongated specialized zygote, the auxospore, occurs). The same two strains were also grown as monoclonal cultures and were collected at the same time points (vegetatively growing controls). Gene expression changes between the sexually reproducing and the vegetatively growing cultures were assessed using RNA-seq.
Cpm values were extracted for the transcripts belonging to the meiotic toolkit (excluding REC-A which is supposed to be chloroplastic) and normalized, after which a heatmap was constructed (Fig. 4). For 37 of the transcripts considered in the present analysis, including RAD21, SPO11-2, RAD51-A, RAD51-B and RAD51-C, expression levels were higher during meiosis when compared to the expression levels at the same time after illumination in vegetatively growing monoclonal cultures. In samples obtained from post-meiotic sexual stages, the expression of meiotic genes decreases, as expected (post-meiosis phase in Fig. 4). Interestingly, the putative mitosis-specific homolog SPO11-3/Top VIA appeared to be more abundant during auxosporulation than in other conditions. The XRCC3 homolog was also more expressed during auxosporulation than during meiosis, similarly to MCM6 and MCM7. For MCM2, MCM4 and RAD1, expression was increased during mating compared to the vegetatively growing samples and remained high during auxosporulation.
Fig. 4.
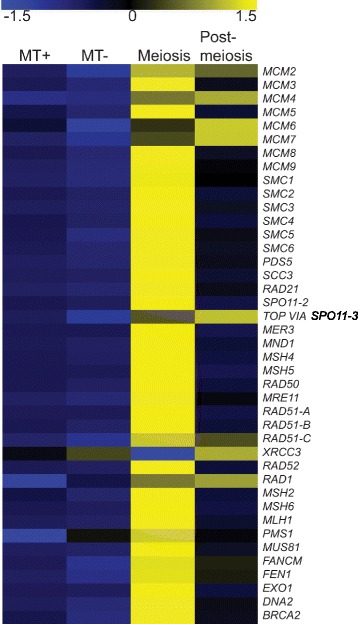
Expression profiles of the meiotic toolkit transcripts found in the transcriptome of Seminavis robusta. Expression values (normalized cpm) were determined for dark-synchronized monoclonal cultures (MT+ and MT-) (7 to 10 h after illumination) and for dark-synchronized mixed MT+ and MT- cultures sampled at the time for meiosis (9-10 h after illumination) and post-meiosis (19–22 h after illumination). Blue colour indicates down-regulation and yellow colour upregulation of expression
Gene expression analyses in P. multistriata
We investigated the gene expression profile of selected meiotic genes at two time points during sexual reproduction in P. multistriata using real-time quantitative PCR (qPCR). Monoclonal cultures of opposite mating type were used as controls. Specifically, RAD21, SPO11 and RAD51 paralogs were selected for expression analyses to assess whether there was an indication of a specific requirement for the only RAD21 homolog and for any of the SPO11 and RAD51 paralogs during meiosis in this species as well. REC8 (the meiotic homolog of RAD21), SPO11 and RAD51 are known to be expressed early in meiotic prophase I [39, 53]. At both time points, the RAD21 transcript was significantly upregulated in the co-cultures with respect to the monoclonal cultures, clearly indicating that this gene functions during meiosis (Fig. 5). Similarly, SPO11-2 showed significant upregulation in its transcript expression in the cultures undergoing sexual reproduction as compared to monoclonal cultures, whereas SPO11-3/TOP VIA did not show any significant change between sexually reproducing cultures and monoclonal cultures (Fig. 5). Although all of the RAD51 homologs examined (RAD51-A1, RAD51-A2, RAD51-B, RAD51-C and XRCC3) showed higher expression during sexual reproduction at both time points investigated (Fig. 5), significant upregulation was observed only for RAD51-A1 and RAD51-C.
Fig. 5.
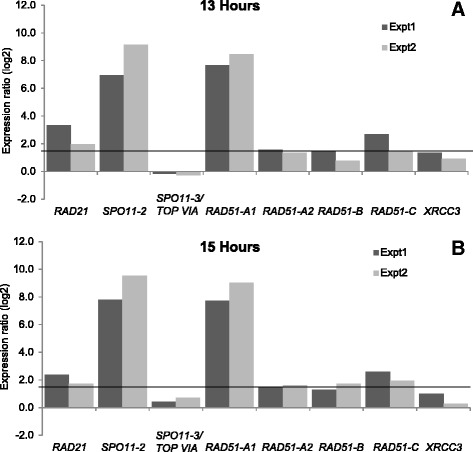
Differential expression analysis of RAD21, SPO11 and RAD51 homologs during meiosis in Pseudo-nitzschia multistriata. Two time points, 13 h (a) and 15 h (b) after strains of opposite mating type were mixed together, were selected for expression analyses. Dark gray bars represent experiment 1 (Expt1, B937 (MT+) with B936 (MT-)) and light gray bars represent experiment 2 (Expt2, B938 (MT+) with B939 (MT-)). Relative fold changes, with respect to vegetatively growing cultures, in log2 scale, are reported on the Y-axis. A gene was considered differentially expressed if its relative expression change is 1.5 fold or greater (horizontal black line)
Discussion
The availability of genomic data from six diatom species with different life cycles has been exploited to define the set of meiotic and meiosis-related genes for this important group of stramenopile microalgae (Additional file 5: Table S1) and guided experiments to define their expression pattern during sexual reproduction in two pennate species. Importantly, a functional differentiation between the two diatom SPO11/TOP VIA homologs can be hypothesized. RAD21 and homologs of RAD51 were also found to be more expressed in cells that were undergoing sexual reproduction in both diatoms, indicating their specific requirement during meiosis.
Genes exclusive to meiosis
Compared to other studies, our list of meiosis-specific genes (Tables 1 and 2) contained three more ZIP (Zipping up meiotic chromosomes protein) genes, ZIP2, ZIP3 and ZIP4 [64], all reported to be required for the synaptonemal complex in budding yeast [47, 64, 65]. Moreover, we list as meiosis-specific both SPO11-1 and SPO11-2, therefore the total number of meiosis-specific genes increases from 11 [9] to 15 (Table 1). Of these 15 genes, five were detected in all the diatom genomes surveyed (Table 2). In the Seminavis robusta dataset, these five genes were all clearly upregulated during mating (Fig. 4), as was the one tested in Pseudo-nitzschia multistriata (SPO11-2, Fig. 5). The role of sex in the ecology and biogeochemical function of natural diatom populations has been challenging to investigate with classical methods as diatom sexual stages are difficult to recognize and preserve poorly [15]. The genes identified in the present study might prove useful as molecular markers to detect events of diatom sexual reproduction in nature.
Ten meiosis-specific genes could not be identified in any of the diatom genomes. The absence of some of these genes has also been reported for other species (Additional file 5: Table S1) and has been correlated with the presence of alternative structures and pathways required during meiotic recombination [66–69].
While duplications of some of the meiotic toolkit genes have been observed in protists and other organisms (Additional file 5: Table S1), the diatom genes were all present in single copies, except for the notable expansion observed in the RAD51 family.
Does Rad21 function as a component of the cohesin complex both during mitosis and meiosis in diatoms and other Stramenopiles?
Although the SMC1, SMC2, SMC3, SMC4, SMC5 and SMC6 genes, whose products are functional components of the cohesin and condensin complexes (required for chromatin organization during cell division), were identified in diatoms, REC8, an important component of cohesin complex and meiosis-specific homolog of RAD21 [50, 70], seemed to be absent in the diatoms investigated (Additional file 5: Table S1). Gene expression analysis of the RAD21 homolog during sexual reproduction in P. multistriata and S. robusta supports the hypothesis that REC8 might be substituted by RAD21, which would function as a component of the cohesin complex both during mitosis and meiosis (Figs. 4 and 5). Indeed, during mammalian meiosis, the mitotic RAD21 cohesin has been shown to perform the role of REC8 [30, 71]. On the contrary, in the ciliate Tetrahymena thermophila that is lacking RAD21, it has been shown that REC8, the meiotic homolog of RAD21, replaces the function of RAD21 during mitosis [72]. The loss of REC8 appears to be a character shared by all Stramenopiles (Additional file 5: Table S1).
SPO11-2 is the meiosis-specific SPO11 paralog
SPO11, encoding a conserved protein involved in DNA DSBs formation and thus in initiation of homologous recombination, was found to have two paralogs in diatoms, SPO11-2 and SPO11-3/TOP VIA (Fig. 2). In many plants, including Arabidopsis thaliana, three paralogs of SPO11 have been reported, of which SPO11-1 and SPO11-2 are meiosis-specific, while SPO11-3/TOP VIA has a topoisomerase function as it interacts with topoisomerase VIB (Top VIB) and is required during vegetative growth [27]. However, most animals, insects and yeasts possess the meiosis-specific SPO11-1 homolog [73] (Additional file 5: Table S1) and lack the other counterpart of topoisomerase assembly, the TOP VIB homolog [73]. In diatoms, red algae and prasinophytes, the SPO11-1 gene seems to be lost, although the SPO11-2 and SPO11-3/TOP VIA homologs are conserved [74, 75]. Although functional differentiation between SPO11-1 and SPO11-2 is not fully resolved in plants, in A. thaliana SPO11-2 mutants the male and female meiosis is severely disrupted, while the mutation does not affect vegetative growth [27]. This suggests that SPO11-2 is involved in meiotic recombination. Similar to many plants, it had been hypothesized that the SPO11-2 homolog in diatoms is involved in meiosis, while the product of SPO11-3/TOP VIA homolog may interact with the Top VIB subunit and might be involved in vegetative growth [27, 73, 74, 76]. The present study demonstrated that SPO11-2 mRNA levels were significantly upregulated during sexual reproduction in P. multistriata and S. robusta, providing the first experimental evidence for the functional distinction between SPO11 paralogs in diatoms. Gene expression studies in the centric diatom Thalassiosira weissflogii also revealed the meiosis-specific role of SPO11-2 (Additional file 6). SPO11-1 is absent in all members of the Stramenopiles analyzed to date, and might have been lost early in the divergence of the alveolate and stramenopile lineages.
Homologous recombination and strand exchange in the absence of DMC1
Archaeal Rad-A homologs Rad51 and Dmc1 work collectively in homology search and strand exchange processes during meiotic recombination, although it is unclear how they cooperate [54, 77]. However, in mitotic cells only Rad51 is induced and carries out recombination [77]. Tsubouchi et al. [78] have proposed two different pathways of homology searching during meiosis. In the first pathway, Dmc1 and Rad51 act together with Hop2 and other accessory proteins to accomplish efficient homology searching. In budding yeast, mutation in the HOP2 gene results in inappropriate homology searching, leading to extensive synaptonemal complex formation between non-homologous chromatids [41, 79]. In this Dmc1-dependent pathway, Hop2 interacts with Mnd1 downstream of Dmc1 and Rad51 homology searching, with Hop2 being a major DNA binding protein and Mnd1 the foremost protein interacting with Rad51 [42, 80]. In the second pathway, only Rad51 is involved in homology searching. The same study [78] also reported that overexpression of RAD51 suppresses defects in DMC1 mutants, indicating Rad51 can carry out effective homology searching independently. Crismani et al. [81] recently reported that in Arabidopsis Rad51 can work together with Mcm8 to repair meiotic double strand breaks when the Dmc1-dependent major repair pathway fails. The Mcm family of DNA helicases consists of nine homologs of which six (Mcm2-7) are conserved across the eukaryotic kingdom and function as heterohexamer helicase in DNA replication [28], whereas the other three (Mcm8-10) are less conserved, with Mcm8-Mcm9 being proved to work during meiotic recombination [82, 83]. Mcm8 and Mcm9 form a stable complex and promote recruitment of Rad51 to the DNA damage sites [83, 84]. Although diatoms lack DMC1 and HOP2 genes, they possess five to six homologs of RAD51 (either one or two homologs of RAD51-A, and one homolog each of RAD51-B, RAD51-C, XRCC3 and REC-A) and MCM8 and MCM9. In A. thaliana, RAD51-C and XRCC3 have been shown to be involved in meiotic recombination [85, 86]. In P. multistriata, we observed that RAD51-A1 and RAD51-C were upregulated during sexual reproduction, and in S. robusta RAD51-A, RAD51-B and RAD51-C appeared to be upregulated during sexual reproduction. Based on the RNA-seq data produced for S. robusta, MCM8 and MCM9 also appeared upregulated during meiosis (Fig. 4). Intriguingly, a homolog of the HOP2 gene, whose protein product forms a heterodimer with Mnd1, could not be found, while MND1 was found in diatoms. Either Hop2 is highly diverged in diatoms and is currently beyond the detection by homology searches or another mechanism involving Mnd1 exists, as the S. robusta homolog is indeed up-regulated during meiosis. Therefore, diatoms may have evolved an alternative meiotic double strand break repair pathway that does not involve Dmc1. Absence of DMC1, HOP2 and MND1 homologs has been reported for certain sexually reproducing organisms such as Caenorhabditis elegans, Drosophila melanogaster and Neurospora crassa (Additional file 5: Table S1) and absence of HOP2 has been reported in Gallus gallus, Phytophthora species, and some fungi [6]. As DMC1 and HOP2 orthologs were identified in the Ectocarpus siliculosus genome but not in sexual diatoms, the losses of a DMC1 dependent DSB repair pathway and of HOP2 might be specific to certain lineages of Stramenopiles and do not correlate with obligate asexuality.
Does cross-over occur without canonical synaptonemal complex (SC) in diatoms?
The synaptonemal complex (SC), a proteinaceous structure, is developed during early prophase I of meiosis and is thought to juxtapose homologous chromatids to enhance crossing over during meiotic recombination [45]. Almost all animals, plants and fungi capable of meiosis possess the SC [87]. The ZMM (Zip, Msh, Mer) group of proteins includes seven functionally similar yet structurally diverse proteins that coordinate recombination events and SC formation during meiosis [45]. Functionally, ZMM proteins can be categorized into three subgroups. Subgroup I includes Mer3 and Msh4-Msh5, subgroup II includes Zip2, Zip3 and Zip4 while subgroup III includes the Zip1 protein. The Zip1 protein produces a stable connection between two homologous chromosomes [47, 88], Zip2, Zip3 and Zip4 facilitate protein-protein interactions [64, 65, 89] and Mer3, Msh4 and Msh5 promote DNA recombination [45, 90]. However, homologs of Zip1 among fungi (Zip1, [47]), animals (Sycp1, [91]) and plants (Zyp1, [88, 92]) are highly divergent. Similarly, the other Zip proteins have functional analogues in fungi (Zip2, Zip3 and Zip4 [64, 65]), animals (Zhp-3/Hei10 [93], Zip4H/Tex11 [94]) and plants (Zip3/Hei10 [95], Zip4 [96]) albeit with very low or no sequence similarity among different taxa. The diatom genomes contained neither identifiable homologs of ZIP genes (Table 2), nor of HOP1 and RED1, the components of lateral elements in SCs (Table 2 and Additional file 5: Table S1) and thus, diatoms may lack canonical SCs. Such a possibility was proposed for ciliates. In ciliate genomes, none of the genes related to canonical SCs were detected [9] and the corresponding absence of a canonical SC in the ciliate Tetrahymena thermophila was also supported by microscopic observations [67, 97]. The presence of a rigid siliceous frustule and dense ring of chromatin around the central spindle at metaphase make it difficult to perform routine cytological studies in diatoms [16, 98], however, in some species SC-like structures have been reported [99, 100], so SC-like structures might involve unidentified proteins that have replaced the Zip and Hop1 protein functions. Alternatively, the homologous proteins are present in diatoms but have diverged so much that they are unrecognizable.
DNA Mismatch repair genes (MSH and MLH gene family)
The MSH gene family comprises homologs of bacterial MutS genes that are important for DNA mismatch recognition and repair (MMR). MSH genes have been reported in all eukaryotes and are fundamentally involved in the initial recognition of nucleotide mismatch during repair [101, 102]. Although seven MSH homologs (MSH1-7) have been identified among eukaryotes, MSH1 and MSH7 are less conserved [102]. Msh proteins form heterodimers, Msh2-Msh6 is principally involved in MMR during mitosis whereas Msh4-Msh5 functions during meiosis [32, 44], stabilizing single strand invasion intermediates formed during early stages of meiotic recombination [90, 103]. Further, it directs Holliday Junction resolution towards crossover formation following an interference sensitive pathway [103, 104]. T-DNA insertional mutation of MSH4 of Arabidopsis exhibits reduced fertility with no effects on normal vegetative growth [103]. MSH2, MSH4, MSH5 and MSH6 were identified in all diatom genomes investigated and they were upregulated during meiosis in S. robusta indicating the presence of a complete and active MMR machinery.
Prokaryotic MutL homologs (Mlh) of DNA MMR proteins are another important group of conserved meiotic genes that work in coordination with Msh homologs. Multiple copies of MLH homologs (MLH1-3 and PMS1-2) are present in eukaryotes [105]. Mlh1 and Pms1/2 form heterodimers and interact with Msh2-Msh4 or Msh4-Msh5 heterodimers to remove DNA mismatches during replication [101, 105, 106]. Msh heterodimers initiate DNA MMR by recognizing and binding to unpaired and impaired bases. In addition, they activate the Mlh complex endonuclease that further incises DNA mismatches [107]. Mlh1-Pms1 is the major heterodimer and in some eukaryotes the Mlh family contains multiple homologs (all of which form heterodimers with Mlh1) [107]. Diatoms contain MLH1 and PMS1 whereas MLH2 and MLH3 were not detected, suggesting that the Mlh1-Pms1 complex plays a major role in MMR in diatoms. The latter hypothesis is supported by the upregulation of both genes during mating in S. robusta. Although MLH2 was not detected in other SAR supergroup members examined, as is the case for diatoms, an MLH3 homolog was detected in E. siliculosus, suggesting that MMR varies among Stramenopiles.
The meiotic toolkit in P. tricornutum and T. pseudonana
Centric and pennate diatoms differ in many aspects of their life cycles, and meiosis also differs in many ways, with a different number of gametes produced in pennates (generally two isogamous gametes) with respect to centrics (one large sized female gamete and many small sized male gametes) [108, 109]. Nevertheless, our results suggest that the molecular machinery employed in meiotic recombination may be shared by all diatom species studied. This includes P. tricornutum and T. pseudonana for which a sexual phase has never been reported. Differences could only be found for the RAD51 family, where P. multistriata, P. multiseries and F. cylindrus appeared to have a duplicated RAD51-A gene, a canonical version of XRCC3 appeared to be present only in P. tricornutum, while T. pseudonana lacked MUS81.
This would suggest that the apparent lack of a sexual phase for P. tricornutum and T. pseudonana is unlikely due to major losses in the meiotic toolkit genes. In spite of the rapid evolution of the genomes of these two diatoms and the presence of a significant amount of transposable elements in their genomes, the meiotic genes are conserved. Since meiotic genes have been reported in the genome of asexual organisms [33, 110], it cannot be excluded that these diatom species are truly asexual, with meiosis-related genes having undergone neo-functionalization and becoming employed in non-meiotic processes such as DNA repair. Moreover, detailed analyses should include an assessment of the integrity of the meiosis-related genes identified, to rule out a recent accumulation of mutations rendering the genes non-functional (i.e., species-specific or even strain-specific loss of sex after isolation in culture). For example, the meiotic SPO11-2 homolog in the T. pseudonana genome appeared to be missing the N-terminal portion of the gene found in other diatoms (Additional file 4). The presence of meiosis related genes in the genome of T. pseudonana, even if some genes may be subject to recent loss-of-function mutations (in the CCMP1335 genome), suggests that some members of the species may have retained this capacity, as seen recently in the coccolithophore Emiliania huxleyi [36]. However, at least for P. tricornutum, which is a pennate diatom (pennate diatoms are generally heterothallic), few strains have been used in laboratories across the world and it could be that the right mating partner has never been used in crossing experiments. This species is both important for fundamental research and promising in biotechnology, and the ability to conduct laboratory breeding would greatly enhance this potential. The isolation of additional wild type P. tricornutum strains should be pursued in order to verify if sexual reproduction can be induced in the laboratory.
Conclusions
Analysis of the meiotic toolkit in diatoms revealed that the majority of meiosis-related genes are present and, in two species tested, showed an expression consistent with their proposed role. However, it seems that not all eukaryotic meiosis-specific genes are required to complete meiosis in diatoms. Specifically, our results suggest the presence of a Dmc1-independent pathway for double strand break repair during meiosis in diatoms. The absence of the genes required for canonical SC formation in diatoms may explain why the SC has not been seen during meiotic divisions. The assignment of specific functional roles to the meiosis-related genes in diatoms, for comparison to roles of homologous proteins in yeasts, plants, and animals, will need further investigation using various approaches, including reverse genetics and protein interaction analyses. More broadly, the presented data refine our knowledge of patterns of evolutionary divergence of meiosis, a fundamental process ancestral to all extant eukaryotes. The SAR supergroup has undergone fundamental modifications to the meiosis process compared to other representatives of both the Diaphoretickes/bikont megaclade (Archaeplastida) and the Amorphea/unikont megaclade (Opisthokonts: animals and fungi). Features common among the SAR members are the absence of Mlh2 and Xrs2/Nbs1 in DNA damage sensing and the loss of components involved in canonical SC formation. Within the Stramenopiles there is also a general trend to lose canonical components in meiotic recombination, some of which have occurred in specific branches, and in some cases the meiosis-specific components may have been replaced by distant homologs with known mitotic functions.
Methods
Culture conditions and strains used
Seminavis robusta strains were grown at 18 °C in a 12 L:12D h (light:dark) regime with cool white fluorescent lamps at approximately 80 μmol photons m−2 s−1. S. robusta strains 85A and 85B used in RNA-seq experiments are publicly available in the diatom culture collection of the Belgian Coordinated Collection of Micro-organisms (BCCM/DCG, http://bccm.belspo.be, accession numbers DCG 0105 and DCG 0107). Pseudo-nitzschia multistriata strains B936 (MT-), B937 (MT+), B938 (MT+) and B939 (MT-) were grown at 18 °C, under 100 μmol photons m−2 s−1 irradiance with 12 L:12D h (light:dark) photoperiod. Cultures were grown in Guillard F/2 medium [111] made with autoclaved filtered natural sea water collected from the North Sea (for S. robusta) or the Gulf of Naples (for P. multistriata) and Guillard’s F/2 solution (Sigma-Aldrich).
Database search for conserved meiotic genes in diatoms
A list of conserved meiotic genes was taken from [6] and expanded with additional genes reported to be involved in meiosis. Meiotic protein sequences of Arabidopsis thaliana or Saccharomyces cerevisiae (Table 2) were used as query sequences for the homology searches. Keyword based searches in the NCBI protein database were made to retrieve the protein sequences. S. cerevisiae proteins were selected when no A. thaliana protein could be found for a given meiotic gene (Table 2). Meiotic protein homologs for four diatom species with publicly available genomes, including Thalassiosira pseudonana v3.0, Phaeodactylum tricornutum v2.0, Fragilariopsis cylindrus v1.0 and Pseudo-nitzschia multiseries v1.0, were retrieved by BLASTp searches from the Joint Genome Institute (JGI) database (http://genome.jgi-psf.org/). In the case of Thalassiosira pseudonana and Phaeodactylum tricornutum genomes, “unmapped sequences” databases (http://genome.jgi.doe.gov/Thaps3_bd/Thaps3_bd.home.html and http://genome.jgi.doe.gov/Phatr2_bd/Phatr2_bd.home.html, respectively) were also searched since a significant portion of their genome sequence is maintained in these additional databases. The diatom homologs were first searched in filtered models and search was further extended to all models only if the respective homolog was not detected in filtered models. Meiotic gene homologs of P. multistriata were retrieved by tBLASTn searches in the v1.4 genome assembly (Ferrante, in preparation). Sequences for the retrieved P. multistriata gene models are given in Additional file 7 and corresponding proteins are given in Additional file 8. The search for the presence of meiotic genes was extended to the de novo transcriptome of S. robusta using tBLASTn searches. Sequences for the S. robusta transcripts are given in Additional file 9. The protein sequences of the resulting transcripts were predicted using Trapid [112] and then manually curated by mapping the transcripts to an in-house draft genome of S. robusta (Vandepoele, De Veylder & Vyverman, in preparation). The resulting protein sequences were blasted (BLASTp) against the Uniprot-Swissprot database to confirm their functional annotation. The resulting protein sequences are given in Additional file 10.
We took into consideration only those sequences showing a BLAST e-value smaller or equal to 1e10−4. The resulting dataset was further manually curated verifying the presence of at least one functional domain using the phmmer search against the UniProtKB sequences with an e-value cutoff of 1e10−4 on the webserver HMMER (http://hmmer.janelia.org/) [113]. For dubious cases, Interpro scan (http://www.ebi.ac.uk/interpro/) and CD-search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?) were performed.
A reciprocal blast was performed to confirm that each retrieved diatom sequence had the corresponding query sequence as top hit when searching against the A. thaliana (or S. cerevisiae) protein database.
An alternative approach to verify absence of genes involved searches using HMMER. Protein sequences (covering major taxa from different eukaryotic groups) from the NCBI protein database were downloaded and aligned using the MUSCLE program [114]. Further, HMM profiles were generated (Additional file 11) using default settings of the HMMBUILD command in the HMMER 3.1b software and these HMM profiles for respective gene families were then used to search against the diatom protein databases (already mentioned above). No additional proteins from any of the diatom genomes could be identified when using this approach. In certain cases Hmmsearch did yield entries but manual inspection of the sequences showed that the resulted protein belonged to other gene families (data not shown).
There is not a defined convention for diatom gene nomenclature, we chose to indicate diatom gene names by capital letters and italics, and proteins in lowercase with a capital first letter, following the convention used for S. cerevisiae.
Phylogenetic analyses
For the phylogenetic analysis of each gene, the corresponding protein sequences from representative taxa of plants, animals, fungi and protists were retrieved from NCBI (http://www.ncbi.nlm.nih.gov/protein/) and JGI with keyword searches and aligned using the sequence alignment software MUSCLE [114]. Maximum likelihood analysis was performed using MEGA 6 (Molecular Evolutionary Genetics Analysis) [115] with appropriate substitution model suggested by the software, specified in the figure legends.
RNA-seq for S. robusta
For the mitotic libraries, S. robusta strains 85A (MT+) and 85B (MT-) with an average cell size below the sexual size threshold (SST) were grown under abovementioned growth conditions and before sampling, the dark period was extended with 12 h to synchronize cells at the G1 phase [116]. After illumination, synchronization was assessed by light microscopy. Pictures were taken using a digital camera connected to a Zeiss Axiovert 40 light microscope and the percentage of dividing cells (distinguished from interphase cells by the newly built cell wall between the two valve-appressed chloroplasts) was counted using cell counter plug-in of the ImageJ software. Cultures were harvested hourly from seven until ten hours post-illumination and cell pellets were frozen in liquid nitrogen and stored at –80 °C until RNA extraction.
For the sexual stages, monoclonal cultures were grown as described above. Three hours before illumination, 85A cell suspensions were added to 85B cultures in dark conditions. Harvesting was done analogous to the vegetative samples at nine and ten hours post-illumination, during which cell-pairing (and thus meiosis) was observed, and at 19, 20 and 21 h, when auxosporulation occurs and thus the meiotic phase is passed.
Total RNA was extracted from each sample using the RNeasy Plant Mini Kit (Qiagen). Cell lysis was achieved by mechanical disruption in 1 mL of RNeasy Lysis buffer (Qiagen) by highest speed agitation with glass/zirconium beads (0.1 mm diameter; Biospec) on a bead mill (Retsch). All other steps for RNA extraction were done according to the manufacturer’s instructions. RNA samples were pooled in equal amounts before sequencing.
Poly-(A) RNA was isolated from 5 μg total RNA using Dynabeads mRNA isolation kit (Invitrogen). Purified RNA was then fragmented using RNA Fragmentation Reagents (Ambion) at 70 °C for 3 mins, targeting fragments range 200–300 bp. Fragmented RNA was purified using Ampure XP beads (Agencourt). Reverse transcription was performed using SuperScript II Reverse Transcription (Invitrogen). Double stranded cDNA fragments were purified and selected for targeted fragments (200–300 bp) using Ampure XP beads. The cDNA was blunt-ended, poly-adenylated, and ligated with library adaptors using Kapa Library Amplification Kit (Kapa Biosystems). Digestion of dUTP was performed using AmpErase UNG (Applied Biosystems) to remove second strand cDNA. Digested cDNA was cleaned up with Ampure XP beads. This was followed by amplification by 10 cycles PCR using Kapa Library Amplification Kit (Kapa Biosystems). The final library was cleaned up with Ampure XP beads. Sequencing was done on the Illumina platform generating paired end reads of 150 bp each.
De novo transcriptome of S. robusta and differential expression analysis
The de novo transcriptome for S. robusta was assembled using RNA-seq data generated in collaboration with the JGI institute (http://www.jgi.doe.gov/) within the project “A deep transcriptomic and genomic investigation of diatom life cycle regulation”. Raw reads are available at http://genome.jgi-psf.org/pages/dynamicOrganismDownload.jsf?organism=SemrobtraphaseII. Libraries used in this study are CYAG (MT+), CYAC (MT-), CYAN (meiosis) and CYAH (post-meiosis).
Raw reads were filtered and trimmed based on quality and adapter inclusion using Trimmomatic [117] with the following parameters: -threads 20 -phred64 ILLUMINACLIP:illumina_adapters.fa:2:40:15 LEADING:5 TRAILING:5 SLIDINGWINDOW:5:20 MINLEN:100. Trimmed and filtered reads were normalized using the normalize_by_kmer_coverage.pl script from the Trinity [118] software (release r2013_08_14) with the following parameters: --seqType fq --JM 240G --max_cov 30 --SS_lib_type RF --JELLY_CPU 24. Assembly was performed using Trinity on the trimmed, filtered and normalized reads with the following parameters: --seqType fq --JM 220G --inchworm_cpu 22 --bflyHeapSpaceInit 22G --bflyHeapSpaceMax 220G --bflyCalculateCPU --CPU 22 --SS_lib_type RF --min_kmer_cov 2 --jaccard_clip. All reads were mapped to the assembled transcriptome using bowtie (version 1) [119] with the following parameters -p 20 -S --chunkmbs 10240 -t --maxins 500 --trim5 20 --trim3 20 --seedlen 20 --tryhard –a. Quantification of the mapping to obtain the number of raw reads mapping on each transcript in each condition was performed using the samtools view, sort, index and idxstats programs with default parameters [120]. Cpm values were calculated for all the genes using R and extracted for the meiosis transcripts, after which a heatmap was constructed using MeV [121].
Experimental set-up for the gene expression studies in P. multistriata
Two experiments were carried out: one (Expt. 1) with P. multistriata strains B936 (MT-) and B937 (MT+) and the other (Expt. 2) with strains B938 (MT+) and B939 (MT-). Exponentially growing cultures were synchronized by incubating them in the dark for 36 h. Monoclonal cultures of MT+ and MT- strains were grown as controls and the same MT+ and MT- strains were mixed together to induce the sexual phase. The timing for collection of samples for RNA was chosen based on earlier observations on the timing of gamete formation: under the specified experimental set up, pairing cells could be observed starting from 10 h after the opposite mating type cells were mixed together and gametes could be observed 24 h after the opposite mating type cells were mixed together (Scalco et al. in press). The samples for RNA were therefore collected from controls and mixed cultures at 13 and 15 h after the start of co-culturing. Mixed cultures and vegetative control samples were collected onto 1.2 μm pore-size membrane filters (RAWP04700 Millipore), placed in Trizol™, flash frozen in liquid nitrogen immediately and stored at -80 °C until RNA extractions. A control plate with the mixed culture was maintained and observed after 24 h of co-culturing to verify that gamete formation had occurred.
RNA was extracted according to the manufacturer’s instructions (Trizol reagent, Invitrogen) and the genomic DNA contamination was removed by DNase I treatment (RNase-Free DNase Set, Qiagen) followed by RNA purification using RNeasy Plant Mini Kit (Qiagen). The quantity of RNA was determined using the Qubit assay (Qubit® 2.0 Fluorometer, Life Technologies) and RNA integrity was assessed by running samples on a 1.5 % agarose gel. One microgram of total RNA was further used for cDNA preparation using the QuantiTect® Reverse Transcription Kit (Qiagen).
RAD21 and homologs of SPO11 and RAD51 genes were retrieved from the genome sequence of P. multistriata and real time qPCR primers were designed manually (Additional file 12). To ensure specificity of the primer to the specific homolog, the homologs were aligned using ClustalX [122] and primers were designed on divergent fragments of the sequence.
The expression profiles of RAD21, SPO11-2, SPO11-3/TopVIA, RAD51-A1, RAD51-A2, RAD51-B, RAD51-C and XRCC3 genes were analyzed using CDK-A and COPA as normalization genes [123]. qPCR amplification was performed as previously described [123]. The results were analyzed and collected in an Excel sheet using the ViiA™ 7 Software. Gene expression analysis was performed on two biological replicates. Each biological sample was run in technical triplicates. Expression analysis was performed using the Relative Expression Software Tool-Multiple Condition Solver (REST-MCS), the calculation software for the relative expression in qPCR, using Pair Wise Fixed Reallocation Randomization Test [124].
Availability of supporting data
The datasets supporting the results of this article are available at http://genome.jgi-psf.org/pages/dynamicOrganismDownload.jsf?organism=SemrobtraphaseII.
Acknowledgements
S.P. has been supported by an Open University PhD fellowship from Stazione Zoologica Anton Dohrn. The project was partially funded by the Marie Curie FP7-PEOPLE-2011-CIG 293887 (GyPSy) to MIF. The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. LDV and WV acknowledge support by grants of the Research Foundation Flanders (G.0288.13) and Ghent University (BOF project GOA 01G01715). M.J.J.H. is a Postdoctoral Fellow of the Research Foundation Flanders. The authors wish to thank Maria Valeria Ruggiero for her help with phylogenetic analyses, Marco Borra for his help with qPCR data analysis, Valerie Devos, Dr. Jeroen Gillard and Dr. M. Vuylsteke for help with sample preparation for the Seminavis RNA-seq experiment and Aurora Storlazzi for her critical comments on the manuscript.
Abbreviations
- DMC1
disrupted meiotic C-DNA1
- DSB
double strand break
- MCM
mini-chromosome maintenance complex
- MMR
DNA mis-match repair
- MT
mating type
- qPCR
quantitative PCR
- SPO11
sporulation-specific protein 11
- SC
synaptonemal complex
- SMC
structural maintenance of chromosomes
- SST
sexual size threshold
- TOP VI
topoisomerase VI
- ZIP
zipping up meiotic chromosomes protein
Additional files
Phylogenetic inferences of meiosis genes searched in diatom genomes. (PDF 862 kb)
Multiple sequence alignment of Rec8/Rad21proteins used in building the phylogenetic tree of Rad21, shown in Fig. 1 . (PDF 894 kb)
Multiple sequence alignment of Spo11 proteins used in building the phylogenetic tree of Spo11, shown in Fig. 2 . (PDF 553 kb)
Multiple sequence alignment of Rad51 proteins used in building the phylogenetic tree of Rad51, shown in Fig. 3 . (PDF 1107 kb)
Overview of phylogenetic distribution of core meiotic proteins among eukaryotes [11, 72, 133–142]. (DOC 235 kb)
Expression profiles of SPO11-2 and SPO11-3/TOP VIA homologs in Thalassiosira weissflogii during spermatogenesis compared to purely asexually-dividing cultures. (PDF 248 kb)
Gene models for meiotic genes from the Pseudo-nitzschia multistriata genome. (DOCX 142 kb)
Protein sequences for meiotic genes from the Pseudo-nitzschia multistriata genome in fasta format. (FASTA 54 kb)
Transcript sequences for meiotic genes in Seminavis robusta . (FAS 108 kb)
Protein sequences for meiotic genes for Seminavis robusta in fasta format. (FAS 37 kb)
HMM profiles used for Hmmsearch. (DOC 90 kb)
List of the genes and corresponding primers tested in Pseudo-nitzschia multistriata with qPCR. (XLSX 13 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SP, MM, PvD, WV and MIF conceived the study, SP, SM and PvD performed the experiments, DM produced the P. multistriata gene models, RS processed the RNA-seq data, SP, SM, MJJH, LvV, RS and MIF analysed the experimental data, SP, PvD and MIF wrote the paper that was discussed and revised by all the authors. All authors read and approved the final manuscript.
Contributor Information
Shrikant Patil, Email: shrikant.patil@szn.it.
Sara Moeys, Email: saramoeys@hotmail.com.
Peter von Dassow, Email: pvondassow@bio.puc.cl.
Marie J. J. Huysman, Email: mahuy@psb.vib-ugent.be
Daniel Mapleson, Email: daniel.mapleson@tgac.ac.uk.
Lieven De Veylder, Email: lieven.deveylder@psb.vib-ugent.be.
Remo Sanges, Email: remo.sanges@szn.it.
Wim Vyverman, Email: wim.vyverman@ugent.be.
Marina Montresor, Email: marina.montresor@szn.it.
Maria Immacolata Ferrante, Phone: +39-081-5833268, Email: mariella.ferrante@szn.it.
References
- 1.Cavalier-Smith T. Origins of the machinery of recombination and sex. Heredity (Edinb) 2002;88:125–41. doi: 10.1038/sj.hdy.6800034. [DOI] [PubMed] [Google Scholar]
- 2.Zimmer C. On the origin of sexual reproduction. Science. 2009;324:1254–6. doi: 10.1126/science.324_1254. [DOI] [PubMed] [Google Scholar]
- 3.Goodenough U, Heitman J. Origins of eukaryotic sexual reproduction. Cold Spring Harb Perspect Biol. 2014;6:a016154. doi: 10.1101/cshperspect.a016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schurko AM, Logsdon JM. Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. BioEssays. 2008;30:579–89. doi: 10.1002/bies.20764. [DOI] [PubMed] [Google Scholar]
- 5.Ramesh MA, Malik SB, Logsdon JM. A phylogenomic inventory of meiotic genes: Evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol. 2005;15:185–91. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Malik SB, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM. An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS One. 2008;3:e2879. doi: 10.1371/journal.pone.0002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parfrey LW, Lahr DJG, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 2011;108:13624–9. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–42. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 9.Chi J, Mahé F, Loidl J, Logsdon J, Dunthorn M. Meiosis gene inventory of four ciliates reveals the prevalence of a synaptonemal complex-independent crossover pathway. Mol Biol Evol. 2014;31:660–72. doi: 10.1093/molbev/mst258. [DOI] [PubMed] [Google Scholar]
- 10.Carr M, Leadbeater BSC, Baldauf SL. Conserved meiotic genes point to sex in the choanoflagellates. J Eukaryot Microbiol. 2010;57:56–62. doi: 10.1111/j.1550-7408.2009.00450.x. [DOI] [PubMed] [Google Scholar]
- 11.Chi J, Parrow MW, Dunthorn M. Cryptic sex in Symbiodinium (alveolata, dinoflagellata) is supported by an inventory of meiotic genes. J Eukaryot Microbiol. 2014;61:322–7. doi: 10.1111/jeu.12110. [DOI] [PubMed] [Google Scholar]
- 12.Field C, Behrenfeld M, Randerson J, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281:237–40. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 13.Nelson DM, Tréguer P, Brzezinski MA, Leynaert A, Quéguiner B. Production and dissolution of biogenic silica in the ocean: Revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem Cycles. 1995;9:359. [Google Scholar]
- 14.Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–93. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chepurnov VA, Mann DG, Sabbe K, Vyverman W. International Review of Cytology. Volume 237. San Diego, CA, USA: Academic; 2004. Experimental studies on sexual reproduction in diatoms; pp. 91–154. [DOI] [PubMed] [Google Scholar]
- 16.Round FE, Crawford RM, Mann DG. The Diatoms: Biology and Morphology of the Genera. Cambridge: Cambridge University Press; 1990. [Google Scholar]
- 17.Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 18.Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–44. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 19.Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C. Transformation of nonselectable reporter genes in marine diatoms. Mar Biotechnol. 1999;1:239–51. doi: 10.1007/pl00011773. [DOI] [PubMed] [Google Scholar]
- 20.Poulsen N, Chesley PM, Kröger N. Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae) J Phycol. 2006;42:1059–65. [Google Scholar]
- 21.Siaut M, Heijde M, Mangogna M, Montsant A, Coesel S, Allen A, et al. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene. 2007;406:23–35. doi: 10.1016/j.gene.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 22.Kooistra WHCF, Gersonde R, Medlin LK, Mann DG. The origin and evolution of the diatoms: Their adaptation to a planktonic existence. In: Falkowski PG, Knoll AH, editors. Evolution of primary producers in the sea/ed. by Paul G. Falkowski, Andrews H. Knoll Amsterdam ; Heidelberg[u.a.] : Elsevier, Academic Press. Burlington: Elsevier Academic Press; 2007. pp. 207–49. [Google Scholar]
- 23.Honda D, Shono T, Kimura K, Fujita S, Iseki M, Makino Y, et al. Homologs of the sexually induced gene 1 (sig1) product constitute the Stramenopile mastigonemes. Protist. 2007;158:77–88. doi: 10.1016/j.protis.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Vanstechelman I, Sabbe K, Vyverman W, Vanormelingen P, Vuylsteke M. Linkage mapping identifies the sex determining region as a single locus in the pennate diatom Seminavis robusta. PLoS One. 2013;8:e60132. doi: 10.1371/journal.pone.0060132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schurko AM, Logsdon JM, Eads BD. Meiosis genes in Daphnia pulex and the role of parthenogenesis in genome evolution. BMC Evol Biol. 2009;9:78. doi: 10.1186/1471-2148-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mlambo G, Coppens I, Kumar N. Aberrant sporogonic development of Dmc1 (a meiotic recombinase) deficient Plasmodium berghei parasites. PLoS One. 2012;7:e52480. doi: 10.1371/journal.pone.0052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stacey NJ, Kuromori T, Azumi Y, Roberts G, Breuer C, Wada T, et al. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 2006;48:206–16. doi: 10.1111/j.1365-313X.2006.02867.x. [DOI] [PubMed] [Google Scholar]
- 28.Lindner K, Gregán J, Montgomery S, Kearsey SE. Essential role of MCM proteins in premeiotic DNA replication. Mol Biol Cell. 2002;13:435–44. doi: 10.1091/mbc.01-11-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strunnikov AV, Jessberger R. Structural maintenance of chromosomes (SMC) proteins: Conserved molecular properties for multiple biological functions. Eur J Biochem. 1999;263:6–13. doi: 10.1046/j.1432-1327.1999.00509.x. [DOI] [PubMed] [Google Scholar]
- 30.Prieto I, Pezzi N, Buesa JM, Kremer L, Barthelemy I, Carreiro C, et al. STAG2 and Rad21 mammalian mitotic cohesins are implicated in meiosis. EMBO Rep. 2002;3:543–50. doi: 10.1093/embo-reports/kvf108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suwaki N, Klare K, Tarsounas M. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin Cell Dev Biol. 2011;22:898–905. doi: 10.1016/j.semcdb.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 32.Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 2003;12:233–46. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 33.Hanson SJ, Schurko AM, Hecox-Lea B, Mark Welch DB, Stelzer CP, Logsdon JM. Inventory and phylogenetic analysis of meiotic genes in monogonont rotifers. J Hered. 2013;104:357–70. doi: 10.1093/jhered/est011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6:1084–97. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carpenter ML, Assaf ZJ, Gourguechon S, Cande WZ. Nuclear inheritance and genetic exchange without meiosis in the binucleate parasite Giardia intestinalis. J Cell Sci. 2012;125:2523–32. doi: 10.1242/jcs.103879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Von Dassow P, John U, Ogata H, Probert I, Bendif EM, Kegel JU, et al. Life-cycle modification in open oceans accounts for genome variability in a cosmopolitan phytoplankton. ISME J. 2015;9:1365–77. doi: 10.1038/ismej.2014.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davidovich NA, Bates SS. Sexual reproduction in the pennate diatoms Pseudo-nitzschia multiseries and P. pseudodelicatissima (Bacillariophyceae) J Phycol. 1998;34:126–37. [Google Scholar]
- 38.D’Alelio D, Amato A, Luedeking A, Montresor M. Sexual and vegetative phases in the planktonic diatom Pseudo-nitzschia multistriata. Harmful Algae. 2009;8:225–32. [Google Scholar]
- 39.Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. In: Egel R, Lankenau D-H, editors. Recombination and Meiosis. Volume 2. Berlin Heidelberg: Springer; 2008. pp. 81–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–84. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 41.Henry JM, Camahort R, Rice DA, Florens L, Swanson SK, Washburn MP, et al. Mnd1/Hop2 facilitates Dmc1-dependent interhomolog crossover formation in meiosis of budding yeast. Mol Cell Biol. 2006;26:2913–23. doi: 10.1128/MCB.26.8.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao W, Saro D, Hammel M, Kwon Y, Xu Y, Rambo RP, et al. Mechanistic insights into the role of Hop2-Mnd1 in meiotic homologous DNA pairing. Nucleic Acids Res. 2014;42:906–17. doi: 10.1093/nar/gkt924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishant KT, Chen C, Shinohara M, Shinohara A, Alani E. Genetic analysis of baker’s yeast Msh4-Msh5 reveals a threshold crossover level for meiotic viability. PLoS Genet. 2010;6:e1001083. doi: 10.1371/journal.pgen.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell. 2004;15:437–51. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 45.Lynn A, Soucek R, Börner GV. ZMM proteins during meiosis: Crossover artists at work. Chromosom Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- 46.Mercier R, Jolivet S, Vezon D, Huppe E, Chelysheva L, Giovanni M, et al. Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3, whereas the other one is not. Curr Biol. 2005;15:692–701. doi: 10.1016/j.cub.2005.02.056. [DOI] [PubMed] [Google Scholar]
- 47.Sym M, Engebrecht JA, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–78. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 48.Rockmill B, Roeder GS. RED1: A yeast gene required for the segregation of chromosomes during the reductional division of meiosis. Proc Natl Acad Sci U S A. 1988;85:6057–61. doi: 10.1073/pnas.85.16.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferdous M, Higgins JD, Osman K, Lambing C, Roitinger E, Mechtler K, et al. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–4. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- 51.Bardhan A. Many functions of the meiotic cohesin. Chromosom Res. 2010;18:909–24. doi: 10.1007/s10577-010-9169-0. [DOI] [PubMed] [Google Scholar]
- 52.Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 53.Shinoharaa A, Shinohara M. Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet Genome Res. 2004;107:201–7. doi: 10.1159/000080598. [DOI] [PubMed] [Google Scholar]
- 54.Kurzbauer M-T, Uanschou C, Chen D, Schlogelhofer P. The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell. 2012;24:2058–70. doi: 10.1105/tpc.112.098459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abdu U, González-Reyes A, Ghabrial A, Schüpbach T. The Drosophila spn-D gene encodes a RAD51C-like protein that is required exclusively during meiosis. Genetics. 2003;165:197–204. doi: 10.1093/genetics/165.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takanami T, Mori A, Takahashi H, Horiuchi S, Higashitani A. Caenorhabditis elegans Ce-rdh-1/rad-51 functions after double-strand break formation of meiotic recombination. Chromosom Res. 2003;11:125–35. doi: 10.1023/a:1022863814686. [DOI] [PubMed] [Google Scholar]
- 57.Lin Z, Kong H, Nei M, Ma H. Origins and evolution of the RecA/RAD51 gene family: Evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci U S A. 2006;103:10328–33. doi: 10.1073/pnas.0604232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–51. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rowan BA, Oldenburg DJ, Bendich AJ. RecA maintains the integrity of chloroplast DNA molecules in Arabidopsis. J Exp Bot. 2010;61:2575–88. doi: 10.1093/jxb/erq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cerutti H, Osman M, Grandoni P, Jagendorf AT. A homolog of Escherichia coli RecA protein in plastids of higher plants. Proc Natl Acad Sci U S A. 1992;89:8068–72. doi: 10.1073/pnas.89.17.8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bendtsen JD, Nielsen H, Von Heijne G, Brunak S. Improved prediction of signal peptides : SignalP 3. 0. J Mol Biol. 2004;340:783–95. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 62.Gruber A, Rocap G, Kroth PG, Armbrust EV, Mock T. Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J. 2015;81:519–28. doi: 10.1111/tpj.12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chepurnov VA, Mann DG, Vyverman W, Sabbe K, Danielidis DB. Sexual reproduction, mating system, and protoplast dynamics of Seminavis (Bacillariophyceae) J Phycol. 2002;38:1004–19. [Google Scholar]
- 64.Perry J, Kleckner N, Börner GV. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc Natl Acad Sci U S A. 2005;102:17594–9. doi: 10.1073/pnas.0508581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsubouchi T, Zhao H, Roeder GS. The Meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev Cell. 2006;10:809–19. doi: 10.1016/j.devcel.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 66.Bähler J, Wyler T, Loidl J, Kohli J. Unusual nuclear structures in meiotic prophase of fission yeast: A cytological analysis. J Cell Biol. 1993;121:241–56. doi: 10.1083/jcb.121.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loidl J, Scherthan H. Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J Cell Sci. 2004;117(Pt 24):5791–801. doi: 10.1242/jcs.01504. [DOI] [PubMed] [Google Scholar]
- 68.Egel-Mitani M, Olson LW, Egel R. Meiosis in Aspergillus nidulans: Another example for lacking synaptonemal complexes in the absence of crossover interference. Hereditas. 2008;97:179–87. doi: 10.1111/j.1601-5223.1982.tb00761.x. [DOI] [PubMed] [Google Scholar]
- 69.Villeneuve AM, Hillers KJ. Whence meiosis? Cell. 2001;106:647–50. doi: 10.1016/s0092-8674(01)00500-1. [DOI] [PubMed] [Google Scholar]
- 70.Strich R. Meiotic DNA Replication. Curr Top Dev Biol. 2004. [DOI] [PubMed]
- 71.Xu H, Beasley M, Verschoor S, Inselman A, Handel MA, McKay MJ. A new role for the mitotic RAD21/SCC1 cohesin in meiotic chromosome cohesion and segregation in the mouse. EMBO Rep. 2004;5:378–84. doi: 10.1038/sj.embor.7400121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Howard-Till RA, Lukaszewicz A, Novatchkova M, Loidl J. A single cohesin complex performs mitotic and meiotic functions in the protist Tetrahymena. PLoS Genet. 2013;9:e1003418. doi: 10.1371/journal.pgen.1003418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Malik SB, Ramesh MA, Hulstrand AM, Logsdon JM. Protist homologs of the meiotic Spo11 gene and topoisomerase VI reveal an evolutionary history of gene duplication and lineage-specific loss. Mol Biol Evol. 2007;24:2827–41. doi: 10.1093/molbev/msm217. [DOI] [PubMed] [Google Scholar]
- 74.Sprink T, Hartung F. The splicing fate of plant SPO11 genes. Front Plant Sci. 2014;5:e00214. doi: 10.3389/fpls.2014.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Worden AZ, Lee J-H, Mock T, Rouzé P, Simmons MP, Aerts AL, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–72. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 76.Hartung F, Puchta H. Molecular characterization of homologues of both subunits A (SPO11) and B of the archaebacterial topoisomerase 6 in plants. Gene. 2001;271:81–6. doi: 10.1016/s0378-1119(01)00496-6. [DOI] [PubMed] [Google Scholar]
- 77.Cloud V, Chan Y-L, Grubb J, Budke B, Bishop DK. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012;337:1222–5. doi: 10.1126/science.1219379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsubouchi H, Roeder GS. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev Cell. 2003;5:915–25. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- 79.Leu JY, Chua PR, Roeder GS. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–86. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- 80.Chi P, San Filippo J, Sehorn MG, Petukhova GV, Sung P. Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 2007;21:1747–57. doi: 10.1101/gad.1563007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crismani W, Portemer V, Froger N, Chelysheva L, Horlow C, Vrielynck N, et al. MCM8 is required for a pathway of meiotic double-strand break repair independent of DMC1 in Arabidopsis thaliana. PLoS Genet. 2013;9:e1003165. doi: 10.1371/journal.pgen.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blanton HL, Radford SJ, McMahan S, Kearney HM, Ibrahim JG, Sekelsky J. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 2005;1:e40. doi: 10.1371/journal.pgen.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lutzmann M, Grey C, Traver S, Ganier O, Maya-Mendoza A, Ranisavljevic N, et al. MCM8- and MCM9-deficient mice reveal gametogenesis defects and genome instability due to impaired homologous recombination. Mol Cell. 2012;47:523–34. doi: 10.1016/j.molcel.2012.05.048. [DOI] [PubMed] [Google Scholar]
- 84.Park J, Long DT, Lee KY, Abbas T, Shibata E, Negishi M, et al. The MCM8-MCM9 complex promotes RAD51 recruitment at DNA damage sites to facilitate homologous recombination. Mol Cell Biol. 2013;33:1632–44. doi: 10.1128/MCB.01503-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abe K, Osakabe K, Nakayama S, Endo M, Tagiri A, Todoriki S, et al. Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis. Plant Physiol. 2005;139:896–908. doi: 10.1104/pp.105.065243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bleuyard JY, White CI. The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 2004;23:439–49. doi: 10.1038/sj.emboj.7600055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Von Wettstein D, Rasmussen SW, Holm PB. The synaptonemal complex in genetic segregation. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- 88.Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FCH. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 2005;19:2488–500. doi: 10.1101/gad.354705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–55. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- 90.Hoffmann ER, Borts RH. Meiotic recombination intermediates and mismatch repair proteins. Cytogenet Genome Res. 2004;107:232–48. doi: 10.1159/000080601. [DOI] [PubMed] [Google Scholar]
- 91.De Vries FAT, de Boer E, van den Bosch M, Baarends WM, Ooms M, Yuan L, et al. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 2005;19:1376–89. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Osman K, Sanchez-Moran E, Higgins JD, Jones GH, Franklin FCH. Chromosome synapsis in Arabidopsis: Analysis of the transverse filament protein ZYP1 reveals novel functions for the synaptonemal complex. Chromosoma. 2006;115:212–9. doi: 10.1007/s00412-005-0042-4. [DOI] [PubMed] [Google Scholar]
- 93.Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 2008;4:e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adelman CA, Petrini JHJ. ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet. 2008;4:e1000042. doi: 10.1371/journal.pgen.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chelysheva L, Vezon D, Chambon A, Gendrot G, Pereira L, Lemhemdi A, et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 2012;8:e1002799. doi: 10.1371/journal.pgen.1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chelysheva L, Gendrot G, Vezon D, Doutriaux M-P, Mercier R, Grelon M. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 2007;3:e83. doi: 10.1371/journal.pgen.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mochizuki K, Novatchkova M, Loidl J. DNA double-strand breaks, but not crossovers, are required for the reorganization of meiotic nuclei in Tetrahymena. J Cell Sci. 2008;121(Pt 13):2148–58. doi: 10.1242/jcs.031799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mann DG, Stickle AJ. Meiosis, nuclear cyclosis, and auxospore formation in Navicula sensu stricto (Bacillariophyceae) Br Phycol J. 1989;24:167–81. [Google Scholar]
- 99.Manton I, Kowallik K, Von Stosch HA. Observations on the fine structure and development of the spindle at mitosis and meiosis in a marine centric diatom (Lithodesmium undulatum). I. Preliminary survey of mitosis in spermatogonia. J Microsc. 1969;89:295–320. doi: 10.1111/j.1365-2818.1969.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 100.Drum RW. Electron microscope observations of diatoms. Österreichische Bot Zeitschrift. 1969;116:321–30. [Google Scholar]
- 101.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–42. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 102.Eisen JA. A phylogenomic study of the MutS family of proteins. Nucleic Acids Res. 1998;26:4291–300. doi: 10.1093/nar/26.18.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Higgins JD, Armstrong SJ, Franklin FCH, Jones GH. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev. 2004;18:2557–70. doi: 10.1101/gad.317504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.De los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The MUS81/MMS4 endonuclease acts independently of double-holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 106.Srivatsan A, Bowen N, Kolodner RD. Mispair-specific recruitment of the Mlh1-Pms1 complex identifies repair substrates of the Saccharomyces cerevisiae Msh2-Msh3 complex. J Biol Chem. 2014;289:9352–64. doi: 10.1074/jbc.M114.552190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marti TM, Kunz C, Fleck O. DNA mismatch repair and mutation avoidance pathways. J Cell Physiol. 2002;191:28–41. doi: 10.1002/jcp.10077. [DOI] [PubMed] [Google Scholar]
- 108.Amato A. Diatom reproductive biology: Living in a crystal cage. Int J Plant Reprod Biol. 2010;2:1–10. [Google Scholar]
- 109.Mizuno M. Evolution of centric diatoms inferred from patterns of oogenesis and spermatogenesis. Phycol Res. 2008;56:156–65. [Google Scholar]
- 110.Flot JF, Hespeels B, Li X, Noel B, Arkhipova I, Danchin EG, et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature. 2013;500:453–7. doi: 10.1038/nature12326. [DOI] [PubMed] [Google Scholar]
- 111.Guillard RRL. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH, editors. Culture of Marine Invertebrate Animals. New York: Plenum Press; 1975. pp. 29–60. [Google Scholar]
- 112.Van Bel M, Proost S, Van Neste C, Deforce D, Van de Peer Y, Vandepoele K. TRAPID: An efficient online tool for the functional and comparative analysis of de novo RNA-Seq transcriptomes. Genome Biol. 2013;14:R134. doi: 10.1186/gb-2013-14-12-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gillard J, Devos V, Huysman MJJ, De Veylder L, D’Hondt S, Martens C, et al. Physiological and transcriptomic evidence for a close coupling between chloroplast ontogeny and cell cycle progression in the pennate diatom Seminavis robusta. Plant Physiol. 2008;148:1394–411. doi: 10.1104/pp.108.122176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Howe EA, Sinha R, Schlauch D, Quackenbush J. RNA-Seq analysis in MeV. Bioinformatics. 2011;27:3209–10. doi: 10.1093/bioinformatics/btr490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 123.Adelfi MG, Borra M, Sanges R, Montresor M, Fontana A, Ferrante MI. Selection and validation of reference genes for qPCR analysis in the pennate diatoms Pseudo-nitzschia multistriata and P. arenysensis. J Exp Mar Bio Ecol. 2014;451:74–81. [Google Scholar]
- 124.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Matsuzaki K, Shinohara A, Shinohara M. Forkhead-associated domain of yeast Xrs2, a homolog of human Nbs1, promotes nonhomologous end joining through interaction with a ligase IV partner protein, Lif1. Genetics. 2008;179:213–25. doi: 10.1534/genetics.107.079236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nakagawa T, Kolodner RD. Saccharomyces cerevisiae Mer3 is a DNA helicase involved in meiotic crossing over. Mol Cell Biol. 2002;22:3281–91. doi: 10.1128/MCB.22.10.3281-3291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, et al. FANCM limits meiotic crossovers. Science. 2012;336:1588–90. doi: 10.1126/science.1220381. [DOI] [PubMed] [Google Scholar]
- 128.Kikuchi K, Taniguchi Y, Hatanaka A, Sonoda E, Hochegger H, Adachi N, et al. Fen-1 facilitates homologous recombination by removing divergent sequences at DNA break ends. Mol Cell Biol. 2005;25:6948–55. doi: 10.1128/MCB.25.16.6948-6955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, et al. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci U S A. 1997;94:7487–92. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, et al. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol. 2009;29:4274–82. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xiong B, Li S, Ai JS, Yin S, Ouyang YC, Sun SC, et al. BRCA1 is required for meiotic spindle assembly and spindle assembly checkpoint activation in mouse oocytes. Biol Reprod. 2008;79:718–26. doi: 10.1095/biolreprod.108.069641. [DOI] [PubMed] [Google Scholar]
- 132.Badie S, Escandell JM, Bouwman P, Carlos AR, Thanasoula M, Gallardo MM, et al. BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol. 2010;17:1461–9. doi: 10.1038/nsmb.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Radakovits R, Jinkerson RE, Fuerstenberg SI, Tae H, Settlage RE, Boore JL, et al. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat Commun. 2012;3:686. doi: 10.1038/ncomms1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schwarzacher T. Meiosis, recombination and chromosomes: a review of gene isolation and fluorescent in situ hybridization data in plants. J Exp Bot. 2003;54:11–23. doi: 10.1093/jxb/erg042. [DOI] [PubMed] [Google Scholar]
- 135.Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FCH. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 2011;190:523–44. doi: 10.1111/j.1469-8137.2011.03665.x. [DOI] [PubMed] [Google Scholar]
- 136.Luo Q, Li Y, Shen Y, Cheng Z. Ten years of gene discovery for meiotic event control in rice. J Genet Genomics. 2014;41:125–37. doi: 10.1016/j.jgg.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 137.Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 138.Andrews J, Bouffard GG, Cheadle C, Lü J, Becker KG, Oliver B. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 2000;10:2030–43. doi: 10.1101/gr.10.12.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Garcia-Muse T, Boulton SJ. Meiotic recombination in Caenorhabditis elegans. Chromosom Res. 2007;15:607–21. doi: 10.1007/s10577-007-1146-x. [DOI] [PubMed] [Google Scholar]
- 140.Winter E. The Sum1/Ndt80 transcriptional switch and commitment to meiosis in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2012;76:1–15. doi: 10.1128/MMBR.05010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shodhan A, Lukaszewicz A, Novatchkova M, Loidl J. Msh4 and Msh5 function in SC-independent chiasma formation during the streamlined meiosis of Tetrahymena. Genetics. 2014;198:983–93. doi: 10.1534/genetics.114.169698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Howard-Till RA, Lukaszewicz A, Loidl J. The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. PLoS Genet. 2011;7:e1001359. doi: 10.1371/journal.pgen.1001359. [DOI] [PMC free article] [PubMed] [Google Scholar]


