In this study, Kim et al. identified WD repeat and SOCS box-containing protein 1 (WSB1) as a novel regulator of pVHL through WSB1's E3 ligase activity. These findings provide important new insights into the understanding of misregulation of the pVHL–HIF pathway in cancer cell invasion and metastasis.
Keywords: WSB1, pVHL, HIFs, metastasis, E3 ligase, degradation
Abstract
The von Hippel-Lindau tumor suppressor pVHL is an E3 ligase that targets hypoxia-inducible factors (HIFs). Mutation of VHL results in HIF up-regulation and contributes to processes related to tumor progression such as invasion, metastasis, and angiogenesis. However, very little is known with regard to post-transcriptional regulation of pVHL. Here we show that WD repeat and SOCS box-containing protein 1 (WSB1) is a negative regulator of pVHL through WSB1's E3 ligase activity. Mechanistically, WSB1 promotes pVHL ubiquitination and proteasomal degradation, thereby stabilizing HIF under both normoxic and hypoxic conditions. As a consequence, WSB1 up-regulates the expression of HIF-1α’s target genes and promotes cancer invasion and metastasis through its effect on pVHL. Consistent with this, WSB1 protein level negatively correlates with pVHL level and metastasis-free survival in clinical samples. This work reveals a new mechanism of pVHL's regulation by which cancer acquires invasiveness and metastatic tendency.
pVHL is ubiquitously expressed in normal tissues and functions as a tumor suppressor (Los et al. 1996; Gossage et al. 2015). Loss of pVHL via gene and germline mutations results in the von Hippel-Lindau (VHL) disease and is characterized by development of various tumors, including renal clear cell carcinomas (RCCs) and other highly vascularized tumors (Kaelin 2007; Bausch et al. 2013; Gossage et al. 2015). pVHL is the substrate recognition component of a cullin RING ubiquitin ligase complex that includes elongin B, elongin C, Rbx1, and Cul2 (Kaelin 2008; Deshaies and Joazeiro 2009). pVHL's main function as an E3 ligase is to target hypoxia-inducible factor-α (HIF-α) for degradation during normoxia (Gossage et al. 2015).
Loss of pVHL function results in constitutive activation of HIF-1α and HIF-2α, which act as important heterodimeric transcription factors (composed of HIF-1α or HIF-2α and HIF-1β) for target genes such as VEGF, GULT1, CAIX, and HK2 (Semenza 2011; Keith et al. 2012; Montagner et al. 2012; Gossage et al. 2015). Constitutive activation of these transcription factors results in the induction of metabolic adaptation and promotes tumor growth, invasion, migration, metastasis, and angiogenesis through the up-regulation of their target genes (Majmundar et al. 2010; Semenza 2010, 2011; Keith et al. 2012). However, the regulation of pVHL at the post-transcriptional level remains underexplored. It has been reported that pVHL may be regulated through the ubiquitin–proteasome pathway (Jung et al. 2006; Chen et al. 2013; Pozzebon et al. 2013), although pVHL's E3 ligase has yet to be identified. Identification of how pVHL is regulated at the post-transcriptional level will further elucidate the mechanism of HIF biology in cancers, especially in cancers with wild-type pVHL.
WSB1 (WD repeat and SOCS box-containing protein 1) has been classified as a substrate recognition subunit of the ECS (Elongin B/C–Cul2/5–SOCS) ubiquitin ligase complexes (Vasiliauskas et al. 1999). WSB1 harbors seven WD40 repeats and a SOCS box, respectively (Choi et al. 2008). WSB1 has been shown to be overexpressed in several cancers, such as pancreatic cancer, hepatocellular carcinoma, and salivary gland tumor (Rhodes and Chinnaiyan 2005; Archange et al. 2008; Silva et al. 2011; Tong et al. 2013). Additionally, WSB1 has been identified as a HIF-1 target gene (Tong et al. 2013). The cellular function of WSB1 is not well studied. WSB1 has been shown to mediate homeodomain-interacting protein kinase 2 (HIPK2) ubiquitination, resulting in its proteasome degradation (Choi et al. 2008). Following DNA damage, WSB1-mediated ubiquitination of HIPK2 is blocked, and HIPK2 is stabilized. HIPK2 in turn phosphorylates p53 at Ser46, which is important for activating proapoptotic gene expression (Puca et al. 2010). WSB1 overexpression has been shown to promote pancreatic cancer cell proliferation (Archange et al. 2008). However, it is unlikely that this effect is due to inactivation of the HIPK–p53 pathway, as the pancreatic cancer cell line used in this study contains mutant p53. This implies that WSB1 could promote cancer cell proliferation through other p53-independent mechanisms.
Here, we identify WSB1 as a pVHL E3 ligase. In normoxic and hypoxic conditions, WSB1 interacts with pVHL and regulates pVHL's stability through proteasome degradation. These results reveal an important missing piece in the regulation of pVHL stability.
Results
WSB1 is associated with metastasis in various tumors
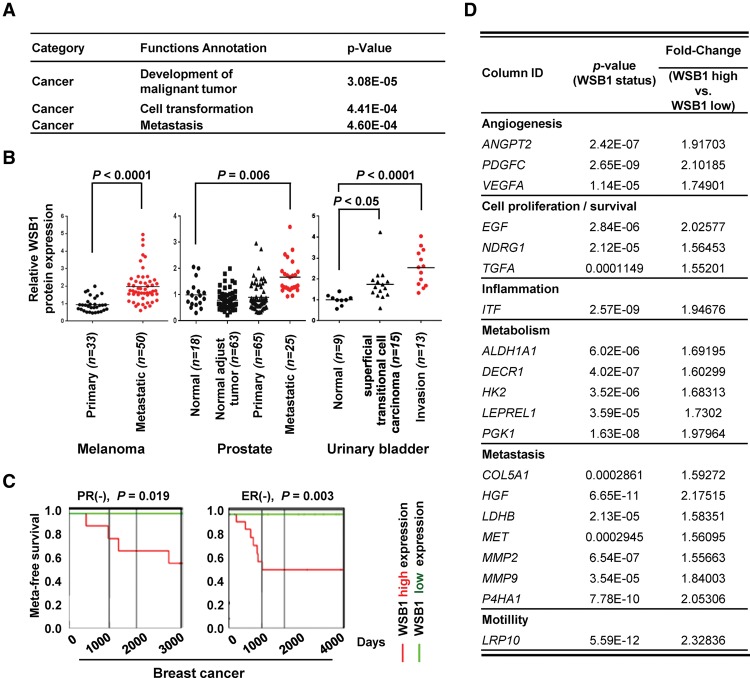
The identification of WSB1 expression as a prognostic marker in tumors prompted us to test its functional role in tumor aggressiveness. We first examined gene expression profiles from 56 pairs of primary lung adenocarcinoma patient samples and analyzed them by WSB1 status using Ingenuity Pathway Analysis (IPA) (Supplemental Figs. S1, S2). Metastasis- or migration-related signaling—such as RhoA (Friedl et al. 2014), TGF-β (Pickup et al. 2013), and actin nucleation by the ARP–WASP complex (Bovellan et al. 2014)—are all top-ranked canonical pathways associated with WSB1 expression (Supplemental Fig. S1). Based on the analysis, WSB1 expression is closely associated with pathways that are involved in metastasis and invasion (Fig. 1A).
Figure 1.
WSB1 positively correlates with metastasis. (A) Analysis of differentially expressed genes by WSB1 expression levels in 56 lung adenocarcinoma patients using IPA. (B) Gene expression of WSB1 in metastatic cancers (Gene Expression Omnibus data sets GDS3966, GDS2545, and GDS1479) (defined in Supplemental Table 1). WSB1 is highly expressed in human metastatic cancers. (C) Kaplan-Meier graph of human breast cancer patients, stratified according to high or low expression levels of WSB1 (PROGgene). (D) Potential HIF-1α target genes associated with WSB1 expression levels in 56 lung adenocarcinoma patients.
To further investigate the relationship between WSB1 and metastasis, we analyzed a cohort of 83 melanoma, 171 prostate cancers, and 37 urinary bladder tumors from three clinically annotated gene expression data sets for WSB1 expression (Supplemental Table 1; Supplemental Material; Dyrskjot et al. 2004; Yu et al. 2004; Chandran et al. 2007; Xu et al. 2008). Within the cohorts, the WSB1 level is significantly higher in metastatic tissues than in normal or primary tissues (melanoma, P < 0.0001; prostate, P = 0.006; urinary bladder, P < 0.0001, respectively) (Fig. 1B). Furthermore, for a subset of breast cancer and colon cancer patients, individuals with high WSB1 expression showed lower metastasis-free survival (breast, P = 0.019 [PR−] and 0.003 [ER−]; colon, P = 0.0455, respectively) (Fig. 1C; Supplemental Fig. 3A–C). Interestingly, for breast cancer, high WSB1 expression is associated with poor survival mostly in ER−, PR−, HER−, or triple-negative subtypes.
To determine the gene expression changes associated with WSB1, we selected 89 cancer metastasis-related genes using IPA. Among these genes, 64 are most differentially expressed on the basis of WSB1 expression status in the 56 fresh-frozen lung adenocarcinoma samples. The expression of prometastasis-related genes such as CXCR4, EGF, MMP2, and MMP9 positively correlates with WSB1 expression, whereas the expression of anti-metastasis genes, including p53, negatively correlates with WSB1 expression (Fig. 1D; Supplemental Table 2). Interestingly, WSB1 expression is also positively associated with the level of HIF target genes such as VEGFA, MMP-9, and HK2 (Fig. 1D). These results suggest that WSB1 is associated with metastasis and might be important for metastasis in malignant tumors.
WSB1 regulates pVHL levels through ubiquitinating pVHL
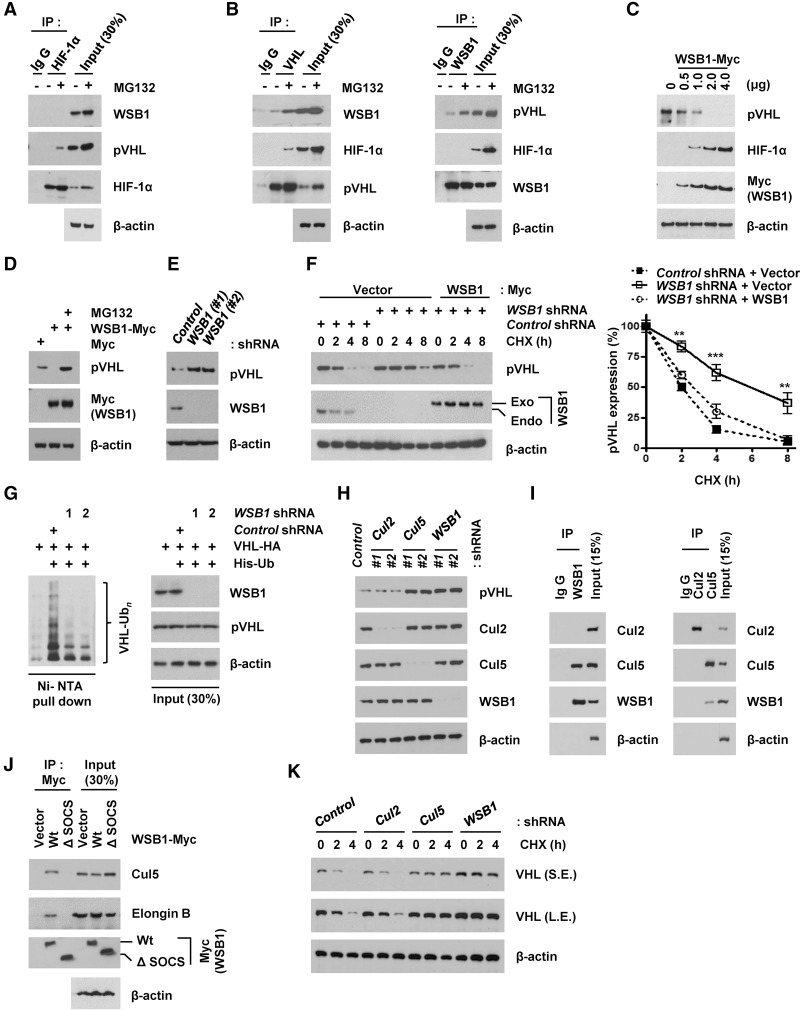
In several tumor types, HIF-1α is a major protein that promotes tumor aggressiveness, metastasis, and patient mortality (Maxwell 2005; Gordan and Simon 2007; Semenza 2010). HIF-1α’s stability can be regulated by E3 ligases such as pVHL and TRAF6 (Sun et al. 2013; Gossage et al. 2015). While pVHL promotes HIF-1α proteasomal degradation, E3 ligase activity of TRAF6 is involved in the increase of HIF-1α’s stability through its K63-linked polyubiquitination. Because WSB1 is known as an E3 ligase (Dentice et al. 2005), we speculated whether HIF-1α is regulated by WSB1. Unexpectedly, we did not find an interaction between WSB1 and HIF-1α (Fig. 2A; Supplemental Fig. S4A). Instead, we found an interaction between WSB1 and pVHL (Fig. 2B; Supplemental Fig. S4A). Therefore, we hypothesized that WSB1 might regulate pVHL. When WSB1 was overexpressed, we found pVHL protein levels decreased and HIF-1α levels increased in a dosage-dependent manner (Fig. 2C).The decrease of pVHL induced by WSB1 overexpression was reversed by the proteasome inhibitor N-carbobenzoxy-l-leucinyl-lleucinyl-l-norleucinal (MG132) (Fig. 2D). Conversely, knockdown of WSB1 resulted in a marked increase of pVHL levels (Fig. 2E). We further found that WSB1 regulated pVHL stability. Depletion of WSB1 increased pVHL stability, while reconstitution of cells with WSB1 restored VHL stability to that of control cells (Fig. 2F). Interestingly, we found high expression of HIF-1 α in several cancer cell lines under normoxic conditions. In addition, we found a negative correlation between WSB1 and pVHL levels and a positive correlation between WSB1 and HIF-1α in pancreatic cancer cell lines, non-small-cell lung cancer (NSCLC) cell lines, and breast cancer cell lines (Supplemental Fig. S4B). Previously, several studies have also shown high HIF-1α protein levels in cancer cells under normoxic conditions (Zhong et al. 1999; Akakura et al. 2001; Jung et al. 2006; Saito et al. 2006). Therefore, WSB1 might be responsible for increased HIF-1α levels under normoxic conditions. We next examined whether WSB1 regulates pVHL stability through promoting pVHL ubiquitination. Indeed, we found that knocking down WSB1 decreased pVHL ubiquitination (Fig. 2G). These results suggest that WSB1 regulates pVHL ubiquitination.
Figure 2.
WSB1 regulates pVHL ubiquitination and degradation. (A,B) Coimmunoprecipitation (co-IP) of endogenous HIF-1α (A), pVHL (B, left), and WSB1 (B, right) from extracts of human embryonic kidney 293T (HEK293T) cells. (C) Regulation of pVHL and HIF-1α protein level by WSB1 in a dose-dependent manner. Cells were transfected with different amounts of WBS1 constructs, and pVHL and HIF-1α levels were examined. (D) Down-regulation of pVHL level by WSB1 was reversed by MG132. Cells transfected with WSB1-Myc were left untreated or were treated with MG132, and pVHL levels were examined. (E) Cells were transfected with WSB1 shRNAs, and the pVHL level was examined. (F) Cells stably expressing control shRNA, WSB1 shRNA, or WSB1 shRNA together with shRNA-resistant WSB1 were treated with 0.1 mg/mL cycloheximide and harvested at the indicated times. The pVHL level was then examined. (Exo) Exogenous; (Endo) endogenous. (Bottom) Quantification of VHL protein expression levels shown in the top panel. (**) P < 0.01; (***) P < 0.001 versus control shRNA by one-way ANOVA. (G) Cells were transfected with the indicated constructs and then treated with MG132. Ubiquitinated proteins were pulled down under denaturing conditions by Ni-NTA agarose and analyzed by immunoblot. (pVHL-Ubn) Polyubiquitinated pVHL. (H) Cells were transfected with Cul2 shRNA, Cul5 shRNA, or WSB1 shRNAs, and pVHL levels were examined. (I) Co-IP of endogenous WSB1 (left) and Cul2 or Cul5 (right) from extracts of HEK293T cells. (J) Cells were transfected with the indicated plasmids. Cells were pretreated with MG132, and the WSB1–Elongin B/C–Cul5 interaction was examined by immunoprecipitation. (Exo) Exogenous; (Endo) endogenous. (K) Cells stably expressing control shRNA, Cul2 shRNA, Cul5 shRNA, or WSB1 shRNA were treated with 0.1 mg/mL cycloheximide and harvested at the indicated times. pVHL levels were then examined by immunoblot. (S.E.) Short exposed; (L.E.) long exposed.
WSB1 acts as a substrate-recognizing subunit of the ECS ubiquitin ligase complex (Dentice et al. 2005). We therefore tested whether Cul2 or Cul5 would regulate the pVHL level. We found that the pVHL level was increased in cells depleted of Cul5 or WSB1 but not in cells depleted of Cul2 (Fig. 2H), suggesting that Cul5 is involved in the regulation of pVHL. Consistent with this, WSB1 interacted with Cul5 but not Cul2 (Fig. 2I). In addition, we found that the SOCS domain of WSB1 was required for its interaction with Cul5 and Elongin B (Fig. 2J). In cells depleted of Cul5, pVHL stability was also significantly increased (Fig. 2K). These results suggest that the Cul5/Elongin/WSB1 ubiquitin ligase regulates pVHL stability.
The SOCS domain of WSB1 is required for pVHL interaction and ubiquitination
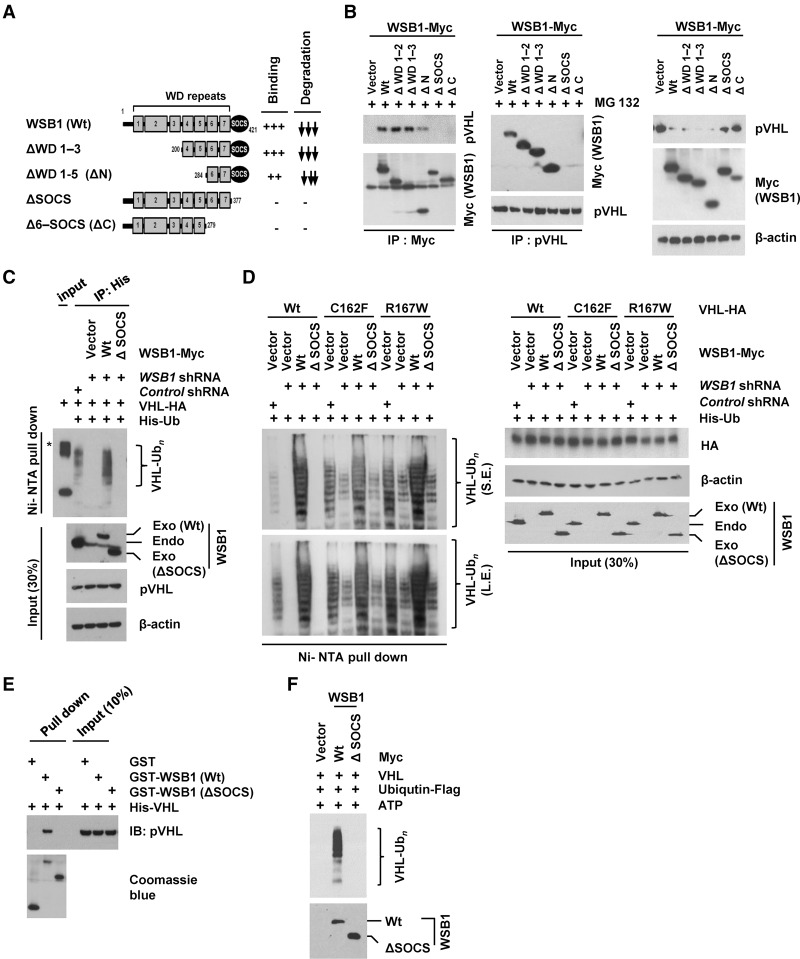
To determine the regions of WSB1 that are required for its interaction with pVHL, we expressed Myc-tagged WSB1 deletion mutants in cells and performed coimmunoprecipitation (co-IP). We found that WSB1's interaction with pVHL also required the SOCS domain. Consistent with the important role of the SOCS domain, down-regulation of pVHL level by WSB1 was dependent on the SOCS domain (Fig. 3A,B). WSB1 fragments that contain the SOCS domain decreased pVHL levels, while fragments lacking the SOCS domain could not do so (Fig. 3B). Furthermore, pVHL polyubiquitination was decreased in cells stably expressing WSB1 shRNA, while reconstitution with shRNA-resistant WSB1 restored pVHL polyubiquitination. However, reconstituting these cells with WSB1 (ΔSOCS) could not do so (Fig. 3C). Because pVHL is also an E3 ligase, to confirm WSB1-mediated ubiquitination of pVHL, we next used E3 ligase-deficient pVHL mutants C162F and R167F (Kamura et al. 2002; Jung et al. 2012). We found that the ubiquitination of E3 ligase-deficient pVHL mutants was promoted by WSB1 but not WSB1 (ΔSOCS) (Fig. 3D). We also found that WSB1 interacted with pVHL directly in vitro (Fig. 3E), and WSB1 promoted pVHL polyubiquitination in vitro (Fig. 3F). These results suggest that WBS1 is an E3 ligase that directly regulates pVHL ubiquitination in cells.
Figure 3.
WSB1 interacts with pVHL and promotes pVHL ubiquitination through the its SOCS domain. (A) Diagrams of wild-type WSB1 and corresponding deletion mutants (ΔWD 1–3, ΔWD 1–5, ΔSOCS, and Δ6-SOCS) used in co-IP experiments with pVHL. Plus and minus symbols indicate the binding affinity of each WSB1 mutants with the pVHL, and arrows indicate the effect of the WSB1 mutants (decreasing pVHL stability). (B) Cells were transfected with the indicated plasmids. (Left) Cells were pretreated with MG132, and the WSB1–pVHL interaction was examined by immunoprecipitation. (Right) pVHL levels were examined without MG132 pretreatment. (C) Deletion of WSB1's SOCS domain inhibits WSB1's E3 ligase activity toward pVHL. Cells were transfected with the indicated constructs and then treated with MG132. Ubiquitinated proteins were pulled down under denaturing conditions by Ni-NTA agarose and analyzed by immunoblot. An asterisk indicates a nonspecific band. (Exo) Exogenous; (Endo) endogenous. (D) HEK293T cells were transfected with the indicated constructs, including wild-type pVHL or loss of E3 ligase function pVHL (C162F and R167W). Cells were then treated with MG132. Ubiquitinated proteins were pulled down under denaturing conditions by Ni-NTA agarose and analyzed by immunoblot. WSB1 antibody was used to detect both endo-WSB1 and exo-WSB1 levels. (S.E.) Short exposed; (L.E.) long exposed; (Exo) exogenous; (Endo) endogenous. (E) In vitro binding assay of recombinant WSB1 with pVHL was performed. (F) WSB1 (wild-type or ΔSOCS) was purified from cells and used in an in vitro ubiquitination reaction using recombinant pVHL.
WSB1 regulates HIF activity
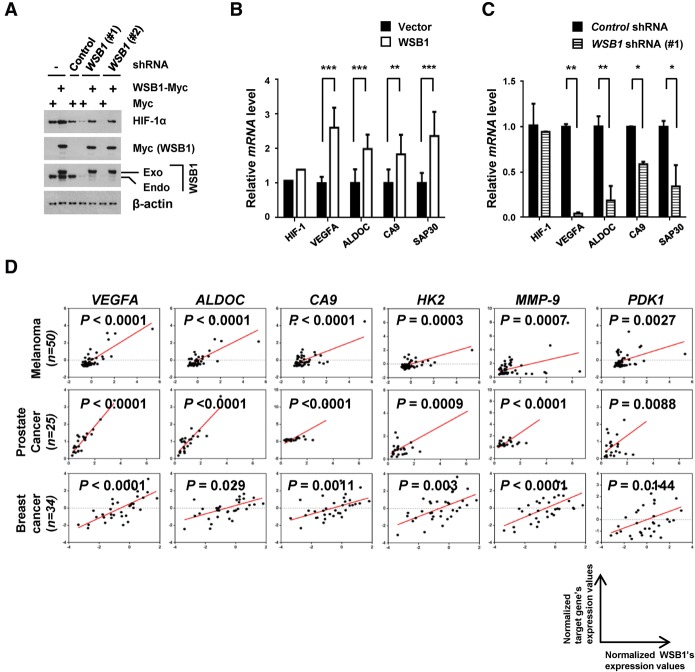
In agreement with the role of WSB1 as a pVHL inhibitor, we found that HIF-1α level increased in cells overexpressing WSB1 (Fig. 2C) and decreased in cells depleted of WSB1 (Fig. 4A; Supplemental Fig. S5A). Reconstitution of WSB1 in cells depleted of WSB1 restored HIF-1α levels, suggesting that WSB1 positively regulates HIF-1α levels. We next examined the mRNA level of HIF-1α’s target genes such as VEGFA, ALDOC, CA9, and SAP30 (Cairns et al. 2011; Montagner et al. 2012; Gilkes et al. 2014) in cells overexpressing WSB1 or depleted of WSB1. We found that WSB1 positively regulated the mRNA level of HIF-1α target genes (Fig. 4B,C; Supplemental Fig. S5B). However, HIF-1α mRNA level was not affected by WSB1. These results suggest that WSB1 positively regulates HIF-1α levels post-transcriptionally. A previous report suggests that WSB1 itself is a HIF-1 target gene (Tong et al. 2013). Therefore, HIF-1 and WSB1 could form a positive feedback loop through WSB1's negative regulation of pVHL. This is consistent with a positive correlation between WSB1 and the HIF targets genes in patient samples from a variety metastatic cancers, including 56 pairs of lung adenocarcinoma patients and five publicly clinically annotated gene expression data sets (Figs. 1D, 4D; Supplemental Fig. S6; Dyrskjot et al. 2004; Yu et al. 2004; Chandran et al. 2007; Boersma et al. 2008; Pantaleo et al. 2008; Xu et al. 2008).
Figure 4.
WSB1 up-regulates HIF-1α activity. (A) HEK293T cells were transfected with the indicated constructs, and HIF-1α level was examined. (Exo) Exogenous; (Endo) endogenous. (B,C) Quantitative PCR (qPCR) analyses of selected HIF-1α targets in HEK293T cells stably transfected with WSB1 cDNA (B) or WSB1 shRNA (C). Expression levels are relative to β-actin; data were normalized to control cells. The results represent the means ± SE of three independent experiments performed in triplicate. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 versus control cells by one-way ANOVA. (D) The corelation between WSB1 and HIF target gene expression in metastatic melanoma (n = 50), prostate cancer (n = 25), and breast cancer (n = 34) patients (Gene Expression Omnibus data set).
WSB1 promotes pVHL proteasomal degradation in hypoxia
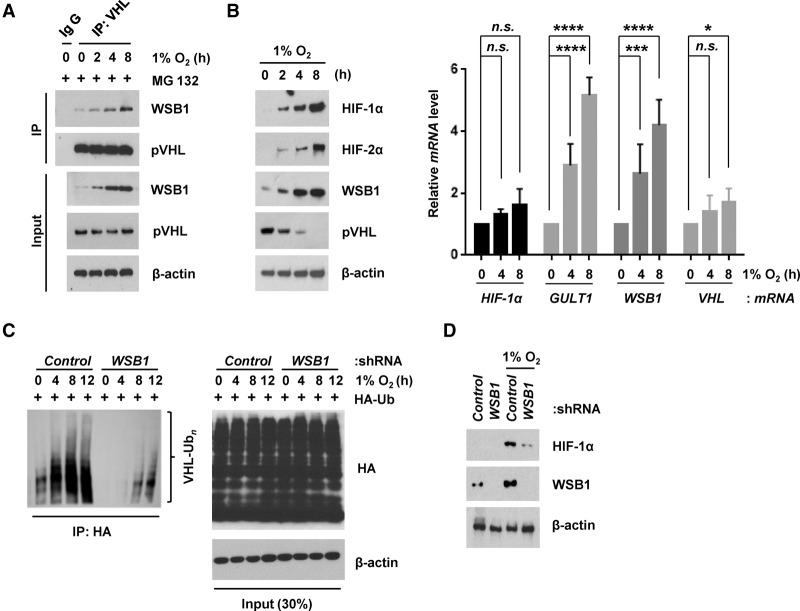
We showed that WSB1 regulates pVHL and HIF-1α levels under normoxic conditions. This raised the question of whether WSB1 also regulates pVHL and HIF-1α levels under hypoxic condition. HIFs have previously been shown to be unstable even under hypoxic conditions (Uchida et al. 2004; Kong et al. 2007; Liu et al. 2011). Furthermore, pVHL level could also decrease in hypoxia (Liu et al. 2011). We found that the interaction between WSB1 and pVHL increased under hypoxic and hypoxia-mimicking conditions (Fig. 5A; Supplemental Fig. S7A). WSB1 level also increased under hypoxia. This increase in the WSB1–pVHL interaction might be due to increased WSB1 expression. The increase of WSB1 under hypoxia is consistent with a previous report suggesting that WSB1 is a HIF-1 target gene (Tong et al. 2013). We also found that, in HIF-1α-depleted cells, hypoxia-induced WSB1 elevation was decreased (Supplemental Fig. S7B), confirming that WSB1 is a HIF-1 target gene. In addition, when cells were cultured under hypoxic conditions, endogenous pVHL level decreased, while HIF-1α, HIF-2α, and WSB1 levels increased (Fig. 5B; Supplemental Fig. S7C). Interestingly, VHL mRNA level was not significantly changed at early time points and was slightly increased at later time points under the same conditions. These results support that WSB1 regulates pVHL at a post-transcriptional level under hypoxic conditions. Furthermore, ubiquitination of pVHL increased under hypoxic conditions, and knocking down WSB1 decreased pVHL polyubiquitination (Fig. 5C). Knocking down WSB1 also decreased the up-regulation of HIF-1α under hypoxic conditions (Fig. 5D). Overall, these results suggest that WSB1 regulates pVHL turnover and contributes to HIF-1α and HIF-2α up-regulation under hypoxic conditions.
Figure 5.
WSB1 regulates pVHL under hypoxic conditions. (A) Endogenous WSB1 was coimmunoprecipitated with pVHL from HEK293T cell extracts under hypoxia for the indicated times. Cells were pretreated with MG132, and the WSB1–pVHL interaction was examined by immunoprecipitation. The 0 time point represents lysates from normoxic cultures that were prepared at the time of transfer to hypoxia. (B) HIF-1α, HIF-2α, and WSB1 protein levels as well as HIF-1α, GULT1, WSB1, and VHL mRNA levels were examined after the indicated times of hypoxia. (Left) Immunoblotting. (Right) qRT–PCR. (*) P < 0.05; (***) P < 0.001; (****) P < 0.0001 versus 0 h by one-way ANOVA. (C) HEK293 cells were transfected with control or WSB1 shRNA, and the ubiquitination of pVHL under hypoxia conditions was examined. (D) Cells were transfected with control or WSB1 shRNA, and HIF-1α expression was examined under hypoxia conditions.
WSB1 promotes cancer cell invasion, migration, and metastasis by inhibiting pVHL
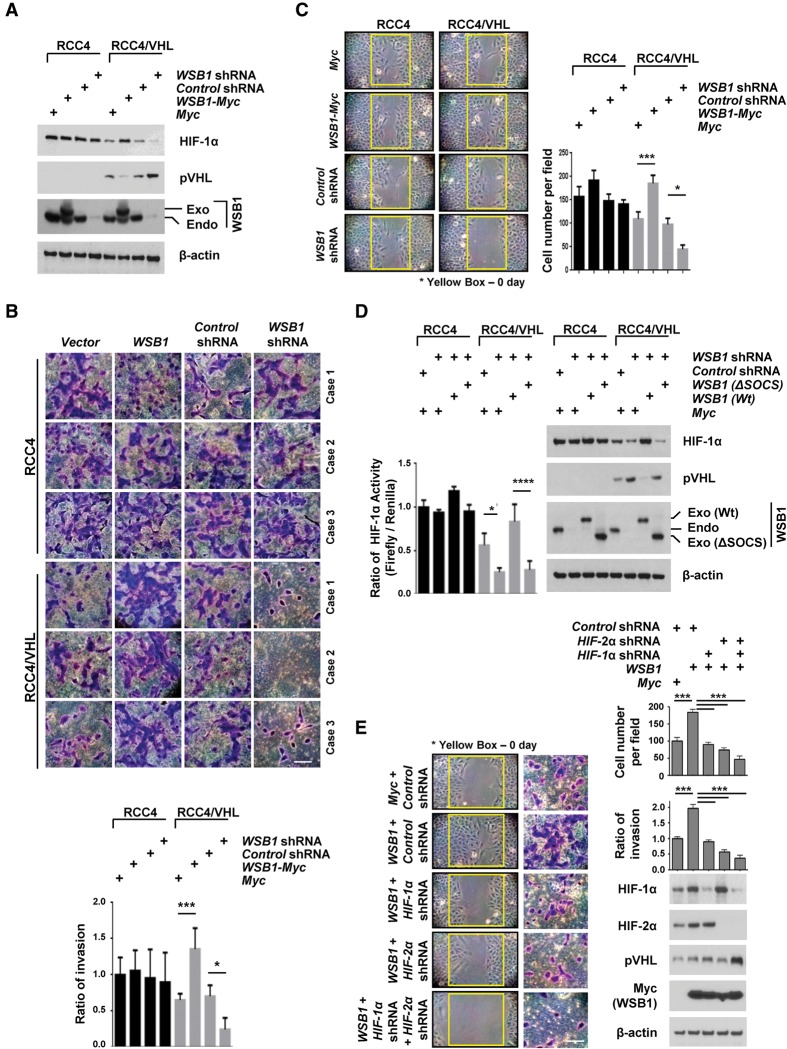
Based on our results, we hypothesized that WSB1 up-regulates HIF-α levels and promotes tumor metastasis through down-regulating pVHL. To confirm this, we used the RCC4 and 786-O renal carcinoma cell lines lacking pVHL as well as their derivatives, reconstituted HA-pVHL (RCC4/VHL and 786-O/VHL) (Hollingsworth et al. 2007). RCC4 expresses both of HIF-1α and HIF-2α, while the 786-O renal carcinoma cell line expresses HIF-2α but not HIF-1α. We discovered that overexpression of WSB1 had no effect on HIF-1α and HIF-2α levels in VHL-deficient RCC4 and 786-O cells (Fig. 6A; Supplemental Fig. S8A). Importantly, WSB1 was able to increase HIF-1α levels only in cells reconstituted with pVHL. Consistent with our hypothesis, overexpression of WSB1 had no significant effect on cell invasiveness and mobility in VHL-deficient RCC4 and 786-O cells, while substantially increased cell motility and invasiveness were observed in cells reconstituted with pVHL (Fig. 6B,C; Supplemental Fig. S8B–D). Furthermore, depletion of WSB1 decreased HIF-1α activity only in RCC4/VHL cells and 786-O/VHL cells but did not affect HIF-1α activity in RCC4 and 786-O cells (Fig. 6D). Cells reconstituted with wild-type WSB1 but not WSB1 (ΔSOCS) in cells depleted of WSB1 restored HIF-1α levels and activity (Fig. 6D). Similar results were obtained using the breast cancer cell line MDA-MB-231 (Supplemental Fig. S8E–G). Next, to determine whether the role of WSB1 in cell motility and invasion was mediated via the pVHL–HIF pathway, we knocked down HIF-1α or HIF-2α in WSB1-overexpressed RCC4/VHL cells. Depleting HIF-1α or HIF-2α inhibited the WSB1-induced increase in cell motility and cell invasion (Fig. 6E). This effect was more apparent in cells codepleted of HIF-1α and HIF-2α. These results establish that WSB1 regulates HIFs, cell motility, and cell invasion through pVHL.
Figure 6.
WSB1 promotes cancer cell invasion and migration by inhibiting pVHL. (A) Cells were transfected with the indicated constructs, and pVHL and HIF-1α levels were examined. (Exo) Exogenous; (Endo) endogenous. (B) Trans-well invasion assay of RCC4 or RCC4/VHL cells stably transfected the indicated shRNAs or plasmids. The plot shows the quantification of the area covered by the invasion cells, relative to the control. The results represent the means ± SE of three independent experiments performed in triplicate. Bar, 10 μm. (*) P < 0.05; (***) P < 0.001 versus control cells by one-way ANOVA. (C) Wound healing assays of RCC4 or RCC4/VHL cell lines stably transfected with the indicated plasmids or shRNA. (Left) Representative images of the assay. (Right) The mean and SD of three independent experiments performed in triplicate are shown. (*) P < 0.05; (***) P < 0.001 versus control cells by one-way ANOVA. (D) RCC4 or RCC4/VHL cells were infected with the indicated constructs, and HIF-1α activity was assayed by a HIF-1α luciferase reporter assay. (Left) The results represent the means ± SE of three independent experiments performed in triplicate. (*) P < 0.05; (****) P < 0.0001 versus control cells by one-way ANOVA. (Right) Cells were blotted with the indicated antibodies. (Exo) Exogenous; (Endo) endogenous. (E) Cells were infected with indicated constructs. Wound healing assays and trans-well invasion assays of HIF-1α, HIF-2α, and pVHL levels were examined. (Left) Representative images of the wound healing assay. (Top right) The mean and SD of three independent experiments performed in triplicate are shown. (Middle) Representative images of the invasion assay. Bar, 10 μm. (Middle right) The mean and SD of three independent experiments performed in triplicate are shown. (Bottom right) Immunoblot with indicated antibodies. (***) P < 0.001 by one-way ANOVA.
WSB1 has previously been shown to down-regulate HIPK2 through its E3 ligase activity (Choi et al. 2008; Tong et al. 2013). Because HIPK2 regulates multiple cellular processes, including p53 activation (Puca et al. 2010; Jin et al. 2012; Rinaldo et al. 2012), we next tested whether HIPK2 regulation by WSB1 plays a role in HIF-1α activity and cell mobility. As shown in Supplemental Figure S9, WSB1 overexpression was able to increase HIF-1α levels and cell mobility in cells depleted of HIPK2, suggesting that WSB1 regulates HIF-1α in a HIPK2-independent manner.
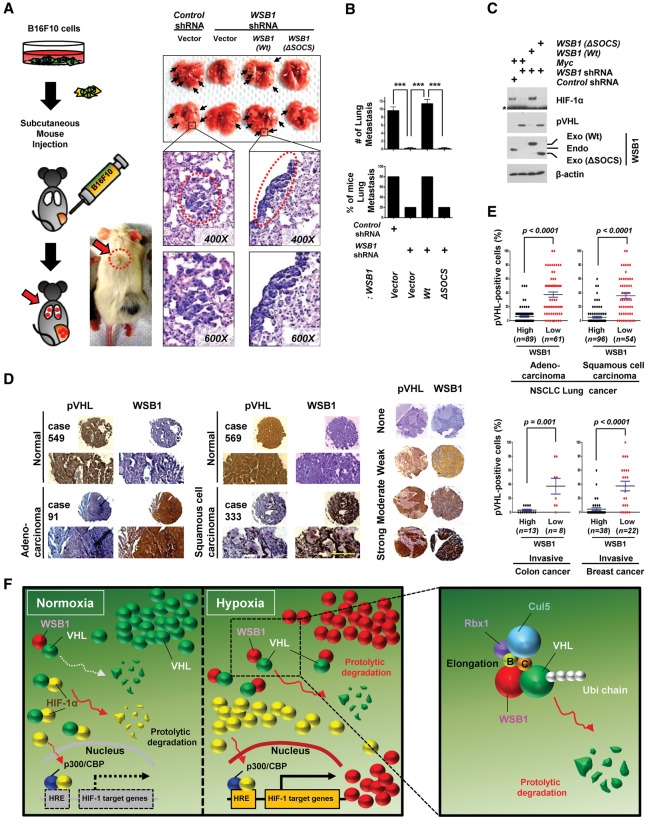
To investigate the functional relevance of WSB1 in malignant behavior in vivo, we performed colonization/metastasis assays in mouse models. We found that depletion of WSB1 resulted in a significant decrease of lung and liver colonization of B16F10 melanoma cells after tail vein injection (Supplemental Fig. S10). These effects were reversed by reconstitution of wild-type WSB1 but not WSB1 (ΔSOCS) (Supplemental Fig. S10). In addition, we examined lung metastasis after subcutaneously implanting B16F10 cells. We found that depletion of WSB1 inhibited lung metastasis (Fig. 7A,B). Reconstitution of cells with wild-type WSB1, but not WSB1 (ΔSOCS), in WSB1-depleted cells restored lung metastasis (Fig. 7A,B). pVHL level was up-regulated while HIF-1 level was down-regulated when WSB1 was depleted. These effects could be reversed by reconstituting wild-type WSB1 but not WSB1 (ΔSOCS) (Fig. 7C). These results suggest that WSB1 is important for cancer metastasis through pVHL degradation and HIF-1α up-regulation.
Figure 7.
WSB1 promotes cancer cell metastasis and negatively correlated with pVHL in various human cancers. (A–C) Lung colonization assay of mice subcutaneously injected with B16F10 cells stably infected with the indicated viral constructs or shRNA. (A, left) Schematic of the experiments. (Top right) Representative lung organs are shown. (Bottom right) H&E stain of mouse lungs with metastasis. B16F10 tumor metastasis (black arrows) is shown with two different magnifications (top panel, 400×; bottom panel, 600×). (B, top) The plot shows the number of metastatic foci per section. (Bottom) The percentage of mice with B16F10 metastasis. n = 5 for B16F10 cells. (***) P < 0.001 versus control cells by one-way ANOVA. (C) Cells as in A were then collected for immunoblot analysis. (Exo) Exogenous; (Endo) endogenous. An asterisk indicates a nonspecific band. (D, left) Representative immunohistochemical images of WSB1 and pVHL in adenocarcinoma and squamous cell carcinoma compared with normal lung tissues. Serial tumor sections from the same patient were processed. (Right) Immunostaining intensities were 0 (negative), 1+ (weak), 2+ (moderate), and +3 (strong). Bar, 50 μm. (E) Quantification of WSB1 and pVHL expression correlation in 400 human NSCLC tissue microarrays and 60 metastatic breast cancer and 21 metastatic colorectal carcinoma patients’ slides. The P-value was calculated by the Student's t-test. (F) Schematic model. WSB1 acts as a negative regulator of pVHL. WSB1 promotes pVHL ubiquitination and proteasomal degradation, thereby stabilizing HIF under both normoxia and hypoxia conditions. WSB1 boosts cancer cell invasion and metastasis through the pVHL–HIF pathway.
Finally, to examine the expression of WSB1 and pVHL in human tumor tissues, we performed immunohistochemical staining of WSB1 and pVHL in 400 lung cancer specimens (n = 300 lung cancer specimens; n = 100 lung normal specimens) spotted on a tissue microarray (TMA) (Fig. 7D,E) and 60 metastatic breast cancer and 21 metastatic colorectal carcinoma patients' slides (Fig. 7E). We found that WSB1 expression is higher in cancer lesions compared with normal adjacent tissues, while pVHL expression negatively correlates with WBS1. These results are consistent with the negative regulation of pVHL by WSB1 in human cancers.
Discussion
In the present study, we define a novel mechanistic link between WSB1 and pVHL (presented in Fig. 7F) in which WSB1 induces pVHL degradation through its E3 ligase function. We propose that the negative regulation of pVHL and up-regulation of HIF is one mechanism by which WSB1 promotes tumor progression.
Several mechanisms have been shown to stabilize pVHL. pVHL is stabilized by Elongin B/C (Schoenfeld et al. 2000). In addition, APC/Ccdh1 has been shown to regulate pVHL in a cell cycle-dependent manner (Liu et al. 2011). However, how this regulation affects pVHL function in tumor initiation and progression is not clear. WSB1 is overexpressed in many cancers, and its expression is positively correlated with cancer progression (Archange et al. 2008; Silva et al. 2011; Tong et al. 2013). However, its cellular function remains unclear. We show that WSB1 regulates pVHL protein stability under not only hypoxic conditions but also normoxia through the ubiquitination–proteasome pathways. In addition, we presented clinical and functional evidence that this pathway is important for metastatic invasion. Our findings provide important clues for understanding WSB1-associated tumorigenesis. Furthermore, the increase of HIF-1α level by WSB1 through pVHL's degradation may explain why high levels of HIF-1α are detected in normoxic regions of human tumors with functional pVHL (Supplemental Fig. S4B; Zhong et al. 1999; Akakura et al. 2001; Saito et al. 2006).
It has been suggested that WSB1 is one of HIF-1α target genes, and its expression is regulated by HIF-1α (Benita et al. 2009; Tong et al. 2013). We also determined that WSB1 expression increased under hypoxic condition, and it is dependent on HIF-1α. Our data suggest that WSB1 could in turn up-regulate HIF-α through the ubiquitination of pVHL. Therefore, HIF-1 and WSB1 form a positive feedback loop in enhancing HIF-1 function. This positive feedback loop ensures robust activation of HIFs.
The expression of HIF-1α and HIF-2α has been positively correlated with a wide variety of human cancers, usually is associated with poor prognosis (Klatte et al. 2007; Keith et al. 2012), and is inversely correlated with pVHL (Gordan et al. 2008). Several studies support the opinion that HIF-1α and HIF-2α promote tumor angiogenesis and growth (Gordan and Simon 2007; Klatte et al. 2007; Keith et al. 2012). Constitutively active HIFs induce the development of ccRCC in mice (Fu et al. 2011, 2013). Consistently, HIF-1α-expressed renal cell carcinomas are also more metastatic (Semenza 2010), and HIF-2α has been suggested as a major protein in VHL-deficient renal carcinogenesis (Frew et al. 2008). Conversely, some studies suggest a tumor suppressor role for HIFα (Shen et al. 2011). The expression of HIF-1α is related to a favorable prognosis in human neuroblastoma and renal cell carcinomas (Xu et al. 2010; Keith et al. 2012; Medina Villaamil et al. 2012). HIF-2α also induces apoptosis in glioma and inhibits oncogenic signaling and activates a tumor suppressor gene in NSCLC (Acker et al. 2005; Mazumdar et al. 2010). However the mechanism by which HIF-1α and HIF-2α function as tumor suppressors remains unclear. Thus, the role of HIF-1α and HIF-2α in cancer biology might be context-dependent and needs to be further studied. As pVHL negatively regulates cell growth and tumor metastatic invasion, WSB1 might act as a main amplifier in the HIF–pVHL pathway. The present study may assist in elucidating the underlying mechanisms by which HIF-1α or HIF-2α affects cancer development.
In this study, we also demonstrate the up-regulation of WSB1 in a high percentage of metastatic lung, breast, colorectal, melanoma, prostate, and urinary bladder cancers. Although the detailed mechanism by which WSB1 is up-regulated in cancer remains to be determined, it is conceivable that both HIF-dependent and HIF-independent mechanisms exist for WSB1 regulation in cancers. For example, CREB and ATF5 have been shown to bind the WSB1 promoter (Rhodes and Chinnaiyan 2005). Additionally, post-translational modifications of WSB1 might also be involved in the regulation of WSB1 level and activity. These types of regulation remain to be studied in the future.
Overall, our study is the first to report a cellular E3 ligase for pVHL that regulates metastasis in animal models and clinical samples. Our study will help to understand how misregulation of the pVHL–HIF pathway contributes to human malignancy and reveal potential targets for therapeutic intervention of human cancers.
Materials and methods
Cells, cell lines, and reagents
All cell lines were sourced from commercial vendors. Human embryonic kidney 293T (HEK293T), HEK293, HeLa cervix carcinoma cells, RCC4, RCC4/VHL, 786-O, and 786-O/VHL human renal carcinoma cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco-Invitrogen). Five human lung cancer cell lines (three adenocarcinoma: H522, H1650, and A549; and two large cell carcinoma: H460 and H1299), six human pancreatic cancer cells (BxPC3, Hup-T3, Mia-Paca, Panc1, Pan 04.03, and ASPC1), four human breast cancer cells (HT-29, HCC1937, HCC1806, and MDA231), and mouse melanoma cells (B16F10) were maintained in Eagle's minimal essential medium (EMEM) or RPMI 1640 (Gibco-Invitrogen). RCC4, RCC4/VHL, 786-O, and 786-O/VHL cells were kindly provided by Dr. William G. Kaelin Jr. (Department of Medicine, Harvard Medical School, Dana-Farber Cancer Institute, Massachusetts). All media contained 10% heat-inactivated FBS (Gibco-Invitrogen), 2 mg/mL sodium bicarbonate (Sigma-Aldrich), 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco-Invitrogen). MG132, cycloheximide, and CoCl2 were purchased from Sigma-Aldrich.
Gene expression profiling from 56 pairs of fresh-frozen primary never-smoked lung adenocarcinomas patients
Gene expression profiling was described previously (Jang et al. 2012). Briefly, after RNA extraction and gene expression profiling by microarray from 56 primary lung tumors, the data for mRNA were processed and normalized through BeadStudio software version 3.0. (Illumina, Inc.) using the quantile normalization method and then log2 transformed and analyzed using the Partek Genomics suite (Partek, Inc.). To identify differentially expressed genes, we applied the one-way ANOVA model for all analyses. mRNA genes with a fold change greater than ±1.5 at a raw P-value of <0.01 and a false- discovery rate (FDR) of <5% were considered significant. We further subclassified fresh-frozen tumors with WSB1 “high” (H) and “low” (L) groups based on the mean of expression after normalization and compared mRNA gene expression differences between the WSB1-H and WSB1-L tumors. We found 2534 mRNA genes (data not shown) differentially expressed based on a fold change of >1.5 and a FDR of <5% using this analysis. We analyzed these 2534 genes using IPA 8.5 software (Ingenuity Systems, http://www.ingenuity.com) to identify biological function in lung adenocarcinoma potentially modified by WSB1 expression level.
Cancer data collection and processing
We retrieved several cancer data sets from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; GDS3966 [Xu et al. 2008], GDS2545 [Yu et al. 2004; Chandran et al. 2007], GDS1479 [Dyrskjot et al. 2004], GDS3501 [Pantaleo et al. 2008], and GDS3096 [Boersma et al. 2008]) and PROGgene (http://watson.compbio.iupui.edu/chirayu/proggene/database/?url=proggene) containing patients’ clinical information and gene expression data (Supplemental Table 1).
Among the various data sets, specifically, we defined two major events; i.e., WSB1 gene levels in metastatic patient samples and the relationship between WSB1 and HIF target genes. To identify the relationship between WSB1 and HIF target genes, we used the classifier described previously (Montagner et al. 2012). Briefly, we defined a classification rule based on summarizing the standardized expression levels of WSB1 and HIF-1α target genes and analyzed log2 expression values of both WSB1 and HIF target genes by the Prism program.
We defined the relationship between WSB1 gene levels and the metastasis-free survival in human breast cancer and human colon cancer patient data sets using the PROGgene Web site (http://watson.compbio.iupui.edu/chirayu/proggene/database/?url=proggene).
In vitro ubiquitination assays
After transfection with Myc or Myc-WSB1 (wild-type and ΔSOCS), cells were collected for immunoprecipitation–immunoblot analysis. WSB1 was immunoprecipitated with Myc antibody and incubated with reaction buffer (50 mM Tris-HCl at pH 7.5, 2.5 mM MgCl2, 0.05% Nonidet P-40, 0.5 mM dithiothreitol), 5 mM Flag-ubiquitin or Myc-ubiquitin, 2 mM ATP, substrate, and recombinant VHL (Abnova) for 90 min at 32°C. Equal volumes of each sample were prepared for the immunoblot. The reaction products were analyzed by immunoblot with pVHL antibody.
In vivo ubiquitination assays
For in vivo ubiquitination, cells were transfected with ubiquitin-his plasmid together with Myc, Myc-WSB1 (wild-type and ΔSOCS), shRNAs, or HA-tagged VHL (wild-type, C162F, and R167W) followed by treatment with 10 μM MG132. Forty-eight hours after transfection, cells were lysed by urea lysis buffer (8 M urea, 0.1 M Na2HPO4, 0.1 M Tris/HCl at pH 8.0, 0.05% Tween 20, 0.01 M imidazole). After centrifugation, the supernatants were collected and incubated with 20 mL of Ni-NTA agarose beads (Qiagen) for 4 h at 4°C. The precipitates were washed three times with urea wash buffer (8 M urea, 0.1 M Na2HPO4, 0.1 M Tris/HCl at pH 8.0, 0.05% Tween 20, 0.02 M imidazole) and Native wash buffer (0.1 M Na2HPO4, 0.1 M Tris/HCl at pH 8.0, 0.05% Tween 20, 0.02 M imidazole), boiled with SDS loading buffer, and then subjected to SDS-PAGE followed by immunoblot analysis.
In vitro binding assay
GST fusion proteins were prepared following standard protocol (Yuan et al. 2010). For in vitro biding assays, WSB1 (wild-type and ΔSOCS) GST fusion proteins bound to GSH Sepharose were incubated with cell lysates. After washing, the bound proteins were separated by SDS-PAGE and immunoblotted with the indicated antibodies.
In vivo assays for animal experiments
Mice were housed in specific pathogen-free (SPF) animal facilities and treated in conformity with approved institutional guidelines (Mayo Clinic). For metastasis assays, B16F10 cells were resuspended in 100 μL of PBS and injected into the tail veins of C57/Bl6 male mice, aged-matched between 5 and 7 wk. We injected seven mice for each sample (3 × 105 cells for each mouse). After 4 wk, animals were sacrificed, and lungs and livers were removed. A subcutaneous metastasis was described previously (Skuli et al. 2009). Briefly, xenograft tumors were generated by injecting B16F10 melanoma cells subcutaneously into the flanks of NOD/SCID mice. After 6 wk, mice were sacrificed, and lung tissues were harvested. Lungs were fixed and serially sectioned to evaluate metastasis formation.
Immunohistochemistry
The tissue arrays included a lung tumor TMA containing 400 pairs of human lung cancer, 21 human invasive colon cancers, and 60 human invasive breast cancers and matched or unmatched normal adjacent tissue. We performed immunohistochemical staining as previously described (Yuan et al. 2010). The primary antibody dilutions used were anti-WSB1 (1:200; Sigma-Aldrich) and anti-pVHL (1:100; Santa Cruz Biotechnology).
Statistical analysis
Each assay was performed in triplicate and independently repeated at least three times. The results were presented as mean ± standard error of the mean. Statistical analyses were performed using GraphPad Prism software version 4.02 (GraphPad Software). One-way ANOVA followed by t-test was used to compare the results. A difference was considered significant if P < 0.05. Statistical significance was defined as P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
Supplementary Material
Acknowledgments
We thank all members of Dr. Lou's laboratory (Dr. K. Luo, Dr. J. Yuan, Dr. H. Pei, Dr. P. Yin, Dr. Y. Li, Dr. F. Yuan, Dr. W. Guo, Dr. L. Zhang, Dr. B. Xu, Dr. H. Shi, S Nowsheen, and Y-H. Lin) for their critical discussion of this work. We thank Dr. M.S. Kim and Dr. C.H. Park for animal experiments. We thank Dr. C.Y. Choi for WSB1 plasmids, and Dr. William G. Kaelin Jr. for RCC4, RCC4/VHL, 786-O, and 786-O/VHL cells. This work was supported by National Institutes of Health grants CA130996, CA148940, and CA189666 to Z.L.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.268128.115.
References
- Acker T, Diez-Juan A, Aragones J, Tjwa M, Brusselmans K, Moons L, Fukumura D, Moreno-Murciano MP, Herbert JM, Burger A, et al. 2005. Genetic evidence for a tumor suppressor role of HIF-2α. Cancer Cell 8: 131–141. [DOI] [PubMed] [Google Scholar]
- Akakura N, Kobayashi M, Horiuchi I, Suzuki A, Wang J, Chen J, Niizeki H, Kawamura K, Hosokawa M, Asaka M. 2001. Constitutive expression of hypoxia-inducible factor-1α renders pancreatic cancer cells resistant to apoptosis induced by hypoxia and nutrient deprivation. Cancer Res 61: 6548–6554. [PubMed] [Google Scholar]
- Archange C, Nowak J, Garcia S, Moutardier V, Calvo EL, Dagorn JC, Iovanna JL. 2008. The WSB1 gene is involved in pancreatic cancer progression. PLoS One 3: e2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch B, Jilg C, Glasker S, Vortmeyer A, Lutzen N, Anton A, Eng C, Neumann HP. 2013. Renal cancer in von Hippel-Lindau disease and related syndromes. Nat Rev Nephrol 9: 529–538. [DOI] [PubMed] [Google Scholar]
- Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ. 2009. An integrative genomics approach identifies hypoxia inducible factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res 37: 4587–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma BJ, Reimers M, Yi M, Ludwig JA, Luke BT, Stephens RM, Yfantis HG, Lee DH, Weinstein JN, Ambs S. 2008. A stromal gene signature associated with inflammatory breast cancer. Int J Cancer 122: 1324–1332. [DOI] [PubMed] [Google Scholar]
- Bovellan M, Romeo Y, Biro M, Boden A, Chugh P, Yonis A, Vaghela M, Fritzsche M, Moulding D, Thorogate R, et al. 2014. Cellular control of cortical actin nucleation. Curr Biol 24: 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. 2011. Regulation of cancer cell metabolism. Nat Rev Cancer 11: 85–95. [DOI] [PubMed] [Google Scholar]
- Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. 2007. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 7: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Chen S, Huang C, Cheng H, Zhou R. 2013. TCTP increases stability of hypoxia-inducible factor 1α by interaction with and degradation of the tumour suppressor VHL. Biol Cell 105: 208–218. [DOI] [PubMed] [Google Scholar]
- Choi DW, Seo YM, Kim EA, Sung KS, Ahn JW, Park SJ, Lee SR, Choi CY. 2008. Ubiquitination and degradation of homeodomain-interacting protein kinase 2 by WD40 repeat/SOCS box protein WSB-1. J Biol Chem 283: 4682–4689. [DOI] [PubMed] [Google Scholar]
- Dentice M, Bandyopadhyay A, Gereben B, Callebaut I, Christoffolete MA, Kim BW, Nissim S, Mornon JP, Zavacki AM, Zeold A, et al. 2005. The Hedgehog-inducible ubiquitin ligase subunit WSB-1 modulates thyroid hormone activation and PTHrP secretion in the developing growth plate. Nat Cell Biol 7: 698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434. [DOI] [PubMed] [Google Scholar]
- Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Moller K, Orntoft TF. 2004. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res 64: 4040–4048. [DOI] [PubMed] [Google Scholar]
- Frew IJ, Thoma CR, Georgiev S, Minola A, Hitz M, Montani M, Moch H, Krek W. 2008. pVHL and PTEN tumour suppressor proteins cooperatively suppress kidney cyst formation. EMBO J 27: 1747–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Wolf K, Zegers MM. 2014. Rho-directed forces in collective migration. Nat Cell Biol 16: 208–210. [DOI] [PubMed] [Google Scholar]
- Fu L, Wang G, Shevchuk MM, Nanus DM, Gudas LJ. 2011. Generation of a mouse model of Von Hippel-Lindau kidney disease leading to renal cancers by expression of a constitutively active mutant of HIF1α. Cancer Res 71: 6848–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Wang G, Shevchuk MM, Nanus DM, Gudas LJ. 2013. Activation of HIF2α in kidney proximal tubule cells causes abnormal glycogen deposition but not tumorigenesis. Cancer Res 73: 2916–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Semenza GL, Wirtz D. 2014. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer 14: 430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Simon MC. 2007. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev 17: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Lal P, Dondeti VR, Letrero R, Parekh KN, Oquendo CE, Greenberg RA, Flaherty KT, Rathmell WK, Keith B, et al. 2008. HIF-α effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell 14: 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossage L, Eisen T, Maher ER. 2015. VHL, the story of a tumour suppressor gene. Nat Rev Cancer 15: 55–64. [DOI] [PubMed] [Google Scholar]
- Hollingsworth JM, Miller DC, Daignault S, Hollenbeck BK. 2007. Five-year survival after surgical treatment for kidney cancer: a population-based competing risk analysis. Cancer 109: 1763–1768. [DOI] [PubMed] [Google Scholar]
- Jang JS, Jeon HS, Sun Z, Aubry MC, Tang H, Park CH, Rakhshan F, Schultz DA, Kolbert CP, Lupu R, et al. 2012. Increased miR-708 expression in NSCLC and its association with poor survival in lung adenocarcinoma from never smokers. Clin Cancer Res 18: 3658–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Ratnam K, Chuang PY, Fan Y, Zhong Y, Dai Y, Mazloom AR, Chen EY, D'Agati V, Xiong H, et al. 2012. A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nat Med 18: 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CR, Hwang KS, Yoo J, Cho WK, Kim JM, Kim WH, Im DS. 2006. E2-EPF UCP targets pVHL for degradation and associates with tumor growth and metastasis. Nat Med 12: 809–816. [DOI] [PubMed] [Google Scholar]
- Jung YS, Lee SJ, Yoon MH, Ha NC, Park BJ. 2012. Estrogen receptor α is a novel target of the Von Hippel-Lindau protein and is responsible for the proliferation of VHL-deficient cells under hypoxic conditions. Cell Cycle 11: 4462–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG. 2007. Von Hippel-Lindau disease. Annu Rev Pathol 2: 145–173. [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr. 2008. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer 8: 865–873. [DOI] [PubMed] [Google Scholar]
- Kamura T, Brower CS, Conaway RC, Conaway JW. 2002. A molecular basis for stabilization of the von Hippel-Lindau (VHL) tumor suppressor protein by components of the VHL ubiquitin ligase. J Biol Chem 277: 30388–30393. [DOI] [PubMed] [Google Scholar]
- Keith B, Johnson RS, Simon MC. 2012. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 12: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatte T, Seligson DB, Riggs SB, Leppert JT, Berkman MK, Kleid MD, Yu H, Kabbinavar FF, Pantuck AJ, Belldegrun AS. 2007. Hypoxia-inducible factor 1α in clear cell renal cell carcinoma. Clin Cancer Res 13: 7388–7393. [DOI] [PubMed] [Google Scholar]
- Kong X, Alvarez-Castelao B, Lin Z, Castano JG, Caro J. 2007. Constitutive/hypoxic degradation of HIF-α proteins by the proteasome is independent of von Hippel Lindau protein ubiquitylation and the transactivation activity of the protein. J Biol Chem 282: 15498–15505. [DOI] [PubMed] [Google Scholar]
- Liu W, Xin H, Eckert DT, Brown JA, Gnarra JR. 2011. Hypoxia and cell cycle regulation of the von Hippel-Lindau tumor suppressor. Oncogene 30: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los M, Jansen GH, Kaelin WG, Lips CJ, Blijham GH, Voest EE. 1996. Expression pattern of the von Hippel-Lindau protein in human tissues. Lab Invest 75: 231–238. [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. 2010. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell 40: 294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH. 2005. The HIF pathway in cancer. Semin Cell Dev Biol 16: 523–530. [DOI] [PubMed] [Google Scholar]
- Mazumdar J, Hickey MM, Pant DK, Durham AC, Sweet-Cordero A, Vachani A, Jacks T, Chodosh LA, Kissil JL, Simon MC, et al. 2010. HIF-2α deletion promotes Kras-driven lung tumor development. Proc Natl Acad Sci 107: 14182–14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina Villaamil V, Aparicio Gallego G, Santamarina Cainzos I, Valladares-Ayerbes M, Anton Aparicio LM. 2012. Searching for Hif1-α interacting proteins in renal cell carcinoma. Clin Transl Oncol 14: 698–708. [DOI] [PubMed] [Google Scholar]
- Montagner M, Enzo E, Forcato M, Zanconato F, Parenti A, Rampazzo E, Basso G, Leo G, Rosato A, Bicciato S, et al. 2012. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature 487: 380–384. [DOI] [PubMed] [Google Scholar]
- Pantaleo MA, Astolfi A, Nannini M, Paterini P, Piazzi G, Ercolani G, Brandi G, Martinelli G, Pession A, Pinna AD, et al. 2008. Gene expression profiling of liver metastases from colorectal cancer as potential basis for treatment choice. Br J Cancer 99: 1729–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup M, Novitskiy S, Moses HL. 2013. The roles of TGFβ in the tumour microenvironment. Nat Rev Cancer 13: 788–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzebon ME, Varadaraj A, Mattoscio D, Jaffray EG, Miccolo C, Galimberti V, Tommasino M, Hay RT, Chiocca S. 2013. BC-box protein domain-related mechanism for VHL protein degradation. Proc Natl Acad Sci 110: 18168–18173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca R, Nardinocchi L, Givol D, D'Orazi G. 2010. Regulation of p53 activity by HIPK2: molecular mechanisms and therapeutical implications in human cancer cells. Oncogene 29: 4378–4387. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Chinnaiyan AM. 2005. Integrative analysis of the cancer transcriptome. Nat Genet 37: S31–S37. [DOI] [PubMed] [Google Scholar]
- Rinaldo C, Moncada A, Gradi A, Ciuffini L, D'Eliseo D, Siepi F, Prodosmo A, Giorgi A, Pierantoni GM, Trapasso F, et al. 2012. HIPK2 controls cytokinesis and prevents tetraploidization by phosphorylating histone H2B at the midbody. Mol Cell 47: 87–98. [DOI] [PubMed] [Google Scholar]
- Saito Y, Sunamura M, Motoi F, Abe H, Egawa S, Duda DG, Hoshida T, Fukuyama S, Hamada H, Matsuno S. 2006. Oncolytic replication-competent adenovirus suppresses tumor angiogenesis through preserved E1A region. Cancer Gene Ther 13: 242–252. [DOI] [PubMed] [Google Scholar]
- Schoenfeld AR, Davidowitz EJ, Burk RD. 2000. Elongin BC complex prevents degradation of von Hippel-Lindau tumor suppressor gene products. Proc Natl Acad Sci 97: 8507–8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. 2010. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 29: 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. 2011. Oxygen sensing, homeostasis, and disease. N Engl J Med 365: 537–547. [DOI] [PubMed] [Google Scholar]
- Shen C, Beroukhim R, Schumacher SE, Zhou J, Chang M, Signoretti S, Kaelin WG Jr. 2011. Genetic and functional studies implicate HIF1α as a 14q kidney cancer suppressor gene. Cancer Discov 1: 222–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AS, Wood SH, van Dam S, Berres S, McArdle A, de Magalhaes JP. 2011. Gathering insights on disease etiology from gene expression profiles of healthy tissues. Bioinformatics 27: 3300–3305. [DOI] [PubMed] [Google Scholar]
- Skuli N, Liu L, Runge A, Wang T, Yuan L, Patel S, Iruela-Arispe L, Simon MC, Keith B. 2009. Endothelial deletion of hypoxia-inducible factor-2α (HIF-2α) alters vascular function and tumor angiogenesis. Blood 114: 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Li XB, Meng Y, Fan L, Li M, Fang J. 2013. TRAF6 upregulates expression of HIF-1α and promotes tumor angiogenesis. Cancer Res 73: 4950–4959. [DOI] [PubMed] [Google Scholar]
- Tong Y, Li QG, Xing TY, Zhang M, Zhang JJ, Xia Q. 2013. HIF1 regulates WSB-1 expression to promote hypoxia-induced chemoresistance in hepatocellular carcinoma cells. FEBS Lett 587: 2530–2535. [DOI] [PubMed] [Google Scholar]
- Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, Clerici C. 2004. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1α and HIF-2α expression in lung epithelial cells: implication of natural antisense HIF-1α. J Biol Chem 279: 14871–14878. [DOI] [PubMed] [Google Scholar]
- Vasiliauskas D, Hancock S, Stern CD. 1999. SWiP-1: novel SOCS box containing WD-protein regulated by signalling centres and by Shh during development. Mech Dev 82: 79–94. [DOI] [PubMed] [Google Scholar]
- Xu L, Shen SS, Hoshida Y, Subramanian A, Ross K, Brunet JP, Wagner SN, Ramaswamy S, Mesirov JP, Hynes RO. 2008. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res 6: 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Ding Q, Fang Z, Zheng J, Gao P, Lu Y, Zhang Y. 2010. Silencing of HIF-1α suppresses tumorigenicity of renal cell carcinoma through induction of apoptosis. Cancer Gene Ther 17: 212–222. [DOI] [PubMed] [Google Scholar]
- Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al. 2004. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol 22: 2790–2799. [DOI] [PubMed] [Google Scholar]
- Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. 2010. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 140: 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. 1999. Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res 59: 5830–5835. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.