Haakonsen et al. find that the essential cell cycle regulator GcrA in Caulobacter crescentus forms a stable complex with RNA polymerase and localizes to almost all active σ70-dependent promoters in vivo but activates transcription primarily at promoters harboring certain DNA methylation sites. GcrA could stabilize RNA polymerase binding and directly stimulate open complex formation to activate transcription.
Keywords: Caulobacter crescentus, DNA methylation, bacterial transcription, cell cycle
Abstract
Cell cycle progression in most organisms requires tightly regulated programs of gene expression. The transcription factors involved typically stimulate gene expression by binding specific DNA sequences in promoters and recruiting RNA polymerase. Here, we found that the essential cell cycle regulator GcrA in Caulobacter crescentus activates the transcription of target genes in a fundamentally different manner. GcrA forms a stable complex with RNA polymerase and localizes to almost all active σ70-dependent promoters in vivo but activates transcription primarily at promoters harboring certain DNA methylation sites. Whereas most transcription factors that contact σ70 interact with domain 4, GcrA interfaces with domain 2, the region that binds the −10 element during strand separation. Using kinetic analyses and a reconstituted in vitro transcription assay, we demonstrated that GcrA can stabilize RNA polymerase binding and directly stimulate open complex formation to activate transcription. Guided by these studies, we identified a regulon of ∼200 genes, providing new insight into the essential functions of GcrA. Collectively, our work reveals a new mechanism for transcriptional regulation, and we discuss the potential benefits of activating transcription by promoting RNA polymerase isomerization rather than recruitment exclusively.
Tightly regulated programs of transcription underlie countless cellular and developmental processes throughout biology. In bacteria, transcription is performed by the multisubunit DNA-dependent RNA polymerase (RNAP). The RNAP core enzyme (composed of α2ββ′ω subunits) is capable of RNA synthesis but cannot initiate promoter-specific transcription. Transcription initiation at promoters requires a σ factor that associates with the core enzyme to form RNAP holoenzyme (Lee et al. 2012). Bacteria typically encode one primary σ factor, such as σ70 in Escherichia coli, that is responsible for most transcription during exponential growth and multiple alternative σ factors that are required for gene expression in different growth or stress conditions (Gruber and Gross 2003). The recognition of conserved promoter elements by the σ subunit leads to binding of RNAP holoenzyme to promoters in an initial, typically unstable, closed complex (RPC). Through a series of isomerization steps, σ mediates strand separation at the −10 element of the promoter, resulting in a transcription-competent, stable, open complex (RPO) that can initiate RNA synthesis (Saecker et al. 2011).
In bacteria, the regulation of transcription occurs primarily at the level of initiation. The predominant mechanism underlying transcription activation involves regulators that bind specific DNA sequences upstream of or within the promoter. These canonical transcription factors typically bind DNA independently, recruiting RNAP to target promoters by contacting the α subunit or domain 4 of σ, which binds the −35 promoter element (Lee et al. 2012). Canonical transcription factors are usually present only at promoters that harbor a close match to their consensus binding motif. In contrast, a few regulators are known to interact tightly with RNAP and are found at virtually all active promoters (Haugen et al. 2008). These regulators, including E. coli DksA (Paul et al. 2004; Rutherford et al. 2009; Lennon et al. 2012) and Mycobacterium tuberculosis CarD (Stallings et al. 2009; Srivastava et al. 2013), do not affect the initial recruitment of RNAP to promoters but instead regulate the transition from the closed to the open complex, although their mechanisms of action are incompletely understood.
Temporal regulation of gene expression is critical for cell cycle progression in the α-proteobacterium Caulobacter crescentus. This organism divides asymmetrically, yielding a sessile, replication-competent “stalked” cell and a motile, replication-incompetent “swarmer” cell, which can then differentiate into a stalked cell (Skerker and Laub 2004; Curtis and Brun 2010). Nearly a third of all genes in Caulobacter show cell cycle-dependent expression (Laub et al. 2000; Fang et al. 2013). These patterns of gene expression are driven by several transcription factors. Some are canonical transcription factors that recognize specific DNA sequences within the promoters of target genes. For example, the response regulator CtrA directly binds to and regulates the expression of ∼100 genes, mostly during late stages of the cell cycle (Laub et al. 2002).
Another important but poorly understood cell cycle transcription factor is GcrA, which accumulates during the swarmer-to-stalked cell transition (Holtzendorff et al. 2004). The precise role of GcrA in cell cycle progression remains unclear, and the direct regulon of GcrA is unknown. The first report on GcrA identified ∼125 genes whose expression changed after depleting GcrA but did not distinguish direct and indirect targets (Holtzendorff et al. 2004). More recently, ChIP-seq (chromatin immunoprecipitation [ChIP] combined with deep sequencing) analysis of GcrA was reported, but there was little overlap between the genes showing the highest GcrA promoter occupancy and those originally reported as GcrA-dependent, and no consensus binding site was identified, although GcrA was shown to bind preferentially to N6-adenine methylated GANTC sites in vitro (Fioravanti et al. 2013).
GcrA was proposed to activate transcription by binding independently to all GANTC sites and recruiting RNAP (Fioravanti et al. 2013). However, little data exist to support a canonical recruitment mechanism. Although some GcrA-bound promoters have nearby GANTC sites, GcrA also associates with many promoters lacking methylation sites, and there are many methylation sites in the genome without GcrA bound. Moreover, the expression of many genes with methylation sites is unaffected by the loss of GcrA. In short, the mechanism by which GcrA affects gene expression is unclear, and the relationship between GcrA and DNA methylation remains ill-defined.
Here, we demonstrate that GcrA forms a stable complex with RNAP holoenzyme through an interaction with the primary σ factor σ73 (hereafter σ70 for consistency with E. coli), associating with nearly all σ70-regulated promoters. However, GcrA does not affect the expression of most genes. Instead, our results show that GcrA primarily boosts expression from promoters harboring an extended recognition element containing certain GANTC methylation sites. GcrA interacts with the domain of σ70 that binds the −10 element during strand separation, and we show that GcrA stimulates transcription at promoters by increasing both binding of RNAP and open complex formation. Collectively, our findings favor a model in which GcrA is brought to promoters primarily via its interaction with RNAP holoenzyme containing σ70, stimulating the expression of genes harboring certain promoter-proximal methylation sites. Unlike many canonical transcription factors, GcrA does not bind only to target promoters and simply recruit RNAP. Informed by these mechanistic studies, we combined ChIP-seq and expression profiling to identify the direct GcrA regulon and define a more precise role for GcrA in cell cycle progression.
Results
GcrA colocalizes genome-wide with RNAP holoenzyme containing σ70
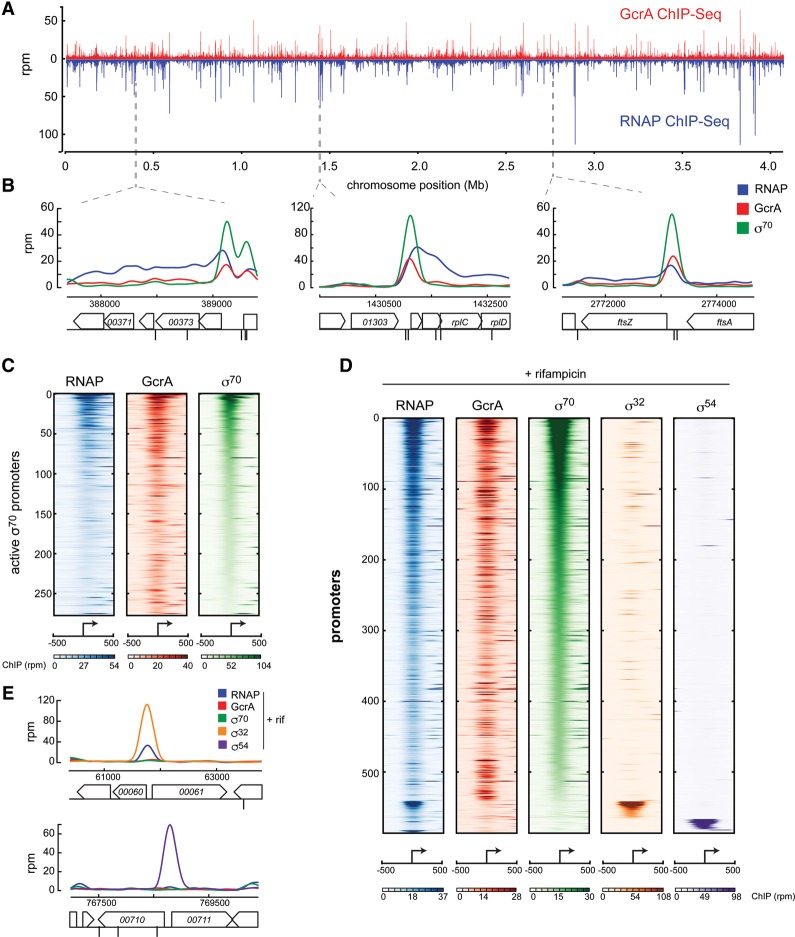
To map the genome-wide binding profile of GcrA, we performed ChIP-seq using an anti-Flag antibody on cells producing GcrA with a 3xFlag tag at its C terminus from either the native gcrA promoter or a xylose-inducible promoter. These two strains produced GcrA at approximately wild-type levels and were morphologically similar to the wild type. The ChIP profiles were highly correlated (R = 0.97) (Supplemental Fig. S1A,B) and similar to a profile produced with a polyclonal antibody (Murray et al. 2013). Comparison of our GcrA-3xFlag ChIP profiles with a control profile generated for cells expressing untagged GcrA indicated widespread enrichment of GcrA-3xFlag across the Caulobacter genome (Supplemental Fig. S1C). GcrA was found primarily in promoter regions, with enrichment above background at >500 different intergenic locations.
To determine whether the promoter regions bound by GcrA were transcriptionally active, we generated ChIP-seq profiles of RNAP and σ70 using an anti-Flag antibody on cells producing a 3xFlag-tagged variant of the β′ subunit of RNAP and an anti-σ70 antibody on wild-type cells, respectively. The profiles for RNAP and σ70 each had striking overall similarity to the GcrA profile (Fig. 1A–C; Supplemental Fig. S1D,E). In general, the σ70 ChIP signal showed high correlation with the GcrA and RNAP ChIP signals; i.e., promoters with high σ70 enrichment tended to also have high GcrA enrichment (Supplemental Fig. S1E), with some exceptions discussed below. However, RNAP was found in promoters and within genes, whereas σ70 and GcrA were found almost exclusively in promoters (Fig. 1B). Collectively, the ChIP-seq profiles indicate that GcrA is localized to virtually all active σ70-dependent promoters. Taken together, our data suggest that (1) σ70 may be the primary determinant of where GcrA binds across the genome, and (2) GcrA associates with RNAP but only during transcription initiation (Fig. 1B).
Figure 1.
GcrA colocalizes genome-wide with RNAP holoenzyme containing σ70. (A) ChIP-seq profiles of GcrA (ΔgcrA, Pxyl-gcrA-3xFlag) (red) and the RNAP β′ subunit (rpoC::rpoC-3xFlag) (blue) across the whole genome. ChIP-seq signals were normalized in reads per million (RPM). (B) ChIP signals for GcrA, RNAP, and σ70 at three genomic regions. Black marks below genes represent locations of GANTC methylation sites. (C) Normalized ChIP-seq signals for GcrA, RNAP, and σ70 at active promoters (see the Supplemental Material) over a 1-kb range around the promoter and sorted based on σ70 signal. Color bars indicate enrichment in RPM. (D) Normalized ChIP-seq signals for the factors indicated from rifampicin-treated cells. (E) ChIP-seq signals for RNAP, GcrA, σ70, σ54, or σ32 from rifampicin-treated cells at representative σ54- or σ32-regulated promoters.
To further test whether GcrA associates specifically with σ70-dependent promoters, we treated cells for 30 min with rifampicin, which traps RNAP at promoters (Campbell et al. 2001). We then performed ChIP-seq for RNAP (β′ subunit), GcrA, σ70, σ54, and σ32. The latter two proteins are alternative σ factors that recognize different promoter consensus sequences. As expected, RNAP was found almost exclusively in intergenic promoter regions. A comparison with the other ChIP profiles for rifampicin-treated cells indicated that GcrA localized to promoters at which σ70, but not σ54 or σ32, was present (Fig. 1D,E). These findings demonstrate that GcrA promoter association in vivo is specific to σ70-dependent promoters, suggesting that GcrA may be targeted to these promoters by interacting with σ70. Additionally, we noted that, in rifampicin-treated cells, GcrA occupied promoters that had not exhibited significant occupancy by GcrA, RNAP, or σ70 in untreated cells (Supplemental Fig. S1F), indicating that GcrA has an even more widespread binding pattern than previously appreciated.
GcrA forms a stable complex with RNAP holoenzyme containing σ70
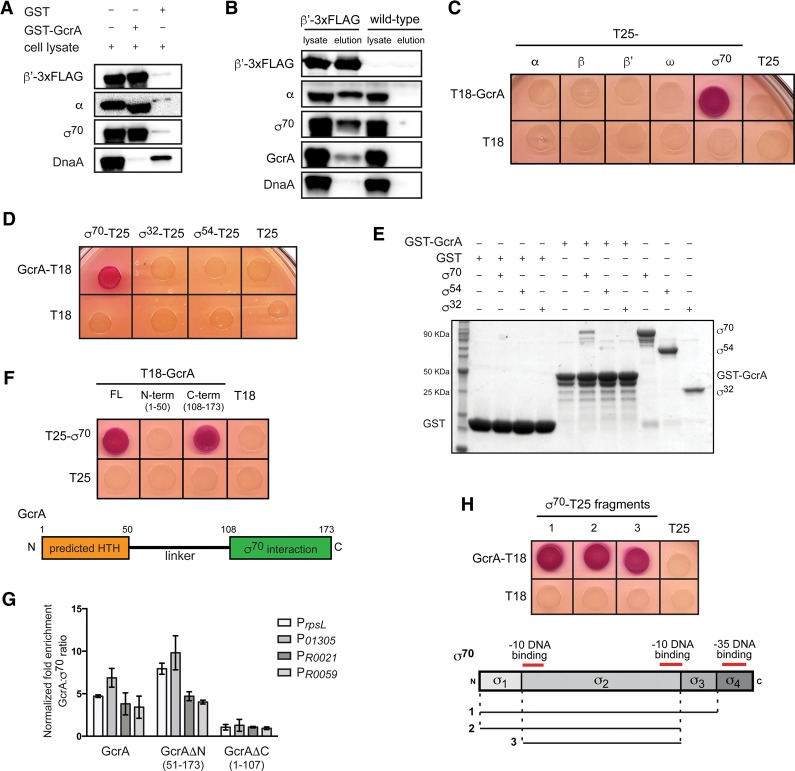
To test whether GcrA is recruited to promoters through a direct interaction with the σ70-containing RNAP holoenzyme, we sought to determine whether GcrA and RNAP holoenzyme interact. First, we mixed Caulobacter cell lysate with purified GST-GcrA immobilized on glutathione beads and then tested for retention of three subunits of RNAP holoenzyme: α, β′, and σ70. We observed clear enrichment of each subunit, indicating that RNAP holoenzyme was pulled down as a complex by GcrA (Fig. 2A). Importantly, the interactions detected were specific, as RNAP holoenzyme subunits were not recovered by GST alone, and GST-GcrA did not pull down other DNA-binding proteins such as DnaA (Fig. 2A). Second, we used an anti-Flag antibody to immunoprecipitate RNAP from cells producing a 3xFlag-tagged variant of the β′ subunit of RNAP; Western blot analysis indicated that GcrA coimmunoprecipitated with RNAP, as did the α subunit of RNAP and σ70, as expected (Fig. 2B). Taken together, these data indicate that GcrA forms a stable complex with RNAP holoenzyme.
Figure 2.
GcrA interacts directly with RNAP through σ70. (A) GST-GcrA pull-down eluates were assessed for the presence of RNAP subunits by immunoblotting. Equal amounts of sample were loaded for each blot. (B) Immunoprecipitated β′-3xFlag was assessed for the presence of coprecipitating RNAP holoenzyme subunits and GcrA by immunoblotting. Equal amounts of sample were loaded for each blot. Identical immunoprecipitation performed on wild-type cells served as a control for specificity. (C) Bacterial two-hybrid analysis. Each RNAP subunit was fused to T25 and tested for interaction with T18-GcrA. Red/purple colonies indicate a positive interaction. (D) Bacterial two-hybrid analysis of the interaction between GcrA-T18 and σ70, σ54, or σ32 fused to T25. (E) Direct interaction of GST-GcrA and σ70. Either GST or GST-GcrA was bound to glutathione sepharose beads and then incubated with the indicated σ factor. Samples were washed, eluted, and then examined by SDS-PAGE and Coomassie staining. (F) Bacterial two-hybrid analysis of the interaction between the N-terminal and C-terminal domains of GcrA and σ70. A schematic of GcrA is shown below. (G) ChIP combined with quantitative PCR (ChIP-qPCR) using an anti-Flag antibody on rifampicin-treated cells expressing GcrA-3xFlag, GcrAΔN-3xFlag, or GcrAΔC-3xFlag from a plasmid in cells depleted of untagged GcrA (ΔgcrA, Pvan-gcrA) for 2 h. Fold enrichment for the GcrA:σ70 ratio is relative to a control strain containing an empty vector. Error bars indicate SD. n = 2 for each of four different promoters. (H) Bacterial two-hybrid analysis of the interaction between GcrA and fragments of σ70 is summarized below.
GcrA is brought to promoters through a direct interaction with domain 2 of σ70
To determine which subunit of RNAP interacts with GcrA, we used a bacterial two-hybrid system based on complementation of the two adenylate cyclase fragments T18 and T25 (Karimova et al. 1998). We fused GcrA to T18 and each subunit of RNAP holoenzyme to T25 and then coexpressed gene fusion pairs in E. coli. We detected an interaction between GcrA and σ70 but not the other subunits of RNAP (Fig. 2C). The interaction was specific to σ70, as we detected no interaction between GcrA and σ54, σ32 (Fig. 2D), or any of the other 16 predicted Caulobacter σ factors (Supplemental Fig. S2A). Although a negative result in the bacterial two-hybrid assay does not completely rule out a potential interaction, these results are consistent with our ChIP-seq data (Fig. 1D). We verified that the interaction between GcrA and σ70 was direct using affinity chromatography in vitro. Again, we observed a stable interaction of purified GST-GcrA with σ70 but not σ32 or σ54 (Fig. 2E).
To determine which domain of GcrA interacts with σ70, we tested truncations of GcrA in the bacterial two-hybrid assay. GcrA has a predicted helix–turn–helix at its N terminus, connected by a long linker to a C-terminal domain of unknown function. We detected an interaction only between the C-terminal domain of GcrA and σ70 (Fig. 2F). To determine whether this interaction is sufficient to localize GcrA to promoters, we performed ChIP combined with quantitative PCR (ChIP-qPCR) using an anti-Flag antibody on cells depleted of full-length GcrA and expressing a 3xFlag-tagged fragment lacking either the N-terminal domain GcrAΔN or the C-terminal domain GcrAΔC. We found that GcrAΔN, but not GcrAΔC, localizes to promoters with σ70 (Fig. 2G), indicating that the N-terminal helix–turn–helix domain is not required to recruit GcrA to promoters. These data strongly support a model in which GcrA is brought to σ70 promoters primarily via an interaction with RNAP holoenzyme not by binding DNA independently.
To determine which domain of σ70 contacts GcrA, we tested the ability of individual domains of σ70 to interact with GcrA in the bacterial two-hybrid system. σ70 has four structural domains, with domains 2 and 4 (σ2 and σ4) involved in binding the −10 and −35 promoter elements, respectively. Transcriptional activators that interact with σ70 typically recruit RNAP to promoters by making contact with σ4 (Lee et al. 2012). However, we found that σ2, not σ4, was sufficient to mediate an interaction with GcrA (Fig. 2H), indicating that GcrA may regulate transcription through an unconventional mechanism. To better define the region of σ2 that interacts with GcrA, we took advantage of the fact that E. coli σ2 does not interact with GcrA (Supplemental Fig. S2B) and tested E. coli–Caulobacter σ2 chimeras for interaction with GcrA. This analysis indicated that GcrA likely interacts with σ2 through the two α helices formed by regions 1.2 and 2.1 as well as the N-terminal and C-terminal ends of the nonconserved region (Supplemental Fig. S2B).
GcrA-binding affinity for methylation sites is sequence-specific
Consistent with GcrA being recruited to σ70-dependent promoters in vivo through a direct interaction with RNAP holoenzyme, our ChIP-seq studies indicated a strong correlation between the binding profiles of GcrA and σ70. However, the correlation was not perfect, perhaps indicating that GcrA exhibits some DNA sequence specificity even though promoter association occurs primarily via σ70. A previous study found that GcrA binds methylated DNA in vitro and proposed that methylated GANTC sites were required for GcrA binding in vivo (Fioravanti et al. 2013). However, many promoters bound by GcrA in vivo do not have methylation sites, and the enrichment of GcrA at promoters with methylation sites varied significantly (Supplemental Fig. S3A).
To examine the DNA sequence and methylation specificity of GcrA, we measured the affinity of GcrA for methylated and unmethylated DNA in vitro by filter binding. Using unmethylated probes derived from the ftsZ and mipZ promoters, we measured a Kd of ∼0.5 μM (Supplemental Fig. S3B). Using methylated PftsZ and PmipZ probes, we observed a 1.4-fold and 1.7-fold reduction in Kd, respectively (Supplemental Fig. S3B). Notably, the Kd of GcrA for methylated PmipZ was lower than for methylated PftsZ, but GcrA occupancy in vivo, as judged by ChIP-seq, was significantly higher at PftsZ (Supplemental Fig. S3C). Thus, DNA-binding affinity and the presence of a GANTC methylation site are insufficient to explain GcrA enrichment at a given promoter.
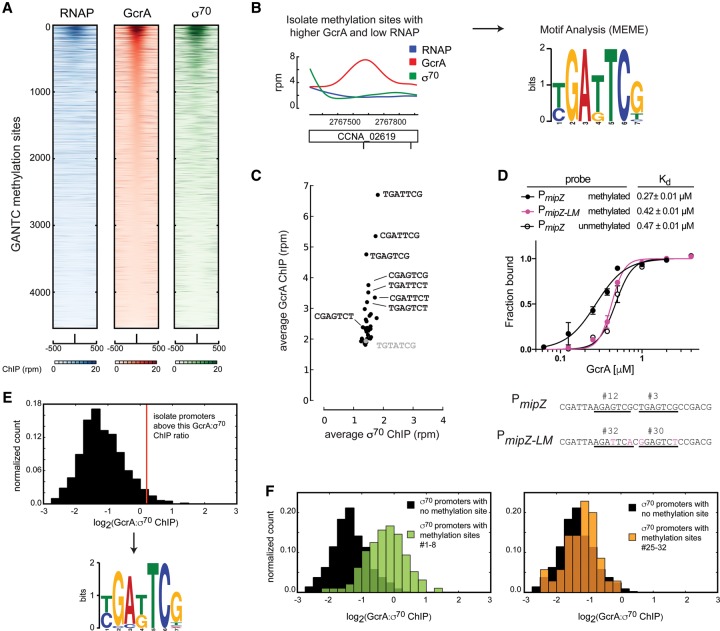
To identify other sequence elements influencing GcrA binding, we examined the ChIP-seq signals for GcrA, σ70, and RNAP at all 4542 GANTC methylation sites across the Caulobacter genome. The majority of sites had little or no GcrA bound (Fig. 3A). Of the methylation sites that were bound by GcrA, most were also bound by σ70, indicating that these sites are in or near promoters (Fig. 3A). However, some methylation sites within genes showed occupancy by GcrA with no enrichment of σ70; the enrichment of GcrA at these sites was substantially less than observed at promoters, supporting the notion that the genomic binding of GcrA is determined primarily by its interaction with σ70. Nevertheless, we hypothesized that intergenic sites bound by GcrA may have sequence elements that strongly favor GcrA binding. MEME analysis of 20 such locations revealed a consensus sequence of TGATTCG or, more broadly, YGAKTCK, where Y = C or T and K = G or T (Fig. 3B).
Figure 3.
GcrA-binding affinity for methylation sites is sequence-specific. (A) ChIP-seq signals for GcrA, RNAP, and σ70 in rifampicin-treated cells plotted over 1-kb ranges centered at each of the 4542 GANTC methylation sites in the Caulobacter genome. Signals were capped at 20 RPM and sorted based on the GcrA signal. (B) Identification of a consensus GcrA-binding motif. ChIP signals for GcrA, RNAP, and σ70 for one of the regions used in the analysis are shown. The motif obtained from the 20 selected sequences is depicted. (C) Plot of the average ChIP signal for σ70 versus GcrA at each of 32 possible NGANTCN variants. Analysis was performed on ChIP signals from rifampicin-treated cells and was restricted to nonpromoter regions. One non-NGANTCN site (gray) is shown for comparison (also see Supplemental Fig. S3D). (D) GcrA-binding curves for the probes indicated: methylated PmipZ, unmethylated PmipZ, and methylated PmipZ mutated to have low-ranking methylation sites [PmipZ(LM)]. Sequences surrounding the methylation sites in PmipZ and PmipZ(LM) and their ranks are indicated. Error bars represent SDs. n = 3. (E) Normalized histogram of the GcrA:σ70 ChIP ratio at σ70 promoters and a motif identified in promoters with high ratios. n = 25. (F) Histograms of the GcrA:σ70 ChIP ratio for promoters lacking methylation sites within 40 base pairs (bp) of the transcription start site (black), containing at least one methylation site ranked 1–8 (green) or 25–32 (orange); ranks were based on C and Supplemental Figure S3D.
To assess the relevance of this motif, we calculated the average GcrA ChIP signal at all intergenic locations that have one of the 32 possible NGANTCN sequences (Fig. 3C; Supplemental Fig. S3D). This analysis revealed a clear hierarchy (or ranking) of the 32 sequences in terms of GcrA binding, with seven of the eight sequences exhibiting the highest GcrA signal included in the consensus YGAKTCK (Fig. 3B,C; Supplemental Fig. S3D). To verify the inferred binding preferences of GcrA, we measured its affinity for a methylated PmipZ probe containing the third and 12th highest-ranked sequences (TGAGTCG and AGAGTCG) and a probe, called PmipZ(LM) (low-ranked methylation sites) in which these sequences were mutated to the 30th and 32nd ranked sequences (GGAGTCT and AGATTCA). The four nucleotide substitutions in PmipZ(LM) reduced the binding affinity of GcrA to a level observed when the PmipZ probe was unmethylated (Fig. 3D). We observed a similar effect when mutating sites in a probe derived from the cckA promoter (Supplemental Fig. S3E).
To test whether the recognition motif YGAKTCK impacts GcrA binding at promoters, we computed the ratio of GcrA:σ70 ChIP signal at each σ70 promoter. MEME analysis of the top 25 promoters revealed a motif (Fig. 3E) almost identical to that identified above. We also found that promoters containing a methylation site ranked 1–8 (i.e., those closely matching the consensus YGAKTCK) typically had much higher GcrA:σ70 ratios than promoters with no methylation site or a low-ranked methylation site (Fig. 3F; Supplemental Fig. S3F). These observations indicate that the DNA-binding specificity of GcrA identified influences the extent of GcrA occupancy at promoters in vivo.
GcrA promotes transcription and RNAP open complex formation at methylated promoters
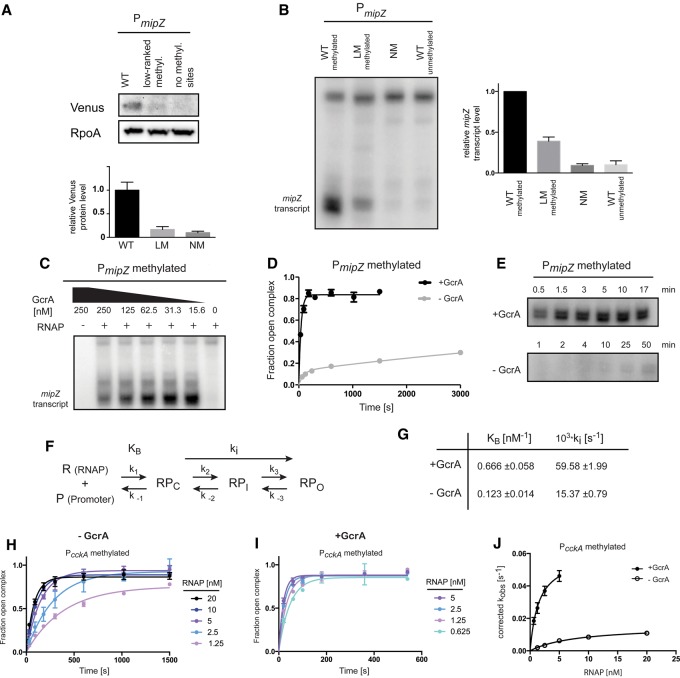
To assess the importance of consensus methylation sites on GcrA-dependent transcription in vivo, we constructed reporters in which a mipZ promoter near the native mipZ chromosomal locus drives expression of the yellow fluorescent protein Venus. Using Western blot analysis, we found that mutating the high-ranked methylation sites in the mipZ promoter to low-ranked sites reduced expression in vivo by nearly 80%, similar to the effect of eliminating the methylation sites completely (Fig. 4A).
Figure 4.
GcrA promotes open complex formation at methylated promoters. (A) Western blot analysis of a Venus reporter expressed from the wild-type PmipZ harboring two high-ranked methylation sites, a variant containing low-ranked methylation sites, or a variant with no methylation sites. (Bottom) Band intensities were quantified. Error bars represent SDs. n = 2. RpoA was used as a loading control. (B) Multiple-round in vitro transcription reactions using the promoter variant indicated as a template. Reactions contained 2 nM purified Caulobacter RNAP holoenzyme and 62.5 nM GcrA. (Right) Band intensities were quantified. Error bars represent SDs. n = 2. (C) Multiple-round in vitro transcription reactions using the methylated wild-type mipZ promoter as a template. Reactions contained 2 nM purified Caulobacter RNAP holoenzyme and varying levels of GcrA. (D) Kinetics of open complex formation using 1.25 nM Caulobacter RNAP holoenzyme ±250 nM GcrA. Error bars represent SDs. n = 2. (E) Single-round in vitro transcription assays using a probe containing the methylated wild-type mipZ promoter as a template. Reactions containing 1.25 nM purified Caulobacter RNAP holoenzyme ±250 nM GcrA and DNA probe were preincubated, and nucleotides, along with heparin, were added at the indicated time points. Samples ±GcrA were imaged on the same gel. (F) Kinetic scheme of transcription initiation highlighting KB (the binding constant of the holoenzyme to promoter DNA) and ki (the aggregate forward rate constant of isomerization from the closed to the open complex). (G) Summary of kinetic parameters for GcrA-dependent transcription from methylated PcckA. (H) Kinetics of open complex formation at the methylated cckA promoter using varying concentrations of Caulobacter RNAP holoenzyme and 1 mM ATP. Error bars represent SDs. n = 2. (I) Kinetics of open complex formation at the methylated cckA promoter using varying concentrations of Caulobacter RNAP holoenzyme, 250 nM GcrA, and 1 mM ATP (see the Supplemental Material). Error bars represent SDs. n = 2. (J) Plot of corrected kobs obtained from each of the curves in H and I versus RNAP concentration and fitted to obtain the constants KB and ki reported in G (see the Supplemental Material).
We also examined the effects of mutating DNA methylation sites using an in vitro transcription assay with purified Caulobacter RNAP holoenzyme containing σ70 and GcrA. Using the wild-type methylated mipZ promoter as a template produced a transcript of the expected size (Fig. 4B; Supplemental Fig. S4). Mutating the two high-ranked methylation sites to low-ranked sites or eliminating methylation sites dramatically reduced transcription in this assay, consistent with our in vivo reporter studies. Importantly, at the concentration of GcrA used, GcrA alone does not stably bind DNA (Fig. 3D). To gain further insight into the binding affinity of GcrA for RNAP holoenzyme, we repeated the in vitro transcription assay with varying levels of GcrA. Transcription was maximal at 15 nM, indicating that GcrA's Kd for RNAP is <15 nM (Fig. 4C). Higher concentrations of GcrA, at which some DNA binding was observed, seemed to partially inhibit the reaction, possibly by preventing binding of RNAP to the promoter. These data strongly support a model in which GcrA is brought to promoters via a tight interaction with RNAP rather than recruiting RNAP through independent DNA binding.
To further examine how GcrA impacts transcription, we implemented a filter-binding assay for measuring open complex formation. We incubated purified Caulobacter RNAP holoenzyme with a radiolabeled, methylated PmipZ probe in the presence or absence of GcrA and then isolated open complexes on nitrocellulose filters after competing away closed complexes with heparin. A time course of open complex formation demonstrated that GcrA strongly promotes open complex formation, with maximal levels of open complex forming within 2–3 min with GcrA while remaining relatively low up to 50 min without GcrA (Fig. 4D). Similarly, we found that GcrA strongly stimulated transcription from the wild-type methylated mipZ promoter in a single-round in vitro transcription assay (Fig. 4E).
The stimulation of open complex formation (Fig. 4D) could indicate that GcrA affects binding of RNAP to promoters, isomerization to the open complex, or both (Fig. 4F). To distinguish between these possibilities, we measured the kinetics of open complex formation using a filter-binding assay while titrating RNAP holoenzyme. For these experiments, we used a probe corresponding to the cckA promoter, as the time-course data for this probe were fit well by a single exponential model, enabling us to disentangle the effects of GcrA on RNAP binding and isomerization (see the Supplemental Material). Our results indicated that GcrA increases KB (the binding constant for RNAP in the closed complex form) by ∼5.4-fold and increases ki (the aggregate forward rate constant for isomerization from the closed to the open complex) by ∼3.9-fold (Fig. 4G–J).
GcrA regulates genes harboring promoter-proximal, near-consensus methylation sites
The composition of the GcrA regulon has been elusive, in large part because ChIP-seq analysis alone is insufficient to pinpoint direct targets, a consequence of the fact that GcrA associates with virtually all σ70-regulated promoters. As a first step to defining the GcrA regulon, we used DNA microarrays to measure RNA levels in synchronized populations of wild-type and GcrA-depleted cells at 30, 45, and 60 min after synchronization, the time window when GcrA normally accumulates to maximal levels (Fig. 5A). We identified 364, 590, and 444 genes at each time point with at least 1.75-fold lower expression in the GcrA-depleted cells relative to wild-type cells (Supplemental Table S1). In total, GcrA appears to positively regulate at least 774 genes either directly or indirectly.
Figure 5.
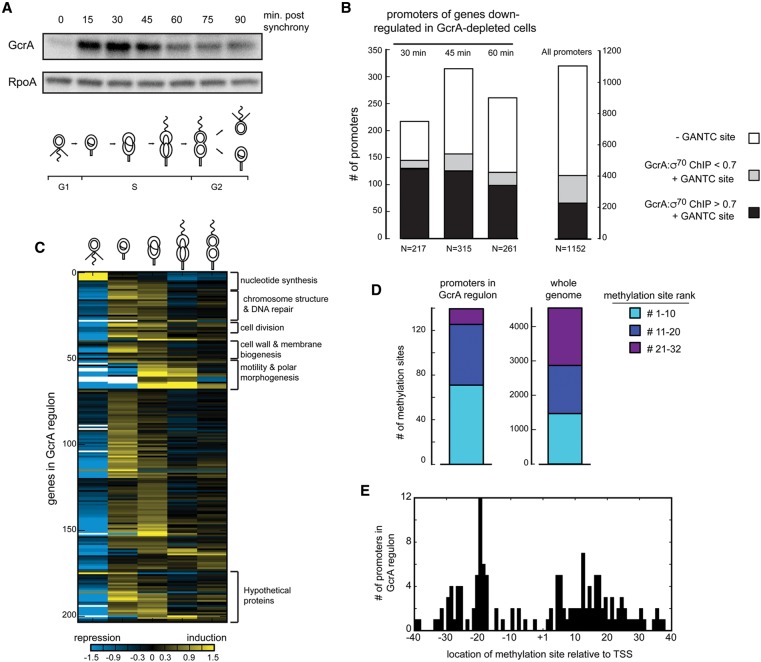
GcrA regulates genes with promoter-proximal, near-consensus methylation sites. (A) Western blot for GcrA and RpoA (loading control) during the cell cycle is summarized below. GcrA, CtrA (not shown), and RpoA were imaged on the same blot. (B) Promoters of genes down-regulated in GcrA-depleted cells at each time point indicated were categorized based on their rifampicin-treated GcrA:σ70 ChIP ratio and the presence of a methylation site within 40 bp of the transcription start site. (Right) Distribution at all promoters is given for comparison. (C) Cell cycle expression patterns of genes directly regulated by GcrA. RNA sequencing data are from Fang et al. (2013). Expression at each time point is relative to a gene's average expression at all time points. (D) Distribution of methylation site ranks for promoters in the GcrA regulon and genome-wide. (E) Histogram for the locations of methylation sites in GcrA-regulated promoters relative to transcription start sites.
We hypothesized that promoters directly regulated by GcrA should have high GcrA:σ70 ChIP enrichment ratios. Although GcrA localizes to promoters mainly via σ70, subsequent interaction with methylation sites within target promoters likely stabilizes GcrA binding or increases the efficiency of ChIP. We found that 78% of genes affected by GcrA depletion at the 30-min time point had promoters with high GcrA:σ70 ratios. The majority of these genes also had at least one methylation site within 40 base pairs (bp) of their transcription start site (Fig. 5B; Supplemental Table S2), consistent with our analysis of the DNA sequence specificity of GcrA.
Combining these observations, we defined the direct GcrA regulon as those genes that had (1) significantly lower expression levels in GcrA-depleted cells at 30 or 45 min, (2) a high GcrA:σ70 ChIP enrichment ratio at their promoters, and (3) a promoter-proximal GANTC site. In total, 140 transcription units encompassing 204 genes satisfied all three criteria (Supplemental Table S3). Notably, most of these genes are cell cycle-regulated (Fig. 5C), with peak expression at or shortly after the time that GcrA levels increase (Fig. 5A), even though cell cycle regulation was not one of our criteria. However, using only cell cycle regulation and lower expression levels in GcrA-depleted cells as criteria to define the GcrA regulon would have identified only 64% of the 140 transcription units that satisfied our three criteria, along with 42 promoters that do not have a high GcrA:σ70 ChIP enrichment ratio and methylation sites (Supplemental Fig. S5B). In sum, our three criteria, chosen based on our mechanistic studies, allowed the precise delineation of the direct GcrA regulon.
Some direct targets of GcrA affect the expression of other genes later in the cell cycle; most of these indirect targets also decrease following depletion of GcrA but lack a promoter-proximal methylation site and have a lower GcrA:σ70 ratio. For example, the gene ctrA, which has a high GcrA:σ70 ratio and a high-ranked methylation site, is likely activated directly by GcrA early in S phase. Almost the entire CtrA regulon is also misregulated in GcrA-depleted cells (Supplemental Fig. S5C); although some may be direct GcrA targets themselves, most are not and instead are indirectly affected via CtrA.
Strikingly, the GANTC sites in promoters called as directly regulated by GcrA were enriched for methylation sites defined above as highly ranked (Fig. 3C; Supplemental Fig. 5D), consistent with our in vitro studies that certain methylation sites better enable GcrA to promote transcription. These methylation sites localize to two regions of target promoters: one narrowly centered around −20 and one more widely distributed from +2 to +30 (Fig. 5E). We observed almost no methylation sites around the −10 element, possibly because this region is bound by σ70 and inaccessible to GcrA. The distribution of methylation sites suggests that GcrA can bind DNA on either side of the −10 element while interacting with domain 2 of σ70, perhaps as a consequence of its predicted long unstructured linker (Fig. 2F). Altogether, our data indicate that it is not simply the presence of a methylation site that dictates whether a promoter is affected by GcrA, but its precise location and flanking nucleotides.
Loss of GcrA delays cytokinesis and desynchronizes DNA replication and cell division
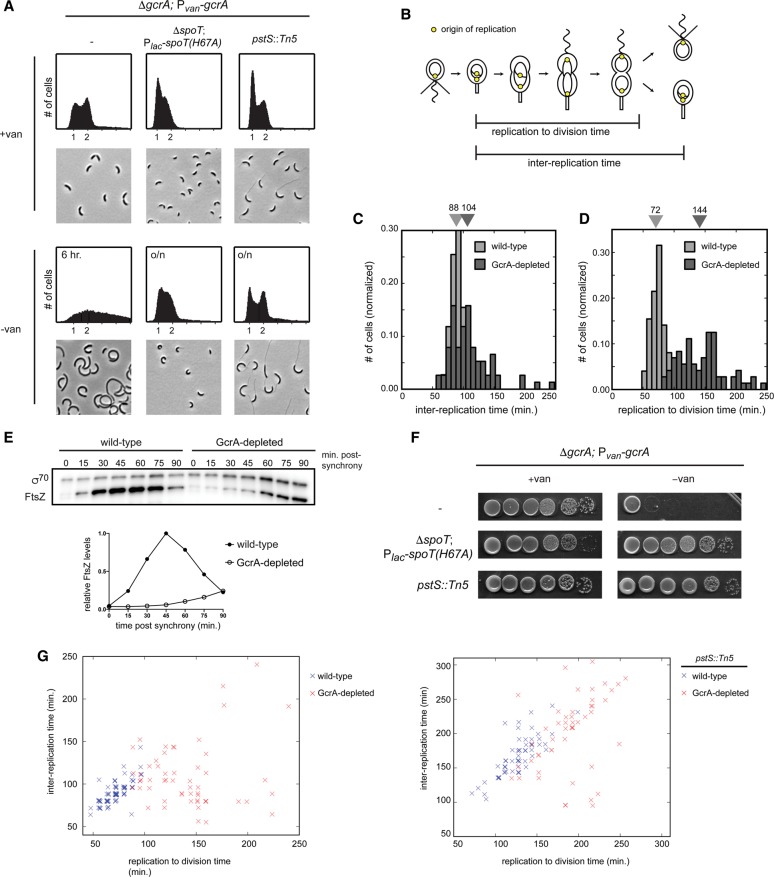
The GcrA regulon spans multiple functional categories (Fig. 5C) and includes several core cell cycle processes: nucleotide synthesis, DNA repair, chromosome organization and segregation, and cell division. Several genes involved in activating the master regulator CtrA were also GcrA targets, including cckA, divL, pleC, podJ, and ctrA itself. The regulon did not include components of the replisome, except for dnaE. The functional categories of direct targets suggest that GcrA helps drive expression patterns critical to S phase and progression through the later stages of the cell cycle. Thus, cells lacking gcrA should exhibit pleiotropic defects. To examine the gcrA phenotype, we built a strain in which the only copy of gcrA is driven by a vanillate-inducible promoter (ΔgcrA; Pvan-gcrA). When grown in the absence of vanillate for 6 h, GcrA-depleted cells grew at approximately the same rate as wild type (Supplemental Fig. S6B) but became elongated and accumulated extra chromosomes (Fig. 6A; Murray et al. 2013), indicating a cell division defect. To quantify the timing of cell division and DNA replication, we performed time-lapse microscopy of GcrA-depleted cells harboring a fluorescent repressor–operator system that labels origins of replication (Fig. 6B; Supplemental Fig. S6A). We found that the interval between successive rounds of DNA replication was delayed by only ∼18% in cells depleted of GcrA (Fig. 6C), whereas the time between DNA replication to cell division increased by 100% (Fig. 6D).
Figure 6.
Loss of GcrA delays cytokinesis and desynchronizes DNA replication and cell division. (A) Flow cytometry analysis of chromosome content and phase-contrast images of a gcrA depletion strain harboring the additional mutations indicated grown in rich medium ± vanillate overnight, except for GcrA-depleted cells that are not viable, for which a 6-h depletion was performed. SpoT(H67A) expression was induced with 500 μM IPTG. (B) Schematic of the cell cycle showing the interreplication and replication to division times. (C,D) Normalized histograms of times between successive replication initiations (C) and times between replication and division (D) in wild-type and GcrA-depleted cells. Median values are listed above. (E) Western blot for FtsZ and σ70 (loading control) during the cell cycle for synchronized wild-type and GcrA-depleted cells. FtsZ levels were normalized to σ70 and are plotted as a fraction of maximum expression. (F) Growth of a gcrA depletion strain harboring the same mutations as in A, assessed by 10-fold serial dilutions on plates ±vanillate. Plates for ΔgcrA; Pvan-gcrA; ΔspoT; Plac-spoTH67A also contained 50 μM IPTG. (G) Scatter plots of the interreplication time and replication to division time in wild-type and GcrA-depleted cells (left) and pstS::Tn5-depleted and pstS::Tn5 GcrA-depleted cells (right). Overlayed data points were slightly shifted by addition of a small random number to facilitate visualization.
These phenotypic analyses indicated that, in cells lacking GcrA, the rate of cell division does not keep pace with the rate of DNA replication. This cell division defect likely results from the misregulated expression of many genes; nearly half of the known division genes are positively regulated by GcrA, with MipZ, FtsE, FtsX, FtsN, and FtsZ being direct targets (Supplemental Fig. S5D). Consistent with the defects of GcrA resulting from a deficiency in expressing multiple genes, we found that the ectopic expression of either ftsN or ftsZ could not substantially restore viability to a strain depleted of GcrA in rich medium (Supplemental Fig. S6B). Increased expression of these genes may marginally increase viability (Murray et al. 2013), but the phenotype of cells lacking GcrA likely stems from its pleiotropic regulation of gene expression.
Importantly, although cell division occurs less frequently in the absence of GcrA, cells do still divide even after GcrA is undetectable (Supplemental Fig. S6C,D). Thus, essential cell division proteins must be transcribed without GcrA but take longer to reach the levels necessary for cytokinesis. Quantitative immunoblotting confirmed that, in GcrA-depleted cells, FtsZ levels do not rise sharply early in the cell cycle as in wild-type cells but do eventually increase (Fig. 6E). These data support a model in which GcrA helps boost the expression of cell division genes so that cells maintain balanced rates of division and DNA replication.
To gain additional insight into the physiological role of GcrA, we sought to identify mutations that restore viability to cells depleted of GcrA. Using transposon mutagenesis, we identified a series of insertions in the phosphate import system comprising PstABC and PstS. The pstS::Tn5 mutation almost completely rescued the plating efficiency of cells lacking GcrA (Fig. 6F), and these cells appeared morphologically similar to cells harboring the pstS::Tn5 mutation alone (Fig. 6A).
Why do mutations in the phosphate import system suppress a gcrA mutant? We suspected that the effect may stem from the fact that pstS mutants grow slowly and are delayed in initiating DNA replication, leading to an increase in G1 cells in flow cytometry analysis (Fig. 6A). As noted above, cells lacking GcrA are delayed for cell division but continue to grow and initiate DNA replication. Hence, the pstS::Tn5, gcrA double mutant has likely rebalanced its rates of growth and DNA replication with rates of cell division; indeed, the intervals between rounds of DNA replication and between replication and division were more closely matched in the pstS::Tn5, gcrA depletion strain than in the gcrA depletion alone (Fig. 6G).
Finally, we wanted to test whether slowing growth and DNA replication in a completely different manner could also rescue the gcrA mutant. We did so by ectopically producing the signaling molecule (p)ppGpp, which is known to slow cell growth and repress DNA replication (Boutte et al. 2012). We used an IPTG-inducible, hydrolase-defective variant of SpoT, the enzyme responsible for the synthesis and hydrolysis of (p)ppGpp in Caulobacter. We observed a nearly complete rescue of plating viability for GcrA-depleted cells expressing this variant of SpoT (Fig. 6F), and the morphology of cells was significantly improved in liquid medium compared with the parent GcrA depletion strain (Fig. 6A). Collectively, our data indicate that the transcription factor GcrA normally helps cell division and potentially other cellular processes keep pace with both cell growth and DNA replication. Consequently, cells lacking GcrA show an uncoupling (or discoordination) of essential cell cycle events, eventually resulting in lethality. However, mutations that specifically slow growth and the rate of replication initiation can largely restore the balance of cell cycle activities, enabling cells to proliferate without GcrA.
Discussion
Mechanism of transcriptional control by GcrA
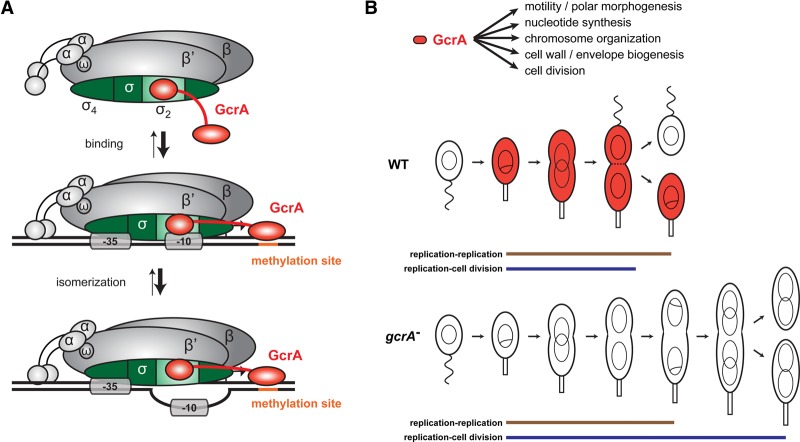
Although critical to cell cycle progression in Caulobacter, the mechanism by which GcrA activates transcription was previously unclear, and, consequently, the extent of its regulon was poorly defined. Our results suggest that GcrA is not a canonical transcription activator that drives gene expression primarily by binding DNA and then recruiting RNAP. Instead, our results favor a model in which GcrA forms a stable complex with RNAP through an interaction with the primary σ factor σ70 and is brought to promoters via this interaction (Fig. 7). Importantly, however, GcrA does not promote transcription from all promoters to which it binds. Rather, GcrA preferentially affects promoters harboring certain GANTC methylation sites, with the identity of the flanking nucleotides and the middle nucleotide of the GANTC site determining GcrA's efficacy as a transcription factor. At such promoters, GcrA may stimulate RNAP binding and open complex formation to activate transcription both in vitro and in vivo. Whether GcrA stimulates transcription from a given promoter likely also depends on whether the rate-limiting step for initiation is the step affected by GcrA.
Figure 7.
Model for the mechanism of transcription control by and cell cycle functions of GcrA. (A) GcrA forms a stable complex with RNAP through an interaction with domain 2 of the primary σ factor σ70 and is brought to promoters via this interaction. The presence of a preferred methylation site near the promoter stabilizes GcrA at the promoter and is required for GcrA-dependent stimulation of transcription. GcrA affects both the binding step and open complex formation. (B) Summary of the role GcrA plays in driving cell cycle progression by regulating gene expression in S phase.
Our results argue strongly against a mechanism in which GcrA binds promoters independently and then recruits RNAP holoenzyme. First, GcrA forms a stable complex with RNAP holoenzyme (Fig. 2A,B), and GcrA is found genome-wide at most σ70-dependent promoters, including those lacking methylation sites (Fig. 1C,D; Supplemental Fig. S3A). Second, we found that a GcrA variant lacking its DNA-binding domain was still highly enriched at σ70-dependent promoters (Fig. 2G). Finally, we found that GcrA promotes transcription in vitro at low nanomolar concentrations (Fig. 4C), where GcrA alone does not stably bind DNA (Fig. 3D), indicating that GcrA's affinity for RNAP is much tighter than its affinity for DNA. GcrA was found at some nonpromoter regions without RNAP, but the ChIP enrichment values at these sites was substantially smaller than that observed at promoters (Fig. 1A; Supplemental Fig. 3C). These observations imply that GcrA binds to promoters together with RNAP holoenzyme rather than independently and prior to RNAP, as previously suggested.
In sum, our results suggest a new model for GcrA as an auxiliary component or cofactor of the σ70-containing RNAP holoenzyme in Caulobacter. GcrA is nearly absent from swarmer cells but is likely very abundant throughout the rest of the cell cycle, with ribosome profiling data (Schrader et al. 2014) indicating synthesis at rates comparable with other components of RNAP. Moreover, because GcrA has a much tighter affinity for RNAP holoenzyme (Kd < 15 nM) (Fig. 4C) than for DNA (Kd ∼ 270 nM for highest affinity sites) (Fig. 3D), most GcrA is likely bound to RNAP. GcrA is then brought to promoters via its interaction with σ70, where our kinetic analyses indicate that, at least for the model cckA promoter, it can affect both KB (the equilibrium binding constant for the closed complex of RNAP) and ki (the composite rate of downstream isomerization steps that ultimately result in strand separation). Whether GcrA affects the two steps differently at other promoters is not clear, as the kinetics of open complex formation for some promoters, like PmipZ, did not exhibit the single exponential behavior needed to elucidate KB and ki.
GcrA's ability to promote isomerization of RNAP through a direct interaction with region 2 of the housekeeping σ factor distinguishes it from other, previously characterized transcription factors. Most transcriptional activators bind DNA independently and recruit RNAP to promoters through direct interactions with the α subunit or domain 4 of σ70. Like GcrA, E. coli DksA and M. tuberculosis CarD affect isomerization from the closed to the open complex, but DksA interacts with the β and β′ subunits, and CarD binds the β subunit. E. coli Crl binds domain 2 of σS but functions differently than GcrA, promoting the association of core RNAP with σS (Banta et al. 2013). In Chlamydia trachomatis, the transcription factor GrgA binds the nonconserved region of domain 2 in the housekeeping σ factor (Bao et al. 2012), but whether GrgA impacts RNAP binding or isomerization is unknown. The actinobacterial protein RbpA also binds domain 2 of a housekeeping σ factor (Tabib-Salazar et al. 2013), but recent data highlighted a role for RbpA in increasing affinity of holoenzyme to promoter DNA (Hubin et al. 2015), so its role, if any, in stimulating isomerization to the open complex is uncertain. Thus, to our knowledge, Caulobacter GcrA is the first transcription factor identified that binds region 2 of a σ factor and can promote the closed-to-open complex step of transcription.
GcrA provides a powerful mechanism for controlling gene transcription. By impacting the isomerization step of transcription initiation, GcrA-regulated genes can still be controlled by other, canonical transcription factors, affording an opportunity for bona fide combinatorial control. Although GcrA-regulated genes in Caulobacter are generally expressed during S phase, the precise timing of their induction varies, possibly the consequence of regulation by other transcription factors. Combined regulation by GcrA and other transcription factors may also increase the dynamic range of expression for target genes, helping to switch them from very low to very high states of expression. For instance, the gene ftsZ is activated by DnaA (Hottes et al. 2005) and GcrA (Fig. 6E). In cells depleted of GcrA, ftsZ is still induced but at significantly reduced levels compared with wild-type cells.
N6-adenine methylation-dependent gene regulation
N6-adenine methylation by orphan methyltransferases affects many processes in bacteria, including gene expression. The most well characterized example involves the γ-proteobacterial methyltransferase Dam (deoxyadenosine methyltransferase), which methylates GATC sequences (Wion and Casadesús 2006). In E. coli, full methylation of the agn43 promoter by Dam activates transcription by preventing binding of the repressor OxyR until DNA replication produces hemimethylated promoters (Haagmans and van der Woude 2000). Similarly, binding of the leucine-responsive regulatory protein Lrp to the traJ promoter in the Salmonella enterica virulence plasmid is blocked when a GATC site is fully methylated. After DNA replication, Lrp can bind to one of the newly replicated hemimethylated plasmids (Camacho and Casadesús 2005). For both OxyR and Lrp, methylation-dependent binding occurs at only a limited number of promoters. In contrast, N6-adenine methylation appears to promote the GcrA-dependent activation of most, if not all, target promoters.
In Caulobacter, the role of CcrM-dependent N6-adenine methylation at GANTC sites in gene regulation has been poorly understood. GANTC sites occur preferentially in intergenic regions, and GcrA was suggested to bind methylated GANTC sites in vitro (Fioravanti et al. 2013). However, GcrA associates with many promoters lacking GANTC sites, and the expression of many genes with promoter-proximal GANTC sites is not affected in cells lacking GcrA. Our results now clarify the relationship between GcrA, CcrM, and promoter methylation. We showed that GcrA is found at virtually all σ70-dependent promoters but primarily affects only promoters containing certain methylation sites, not any GANTC (Fig. 3). In particular, the two bases flanking a GANTC site and the middle “N” base strongly influenced GcrA binding and GcrA-dependent activation both in vitro and in vivo.
Whether N6-adenine methylation affects GcrA binding while RNAP is in the open or the closed complex remains to be determined. Promoter methylation strongly affected GcrA-dependent transcription both in vitro and in vivo (Fig. 4A,B) but had only a modest (approximately twofold) effect on binding to dsDNA (Fig. 3D; Supplemental Fig. S3E), perhaps indicating an effect after strand separation. Further structural studies of GcrA will help reveal how methylation impacts DNA binding and transcription.
Whether GcrA's transcriptional activity differs for fully versus hemimethylated promoters is also not yet clear. CcrM is cell cycle-regulated and accumulates only after DNA replication is nearly complete (Wright et al. 1996). Thus, many GANTC sites, particularly those close to the origin, remain hemimethylated for significant periods of time after passage of the replication fork. The ctrA P1 promoter, which is GcrA-regulated, was postulated to be more active in the hemimethylated state such that ctrA expression was coupled to passage of the replication fork (Reisenauer and Shapiro 2002). However, the induction of ctrA P1 transcription may simply be driven by the cell cycle-dependent accumulation of GcrA (Fig. 7). Moreover, the constitutive production of CcrM, leading to constitutive methylation, only affects the expression of a few cell cycle genes (Gonzalez et al. 2014). Thus, a role for changes in DNA methylation as a “trigger” for GcrA-dependent gene expression remains uncertain.
Role of GcrA in the Caulobacter cell cycle
Collectively, our findings on GcrA also force a significant reconsideration of its role in the Caulobacter cell cycle. First, because GcrA does not promote transcription at all of the promoters to which it binds, ChIP-based studies alone are inherently unable to accurately define GcrA target genes. By developing a mechanistic understanding of GcrA, we were able to then better delineate the set of genes whose transcription is affected by GcrA. These findings indicate that GcrA directly affects at least 204 genes, with many additional indirect targets (Fig. 5). These genes participate in numerous cell cycle processes, many of which help cells progress through S phase, including nucleotide synthesis, DNA repair, chromosome organization and segregation, and cell division. Notably, however, and in contrast to prior suggestions, the GcrA regulon does not include dnaA or, with one exception, core components of the replisome. Consistent with this finding, cells lacking GcrA continue to periodically initiate new rounds of DNA replication, often without an intervening cell division (Fig. 6). A similar phenomenon occurs with mutants in the CtrA phosphorylation pathway (Jonas et al. 2011). Together, these results underscore the fact that oscillations in DnaA (and DNA replication) occur independently of CtrA and GcrA, meaning that these three cell cycle regulators do not comprise a single genetic oscillator.
Second, GcrA-regulated genes are still expressed in cells depleted of GcrA, albeit at lower rates, leading to a delay in processes such as cell division, not a complete disruption. The magnitude of expression changes following GcrA depletion is often only twofold to threefold. In contrast, strains depleted of a canonical transcription factor like CtrA are almost completely blocked for cell division, motility, and other CtrA-dependent processes, with target genes typically decreasing 10-fold or more (Laub et al. 2002). The differences in gene expression following a loss of GcrA or CtrA are mirrored in their phenotypic differences. Most notably, gcrA mutants are viable in mutant backgrounds that slow growth and DNA replication initiation, which likely rebalances cell cycle events such that the periodicity of DNA replication better matches the periodicity of cell division. Growth in minimal medium also partially rescues the essentiality of GcrA (Murray et al. 2013) but not nearly as well as slowing growth and replication initiation, supporting the notion that cells lacking GcrA have imbalanced replication and division cycles (Fig. 6C,D; Supplemental Fig. 7B). In contrast to GcrA, cells lacking CtrA are inviable in most growth conditions and are not rescued by mutations that slow replication and growth.
In sum, we propose that GcrA is a novel accessory factor of RNAP in Caulobacter that stimulates the transcription of target genes through an unconventional interaction with region 2 of the housekeeping σ factor σ70. In contrast to classical activators that typically bind DNA independently to recruit RNAP, GcrA travels with RNAP to promoters, where it can stabilize binding and promote isomerization. Because GcrA interfaces with region 2, other transcription factors can still target region 4 of σ70 or other RNAP subunits, enabling combinatorial control and synergistic activation of gene expression. GcrA is extremely well conserved among α-proteobacteria, where it may play a role in regulating gene expression similar to that elucidated here. Additionally, Caulobacter and many α-proteobacteria encode paralogs of GcrA, and homologs have been found in bacteriophage genomes (Gill et al. 2012). Each of these proteins may, like GcrA, interact directly with RNAP to globally regulate gene expression.
Materials and methods
For details on strain construction, growth conditions, and detailed experimental procedures, see the Supplemental Material and Supplemental Tables S4–S6.
ChIP-seq and data analysis
For details on ChIP, library generation, and data analysis, see the Supplemental Material. For a summary of ChIP-seq analyses performed, see Supplemental Table S7 and Gene Expression Omnibus GSE73925.
Protein purification
The RNAP holoenzyme was purified by Ni-NTA affinity chromatography from Caulobacter cells producing RpoC with a His10 tag at the C terminus, followed by gel filtration and ion exchange chromatography. His6-GcrA, His6-CcrM, His6-σ70, His6-σ54, and His6-σ32 were purified by Ni-NTA affinity chromatography and gel filtration chromatography for His-GcrA. GST and GST-GcrA were purified by affinity chromatography using glutathione sepharose beads.
Filter-binding and open complex assays
DNA probes were generated by PCR; gel-purified; labeled with T4-polynucleotide kinase in 10-μL reactions containing 7 μL of DNA, 1 μL of radiolabeled γ-P32ATP, 1 μL of T4-PNK, and 1 μL of T4-PNK buffer; and incubated for 90 min at 37°C. Kinetic open complex assays were performed with Caulobacter RNAP holoenzyme diluted in transcription buffer (40 mM Tris-HCl at pH 8.0, 100 mM NaCl, 10 mM MgCl2, 1.4% Tween-20, 50 μg/mL BSA [Ambion], 1 mM DTT) and ±250 nM GcrA. This was incubated for 10 min at 30°C prior to addition of 0.1–0.25 nM labeled DNA diluted in 10 mM Tris-HCl (pH 8.5) and pre-incubated for 10 min at 30°C. Reactions were challenged at the indicated times with 50 μg/mL heparin for 20 sec, bound to prewashed nitrocellulose filters, and immediately washed. Radioactive signal was quantified by scintillation counting. Kinetic parameters were fitted as in Saecker et al. (2002) (see the Supplemental Material).
Supplementary Material
Acknowledgments
We thank K. Schmitz, D. Barthelme, V. Baytshtok, and C. Aakre for reagents and discussions, and C. Haakonsen for help with ChIP-seq analysis. This work was supported by a Howard Hughes Medical Institute International Predoctoral Fellowship to D.L.H., and a National Institutes of Health grant (R01GM082899) to M.T.L., who is also an Investigator at the Howard Hughes Medical Institute. A.H.Y. designed and performed the GcrA suppressor screen (Fig. 6A). All other experiments were performed by D.L.H. D.L.H. and M.T.L. designed experiments, analyzed data, and wrote the paper.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.270660.115.
References
- Banta AB, Chumanov RS, Yuan AH, Lin H, Campbell EA, Burgess RR, Gourse RL. 2013. Key features of σS required for specific recognition by Crl, a transcription factor promoting assembly of RNA polymerase holoenzyme. Proc Natl Acad Sci 110: 15955–15960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Nickels BE, Fan H. 2012. Chlamydia trachomatis protein GrgA activates transcription by contacting the nonconserved region of σ66. Proc Natl Acad Sci 109: 16870–16875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutte CC, Henry JT, Crosson S. 2012. ppGpp and polyphosphate modulate cell cycle progression in Caulobacter crescentus. J Bacteriol 194: 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho EM, Casadesús J. 2005. Regulation of traJ transcription in the Salmonella virulence plasmid by strand-specific DNA adenine hemimethylation. Mol Microbiol 57: 1700–1718. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104: 901–912. [DOI] [PubMed] [Google Scholar]
- Curtis PD, Brun YV. 2010. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev 74: 13–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Passalacqua KD, Hocking J, Llopis PM, Gerstein M, Bergman NH, Jacobs-Wagner C. 2013. Transcriptomic and phylogenetic analysis of a bacterial cell cycle reveals strong associations between gene co-expression and evolution. BMC Genomics 14: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioravanti A, Fumeaux C, Mohapatra SS, Bompard C, Brilli M, Frandi A, Castric V, Villeret V, Viollier PH, Biondi EG. 2013. DNA binding of the cell cycle transcriptional regulator GcrA depends on N6-adenosine methylation in Caulobacter crescentus and other alphaproteobacteria. PLoS Genet 9: e1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill JJ, Berry JD, Russell WK, Lessor L, Escobar-Garcia DA, Hernandez D, Kane A, Keene J, Maddox M, Martin R, et al. 2012. The Caulobacter crescentus phage phiCbK: genomics of a canonical phage. BMC Genomics 13: 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D, Kozdon JB, McAdams HH, Shapiro L, Collier J. 2014. The functions of DNA methylation by CcrM in Caulobacter crescentus: a global approach. Nucleic Acids Res 42: 3720–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. 2003. Multiple σ subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 57: 441–466. [DOI] [PubMed] [Google Scholar]
- Haagmans W, van der Woude M. 2000. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol Microbiol 35: 877–887. [DOI] [PubMed] [Google Scholar]
- Haugen SP, Ross W, Gourse RL. 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol 6: 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzendorff J, Hung D, Brende P, Reisenauer A, Viollier PH, McAdams HH, Shapiro L. 2004. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science 304: 983–987. [DOI] [PubMed] [Google Scholar]
- Hottes AK, Shapiro L, McAdams HH. 2005. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol Microbiol 58: 1340–1353. [DOI] [PubMed] [Google Scholar]
- Hubin EA, Tabib-Salazar A, Humphrey LJ, Flack JE, Olinares PDB, Darst SA, Campbell EA, Paget MS. 2015. Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA. Proc Natl Acad Sci 112: 7171–7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Chen YE, Laub MT. 2011. Modularity of the bacterial cell cycle enables independent spatial and temporal control of DNA replication. Curr Biol 21: 1092–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci 95: 5752–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290: 2144–2148. [DOI] [PubMed] [Google Scholar]
- Laub MT, Chen SL, Shapiro L, McAdams HH. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci 99: 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Minchin SD, Busby SJW. 2012. Activating transcription in bacteria. Annu Rev Microbiol 66: 125–152. [DOI] [PubMed] [Google Scholar]
- Lennon CW, Ross W, Martin-Tumasz S, Toulokhonov I, Vrentas CE, Rutherford ST, Lee J-H, Butcher SE, Gourse RL. 2012. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev 26: 2634–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SM, Panis G, Fumeaux C, Viollier PH, Howard M. 2013. Computational and genetic reduction of a cell cycle to its simplest, primordial components. PLoS Biol 11: e1001749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118: 311–322. [DOI] [PubMed] [Google Scholar]
- Reisenauer A, Shapiro L. 2002. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J 21: 4969–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Villers CL, Lee J-H, Ross W, Gourse RL. 2009. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev 23: 236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saecker RM, Tsodikov OV, McQuade KL, Schlax PE, Capp MW, Record MT. 2002. Kinetic studies and structural models of the association of E. coli σ70 RNA polymerase with the λPR promoter: large scale conformational changes in forming the kinetically significant intermediates. J Mol Biol 319: 649–671. [DOI] [PubMed] [Google Scholar]
- Saecker RM, Record MT, Dehaseth PL. 2011. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol 412: 754–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader JM, Zhou B, Li G-W, Lasker K, Childers WS, Williams B, Long T, Crosson S, McAdams HH, Weissman JS, et al. 2014. The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS Genet 10: e1004463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Laub MT. 2004. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat Rev Microbiol 2: 325–337. [DOI] [PubMed] [Google Scholar]
- Srivastava DB, Leon K, Osmundson J, Garner AL, Weiss LA, Westblade LF, Glickman MS, Landick R, Darst SA, Stallings CL, et al. 2013. Structure and function of CarD, an essential mycobacterial transcription factor. Proc Natl Acad Sci 110: 12619–12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. 2009. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138: 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabib-Salazar A, Liu B, Doughty P, Lewis RA, Ghosh S, Parsy M-L, Simpson PJ, O'Dwyer K, Matthews SJ, Paget MS. 2013. The actinobacterial transcription factor RbpA binds to the principal σ subunit of RNA polymerase. Nucleic Acids Res 41: 5679–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wion D, Casadesús J. 2006. N6-methyl-adenine: an epigenetic signal for DNA–protein interactions. Nat Rev Microbiol 4: 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R, Stephens C, Zweiger G, Shapiro L, Alley MR. 1996. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev 10: 1532–1542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.