The host chromatin-binding factor LEDGF/p75 interacts with HIV-1 integrase and directs integration to active transcription units. Singh et al. show that LEDGF/p75 interacts with splicing factors, contributes to exon choice, and directs HIV-1 integration to transcription units that are highly spliced.
Keywords: HIV-1, integration, mRNA splicing, LEDGF, p75, retrovirus
Abstract
The host chromatin-binding factor LEDGF/p75 interacts with HIV-1 integrase and directs integration to active transcription units. To understand how LEDGF/p75 recognizes transcription units, we sequenced 1 million HIV-1 integration sites isolated from cultured HEK293T cells. Analysis of integration sites showed that cancer genes were preferentially targeted, raising concerns about using lentivirus vectors for gene therapy. Additional analysis led to the discovery that introns and alternative splicing contributed significantly to integration site selection. These correlations were independent of transcription levels, size of transcription units, and length of the introns. Multivariate analysis with five parameters previously found to predict integration sites showed that intron density is the strongest predictor of integration density in transcription units. Analysis of previously published HIV-1 integration site data showed that integration density in transcription units in mouse embryonic fibroblasts also correlated strongly with intron number, and this correlation was absent in cells lacking LEDGF. Affinity purification showed that LEDGF/p75 is associated with a number of splicing factors, and RNA sequencing (RNA-seq) analysis of HEK293T cells lacking LEDGF/p75 or the LEDGF/p75 integrase-binding domain (IBD) showed that LEDGF/p75 contributes to splicing patterns in half of the transcription units that have alternative isoforms. Thus, LEDGF/p75 interacts with splicing factors, contributes to exon choice, and directs HIV-1 integration to transcription units that are highly spliced.
Gene therapy is being used with increasing success in the treatment of genetic disorders and is showing particular promise for immunotherapy of cancer. These therapies commonly use retroviral vectors to stably integrate the corrective/therapeutic sequences in the genomes of the patient's cells. The first successful retroviral gene therapies used vectors derived from the γ retroviruses to correct disorders such as X-linked severe combined immunodeficiency (SCID-X1) (Hacein-Bey-Abina et al. 2003, 2010). However, there was a significant risk associated with the use of γ retrovirus-based vectors because of their potential to activate, by integration, proto-oncogenes. This was a problem for some of the SCID-X1 patients who had vector-induced clonal proliferation of their T cells that progressed to leukemia (Hacein-Bey-Abina et al. 2003, 2010).
The γ retroviruses have a strong preference for integrating near-active enhancers (Wu et al. 2003; De Rijck et al. 2013; Gupta et al. 2013; Sharma et al. 2013; De Ravin et al. 2014; LaFave et al. 2014). This bias increases the risk of activating proto-oncogenes and has led to the use of lentivirus vectors for recent gene therapy trials. HIV-1-based vectors are thought to be less likely to cause malignancies because integration is not directed to active enhancers (De Rijck et al. 2013). However, HIV-1 integration occurs throughout the bodies of active transcription units (Schroder et al. 2002; Wu et al. 2003; Wang et al. 2007; Ferris et al. 2010), raising the concern that HIV integrations also have the potential to induce the expression of proto-oncogenes. This concern was heightened by recent studies of HIV-1 patients undergoing suppressive combination anti-retroviral therapy, which indicated that integration in specific genes can cause clonal expansion and persistence of the infected cells (Maldarelli et al. 2014; Wagner et al. 2014).
Structural and biochemical data show that HIV-1 integrase interacts with the host factor LEDGF/p75 (Cherepanov et al. 2003, 2004, 2005; Maertens et al. 2003; Llano et al. 2004; Busschots et al. 2005; Shun et al. 2007), and this interaction favors integration in active transcription units, which is the portion of genes that are transcribed (Ciuffi et al. 2005; Llano et al. 2006a; Vandekerckhove et al. 2006; Shun et al. 2007; Ferris et al. 2010; Wang et al. 2012; Koh et al. 2013). Alternative splicing of LEDGF produces a full-length isoform (p75) and a truncated isoform (p52). Both isoforms contain the chromatin-binding domains (PWWP and AT-hook elements), the major determinants for high-affinity and site-specific binding to chromatin (Llano et al. 2006b; Turlure et al. 2006; Meehan et al. 2009; Ferris et al. 2010; Gijsbers et al. 2010, 2011); however, only LEDGF/p75 contains the integrase-binding domain (IBD), and only this isoform mediates HIV-1 integration.
Although it is clear that LEDGF/p75 helps direct HIV-1 integration to active transcription units, there are important questions about the role of LEDGF/p75 that remain to be addressed. It is not clear how LEDGF/p75 recognizes active transcription units (Schroder et al. 2002; Ferris et al. 2010; Wang et al. 2012). It is not known whether LEDGF/p75 couples integration with transcription or merely interacts with features of transcription units. An important reason these questions are unanswered is that relatively little is known about the cellular functions of LEDGF/p75.
In order to understand integration targeting and its biological impact, we sought to measure integration at the level of individual genes. To obtain the largest possible number of integration events, we generated maps of integration sites with a single-round HIV-1 vector in HEK293T cells. This system and improvements in sequencing methods allowed us to map 961,274 independent integration sites; of the sites in transcription units, 82% occurred in just 4000 genes. Importantly, the 1000 transcription units with the highest numbers of integration sites were highly enriched for cancer-associated genes. Analysis of the integration site densities per transcription unit (integration sites per kilobase) revealed a striking bias that favored transcription units with high numbers of introns and those that produced multiple spliced mRNAs. Our analysis of published profiles of HIV-1 integration demonstrated that LEDGF is required for targeting highly spliced transcription units. Affinity purification of LEDGF/p75 coupled with tandem mass spectrometry (MS/MS) identified a variety of associated splicing factors, including components of snRNPs, hnRNPs, and helicases. Biallelic deletion of the PSIP1 region encoding the IBD of LEDGF/p75 resulted in significant changes in the splicing patterns of the mRNAs from >5000 genes. Together, these results show that LEDGF/p75 specifically interacts with splicing machinery and is required for targeting HIV-1 integration to highly spliced genes.
Results
Approximately 75% of HIV-1 integration occurs in RNA polymerase II (Pol II) transcription units (Schroder et al. 2002; Wang et al. 2007). To improve our understanding of HIV-1 integration, we generated a high-density map of HIV-1 integration sites in cultured human cells (Materials and Methods). A single-round replication-defective virus was used to infect HEK293T cells. Both the ligation reactions and PCR were performed in multiplexed format, which made it possible to generate complex integration site libraries that were directly sequenced without nested PCR amplification or additional rounds of adaptor ligation. A total of 961,274 unique virus–host junctions were obtained. This profile is large enough to provide integration frequencies in individual transcription units that are highly reproducible, as shown by pairwise comparisons of eight independently generated sublibraries (R2 values between 0.89 and 0.98) (Supplemental Table S1 for RefSeq genes).
We counted the total number of integrations within each transcription unit and found that the number of integrations per transcription unit was high in a relatively small number of transcription units (Supplemental Fig. S1); 82% of all of the integration sites in transcription units mapped to just 4000 transcription units. Analysis of the 1000 transcription units with the highest number of total integration sites revealed significant enrichment associated with mRNA splicing and histone methylation (Table 1). An extended list of gene ontologies enriched in this group of the top 1000 transcription units is given in Supplemental Table S2.
Table 1.
The 1000 transcription units with the highest total number of integration sites in HEK293T cells are enriched with functions associated with histone methylation and cancer
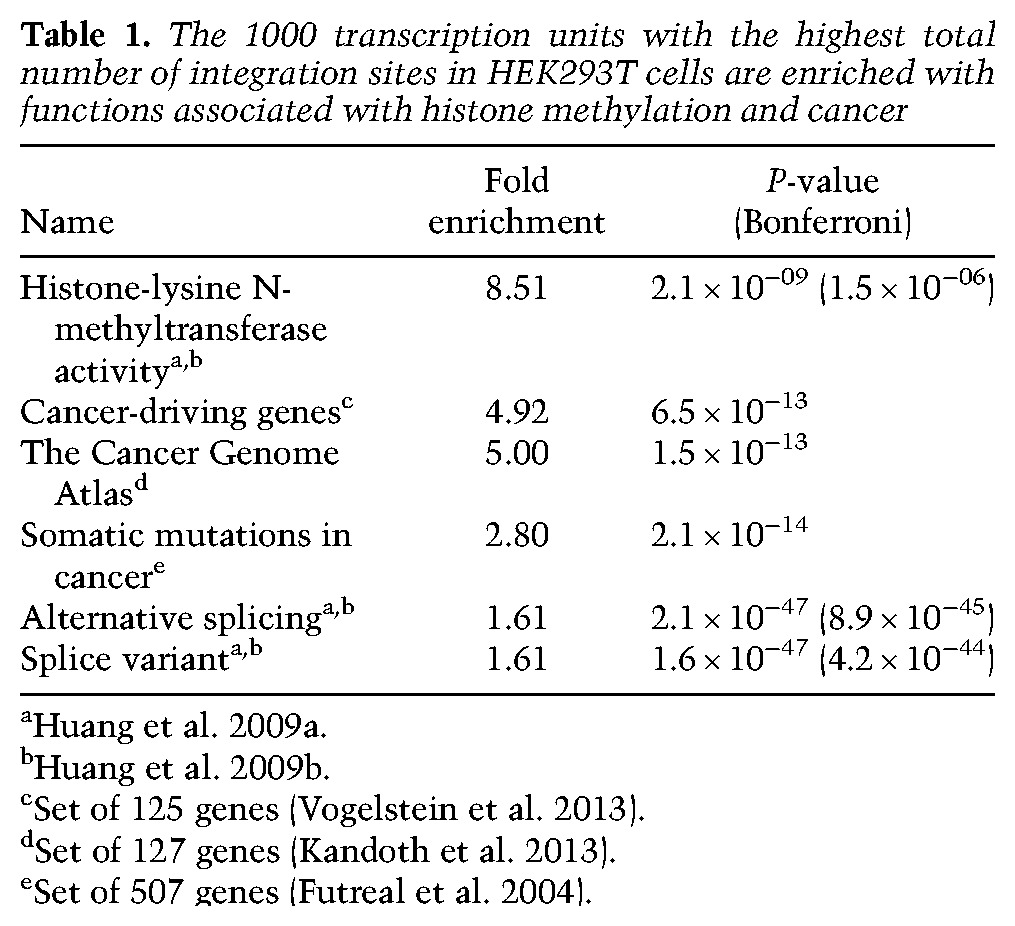
Recent identification of HIV-1 integration sites in peripheral blood lymphocytes of infected patients undergoing antiretroviral therapy indicated that integration into some transcription units may cause clonal expansion of the infected cells (Maldarelli et al. 2014; Wagner et al. 2014). We asked what fraction of the HEK293T integration sites was within transcription units that are known to be associated with an increased risk of cancer. By evaluating independently curated lists of cancer-associated genes, we found that the group of 1000 transcription units with the highest numbers of total integration sites included three to five times more cancer genes than would be predicted based on their genome-wide prevalence (Table 1; Kandoth et al. 2013; Vogelstein et al. 2013; Forbes et al. 2014). These observations indicate that HIV-1 integration favors cancer genes.
To determine whether the high number of integration sites in specific groups of transcription units such as cancer-associated genes was due to targeted integration, we normalized the number of integration sites in each transcription unit by the length. By analyzing the top 1000 transcription units based on integration sites per kilobase, cancer-associated genes, histone methyltransferases, and splicing-associated genes were still favored (Supplemental Tables S3, S4). This shows that HIV-1 integration favors cancer-associated genes with enrichment levels as much as fourfold.
LEDGF causes integration to favor the 5′ end of transcription units
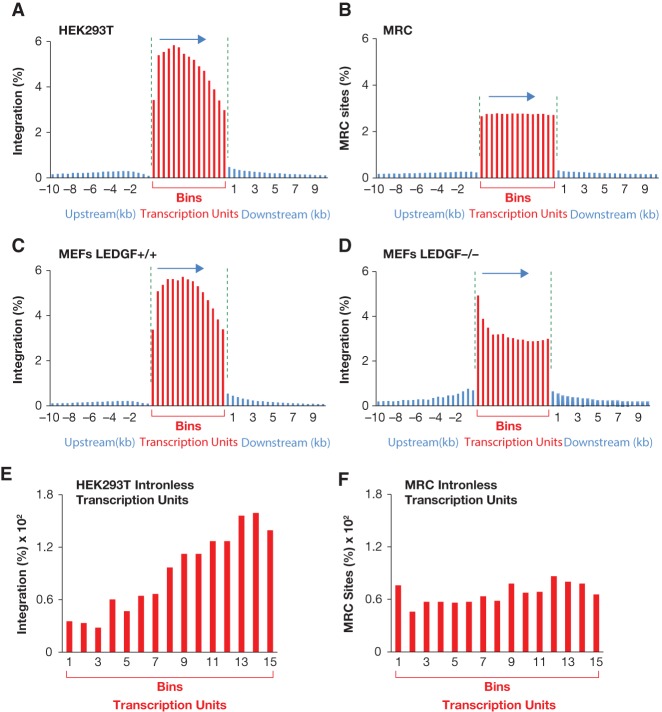
We examined how HIV-1 integration sites were distributed within transcription units by dividing RefSeq transcription units into 15 equal segments (bins) and tabulating the integration sites within each bin (Fig. 1A). Intergenic integration sites were displayed in 500-base-pair (bp) segments either upstream of or downstream from transcription units, depending on whether the integration sites were nearer to the 5′ or 3′ ends of transcription units. Seventy-two percent of the 961,274 integration sites mapped within transcription units and the integration sites had a strong 5′ bias. No such bias was observed in a matched random control (MRC) set of insertions generated in silico (Materials and Methods) to simulate random insertions (Fig. 1B; Berry et al. 2006). Because the integration site libraries were generated from MseI-digested DNA, this random library was designed so that the sites matched the distances to MseI sites seen in the actual integration site library. Forty-one percent of randomly generated MRC insertions occurred in transcription units, a number similar to the fraction of the genome that lies within the known Pol II transcription units. In the HEK293T library, there was an obvious reduction in the number of integrations in the region between 500 and 1500 bp upstream of transcription units, which was not seen in the MRC library. (Fig. 1, A vs. B).
Figure 1.
Distribution of HIV-1 integration sites within transcription units. Each transcription unit was divided into 15 equal parts (bins), and HIV-1 integration sites or MRC insertion sites were counted for each bin (shown as red bars). Outside of transcription units, the genome was divided into 500-bp bins, and the HIV-1 integration sites or MRC insertion sites were counted in each bin (shown as blue bars). The percentages of all of the HIV-1 integration sites and MRC insertion sites are shown on the Y-axis. The horizontal arrow shows the direction of transcription. The green vertical lines separate the transcription units from the upstream and downstream regions to indicate that the bins within the transcription units are not equivalent in size to the 500-bp bins outside of the transcription units. (A) Distribution of HIV-1 integration sites in human HEK293T cells. (B) Distribution of MRC insertion sites in human transcription units. (C) Distribution of HIV-1 integration sites in transcription units in mouse fibroblast cells that contain the wild-type LEDGF gene. (D) Distribution of HIV-1 integration sites in mouse fibroblast cells that lack the LEDGF gene. (E,F) Distribution of HIV-1 integration sites (E) and MRC insertion sites (F) within intronless transcription units.
The availability of previously published HIV-1 integration data in mouse embryonic fibroblasts (MEFs) allowed us to compare the data that we obtained from infected HEK293T cells with data for HIV-1 integration in cells from a different species to see whether there was the same strong bias for the 5′ end of transcription units (Wang et al. 2012). The bias for preferential integration near the 5′ end of Pol II transcription units was apparent in the MEF cells (Fig. 1C) and, more importantly, was absent in cells lacking LEDGF (Fig. 1D). Broadly speaking, the distribution of integration sites in MEF cells lacking LEDGF was similar to the MRC sites (Fig. 1B); however, there was a strong bias for integration in the extreme 5′ end of transcription units. Very similar integration site distribution patterns were seen in two other independently generated HIV-1 integration site libraries generated from infected MEFs that did or did not express LEDGF (Supplemental Figs. S2, S3; Wang et al. 2012; Koh et al. 2013).
Efforts to identify the features of transcription units that were responsible for preferential integration near the 5′ ends of transcription units included tests of whether introns might be involved. In transcription units that lacked introns, integration favored the 3′ end of the transcription units (Fig. 1E). There was no such bias when the MRC insertions were mapped to intronless transcription units (Fig. 1F).
Integration is targeted to highly spliced transcription units by a mechanism that requires LEDGF
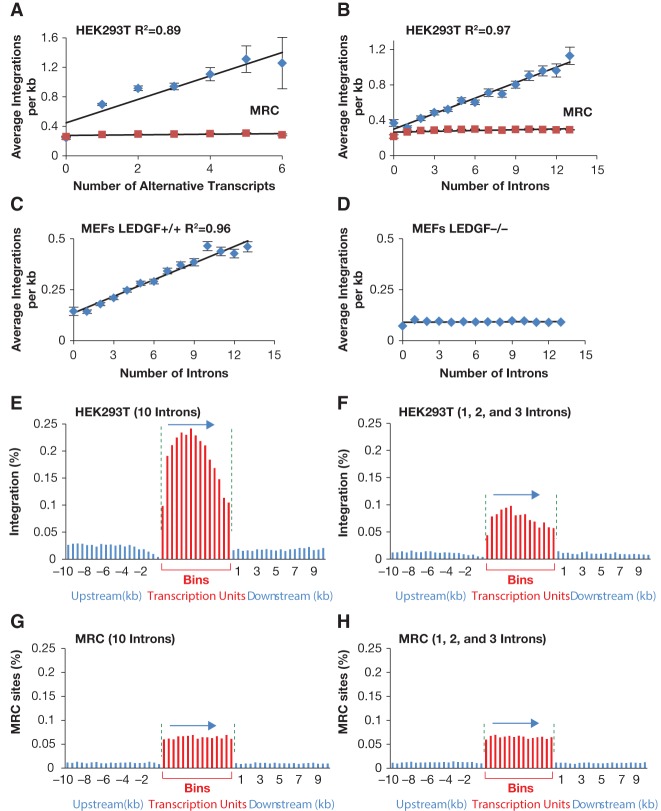
The marked difference in the distribution of integration sites in transcription units that have and lack introns led us to ask whether transcription units with more mRNA splicing had higher levels of integration. We used the number of alternatively spliced transcripts produced per transcription unit as determined with RNA sequencing (RNA-seq) data from our HEK293T cells as a measure of mRNA splicing (analyzed with Cufflinks) (Supplemental Table S5). We observed a strong correlation between the numbers of spliced isoforms per transcription unit and the density of integration sites (integration sites per kilobase) that was absent in the MRC (Fig. 2A). We also used, as an independent measure of splicing within a transcription unit, the total number of introns as annotated by RefSeq (Supplemental Table S6). We grouped all transcription units by the number of introns they contain and calculated the average integration site density (integration sites per kilobase) for each group. This measure revealed a striking correlation between the total numbers of introns and the density of integration sites in the transcription unit (Fig. 2B). This relationship is absent in the MRC. We also observed a strong correlation between the number of introns and integration site density when we analyzed integration sites obtained from HIV-1-infected MEF cells (Fig. 2C; Wang et al. 2012). Importantly, these correlations were not seen in an analysis of integration sites in MEF cells lacking LEDGF (Fig. 2D). The strong correlation between intron number and integration site density was also seen in the two other independent sets of integration data generated in MEFs (Supplemental Figs. S4, S5; Wang et al. 2012; Koh et al. 2013).
Figure 2.
Integration density correlates strongly with the level of splicing. (A) Correlation between HIV-1 integration density and the number of alternative transcripts produced by transcription units in human HEK293T cells. Each transcription unit was assigned to a group based on the number of alternatively spliced transcripts that originated from it. The X-axis shows the number of alternative transcripts per transcription unit, and the Y-axis shows the average HIV-1 integration density for all transcription units that produce the same number of alternative transcripts. The blue diamonds represent data for HIV-1 integration sites in HEK293T, whereas the red rectangles are for the MRC insertion sites. The vertical bar for each data point is the standard error. R2 is the Pearson correlation. (B–D) Correlation between average HIV-1 integration density and the number of introns in the transcription units. The X-axis shows the number of introns per transcription unit, and the Y-axis shows the average HIV-1 integration density (integration sites per kilobase) for all transcription units that have the same number of introns. The blue diamonds represent data for HIV-1 integrations, and the red rectangles are for the MRC insertion sites. (B) HIV-1 integration sites in human HEK293T cells. C and D are based on HIV-1 integration sites in mouse fibroblast cells that either have or lack the LEDGF gene, respectively. (E–H) Distribution of HIV-1 integration sites (E,F) or MRC insertion sites (G,H) within transcription units that contain either 10 introns (E,G) or one to three introns (F,H), based on the data from HEK293T cells. For each transcription unit with 10 introns, a partner transcription unit of equal size was selected from the group that contains transcription units with one to three introns, so the total length of all transcription units with 10 introns is equal to the total length of all transcription units with one to three introns. As in Figure 1, each transcription unit was divided into 15 equal parts (shown in red), and HIV-1 integration sites or MRC sites were counted in each segment.
One possible reason why the integration density correlated with the number of introns is that intronic sequences could have higher numbers of integration sites per kilobase than exons. However, we found that the average integration density for all introns is 0.57 per kilobase, a number that is very similar to the average integration density in exons (0.54 per kilobase) (Supplemental Table S7). It is also possible that transcripts with more introns have a higher density of integration sites because they are larger and, as a result of an unknown mechanistic property, better able to recruit preintegration complexes. To test this possibility, we compared two sets of 673 transcription units that were matched to have equal lengths. The first set of transcription units had 10 introns, and the size-matched transcription units in the other set had one, two, or three introns. The size-matched transcription units with 10 introns had substantially more integrations than the transcription units with one, two, or three introns (Fig. 2E,F). Moreover, the distribution of integration sites throughout the transcription units with one to three introns showed that the 5′ bias was much weaker compared with the integration sites in transcription units with 10 introns (Figs. 1A,C, 2, F vs. E). The high level of integration and the 5′ bias seen in transcription units with 10 introns was not seen with the MRC sites (Fig. 2G). Interestingly, the integration density in transcription units with one, two, and three introns was similar to the MRC set for these transcription units, suggesting that, in this group, the integration was similar to what would occur if integration were random (Fig. 2, cf. F and H).
Previous studies reported that HIV-1 integration favors highly expressed genes (Schroder et al. 2002; Wang et al. 2007; Ferris et al. 2010). We determined average integration site densities from the HEK293T data for transcription units sorted by levels of transcription and observed, as reported by others, that integration site density in genes correlates with the level of expression (Supplemental Fig. S6). However, this tendency was not responsible for the high integration densities that we observed in highly spliced transcription units because transcripts with higher numbers of introns have, on average, lower expression levels (Supplemental Fig. S7). Thus, the observed increase in the density of integration sites in transcripts with higher numbers of introns is more pronounced when the integration site densities are normalized for the average levels of transcription (Supplemental Fig. S8).
The results presented above indicate that splicing is a key determinant of HIV-1 integration site selection. With this knowledge, we asked whether levels of splicing might explain the high numbers of integrations that we observed in cancer genes. The average number of introns per transcription unit for all RefSeq genes is 8.6. Of the 1000 transcription units with the highest integration site densities, cancer genes had significantly higher numbers of introns, averaging 24.6, 17.7, and 18.4, depending on which specific set of cancer genes was analyzed (Supplemental Table S3). The average number of introns for the other types of genes in the top 1000 HIV-1 targeted transcription units was also high (29 for histone methyltransferases and 15 for alternative splicing factors) (Supplemental Table S3), indicating that genes with a high number of introns were favored for HIV-1 integration.
LEDGF interacts with mRNA splicing machinery
Several lines of evidence demonstrate that mRNA splicing is highly coordinated with transcription and, in some cases, functionally coupled to transcription (Cramer et al. 1999; de la Mata et al. 2003; Reed 2003; Kornblihtt et al. 2004; Listerman et al. 2006; Munoz et al. 2010; Brody et al. 2011; David and Manley 2011; Moehle et al. 2014). As a result, it is possible that LEDGF/p75 not only tethers HIV-1 integrase to chromatin of active transcription units but also interacts with mRNA splicing factors. Such an interaction could result in higher densities of integration in highly spliced transcription units.
A survey of the cellular binding partners of LEDGF could reveal its role in biological processes. Therefore, we used MS/MS to identify cellular proteins from nuclear extracts of HEK293T cells that interacted with GST-LEDGF/p75 or GST-LEDGF/p52. A total of 285 and 293 proteins were detected in the GST-LEDGF/p75 and GST-LEDGF/p52 samples, respectively, that were not present in GST control fractions. To characterize the interacting partners, the biological processes of the proteins were assigned using the UniProt ID mapping tool. One-hundred-ninety-two proteins had at least one identified biological process in each of the GST-LEDGF/p75 and GST-LEDGF/p52 samples. The most represented biological process in the LEDGF/p75 data set was mRNA processing (80 protein hits), and the second most abundant group was the 68 proteins that are involved in transcription (Supplemental Fig. S9). Of the LEDGF/p52-binding partners, 80 proteins are involved in transcription, and 79 proteins are involved in mRNA processing (Supplemental Fig. S10). These findings strongly suggest a role for LEDGF in mRNA processing and transcription.
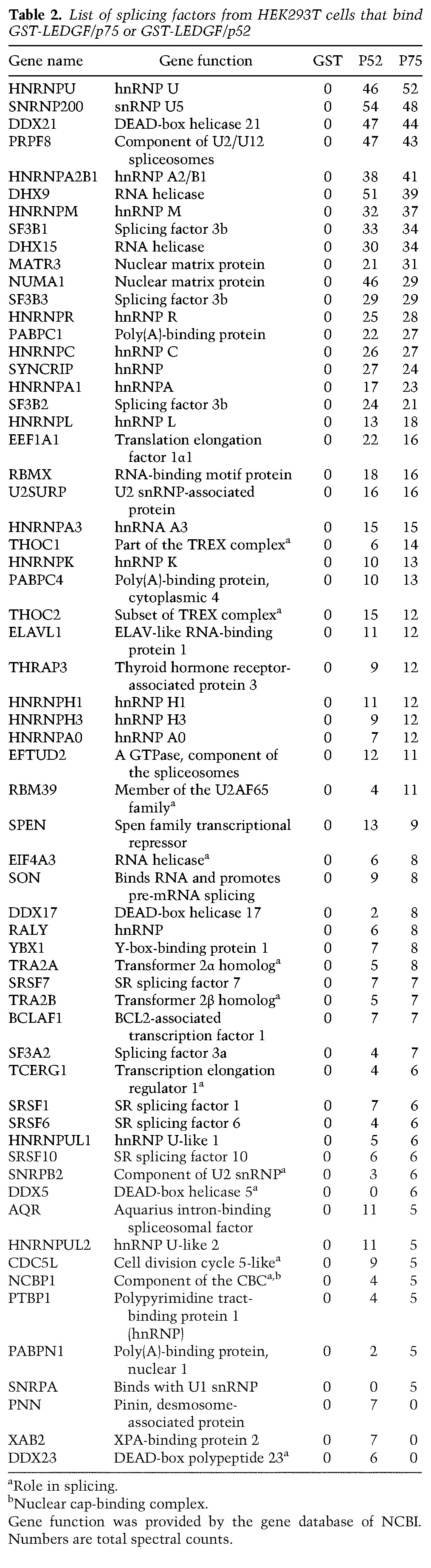
To help determine which of the proteins identified in the LEDGF/p75 and LEDGF/p52 data sets are involved in mRNA processing, the proteins were cross-referenced against the Spliceosome Database (http://spliceosomedb.ucsc.edu), which identified a large number of splicing factors (Table 2). The vast majority of the splicing factors that were detected were found in both the LEDGF/p75 and LEDGF/p52 samples; however, the splicing factors DDX5 and SNRPA were found only in the LEDGF/p75 data set. Two proteins, KMT2B and Men1, which are known to interact selectively with LEDGF/p75 but not with LEDGF/p52, yielded total spectrum counts of 26 and seven, respectively, in the LEDGF/p75 data set, whereas both proteins had a count of zero for GST and GST-LEDGF/p52 (data not shown).
Table 2.
List of splicing factors from HEK293T cells that bind GST-LEDGF/p75 or GST-LEDGF/p52
Our MS data showed that both GST-LEDGF/p75 and GST-LEDGF/p52 can interact with a number of cellular proteins that play a role in splicing. Because there were high spectral counts and because of the previous report of interactions with LEDGF/p52, we tested SRSF1, SF3B2, and hnRNP M for their ability to interact with both LEDGF isoforms in affinity pull-down experiments. The results showed that all three proteins (SRSF1, SF3B2, and hnRNP M) bind to LEDGF/p75 and LEDGF/p52 (Fig. 3).
Figure 3.
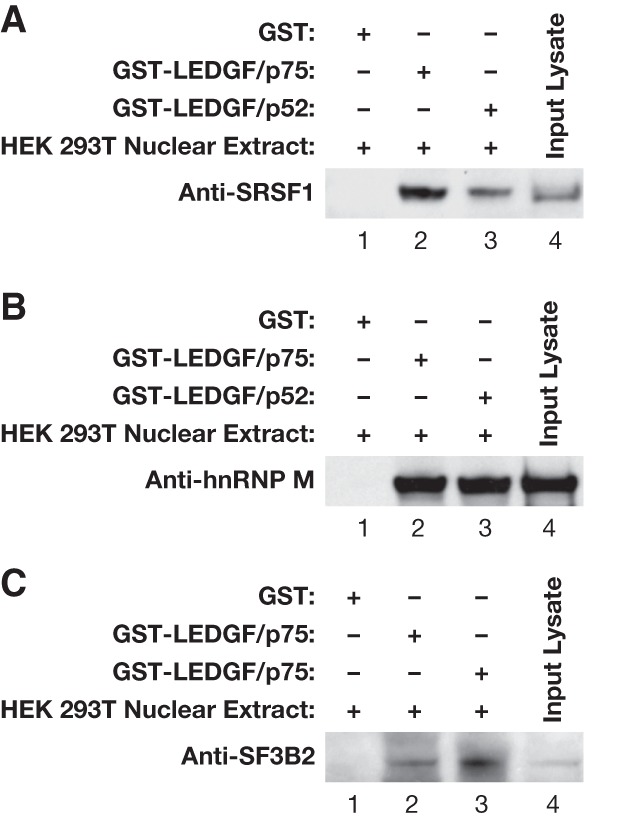
The interactions of representative splicing proteins with LEDGF/p75 and LEDGF/p52. GST pull-down of HEK293T nuclear lysate with GST, GST-LEDGF/p75, or GST–LEDGF/p52 followed by Western blotting to detect splicing factors SRSF1 (A), hnRNP M (B), and SF3B2 (C).
Deletion of the IBD of LEDGF/75 caused substantial changes in splicing patterns
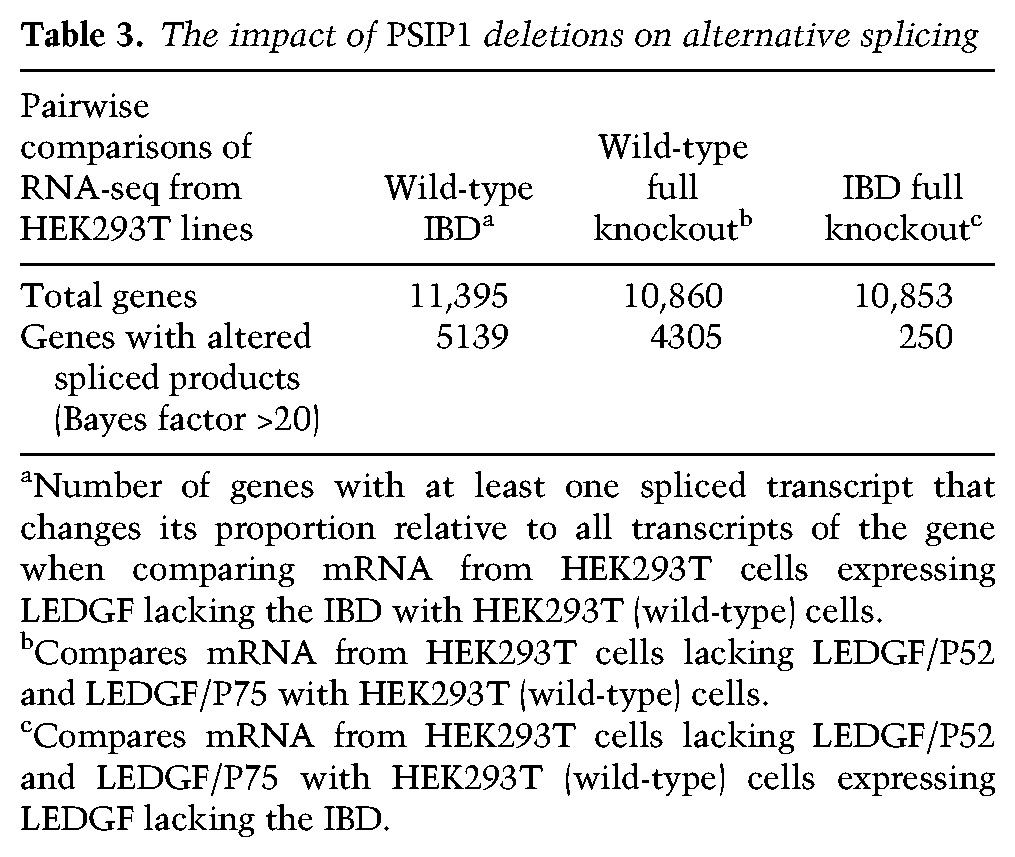
Deletion of PSIP1, the gene encoding LEDGF in MEF cells, was previously reported to cause changes in splicing patterns. The absence of LEDGF was shown to alter inclusion of alternative exons in 90 genes (Pradeepa et al. 2012), and the changes in the splicing patterns of these genes were attributed to the loss of the LEDGF/p52 isoform. To test the role of LEDGF/p75 in splicing, we performed RNA-seq on HEK293T cells that were altered with TALEN endonucleases to express LEDGF/p52 and a truncated form of LEDGF/p75 that lacked the IBD (Fadel et al. 2014). We also analyzed a line of HEK293T cells that had been modified by a TALEN-generated deletion so that it lacked both LEDGF/p75 and LEDGF/p52. For each cell line, four independent RNA samples were subjected to RNA-seq analysis, and the data were used to quantitate statistically significant differences in the amounts of spliced RNAs in the cells. The analysis included ∼11,000 transcription units that produced two or more spliced mRNA products. When the RNA from the wild-type HEK293T cells was compared with RNA from cells with a complete deletion of PSIP1, which encodes both LEDGF/p75 and LEDGF/p52, there were significant changes in the ratio of spliced products of 4305 transcription units (Table 3, Bayes factor >20). Analysis of the RNA species expressed by cells that produce LEDGF lacking the IBD showed that there were significant changes in spliced products of 5139 genes (Table 3, Bayes factor >20). These results show that LEDGF/p75 and the IBD contribute significantly to the pattern of alternative splicing. The changes in alternative splicing seen in cells lacking the entire LEDGF protein verses just the IBD were very similar, indicating that, in the absence of an intact LEDGF/p75, LEDGF/p52 alone had little effect on the pattern of alternate splicing (Table 3, IBD vs. LEDGF).
Table 3.
The impact of PSIP1 deletions on alternative splicing
Intron density of transcription units is the strongest predictor of integration
To evaluate which features of transcription units best predict the level of integration, we developed regression models using factors previously shown to correlate with integration in genome-wide analyses (Schroder et al. 2002; Berry et al. 2006; Wang et al. 2007; Craigie and Bushman 2012). For each of the 21,188 transcription units analyzed, we tabulated values for intron density, histone H3K4 trimethylation (H3K4me3), transcription level (FPKM [fragments per kilobase per million mapped fragments]), DNase I cleavage sites, and percent GC base pairs. The transcription units were grouped into sets of 100 based on integration density, and, for each group of transcription units, the values of each factor were averaged. Natural log values for FPKM, DNase I sites, and integration density were used because this resulted in higher correlations. An examination of each factor against the log(integration density) showed virtually no correlation of the integration density with the percent GC content (Supplemental Material). A weak, negative correlation was observed with log(DNAse1) (r = −0.387), and strong correlations were obtained for log(FPKM) (r = 0.808) and histone H3 trimethylation (r = 0.886), a histone modification that is strongly associated with the level of gene expression. The strongest correlation was with intron density (r = 0.903). This means that there is a strong linear relationship between intron density and log(integrations density), and therefore it is the best single predictor of integration density in transcription units.
To identify the strongest predictor of integration density in transcription units, we performed five-factor multivariate analysis. When all five factors are included in multivariate analysis (Supplemental Material), the GC content is ignored because it does not contribute to the fit. The importance of the remaining four factors reflects the same ordering as found in the individual correlations with log(integration density). The intron density is the most important factor, followed by H3K4me3, log(FPKM), and then log(DNase1). By removing each factor individually and performing the multivariate fit, we found that the percent GC content and the log(DNase1) made small contributions. When log(FPKM) was removed, the best multivariate fit of the linear model included just intron density and H3K4me3 (r2 of 0.875). Finally, removing intron density had by far the largest effect on the multivariate fit, decreasing the r2 to 0.812. All of these results are consistent with the conclusion that, of the five factors considered, intron density is the strongest predictor of log(integration density) in transcription units, followed by H3K4me3.
Discussion
Over 2000 gene therapy clinical trials have been conducted, and this number is growing rapidly (Kaufmann et al. 2013). The majority of the gene therapies are designed to treat disorders of proliferating cells such as leukemia and lymphoma, in large part because of progress in developing protocols to isolate haematopoietic stem cells, introducing therapeutic genes into these cells, and transplanting the altered stem cells back to the patient. Of particular note are the recent successes in the treatment of B-cell leukemia by introducing chimeric antigen receptors (CARs) into T cells. The bulk of these clinical protocols use lentivirus vectors to achieve stable gene transfer.
The data we present here show that HIV-1 integration disproportionately targets cancer genes and raises concerns about whether using lentivirus vectors for gene therapy could cause hyperplasia. Although our experiments were conducted in cultured cells, recent studies showed that integration sites in HeLa cells closely paralleled integration sites in human CD34+ cells (Maldarelli et al. 2014). The possibility that lentivirus vectors could target cancer genes and induce hyperplasia is consistent with the observation that, in HIV-1-infected individuals, integration in certain oncogenes (MKL2 and BACH2) was associated with clonal expansion (Maldarelli et al. 2014; Wagner et al. 2014; Cohn et al. 2015). These results from HIV-1-infected patients, taken together with our high-density integration site data, argue that patients who have been treated with cells infected with lentivirus vectors should be carefully monitored for evidence of clonal outgrowth of any of the infected cells and the possibility that this might lead to associated cancers.
In HEK293T cells, we showed that integration favored the 5′ regions of transcription units. A preference for the 5′ ends of transcription units was also present in published data sets, which allowed us to show that LEDGF was required for this preference. In addition, integration densities were low upstream of transcription start sites (Fig. 1A,C). This is likely due to the low levels of nucleosomes upstream of start sites. The PWWP domain of LEDGF/p75 binds to modified histone tails, and the prototype foamy virus preintegration complex is known to preferentially bind to bent DNA (Maertens et al. 2010; Pradeepa et al. 2012; Eidahl et al. 2013). There is also evidence that HIV-1 integration favors regions that have nucleosomes (Pryciak and Varmus 1992; Wang et al. 2007). These factors direct integration to nucleosomes; conversely, regions that lack nucleosomes have low numbers of integrations. As expected, in cells lacking LEDGF, integration upstream of transcription units was significantly increased.
Structural, biochemical, and genetic evidence shows that LEDGF/p75 interacts with HIV-1 integrase and helps to direct the preintegration complex to transcription units (Cherepanov et al. 2003, 2004, 2005; Maertens et al. 2003; Busschots et al. 2005; Ciuffi et al. 2005; Llano et al. 2006a; Shun et al. 2007; Ferris et al. 2010; Wang et al. 2012; Koh et al. 2013). However, much less is known about the cellular functions of LEDGF/p75. Previous studies of LEDGF/p52, the short isoform of LEDGF, revealed significant interactions with several splicing-associated factors, including SR proteins such as SRSF1 and proteins of the spliceosomal complex C (Ge et al. 1998; Pradeepa et al. 2012). In addition, LEDGF/p75 was previously reported to interact with NOVA1, an RNA-binding protein that regulates splicing (Morchikh et al. 2013). Our proteomic experiments suggest that the LEDGF proteins have a role in mRNA processing. In particular, we found that LEDGF/p75 and LEDGF/p52 interact with many components of the splicing machinery, including U2 snRNP (SF3B1, SF3B2, and SF3B3), U2-associated proteins (PRPF8 and U2SURP), a factor of the U5 snRNP (SNRNP200), and many hnRNPs that are associated with alternative splicing (Table 2). While a subset of these splicing factors was previously reported to interact with LEDGF/p52 (Pradeepa et al. 2012), our data identify a much wider range of splicing-associated interactions. We note that some of the interactions with splicing machinery could be due to a small number of direct contacts with large nucleoprotein complexes that are themselves held together by protein–protein and protein nucleic acid interactions.
The interactions between LEDGF/p75 and splicing components suggested that LEDGF may contribute to intron/exon choices. This idea is supported by our finding that deletion of either the LEDGF IBD or all of LEDGF caused widespread changes in splicing patterns. This result provides new insight into the cellular function of LEDGF; however, further study is necessary to determine how LEDGF contributes to alternative splicing choices.
Detailed analyses of our data revealed a striking correlation between the transcription units that produce highly spliced mRNAs and the density of HIV-1 integration. Integration densities correlated with the numbers of introns in a transcript and the numbers of spliced RNAs produced by transcription units. The correlation with the numbers of introns was supported by an analysis of several sets of previously published integration site data. Significantly, we found that integration in highly spliced transcription units was dependent on LEDGF. The high level of LEDGF/p75-mediated integration in highly spliced transcription units, taken together with LEDGF interactions with splicing components, provides strong support for a model in which LEDGF/p75 interacts with splicing machinery, and this interaction helps to direct integration to highly spliced transcription units (Fig. 4). This model is also supported by multivariate regression analyses that predict which factors best correlate with the integration densities of transcription units. Considering the factors previously found to correlate best with integration (transcription levels, histone H3K4 trimethylation, and DNase I cleavage sites) we found intron density was the strongest predictor.
Figure 4.
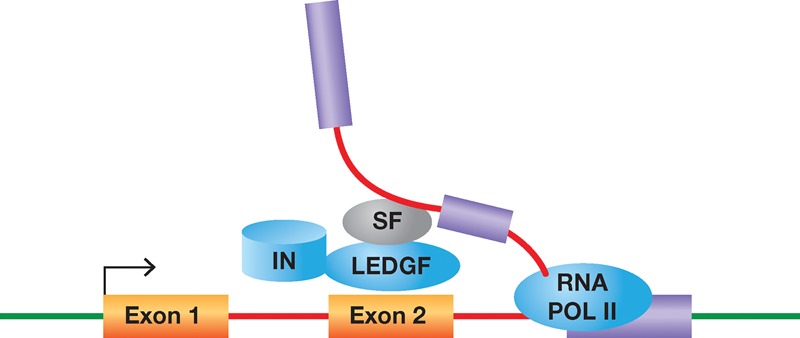
Model for HIV-1 integration in highly spliced transcription units. RNA Pol II transcription and splicing of introns are concurrent. Splicing factors (SF) interact with LEDGF/p75, which in turn binds HIV-1 integrase (IN). These interactions help direct HIV-1 integration to transcription units with a large number of introns.
The significance of the preferential integration of HIV-1 in highly spliced genes is underscored by our finding that cancer-related genes are highly targeted by HIV-1. The high number of introns in cancer-related genes indicates that the recognition of the splicing machinery by LEDGF/p75 directs HIV-1 integration to this collection of medically relevant targets (Table 1). Therefore, the number of introns is a relevant issue when evaluating whether the integration of lentivirus vectors is linked to clonal expansion in gene therapy patients.
Materials and methods
The Supplemental Material contains detailed information about the sequencing of integration sites, the oligonucleotides and barcodes (Supplemental Tables S8, S9), the mapping of integration sites in transcription units, analysis of alternatively spliced transcription units, reproducibility of RNA-seq data sets (Supplemental Fig. S11), MS-based proteomics (Supplemental Table S10), the multivariate regression analyses (Supplemental Table S11), and analysis of integration in cancer genes. All sequences of integration sites and RNA-seq reads have been deposited with the Sequence Read Archive accession number SRP065157.
Cell culture and virus production
HEK293T cells lacking LEDGF/p75 or the LEDGF IBD have been described (Fadel et al. 2014). HEK293T cells (wild type, LEDGF−/−, and IBD−/−) were maintained in DMEM supplemented with 5% fetal bovine serum, 5% newborn calf serum, and 50 U/mL penicillin plus 50 Ug/mL streptomycin. Replication-defective HIV-1 virus was produced in wild-type HEK293T cells and quantified as described (Ferris et al. 2010).
Infections and nucleic acid isolation
HEK293T cells were infected with 0.5–1 µg of VSVg pseudotyped pNLNgoMIVR-Emodluc, and genomic DNA was isolated ∼4 d following infection (Ferris et al. 2010). Total RNA was isolated from uninfected cells using a Qiagen RNeasy minikit.
The derivation of the PSIP1−/− and IBD−/− segment 293T cells by site-specific gene editing has been described elsewhere (Fadel et al. 2014).
Analysis of spliced transcripts
Sequence read densities obtained by RNA-seq were analyzed by MISO as previously described (Katz et al. 2010). Briefly, read densities for each transcript relative to all reads for a gene were defined as ψ. Δψ values were the differences in the relative read densities of a transcript between two RNA-seq experiments. ψ and Δψ values were evaluated statistically using the Bayes factor, which quantifies the odds of differential regulation occurring. A Bayes factor of 20 indicates that the probability that a transcript is differentially expressed between two data sets is 20 times greater than if the densities occurred by chance.
Supplementary Material
Acknowledgments
P.K.S. dedicates this article to his parents, Shri Dhurandhar Singh and Smt. Sushila Devi. We thank Anthony Hickey for computational assistance. We are grateful to Allen Kane for help in preparing the figures. This research was supported by the Intramural Research Programs of the National Institutes of Health from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (H.L.L.) and the National Cancer Institute (S.H.H.). This research also received support from the National Institutes of Health Intramural AIDS Targeted Antiviral Program (H.L.L. and S.H.H.). The present study was also supported by National Institutes of Health grants AI062520 (to M.K.) and AI77344 (to E.M.P.). Funds from the National Cancer Institute under contract number HHSN261200800001E to the Frederick National Laboratory for Cancer Research (B.T.L.) were also used. P.K.S., M.R.P., A.L.F., and H.J.F. designed and performed experiments and analyzed data. J.R.I., C.E., and X.W. provided computational expertise and analysis. E.M.P., S.H.H., M.K., and H.L.L. designed, supervised, and analyzed experiments. B.T.L. and P.K.S. conducted the multivariate analyses. P.K.S. and H.L.L. conceived the study and wrote the paper. All authors contributed to editing of the paper.
Footnotes
Supplemental Material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.267609.115.
References
- Berry C, Hannenhalli S, Leipzig J, Bushman FD. 2006. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol 2: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y. 2011. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol 9: e1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busschots K, Vercammen J, Emiliani S, Benarous R, Engelborghs Y, Christ F, Debyser Z. 2005. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J Biol Chem 280: 17841–17847. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem 278: 372–381. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Devroe E, Silver PA, Engelman A. 2004. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem 279: 48883–48892. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. 2005. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci 102: 17308–17313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med 11: 1287–1289. [DOI] [PubMed] [Google Scholar]
- Cohn LB, Silva IT, Oliveira TY, Rosales RA, Parrish EH, Learn GH, Hahn BH, Czartoski JL, McElrath MJ, Lehmann C, et al. 2015. HIV-1 integration landscape during latent and active infection. Cell 160: 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R, Bushman FD. 2012. HIV DNA integration. Cold Spring Harb Perspect Med 2: a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Caceres JF, Cazalla D, Kadener S, Muro AF, Baralle FE, Kornblihtt AR. 1999. Coupling of transcription with alternative splicing: RNA Pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell 4: 251–258. [DOI] [PubMed] [Google Scholar]
- David CJ, Manley JL. 2011. The RNA polymerase C-terminal domain: a new role in spliceosome assembly. Transcription 2: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532. [DOI] [PubMed] [Google Scholar]
- De Ravin SS, Su L, Theobald N, Choi U, Macpherson JL, Poidinger M, Symonds G, Pond SM, Ferris AL, Hughes SH, et al. 2014. Enhancers are major targets for murine leukemia virus vector integration. J Virol 88: 4504–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijck J, de Kogel C, Demeulemeester J, Vets S, El Ashkar S, Malani N, Bushman FD, Landuyt B, Husson SJ, Busschots K, et al. 2013. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep 5: 886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidahl JO, Crowe BL, North JA, McKee CJ, Shkriabai N, Feng L, Plumb M, Graham RL, Gorelick RJ, Hess S, et al. 2013. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res 41: 3924–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel HJ, Morrison JH, Saenz DT, Fuchs JR, Kvaratskhelia M, Ekker SC, Poeschla EM. 2014. TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism. J Virol 88: 9704–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun MC, Allis CD, Engelman A, et al. 2010. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc Natl Acad Sci 107: 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. 2014. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 43: D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. 2004. A census of human cancer genes. Nat Rev Cancer 4: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Si Y, Wolffe AP. 1998. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol Cell 2: 751–759. [DOI] [PubMed] [Google Scholar]
- Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M, Bushman FD, Debyser Z. 2010. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol Ther 18: 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijsbers R, Vets S, De Rijck J, Ocwieja KE, Ronen K, Malani N, Bushman FD, Debyser Z. 2011. Role of the PWWP domain of lens epithelium-derived growth factor (LEDGF)/p75 cofactor in lentiviral integration targeting. J Biol Chem 286: 41812–41825. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gupta SS, Maetzig T, Maertens GN, Sharif A, Rothe M, Weidner-Glunde M, Galla M, Schambach A, Cherepanov P, Schulz TF. 2013. Bromo and ET domain (BET) chromatin regulators serve as co-factors for murine leukemia virus integration. J Virol 87: 12721–12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302: 415–419. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Hauer J, Lim A, Picard C, Wang GP, Berry CC, Martinache C, Rieux-Laucat F, Latour S, Belohradsky BH, et al. 2010. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med 363: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009a. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009b. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann KB, Büning H, Galy A, Schambach A, Grez M. 2013. Gene therapy on the move. EMBO Mol Med 5: 1642–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh Y, Wu X, Ferris AL, Matreyek KA, Smith SJ, Lee K, KewalRamani VN, Hughes SH, Engelman A. 2013. Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J Virol 87: 648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. 2004. Multiple links between transcription and splicing. RNA 10: 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFave MC, Varshney GK, Gildea DE, Wolfsberg TG, Baxevanis AD, Burgess SM. 2014. MLV integration site selection is driven by strong enhancers and active promoters. Nucleic Acids Res 42: 4257–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM. 2006. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 13: 815–822. [DOI] [PubMed] [Google Scholar]
- Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. 2004. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol 78: 9524–9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. 2006a. An essential role for LEDGF/p75 in HIV integration. Science 314: 461–464. [DOI] [PubMed] [Google Scholar]
- Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM. 2006b. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J Mol Biol 360: 760–773. [DOI] [PubMed] [Google Scholar]
- Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem 278: 33528–33539. [DOI] [PubMed] [Google Scholar]
- Maertens GN, Hare S, Cherepanov P. 2010. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468: 326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, et al. 2014. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345: 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan AM, Saenz DT, Morrison JH, Garcia-Rivera JA, Peretz M, Llano M, Poeschla EM. 2009. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog 57: e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Braberg H, Krogan NJ, Guthrie C. 2014. Adventures in time and space: splicing efficiency and RNA polymerase II elongation rate. RNA Biol 11: 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morchikh M, Naughtin M, Di Nunzio F, Xavier J, Charneau P, Jacob Y, Lavigne M. 2013. TOX4 and NOVA1 proteins are partners of the LEDGF PWWP domain and affect HIV-1 replication. PLoS One 8: e81217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz MJ, de la Mata M, Kornblihtt AR. 2010. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem Sci 35: 497–504. [DOI] [PubMed] [Google Scholar]
- Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. 2012. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet 8: e1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak PM, Varmus HE. 1992. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell 69: 769–780. [DOI] [PubMed] [Google Scholar]
- Reed R. 2003. Coupling transcription, splicing and mRNA export. Curr Opin Cell Biol 15: 326–331. [DOI] [PubMed] [Google Scholar]
- Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110: 521–529. [DOI] [PubMed] [Google Scholar]
- Sharma A, Larue RC, Plumb MR, Malani N, Male F, Slaughter A, Kessl JJ, Shkriabai N, Coward E, Aiyer SS, et al. 2013. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci 110: 12036–12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev 21: 1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. 2006. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res 34: 1653–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekerckhove L, Christ F, Van Maele B, De Rijck J, Gijsbers R, Van den Haute C, Witvrouw M, Debyser Z. 2006. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J Virol 80: 1886–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. 2013. Cancer genome landscapes. Science 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. 2014. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345: 570–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. 2007. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res 17: 1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jurado KA, Wu X, Shun MC, Li X, Ferris AL, Smith SJ, Patel PA, Fuchs JR, Cherepanov P, et al. 2012. HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res 40: 11518–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XL, Li Y, Crise B, Burgess SM. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300: 1749–1751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.