Brien et al. show that while Polycomb-like proteins PCL2 and PCL3 are E2F-regulated genes expressed in proliferating cells, PCL1 is a p53 target gene predominantly expressed in quiescent cells. PCL1 binds to and stabilizes p53 to block cellular proliferation, and depletion of PCL1 phenocopies the defects in maintaining cellular quiescence associated with p53 loss.
Keywords: Polycomb-like, cellular senescence, p53, cellular quiescence, PHD reader domain, neofunctionalization
Abstract
Polycomb-like proteins 1–3 (PCL1–3) are substoichiometric components of the Polycomb-repressive complex 2 (PRC2) that are essential for association of the complex with chromatin. However, it remains unclear why three proteins with such apparent functional redundancy exist in mammals. Here we characterize their divergent roles in both positively and negatively regulating cellular proliferation. We show that while PCL2 and PCL3 are E2F-regulated genes expressed in proliferating cells, PCL1 is a p53 target gene predominantly expressed in quiescent cells. Ectopic expression of any PCL protein recruits PRC2 to repress the INK4A gene; however, only PCL2 and PCL3 confer an INK4A-dependent proliferative advantage. Remarkably, PCL1 has evolved a PRC2- and chromatin-independent function to negatively regulate proliferation. We show that PCL1 binds to and stabilizes p53 to induce cellular quiescence. Moreover, depletion of PCL1 phenocopies the defects in maintaining cellular quiescence associated with p53 loss. This newly evolved function is achieved by the binding of the PCL1 N-terminal PHD domain to the C-terminal domain of p53 through two unique serine residues, which were acquired during recent vertebrate evolution. This study illustrates the functional bifurcation of PCL proteins, which act in both a chromatin-dependent and a chromatin-independent manner to regulate the INK4A and p53 pathways.
Polycomb group proteins are chromatin-associated gene repressors with essential roles in embryonic development, stem cell differentiation, and cellular proliferation (Sauvageau and Sauvageau 2010; Lanzuolo and Orlando 2012). They were identified in Drosophila melanogaster based on their requirement for maintaining the repressed state of homeotic genes during embryonic development (Di Croce and Helin 2013). Subsequent genome-wide binding studies in both D. melanogaster and mammalian cells confirmed that Polycombs directly bind homeotic gene loci in addition to a broader cohort of developmental genes (Boyer et al. 2006; Bracken 2006; Lee et al. 2006; Tolhuis et al. 2006). Polycombs form multiprotein complexes, the best characterized of which are Polycomb-repressive complex 1 (PRC1) and PRC2 (Simon and Kingston 2013). The PRC1 complex is defined by a heterodimeric RING-PCGF core that mediates the monoubiquitination of histone H2A at Lys119 (Gao et al. 2012; Tavares et al. 2012), while the PRC2 complex is composed of a trimeric core of SUZ12, EED, and EZH1/2 and methylates histone H3 at Lys27 (Margueron and Reinberg 2011; Ferrari et al. 2014). Recent genetic evidence indicates that the function of the PRC2 complex is deregulated in multiple cancer types (Hock 2012; Helin and Dhanak 2013). However, it is unclear whether PRC2 deregulation contributes to cancer progression by disrupting stem cell differentiation, cellular proliferation, or both.
PRC1 and PRC2 have a well-established role in the regulation of cellular proliferation via their ability to confer chromatin-mediated repression of the INK4A gene locus (Jacobs et al. 1999; Lessard and Sauvageau 2003; Park et al. 2003; Gil et al. 2004; Bracken et al. 2007; Maertens et al. 2009). The INK4A gene encodes the p16 protein, a cyclin-dependent kinase (CDK) inhibitor and key upstream regulator of cellular senescence (Collado et al. 2007; Lanigan et al. 2011). The ectopic expression of the PRC1 complex components BMI1 (Jacobs et al. 1999; Bracken et al. 2007), CBX7 (Gil et al. 2004), CBX8 (Dietrich et al. 2007), and KDM2B (Tzatsos et al. 2009) confers a proliferative advantage to primary cells due to sustained repression of INK4A. Accordingly, deletion of Bmi1, Eed, or Ezh1 results in derepression of Ink4a and consequent stem and progenitor cell senescence (Lessard and Sauvageau 2003; Molofsky et al. 2003; Park et al. 2003; Hidalgo et al. 2012; Xie et al. 2014). However, it is also clear that Polycombs can regulate cellular proliferation independently of INK4A repression (Bruggeman et al. 2007; Datta et al. 2007; Douglas et al. 2008; Piunti et al. 2014).
Polycombs have emerging, albeit less well-characterized, roles in the control of cellular proliferation independent of chromatin association. For example, in D. melanogaster, the mammalian PCGF homolog Psc ubiquitinates the mitotic regulator Cyclin-B, thereby mediating its proteasomal degradation (Mohd-Sarip et al. 2012). This interaction is essential for normal cell cycle progression, as loss of Psc leads to an accumulation of cells undergoing abnormal mitoses. In mammals, the PCGF1–6 proteins have not been ascribed a similar chromatin-independent role, likely because they lack the extended C-terminal region of Psc needed to interact with Cyclin-B. Interestingly, the mammalian SCML2 gene encodes two isoforms: the chromatin- and PRC1-associated SCML2A and the predominantly nucleoplasmic SCML2B. SCML2B associates with CDK/CYCLIN/p21 and p27 complexes to enhance the function of p21/p27, thereby reducing progression through the cell cycle (Lecona et al. 2013). These observations indicate that Polycombs may play a broad role in cellular proliferation control beyond their canonical function as chromatin-associated gene repressors. However, currently, no component of the PRC2 complex has been linked with a chromatin-independent role in the control of cellular proliferation.
The Polycomb-like (PCL) proteins PCL1–3 (also known as PHF1, MTF2, and PHF19, respectively) are substoichiometric PRC2 complex components that are required for the maintenance and de novo recruitment of PRC2 to chromatin (Ballaré et al. 2012; Brien et al. 2012; Di Croce and Helin 2013). We and others demonstrated that the Tudor domains of PCL1–3 bind to H3K27me3 and H3K36me2/3 in vitro and are essential for the maintenance of PRC2 on chromatin in embryonic stem cells and its recruitment to de novo sites during differentiation (Ballaré et al. 2012; Brien et al. 2012; Musselman et al. 2012; Cai et al. 2013). However, despite being indispensible for PRC2 function, the potential roles of PCL proteins in the control of cellular proliferation have yet to be addressed.
Here we demonstrate the functional bifurcation of PCL1–3 in regulating cellular proliferation. First, we show that while PCL2 and PCL3 are transcriptionally regulated by the E2F pathway, PCL1 is a direct target gene of p53 and is highly expressed in noncycling, quiescent cells. While all three PCL proteins recruit PRC2 to directly repress INK4A, only PCL2 and PCL3 confer an INK4A-dependent proliferative advantage. In contrast to PCL2 and PCL3, we show that PCL1 has an INK4A-, PRC2-, and chromatin-independent role in mediating a p53-dependent growth arrest. We also show that the N-terminal PHD domain of PCL1 has diverged functionally from that of PCL2 and PCL3 by using two unique serine residues to physically interact with the p53 C-terminal domain (CTD). As reported previously, the interaction of PCL1 with p53 leads to stabilization of p53 protein levels and increased expression of p53 target genes (Yang et al. 2013); however, we further show that this increase in p53 levels is associated with induction of cellular quiescence. Conversely, depletion of PCL1 in quiescent cells phenocopies p53 loss, leading to a failure to maintain cellular quiescence. Taken together, these data reveal that, while all three PCL proteins regulate the INK4A pathway via a chromatin-dependent mechanism, PCL1 has evolved a PRC2- and chromatin-independent role in the regulation of the p53 pathway and cellular quiescence.
Results
PCL gene expression is divergently regulated in cycling and noncycling cells
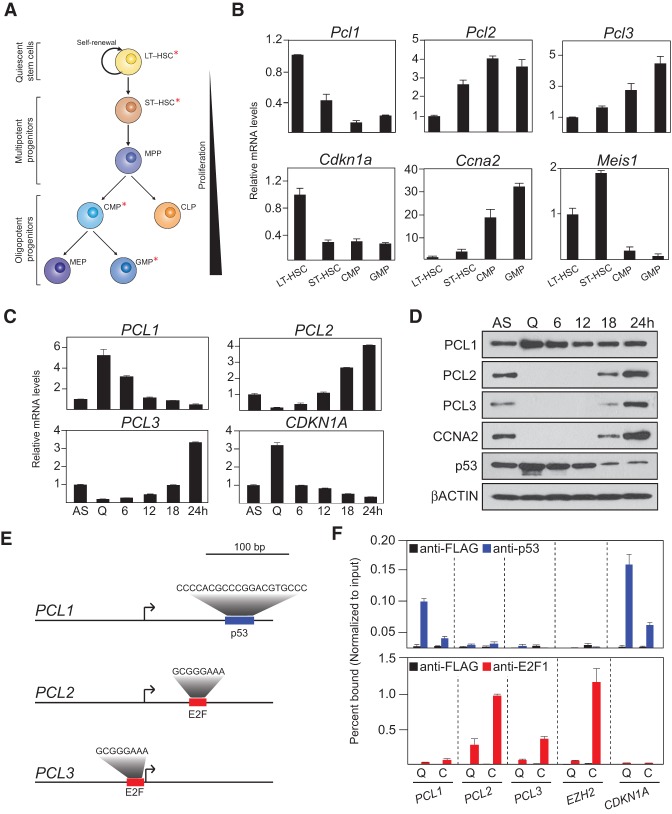
To begin to understand why three PCL genes exist in mammals, we examined the mRNA expression of all three genes in FACS-purified populations from the mouse hematopoietic system (Fig. 1A,B). This demonstrated that Pcl1 is most highly expressed in quiescent (noncycling) long-term hematopoietic stem cells (LT-HSCs), while its expression is sharply reduced in the proliferating common myeloid progenitor (CMP) and granulocyte macrophage progenitor (GMP) populations (Fig. 1B). Conversely, Pcl2 and Pcl3 are both expressed at low levels in LT-HSCs and are induced in the proliferating progenitor populations. Importantly, the proliferative status and identity of these sorted cells were confirmed by analyzing the expression of the cell cycle regulators Cdkn1a and Ccna2 and the HSC marker Meis1 (Fig. 1B). Next, we sought to ascertain whether the expression of human PCL1–3 is similarly associated with cellular proliferation. To do this, we monitored the expression of PCL1–3 in asynchronously growing and quiescent human diploid fibroblasts (HDFs). HDFs were serum-starved for 96 h, resulting in cell cycle exit, as evidenced by the absence of Cyclin A2 protein and increased CDKN1A mRNA levels (Fig. 1C,D). Quantitative RT–PCR and Western blot analyses demonstrated that PCL1, but not PCL2/3, is present at both the mRNA and protein level in quiescent cells. Stimulation of quiescent HDFs to re-enter the cell cycle led to increases in the mRNA and protein levels of PCL2/3 and decreases in the mRNA levels of PCL1 and CDKN1A, accompanied by a decrease in the protein levels of p53 protein (Fig. 1C,D). Taken together, these data indicate that the mammalian PCL proteins are divergently expressed in cycling and noncycling cells.
Figure 1.
PCL1 is a p53 target gene highly expressed in quiescent cells. (A) Schematic representation of the mouse hematopoietic differentiation hierarchy. Asterisks indicate the largely quiescent long-term and short-term HSC (LT/ST-HSC) populations and the highly proliferative CMP and GMP cell populations. (B) Quantitative RT–PCR analyses of the indicated mRNA transcripts in the highlighted (*) in vivo FACS-purified populations. (C) Quantitative RT–PCR analyses of the indicated mRNA transcripts in asynchronously (AS) growing HDFs or the same cells following 96 h of serum starvation (quiescent [Q]). Starved cells were further induced to re-enter the cell cycle following addition of 20% serum for the indicated time points up to 24 h. (D) Western blot analyses using the indicated antibodies on whole-cell protein lysates from the cells in C. (E) Schematic representations of the PCL1, PCL2, and PCL3 promoters indicating the presence of a p53 consensus binding site in the PCL1 promoter and consensus E2F-binding sites in the PCL2 and PCL3 promoters. The Jaspar program (Sandelin et al. 2004) identified consensus E2F sites in the PCL2 and PCL3 promoters but not in the PCL1 promoter. The TRANSFAC program (Matys et al. 2006) identified a consensus p53 site in the PCL1 promoter but not in the PCL2 or PCL3 promoters. (F) Quantitative chromatin immunoprecipitation (ChIP) analyses using the indicated antibodies in HDFs following 96 h of serum starvation (quiescent [Q]) or the same cells following treatment with 20% serum for 24 h (C). Precipitated DNA was analyzed by quantitative PCR using primers directed toward the promoters of the indicated genes. ChIP enrichments are presented as the percentage of protein bound normalized to input. The data represent the average of three biological replicates.
Next, transcription factor-binding motif analysis identified consensus E2F-binding sites in the PCL2 and PCL3 gene promoters and a motif with a high degree of similarity to the consensus p53-binding site in the PCL1 promoter, which could potentially explain their divergent transcriptional regulation (Fig. 1E). Chromatin immunoprecipitation (ChIP) analyses for E2F factors and p53 in quiescent and cycling HDFs demonstrated that the promoters of PCL2 and PCL3 were strongly bound by E2F1 (an activating E2F transcription factor) in cycling HDFs and E2F4 (a repressive E2F) in quiescent HDFs (Fig. 1F; Supplemental Fig. S1), comparable with EZH2, which we previously reported as an E2F-regulated gene (Bracken et al. 2003). In contrast, the PCL1 promoter was bound by p53 in quiescent HDFs, albeit to a lesser extent than the CDKN1A promoter, which contains a perfect consensus p53-binding site. Taken together, these data demonstrate that the divergent expression of PCL1, compared with PCL2–3 in primary cells, is regulated by the p53 and E2F pathways, respectively.
PCL proteins execute divergent roles in cellular proliferation control
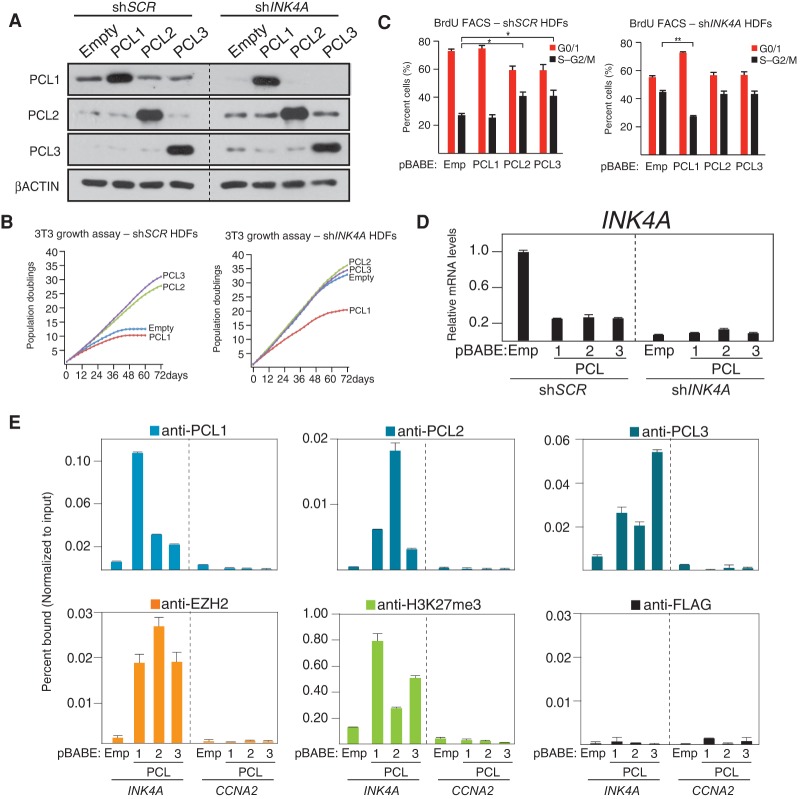
To investigate the potential divergent roles of PCL proteins in cell growth control, we ectopically expressed all three proteins in HDFs. Interestingly, while ectopic expression of PCL2 and PCL3 promoted cellular proliferation and delayed the onset of cellular senescence, ectopic expression of PCL1 did not (Supplemental Fig. S2; data not shown). In order to ascertain whether the observed growth advantage in PCL2- and PCL3-expressing cells was dependent on p16INK4A, we ectopically expressed PCL1–3 in HDFs expressing either scrambled (shSCR)-specific or INK4A-specific (shINK4A) shRNAs (Fig. 2A). This demonstrated that the growth advantage conferred by PCL2/3 was completely dependent on p16INK4A (Fig. 2B). Strikingly, ectopic expression of PCL1 in shINK4A HDFs led to a significant reduction in the rate of cellular proliferation (Fig. 2B), which was associated with an increased proportion of cells in G0/G1 phase of the cell cycle (Fig. 2C). This suggests that PCL1 plays a role independent of INK4A to negatively affect cellular proliferation. Despite this difference, the ectopic expression of all three PCL proteins led to repression of p16INK4A (Fig. 2D), and ChIP analyses demonstrated that all three proteins localized to the INK4A gene promoter, which correlated with recruitment of EZH2 and increased levels of H3K27me3 (Fig. 2E). Taken together, these data suggest that while all three PCL proteins recruit PRC2 to repress INK4A, PCL1 has a divergent role in the control of cellular proliferation.
Figure 2.
An anti-proliferative role for PCL1 independent of INK4A. (A) Western blot analyses using the indicated antibodies of whole-cell protein lysates prepared from HDFs infected with the indicated pBABE retrovirus at population doubling 32 (PD32), where cells were previously infected with either control (shSCR) or INK4A targeting (shINK4A) pLKO lentivirus at PD28. Westerns were performed at PD48. (B) 3T3 growth assays performed on cells from A. Assays were initiated at PD36 and continued for 72 d or until the control shSCR cells became senescent. (C) Quantitative cell cycle BrdU FACS analyses of cells from B, performed at day 48 of the 3T3 assay. Analyses were performed on two independent biological replicates. (D) Quantitative RT–PCR analysis of the INK4A mRNA in cells from C. (E) Quantitative ChIP analyses using the indicated antibodies on cells from C. ChIPs were performed at PD48. Precipitated DNA was analyzed by quantitative PCR using primers directed toward the promoters of the indicated genes. ChIP enrichments are presented as the percentage of protein bound normalized to input.
PCL1 specifically binds and stabilizes p53 to block cellular proliferation
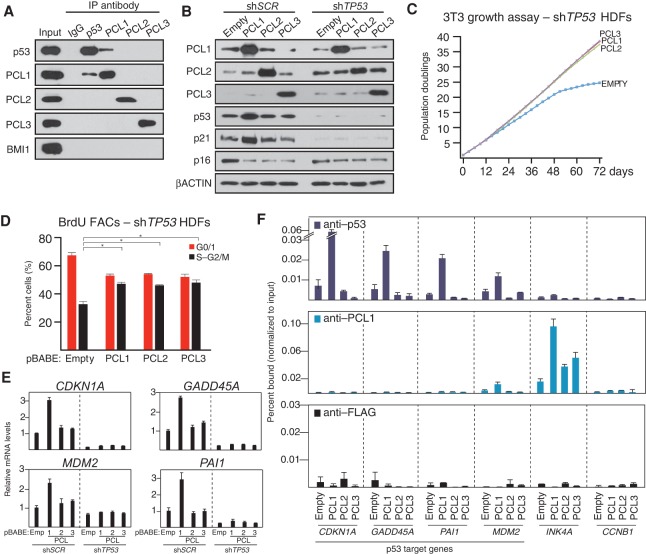
We next sought to explore the mechanism by which PCL1 confers its specific, negative effect on cellular proliferation. A recent study established that PCL1 interacts with and stabilizes p53 by blocking MDM2-mediated ubiquitination (Hong et al. 2008; Yang et al. 2013). To determine whether this interaction is specific to PCL1 and not PCL2 and PCL3, we performed endogenous coimmunoprecipitations (co-IPs) of p53 and PCL1–3 in HDFs (Fig. 3A), which demonstrated that PCL1 is the only PCL capable of coimmunoprecipitating p53. Moreover, in the reciprocal co-IP, PCL1 was the only PCL that coimmunoprecipitated with p53. Interestingly, the PCL1–p53 interaction is independent of the PRC2 complex and MDM2, as p53 did not copurify any other PRC2 components (Fig. 3A; Supplemental Fig. S3A), and MDM2 does not immunoprecipitate PCL1.
Figure 3.
PCL1 is the only PCL that interacts with and stabilizes p53 to block cellular proliferation. (A) Western blot analyses using the indicated antibodies of immunoprecipitations of the indicated endogenous proteins performed on nuclear protein lysate HDFs at PD38. (B) Western blot analyses using the indicated antibodies of whole-cell proteins prepared from HDFs infected with the indicated pBABE retrovirus at PD32, where cells were previously infected with a control (shSCR) or TP53 targeting (shTP53) pLKO lentivirus at PD28. Western blots were performed at PD48. (C) 3T3 growth assays performed in cells from B. Assays were initiated at PD36 and continued for 72 d. (D) Quantitative cell cycle BrdU FACS analyses of cells from B. Analyses were performed on two independent biological replicates. (E) Quantitative RT–PCR analyses of the indicated mRNA transcripts in cells from B. (F) Quantitative ChIP analyses using the indicated antibodies in HDFs infected with the indicated pBABE retrovirus at PD32; ChIPs were performed at PD48. Precipitated DNA was analyzed by quantitative PCR using primers directed toward the promoters of the indicated genes. ChIP enrichments are presented as the percentage of protein bound normalized to input.
We next examined whether the ability of PCL1 to negatively affect cellular proliferation is dependent on p53. To do this, we ectopically expressed PCL1–3 in HDFs expressing either a scrambled control shRNA (shSCR) or a TP53-specific (shTP53) shRNA (Fig. 3B). Strikingly, 3T3 growth assays and BrdU FACS analyses of these cells revealed that, in the absence of p53, ectopic expression of PCL1 confers a proliferative advantage, comparable with that conferred by PCL2 and PCL3 (Fig. 3C,D). Furthermore, ectopic expression of PCL1, but not PCL2 or PCL3, in shSCR cells stabilized the protein levels of p53 (Fig. 3B). This increase of p53 protein levels correlated with increased CDKN1A mRNA and p21 protein levels (Fig. 3B,E). Additional p53 target genes with established roles in the negative regulation of cellular proliferation, including GADD45A, MDM2, and PAI1, also had elevated mRNA levels in HDFs ectopically expressing PCL1 but not PCL2 or PCL3 (Fig. 3E). Furthermore, ChIP analyses demonstrated that increased transcription of these genes correlated with increased p53 occupancy at their promoters (Fig. 3F). ChIPs of PCL1 revealed that it was absent from p53 target gene promoters, suggesting that the PCL1–p53 interaction takes place “off chromatin.” Interestingly, the mRNA levels of a number of p53 target genes with roles in promoting apoptosis, such as PUMA, BAX, and NOXA, were not affected by ectopic expression of PCL1 (Supplemental Fig. S3B), suggesting that PCL1-mediated p53 stabilization leads to context-dependent downstream transcriptional changes. Taken together, these data suggest that PCL1 has diverged functionally from PCL2/3 to mediate its specific negative affects on cellular proliferation by binding to and stabilizing the p53 protein off chromatin and in a PRC2-independent manner.
PCL1 activates p53 target genes and blocks proliferation independently of chromatin association
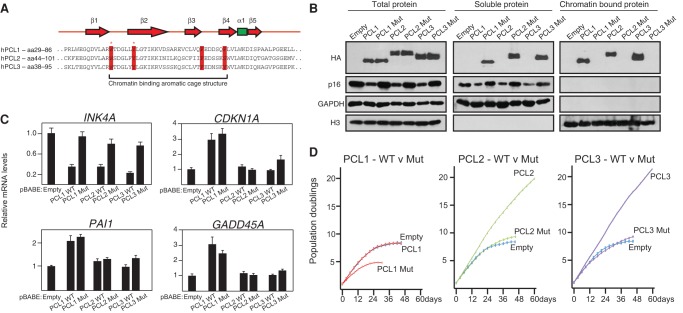
We next wished to confirm that the ability of PCL1 to activate p53 target genes and negatively affect cellular proliferation is independent of its ability to bind chromatin. To do this, we took advantage of two conserved residues within the Tudor domain of the PCL proteins (Fig. 4A), which we and others demonstrated are required for their ability to bind chromatin (Ballaré et al. 2012; Brien et al. 2012; Hunkapiller et al. 2012; Musselman et al. 2012; Cai et al. 2013). We generated mutant forms of each PCL protein in which these conserved tryptophan and tyrosine residues were mutated to cysteine and alanine, respectively. Western blotting of fractionated whole-cell, soluble, and chromatin-bound lysates confirmed that these Tudor domain mutations were sufficient to render ectopically expressed HA-tagged PCL1–3 incapable of binding chromatin (Fig. 4B). ChIP analyses confirmed that ectopic expression of wild-type, but not mutant, PCL1–3 led to the recruitment of EZH2 and PRC2 activity to the INK4A promoter (Supplemental Fig. S4A). Consistent with this, the ectopic expression of wild-type, but not mutant, PCL1–3 led to decreases in p16INK4A protein levels (Fig. 4B), which correlated with decreases in INK4A mRNA levels (Fig. 4C). As expected, the PCL2 and PCL3 Tudor domain mutants did not affect the rate of cellular proliferation, compared with the empty vector control (Fig. 4D). Strikingly, the removal of the ability of PCL1 to bind chromatin and repress INK4A led to a pronounced negative effect on cellular proliferation (Fig. 4D). Interestingly, the ectopic expression of wild-type and mutant PCL1 led to comparable increases in the levels of several p53 target genes, including CDKN1A, PAI1, and GADD45A (Fig. 4C; Supplemental Fig. S4B). This suggests that the differential effect on cellular proliferation between wild-type and mutant PCL1 is due to the ability of wild-type PCL1 to associate with chromatin and repress the INK4A gene. Taken together, these data establish that PCL1 blocks cellular proliferation by stabilizing p53 independently of INK4A repression and chromatin association.
Figure 4.
PCL1 activates the p53 pathway independently of its ability to bind chromatin. (A) Amino acid sequence alignments of the Tudor domains of human PCL1–3. The residues representing the aromatic cage motif are highlighted in red, and asterisks represent the tryptophan (W) and tyrosine (Y) residues that were mutated to cysteine (C) or alanine (A), respectively. Secondary structural elements of the PCL Tudor domain are shown above the alignment; red represents β sheets, and green represents an α helix. (B) Western blot analyses using the indicated antibodies of total, soluble, and chromatin-bound proteins prepared from HDFs infected with the indicated pBABE-HA-tagged retroviruses at PD32. Western blots were performed at PD48. (C) Quantitative RT–PCR analysis for the mRNA levels of INK4A and three p53 target genes in cells from B. (D) 3T3 growth assays performed on cells from B. Assays were initiated at PD36 and continued for 60 d or until the cells became senescent.
Two divergent domains in PCL1 are required and sufficient for binding p53
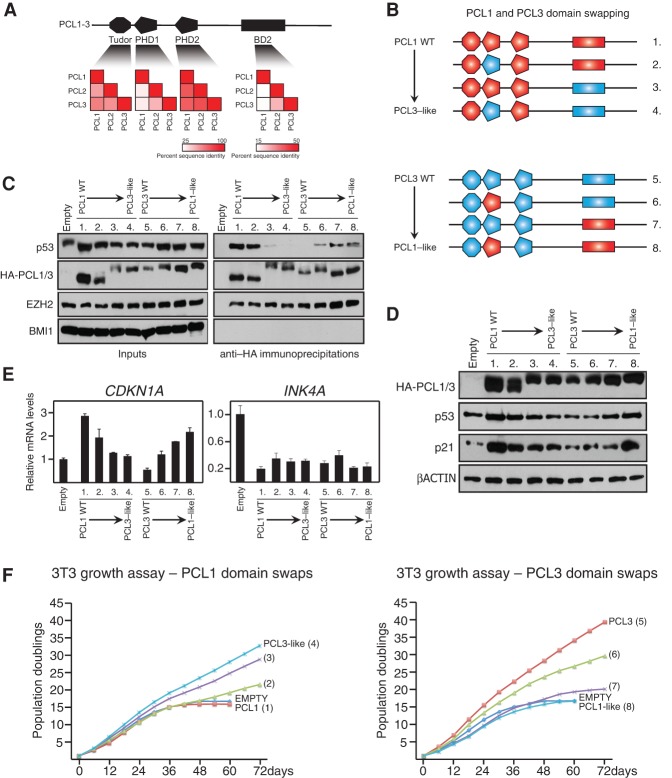
Previous work demonstrated that an N-terminal region encompassing the first PHD domain (PHD1) and a C-terminal region termed “BD2” mediate the ability of PCL1 to bind p53 and block MDM2-mediated ubiquitinylation of p53 (Yang et al. 2013). A comparison of the amino acid sequences of all three PCL proteins revealed that the PHD1 and BD2 regions of PCL1 have diverged from the equivalent regions in PCL2 and PCL3 (Fig. 5A). In contrast, the Tudor and PHD2 domains of all three proteins exhibit a broadly conserved sequence identity.
Figure 5.
Two divergent domains within PCL1 are required and sufficient for its association with p53 and ability to block cellular proliferation. (A, top panel) Representation of the domain structure of human PCL1–3. The PHD1 and BD2 regions of PCL1 were previously shown to interact with p53. (Bottom panel) Heat map representations of the level of amino acid sequence conservation in the indicated domains of PCL1–3. The bar indicates the percentage of sequence conservation. (B) Schematic representation of the PCL1 and PCL3 domain swap strategy used in this study. (C) Western blot analyses using the indicated antibodies of anti-HA immunoprecipitations of nuclear protein lysates prepared from HDFs infected with the indicated pBABE expression constructs. (D) Western blot analyses using the indicated antibodies of whole-cell proteins prepared from cells in C. (E) Quantitative RT–PCR analyses of the indicated mRNA transcripts in cells from C. (F) 3T3 growth assays performed in cells from C. Assays were initiated at PD36 and continued for 60 d.
To determine whether the PHD1 and BD2 domains of PCL1 are sufficient and/or required for its divergent ability to bind p53, we performed domain swap experiments, for which we generated six chimeric versions of PCL1 and PCL3 (Fig. 5B). Next, we ectopically expressed these chimeric proteins and wild-type PCL1 and PCL3 containing a HA-tag in HDFs. We performed co-IPs using anti-HA agarose beads and confirmed that all proteins were capable of associating with EZH2 to a comparable level, suggesting that they all incorporated into the PRC2 complex (Fig. 5C). Importantly, the PCL1 chimera containing the PHD1 and BD2 regions of PCL3 was incapable of interacting with p53, indicating that these regions are required for mediating the PCL1–p53 interaction. Conversely, all three PCL3 chimeras, which contained either one or both of the PHD1 and BD2 regions of PCL1, were capable of interacting with p53, indicating that both regions are sufficient for mediating the interaction. Consistent with these results, ectopic expression of the PCL1 hybrids had little or no affect on p53 protein levels or target gene expression when compared with wild-type PCL1, while ectopic expression of the PCL3 hybrids led to an increase in p53 protein levels and CDKN1A mRNA expression compared with wild-type PCL3 (Fig. 5D,E). Finally, 3T3 growth assays in these cells indicated that the PCL1–p53 interaction is essential for the ability of PCL1 to negatively affect proliferation, since ectopic expression of the “PCL3-like” hybrid containing both the PHD1 and BD2 regions of PCL3, which does not interact with p53, led to a proliferative advantage in HDFs (Fig. 5F). Moreover, ectopic expression of the equivalent “PCL1-like” hybrid did not lead to a proliferative advantage despite a reduction in INK4A levels, most likely owing to increased p53 protein levels and CDKN1A expression (Fig. 5D–F).
The ability of the PCL1 PHD domain to bind the p53 CTD is a newly evolved function
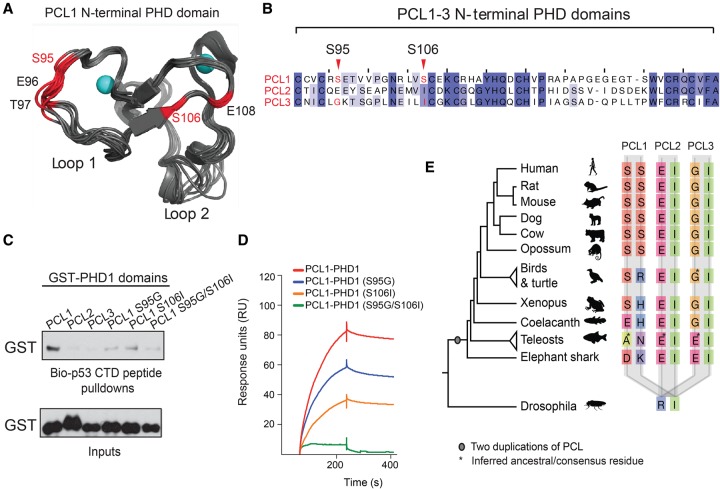
We next sought to further explore how PCL1 can specifically interact with p53 and why PCL2 and PCL3 lack this ability. To do this, we examined the structural determinants required for the binding specificity of PCL1 versus PCL2/3. Since predictions indicated that the BD2 region is likely to be largely unstructured (Supplemental Fig. S5A), we decided to focus our analysis on the PCL1 PHD1 region. This PHD domain shares a 38% sequence identity with the PHD domain of TRIM24, which has been cocrystalized with its H3(1–10)K4 substrate peptide (Tsai et al. 2010). Interestingly, the CTD of p53, which is the minimal region of p53 required for interaction with PCL1 (Yang et al. 2013), also bears sequence similarities to H3(1–10). Molecular dynamics simulations of a homology model of the PCL1 PHD1 based on the TRIM24 PHD-H3(1–10)K4 complex suggest that two serine residues (S95 and S106) could be essential for determining the specificity of PCL1 for p53 (Fig. 6A). In contrast, PCL2 and PCL3 lack these serines (Fig. 6B). We next wished to validate the potential importance of these two serine residues in the PCL1 PHD1 domain for binding to p53. To do this, we purified recombinant glutathione S-transferase (GST) fusion protein fragments representing the PHD1 domains of PCL1–3. Moreover, we also generated single-point mutant PCL1 PHD1 GST fusions in which we converted the serine residues to their equivalent residues in PCL3 (S95G and S106I) in addition to a double mutant in which both residues were mutated (S95G/S106I). Next, we performed peptide pull-downs using a biotinylated peptide representing all 30 amino acids of the unmodified p53 CTD and analyzed these by Western blot (Fig. 6C). This revealed that the wild-type PHD1 domain of PCL1 bound the unmodified p53 CTD peptide more strongly than the same domain in PCL2 and PCL3. Since these experiments were conducted with an unmodified p53 CTD peptide, they suggest that PCL1 binding to p53 is not reliant on the presence of specific post-translational modifications within this region. Importantly, the binding of the PCL1 containing point mutations in the PHD1 domain to the p53 CTD peptide was impaired, directly implicating these residues as essential mediators of the PCL1–p53 interaction. Next, we performed surface plasmon resonance (SPR) to accurately measure the in vitro binding of the recombinant GST proteins to the p53 CTD peptide (Fig. 6D). This validated the peptide pull-down analysis, confirming that both serines are required for the in vitro interaction between GST-PCL1 PHD1 and the p53 CTD peptide (Fig. 6D; Supplemental Fig. S5B). It also revealed that the binding of GST-PCL1-PHD1 to the p53 CTD peptide was highly reproducible, with an apparent Kd of 26.4 nM, as determined from independent measurements at four different concentrations of the protein (Supplemental Fig. S5C; data not shown). Taken together, these data revealed that the two serine residues at positions 95 and 106 of the N-terminal PHD domain of PCL1, which have diverged from PCL2/3, are necessary for the specific, high-affinity binding of PCL1 to the p53 CTD.
Figure 6.
The N-terminal PHD domain of mammalian PCL1 requires two divergent serine residues to associate with the p53 CTD. (A) Identification of a set of critical residues (hot spots) for the PCL1 PHD1 interaction with the p53 CTD. Ten of the dominant conformations of PCL1 PHD1 (shown in gray), identified from 2-μsec molecular dynamics (MD) simulations. The PCL1 PHD1 structure had been determined by homology modeling based on the TRIM24 PHD domain in complex with an unmodified H3(1–10)K4 peptide (Protein Data Bank [PDB] ID 3o37). The hot spots are labeled by residue type and number, and Zn atoms are shown in cyan. (B) Multiple alignment of the PHD1 domain of human PCL1, PCl2, and PCL3 proteins, highlighting the divergent serine residues at positions 95 and 106 in PCL1. (C) Peptide pull-downs of recombinant GST-tagged PCL1–3 PHD1 domains and mutant PCL1 PHD1 domains, with a biotin-tagged 30-amino-acid peptide representing the p53 CTD peptide. (D) Representative SPR sensorgrams for the wild-type and mutant PCL1 PHD1 domains binding to the p53 CTD peptide. (E) Segment of alignment of vertebrate PCL proteins showing residues aligned to Homo sapiens PCL1 Ser95 (first column for each PCL) and Ser106 (second column for each PCL). There is a single PCL in nonvertebrates, as represented by Drosophila. The phylogenetic tree indicates the species relationships; branch lengths are not to scale. Where there was more than one representative of a group and where they did not have identical sequences, the ancestral residue was inferred as the one requiring the fewest evolutionary changes. Such cases are indicated with an asterisk.
In order to understand the evolutionary origins of the two PCL1 serine residues, we performed a multiple sequence alignment and evolutionary tree analysis of the vertebrate PCL proteins (Fig. 6E). This confirmed that the three PCL genes are present in all vertebrates and are related through two whole-genome duplications at the base of the vertebrate tree (Makino and McLysaght 2010). We were specifically interested in whether the ability to bind p53 is a newly evolved function of PCL1 (that is, a so-called “neofunctionalization” event) or an ancestral function that was lost from the other two PCLs. The pattern of sequence divergence at sites corresponding to human Ser95 and Ser106 clearly indicates that the substitutions giving rise to each of these residues occurred uniquely in PCL1 after the duplication events. Importantly, these two serine residues are completely conserved across all mammalian PCL1 genes and, furthermore, uniquely co-occur in mammals (Fig. 6E). This suggests that PCL1 has undergone neofunctionalization during evolution, acquiring the ability to bind the p53 CTD in mammals.
Endogenous PCL1 stabilizes p53 to underpin cellular quiescence
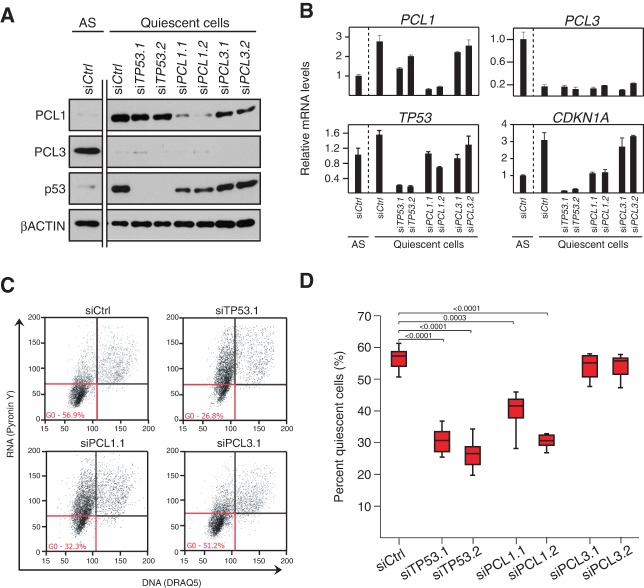
We next wished to further explore the potential biological significance of the PCL1–p53 interaction. Interestingly, despite the dramatic increase in p53 protein levels in quiescent cells, MDM2 mRNA and protein levels are unchanged between proliferating and nonproliferating cells (Supplemental Fig. S6A,B). This observation, taken together with the work of Yang et al. (2013), who demonstrated that PCL1 blocks MDM2-mediated ubiquitinylation of p53, suggested that the changes in PCL1 protein levels between quiescent and growing cells may be key to stabilizing p53 in these cells, thereby maintaining cells in a nonproliferative state. To test this possibility, we evaluated the functional consequences of transiently knocking down PCL1, TP53, or PCL3 (as a negative control) in serum-starved quiescent HDFs (Fig. 7A). The depletion of PCL1 led to a decrease in the levels of p53 protein and CDKN1A mRNA (Fig. 7A,B). In contrast, the depletion of PCL3 had no affect on p53 protein levels or CDKN1A expression, further supporting the idea that PCL1 plays a specific role in stabilizing p53.
Figure 7.
PCL1 stabilizes p53 to maintain cellular quiescence. (A) Western blot analyses using the indicated antibodies on whole-cell protein lysates from HDF cells transfected with siRNAs targeting TP53, PCL1, or PCL3 and then placed in serum-free medium for a further 72 h. (B) Quantitative RT–PCR analyses of the indicated mRNA transcripts on cells from C. (C) Representative data from a multicolor flow cytometry experiment used to determine the percentage of quiescent cells in serum-starved HDFs from C. Quiescent cells are defined as having low levels of Pyronin Y (RNA) and DRAQ5 positivity (DNA). (D) Quantification of the percentage of serum-starved HDFs in quiescence in cells from C. n = 6.
Since the p53 protein plays an essential role in maintaining cellular quiescence (Itahana et al. 2002; Meletis et al. 2006; Liu et al. 2009; Cheung and Rando 2013), we next asked whether depletion of PCL1 would phenocopy loss of p53 in our in vitro model of cellular quiescence. To do this, we quantified the proportion of quiescent cells following siRNA-mediated depletion of TP53, PCL1, and PCL3 in serum-starved quiescent HDFs (Fig. 7C,D). Remarkably, this demonstrated that depletion of PCL1 led to a similar, approximately twofold reduction in the overall quiescent population, similar to p53 depletion in these cultures, whereas depletion of PCL3 had no effect (Fig. 7C,D). These effects on quiescence maintenance are comparable with previous in vivo results, where p53 knockout in quiescent HSCs results in a 1.5-fold to twofold reduction in quiescent HSCs (Liu et al. 2009). This is particularly remarkable because these cells do not have serum or growth factors that would normally be required for quiescent cells to exit the G0 block and enter the cell cycle. Taken together, these data indicate that PCL1 specifically stabilizes the p53 protein in quiescent cells and, like p53, is required for the maintenance of the quiescent cell state.
Discussion
Our initial observation that, unlike PCL2 and PCL3, PCL1 is expressed in quiescent cells suggested that it might have a unique function in nondividing cells. We pursued this and unraveled an essential PCL1–p53 regulatory axis. The PCL1 gene is transcriptionally regulated by p53 in quiescent cells, while the PCL1 protein physically interacts with the p53 protein to stabilize it. Remarkably, mammalian PCL1 has only recently evolved the ability to bind the p53 CTD through the acquisition of two unique serine residues in its N-terminal PHD domain. We show here that this divergent ability of PCL1 is both PRC2- and chromatin-independent and is essential in quiescent cells to maintain their nonproliferative state.
PCL1 as an upstream regulator of cellular quiescence
Our initial demonstration that the PCL1 gene is highly expressed in quiescent cells prompted us to investigate its potential role as a regulator of cellular quiescence. We observed that ectopic expression of PCL1 was sufficient to mediate stabilization of p53 protein levels, leading to increased transactivation of p53 target genes such as CDKN1A and ultimately to an accumulation of G0/quiescent cells. Moreover, transient loss of PCL1 was sufficient to phenocopy p53 depletion in an in vitro model of cellular quiescence. This is a particularly interesting finding, since no Polycomb group protein has previously been linked with a direct role in the regulation of cellular quiescence, and raises the possibility that, like p53, PCL1 could be required to maintain the quiescent state of tissue-specific stem cells in vivo (Itahana et al. 2002; Meletis et al. 2006; Liu et al. 2009; Cheung and Rando 2013). This possibility will require additional study and the generation of suitable in vivo genetic models for the study of PCL1 function in the context of cellular quiescence. A challenge here will be the fact that a straightforward conditional deletion of Pcl1 in quiescent stem cells would likely also disrupt its other function within the PRC2 complex. In this regard, it is worth noting that loss of PRC2 components such as Ezh1 or Eed in HSCs leads to the aberrant activation of Polycomb target genes, including Ink4a, and consequent downstream phenotypic changes (Hidalgo et al. 2012; Xie et al. 2014). Therefore, to delineate the two functions of PCL1, it will be necessary to generate a conditional PCL1 allele, producing a protein incapable of interacting with p53 but still capable of recruiting and maintaining the PRC2 complex to target genes. Therefore, a more extensive molecular characterization of the PCL1–p53 interaction will be necessary to guide the generation of such an allele and thereby help further decipher the molecular mechanisms regulating adult stem cell quiescence.
PCL1 as a new example of neofunctionalization
We provide evidence that the PCL1 gene has undergone neofunctionalization, acquiring the unique ability among the three PCL proteins to bind the CTD of p53. However, we also show that the PCL1 protein retains its ancestral function within the PRC2 complex. The divergent N-terminal PHD domain and C-terminal “BD2” region of PCL1 are both required and sufficient for this unique ability. We further characterized two unique serines in the N-terminal PHD domain of PCL1 that have emerged during vertebrate evolution and are essential for its interaction with p53. This functional divergence of PCL1 goes some way to explaining the acquisition and ultimate maintenance of multiple PCL genes in vertebrate genomes. They are not redundant copies, as might have seemed to be the case, but have a novel functionality that evolved alongside the ancestral one. The idea of neofunctionalization of duplicated genes has been popular for >40 years (Ohno 1970), but there are relatively few examples such as this, where the sequence substitutions involved have been clearly described (Conant and Wolfe 2008). Therefore, PCL1 represents an archetypal example of neofunctionalization, and further studies into its functional interplay with p53 will likely shed new light on the evolution of the molecular mechanisms regulating the process of cellular quiescence.
Roles of PCL proteins in cancer
Our results have implications for how the deregulation of PCL protein function could contribute to cancer. We show that ectopic expression of PCL1–3 leads to an increase in the recruitment of the PRC2 complex and H3K27me3 deposition on the INK4A tumor suppressor gene locus. Therefore, the elevated levels of PCL3 observed in multiple cancer types (Wang et al. 2004; Li et al. 2013) could directly contribute to their sustained proliferation by increasing the function of the PRC2 complex on Polycomb target genes such as INK4A. It is interesting to note that no other PRC2 component has been reported to repress INK4A when ectopically expressed, despite their requirement to maintain its repression (Bracken et al. 2007). This is likely related to the stoichiometry of the PRC2 complex and the fact that the core components—EZH1/2, SUZ12, and EED—exist at a ratio of 1:1:1 in vivo (Smits et al. 2013), which is essential for its activity (Kuzmichev et al. 2002), making it unlikely that overexpression of a single core PRC2 component would lead to increased PRC2 activity. However, since the PCL1–3 proteins exist in less than one in five PRC2 complexes (Smits et al. 2013), it follows that overexpression of any one PCL protein would stabilize the binding of PRC2 on its target genes. Furthermore, since PCL proteins require their Tudor domain to associate with chromatin, targeting this domain could be a novel therapeutic strategy to reactivate INK4A in cancer cells, particularly in cancers with deregulated PRC2 function.
The PCL1 gene is frequently translocated in endometrial stromal sarcomas, and the PCL1 fusion products have been proposed to contribute directly to the pathogenesis of these cancers (Micci et al. 2006; Sauvageau and Sauvageau 2010). However, the mechanisms of how PCL1 fusion gene products could contribute to oncogenesis are unexplored. Interestingly, while these PCL1 fusion genes retain the vast majority of the PCL1-coding sequence, the promoter region is lost in the translocation event, implying that the normal transcriptional control of PCL1 is lost in the case of the fusion genes. Therefore, we speculate that ectopic expression of these PCL1 fusions may contribute to cancer by increasing PRC2 complex function, potentially leading to the repression of Polycomb target genes such as INK4A. Intriguingly, our study and the nature of the PCL1 fusion genes would suggest that the PCL1 fusion gene products should retain the ability of wild-type PCL1 to interact with p53, raising questions about the status of the TP53 gene in these cancers. Finally, strategies to block the PCL1–p53 interaction could potentially be exploited for therapeutic gain in cancer. For example, it will be important to examine the relationship of PCL1 and wild-type p53 in quiescent, slowly proliferating cancer stem cells in vivo, since disrupting the interaction could force these cells into the cell cycle, potentially rendering them more sensitive to chemotherapy.
In conclusion, our study not only establishes critical functions for PCL1–3 in positively and negatively regulating cellular proliferation but also highlights potential opportunities to exploit this knowledge for cancer therapy.
Materials and methods
Cell culture and RNAi
HDFs and viral packaging cells were grown in DMEM (Gibco) with 10% (v/v) FBS (Gibco), 100 U/mL penicillin, and 100 U/mL streptomycin (Gibco). For serum starvation experiments, serum was removed for 96 h, and cells were stimulated to re-enter the cell cycle by addition of medium containing 20% (v/v) FBS for 24 h. For RNAi experiments, HDFs were transfected at 30%–50% confluence with 20 μM siRNA using Lipofectamine RNAi MAX (Invitrogen).
3T3 growth assays and cell cycle analyses
For 3T3 assays, 7.5 × 105 cells were plated on 100-mm plates. Three days later, the total number of cells was counted, and 7.5 × 105 cells were plated again. The cumulative increase in cell number was calculated according to the formula log(Nf/Ni)/log2, where Ni and Nf are the initial and final numbers of cells plated and counted after 3 d, respectively. For BrdU FACS analyses, cells were pulsed with 33 μM BrdU for 45 min. BrdU incorporation was measured using an anti-BrdU antibody followed by FACS analysis. DNA content was measured by propidium iodide staining. For G0, FACS cells were stained with 10 μM DRAQ5 (Biostatus) and 5 μg/mL Pyronin Y at 37°C.
Immunoprecipitations
Immunoprecipitations were performed as previously described (Van Den Berg et al. 2010). Briefly, nuclear pellets were lysed in buffer C containing protease inhibitors (20 mM HEPES at pH 7.6, 20% [v/v] glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, aprotinin 1 μg mL−1, leupeptin 10 μg mL−1, PMSF 1 mM) and subsequently dialyzed against buffer C-100 (20 mM HEPES at pH 7.6, 20% [v/v] glycerol, 0.2 mM EDTA, 100 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA). Antibody-coupled beads were incubated with dialyzed nuclear extracts containing 250 U of benzonase (Sigma) for 3 h at 4°C. Beads were then washed, and elutions were performed with 2× SDS loading dye or 250 μg/mL HA peptide (Sigma).
ChIP
ChIP analyses were performed as described (Brien et al. 2012). Briefly, formaldehyde cross-linked chromatin was sheared to 200–1000 base pairs by sonication. Chromatin was incubated overnight at 4°C with the indicated antibodies, and immune complexes were extracted using protein A or G Sepharose beads (Sigma). ChIP samples were eluted, and DNA was purified by phenol/chloroform extraction and ethanol precipitation.
Quantitative real-time PCRs
Total RNA was extracted from cells using the RNeasy kit (Qiagen) and was used to generate cDNA by RT–PCR using the TaqMan reverse transcription kit (Applied Biosystems). Relative mRNA expression levels were determined using the SYBR Green I detection chemistry (Applied Biosystems).
Purification of recombinant GST fusion proteins
The PCL1/2/3-PHD1 fragments were cloned into pGEX-6P1 and expressed in BL21-DE3 Escherichia coli. Protein expression was induced by addition of 0.5 mM IPTG to bacteria cultures, and GST fusion proteins were purified over GSH-agarose beads (Pierce).
In vitro peptide pull-down
Biotinylated p53 CTD peptide (residues 363–393), bound to strepavidin-coated agarose beads (Invitrogen), was incubated for 30 min with GST fusion proteins. Beads were washed extensively in binding buffer, and bound protein was eluted using 2× Laemlli dye.
SPR
Biotinylated p53 C-terminal peptide was immobilized on a streptavidin-coated SPR surface, and concentration series of each GST-PCL-PHD1 fusion proteins were subsequently injected to determine interaction affinities (Kd).
Supplementary Material
Acknowledgments
We thank Peter Verrijzer, Bill Keyes, Jean-Christophe Marine, and members of the Bracken laboratory for helpful discussions and critical reading of the manuscript. We also thank Barry Moran of the Trinity College Dublin FACS core facility and Conor Henry for assistance with FACS experiments. We thank the Irish Centre for High-End Computing (ICHEC) for the generous allocation of computational resources. A.M.R. and A.M. are supported by funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013)/European Research Council grant agreement 309834. Work in the Bracken laboratory is supported by Science Foundation Ireland (SFI) under the Principal Investigator Career Advancement Award (SFI PICA SFI/10/IN.1/B3002), the Health Research Board under the Health Research Awards 2010 (HRA_POR/2010/124), the Irish Research Council, St. Vincent's Foundation, the Irish Cancer Society Collaborative Cancer Research Centre, and BREAST-PREDICT grant CCRC13GAL (http://www.breastpredict.com).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.267930.115.
References
- Ballaré C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, et al. 2012. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol 19: 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353. [DOI] [PubMed] [Google Scholar]
- Bracken AP. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 22: 5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Mönch K, Minucci S, Porse BT, Marine J-C, et al. 2007. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 21: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien GL, Gambero G, O'Connell DJ, Jerman E, Turner SA, Egan CM, Dunne EJ, Jurgens MC, Wynne K, Piao L, et al. 2012. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol 19: 1273–1281. [DOI] [PubMed] [Google Scholar]
- Bruggeman SWM, Hulsman D, Tanger E, Buckle T, Blom M, Zevenhoven J, van Tellingen O, van Lohuizen M. 2007. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell 12: 328–341. [DOI] [PubMed] [Google Scholar]
- Cai L, Rothbart SB, Lu R, Xu B, Chen W-Y, Tripathy A, Rockowitz S, Zheng D, Patel DJ, Allis CD, et al. 2013. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol Cell 49: 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, Rando TA. 2013. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 14: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M. 2007. Cellular senescence in cancer and aging. Cell 130: 223–233. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950. [DOI] [PubMed] [Google Scholar]
- Datta S, Hoenerhoff MJ, Bommi P, Sainger R, Guo WJ, Dimri M, Band H, Band V, Green JE, Dimri GP. 2007. Bmi-1 cooperates with H-Ras to transform human mammary epithelial cells via dysregulation of multiple growth-regulatory pathways. Cancer Res 67: 10286–10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L, Helin K. 2013. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 20: 1147–1155. [DOI] [PubMed] [Google Scholar]
- Dietrich N, Bracken AP, Trinh E, Schjerling CK, Koseki H, Rappsilber J, Helin K, Hansen KH. 2007. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J 26: 1637–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas D, Hsu JHR, Hung L, Cooper A, Abdueva D, Van Doorninck J, Peng G, Shimada H, Triche TJ, Lawlor ER. 2008. BMI-1 promotes Ewing sarcoma tumorigenicity independent of CDKN2A repression. Cancer Res 68: 6507–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stützer A, Fischle W, Bonaldi T, Pasini D. 2014. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell 53: 49–62. [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45: 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Bernard D, Martínez D, Beach D. 2004. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol 6: 67–72. [DOI] [PubMed] [Google Scholar]
- Helin K, Dhanak D. 2013. Chromatin proteins and modifications as drug targets. Nature 502: 480–488. [DOI] [PubMed] [Google Scholar]
- Hidalgo I, Herrera-Merchan A, Ligos JM, Carramolino L, Nuñez J, Martinez F, Dominguez O, Torres M, Gonzalez S. 2012. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell 11: 649–662. [DOI] [PubMed] [Google Scholar]
- Hock H. 2012. A complex Polycomb issue: the two faces of EZH2 in cancer. Genes Dev 26: 751–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Jiang J, Lan L, Nakajima S, Kanno S, Koseki H, Yasui A. 2008. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic Acids Res 36: 2939–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller J, Shen Y, Diaz A, Cagney G, McCleary D, Ramalho-Santos M, Krogan N, Ren B, Song JS, Reiter JF. 2012. Polycomb-like 3 promotes polycomb repressive complex 2 binding to CpG islands and embryonic stem cell self-renewal. PLoS Genet 8: e1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Dimri GP, Hara E, Itahana Y, Zou Y, Desprez PY, Campisi J. 2002. A role for p53 in maintaining and establishing the quiescence growth arrest in human cells. J Biol Chem 277: 18206–18214. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, Van Lohuizen M. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397: 164–168. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanigan F, Geraghty JG, Bracken AP. 2011. Transcriptional regulation of cellular senescence. Oncogene 30: 2901–2911. [DOI] [PubMed] [Google Scholar]
- Lanzuolo C, Orlando V. 2012. Memories from the polycomb group proteins. Annu Rev Genet 46: 561–589. [DOI] [PubMed] [Google Scholar]
- Lecona E, Rojas LA, Bonasio R, Johnston A, Fernández-Capetillo O, Reinberg D. 2013. Polycomb protein SCML2 regulates the cell cycle by binding and modulating CDK/CYCLIN/p21 complexes. PLoS Biol 11: e1001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. 2003. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423: 255–260. [DOI] [PubMed] [Google Scholar]
- Li G, Warden C, Zou Z, Neman J, Krueger JS, Jain A, Jandial R, Chen M. 2013. Altered expression of polycomb group genes in glioblastoma multiforme. PLoS One 8: e80970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, et al. 2009. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 4: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, El Messaoudi-Aubert S, Racek T, Stock JK, Nicholls J, Rodriguez-Niedenführ M, Gil J, Peters G. 2009. Several distinct polycomb complexes regulate and co-localize on the INK4a tumor suppressor locus. PLoS One 4: e6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T, McLysaght A. 2010. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci 107: 9270–9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletis K, Wirta V, Hede S-M, Nistér M, Lundeberg J, Frisén J. 2006. P53 suppresses the self-renewal of adult neural stem cells. Development 133: 363–369. [DOI] [PubMed] [Google Scholar]
- Micci F, Panagopoulos I, Bjerkehagen B, Heim S. 2006. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Res 66: 107–112. [DOI] [PubMed] [Google Scholar]
- Mohd-Sarip A, Lagarou A, Doyen CM, van der Knaap JA, Aslan U, Bezstarosti K, Yassin Y, Brock HW, Demmers JAA, Verrijzer CP. 2012. Transcription-independent function of Polycomb group protein PSC in cell cycle control. Science 336: 744–747. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park I-K, Clarke MF, Morrison SJ. 2003. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425: 962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde M-E, Hong Z, Allen C, Roy S, Nuñez JK, Nickoloff J, et al. 2012. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol 19: 1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Springer, New York. [Google Scholar]
- Park I, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302–305. [DOI] [PubMed] [Google Scholar]
- Piunti A, Rossi A, Cerutti A, Albert M, Jammula S, Scelfo A, Cedrone L, Fragola G, Olsson L, Koseki H, et al. 2014. Polycomb proteins control proliferation and transformation independently of cell cycle checkpoints by regulating DNA replication. Nat Commun 5: 3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. 2004. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32: D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. 2010. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7: 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49: 808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits AH, Jansen PWTC, Poser I, Hyman AA, Vermeulen M. 2013. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res 41: e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148: 664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, De Wit E, Muijrers I, Teunissen H, Talhout W, Van Steensel B, Van Lohuizen M. 2006. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet 38: 694–699. [DOI] [PubMed] [Google Scholar]
- Tsai W-W, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai C-Y, Shi X, Schwarzer D, Plunkett W, et al. 2010. TRIM24 links a non-canonical histone signature to breast cancer. Nature 468: 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzatsos A, Pfau R, Kampranis SC, Tsichlis PN. 2009. Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc Natl Acad Sci 106: 2641–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Berg DLC, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. 2010. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Robertson GP, Zhu J. 2004. A novel human homologue of Drosophila polycomblike gene is up-regulated in multiple cancers. Gene 343: 69–78. [DOI] [PubMed] [Google Scholar]
- Xie H, Xu J, Hsu JH, Nguyen M, Fujiwara Y, Peng C, Orkin SH. 2014. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell 14: 68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang C, Zhang P, Gao K, Wang D, Yu H, Zhang T, Jiang S, Hexige S, Hong Z, et al. 2013. Polycomb group protein PHF1 regulates p53-dependent cell growth arrest and apoptosis. J Biol Chem 288: 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.