Hammoud et al. conducted extensive genomic profiling and classified three broad spermatogonial stem cell (SSC) populations in juveniles: (1) epithelial-like SSCs, (2) more abundant mesenchymal-like SSCs, and (3) (in older juveniles) abundant cells committing to gametogenesis. Mesenchymal-like SSCs lacked imprinting specifically at paternally imprinted loci but fully restored imprinting prior to puberty. Mesenchymal-like SSCs also displayed developmentally linked DNA demethylation at meiotic genes and also at certain monoallelic neural genes.
Keywords: germline, stem cells, imprinting, spermatogonia, DNA methylation, monoallelic
Abstract
Postnatal spermatogonial stem cells (SSCs) progress through proliferative and developmental stages to populate the testicular niche prior to productive spermatogenesis. To better understand, we conducted extensive genomic profiling at multiple postnatal stages on subpopulations enriched for particular markers (THY1, KIT, OCT4, ID4, or GFRa1). Overall, our profiles suggest three broad populations of spermatogonia in juveniles: (1) epithelial-like spermatogonia (THY1+; high OCT4, ID4, and GFRa1), (2) more abundant mesenchymal-like spermatogonia (THY1+; moderate OCT4 and ID4; high mesenchymal markers), and (3) (in older juveniles) abundant spermatogonia committing to gametogenesis (high KIT+). Epithelial-like spermatogonia displayed the expected imprinting patterns, but, surprisingly, mesenchymal-like spermatogonia lacked imprinting specifically at paternally imprinted loci but fully restored imprinting prior to puberty. Furthermore, mesenchymal-like spermatogonia also displayed developmentally linked DNA demethylation at meiotic genes and also at certain monoallelic neural genes (e.g., protocadherins and olfactory receptors). We also reveal novel candidate receptor–ligand networks involving SSCs and the developing niche. Taken together, neonates/juveniles contain heterogeneous epithelial-like or mesenchymal-like spermatogonial populations, with the latter displaying extensive DNA methylation/chromatin dynamics. We speculate that this plasticity helps SSCs proliferate and migrate within the developing seminiferous tubule, with proper niche interaction and membrane attachment reverting mesenchymal-like spermatogonial subtype cells back to an epithelial-like state with normal imprinting profiles.
The pool of spermatogonial stem cells (SSCs), which ensures male fertility throughout adult life, is established early after birth by prospermatogonia (gonocytes). The precursors of prospermatogonia, the primordial germ cells (PGCs), originate at 5.5 d post-coitum (dpc) in mice (Ohinata et al. 2009). During their migration toward the gonads, PGCs undergo a global DNA demethylation (which includes complete erasure of parental imprints) (Seki et al. 2005; Dawlaty et al. 2011; Hackett et al. 2012, 2013; Seisenberger et al. 2012) by both active and passive mechanisms (Seki et al. 2005; Dawlaty et al. 2011; Hackett et al. 2012, 2013; Seisenberger et al. 2012; Ohno et al. 2013). Subsequently (between embryonic day 13.5 [E13.5] and E16.5), DNA methylation (DNAme) is gradually restored, and both maternal- and paternal-specific imprints are thought to be fully established by birth in the male prospermatogonia.
At birth, these prospermatogonia are mitotically arrested but start cycling at postnatal days 1–2 (P1–P2) (Culty 2009). During the subsequent week (P3–P10), cycling spermatogonia proliferate and populate the seminiferous tubule; here, a portion gives rise to the self-renewing SSCs, and the remainder proceeds to differentiation (becoming highly KIT+) without self renewal, originating the first wave of spermatogenesis (Kluin and de Rooij 1981; Yoshida et al. 2006), or instead commits apoptosis. This phase of germ cell development is therefore crucial to initiate and maintain male fertility throughout adult life.
Following puberty, adult SSCs either self-renew (forming two single unpaired cells [As]) or divide into paired cells (Apr) connected by an intracellular cytoplasmic bridge. This is the first step toward differentiation, which culminates in the production of mature sperm (Hess and Renato de Franca 2008). These sequential transitions coincide with global changes in the epigenome (Khalil et al. 2004; Delaval et al. 2007; Oakes et al. 2007; Turner 2007; Soumillon et al. 2013; Hammoud et al. 2014), which simultaneously reflect the cellular developmental path, its current transcriptional program, and its future commitment to differentiate.
We provide here an in-depth epigenomic and transcriptomic analysis of male germline development that suggests three broad SSC populations in juveniles: epithelial-like cells, mesenchymal-like cells, and cells committing to gametogenesis, which were defined by different signatures related to known SSC markers, cell adhesion/migration markers, and SSC differentiation markers. We also reveal novel candidate receptor–ligand networks involving SSCs and the niche. Curiously, we reveal novel and unexpected DNAme/imprinting dynamics in the mesenchymal-like population. Together, this study and data sets provide foundational new information about spermatogonial cell development.
Results
Genomic profiling of developing SSCs
THY1 and KIT are useful markers for distinguishing self-renewing/transplantable SSCs (THY1+-enriched) from nontransplantable cells committing to gametogenesis/meiosis (KIT+-enriched) (Kubota et al. 2004; Oatley et al. 2009; Hammoud et al. 2014). Although proportions change during postnatal development, a combination of immunostaining and FACS analyses (at P7, for example) reveals that most postnatal cells are KIT+ (∼50%–60%), a somewhat smaller proportion is THY1+ (30%–40%), and ∼10%–20% of cells appear both KIT+ and THY1+ (data not shown). To examine THY1+- or KIT+-enriched cells, we initially implemented antibody-based magnetic cell sorting (MACS) from the testes of newborn (P0; THY1+-only) or juvenile (P7, P12, and P14; THY1+, KIT+, or combined GFRa1+ and THY1+) mice (Fig. 1A; Supplemental Fig. 1A). THY1+ selections provide a population enriched for cells yielding successful transplantation or culturing. Here, our MACS procedure provided a population that was ∼86% positive for PLZF (Supplemental Fig. 1C) but still heterogeneous for other SSC markers, prompting additional isolations of less abundant subpopulations such as those with high OCT4, GFRa1, or ID4, as they may be more stem-like (Chan et al. 2014). These isolations involved fluorescence-activated cell sorting (FACS) and isolation of high-GFP+ cells from newborn (P0) transgenic animals (Oct4-GFP) (Yoshimizu et al. 1999) and P7 transgenic animals (Oct4-GFP and Id4-GFP). Here, we note that GFP+ cells from Oct4-GFP transgenics represent only a subset of the total OCT4+ cells (by immunohistochemistry) in postnatal SSCs, typically with the highest OCT4 (Supplemental Fig. 1B). We also isolated the high-VASA (high-GFP+) spermatogonia at P0 and P7. We typically isolated two biological replicates of each sample type, which were processed separately. We note that although MACS and FACS highly enrich, they do not fully purify cell populations; however, we chose stringent parameters to gate FACS populations, isolating only cells with relatively high levels of GFP (see the Materials and Methods).
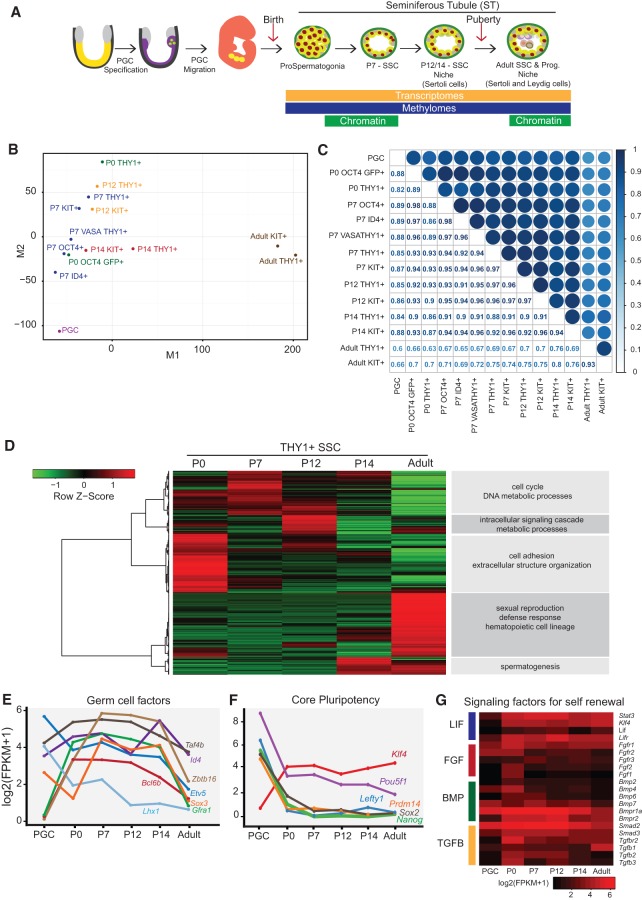
Figure 1.
Transcriptional changes accompanying SSC development. (A) Graphical summary of the biology of germline stem cell specification, transitions, and data sets generated in this study. (B) Multidimensional scaling (MDS) plot comparing transcriptional profiles of PGCs, undifferentiated SSCs (THY1+, high-ID4, OCT4, or VASA), and differentiating SSCs (KIT+) from all tested developmental stages. (C) Pairwise RNA sequencing (RNA-seq) correlation matrix plot of all data sets generated. The color intensity and the size of the circle reflect the correlation between the data sets. (D) RNA-seq hierarchical clustering of developing THY1+-enriched SSCs, with enriched gene ontology terms at the right. Note that all cell purifications were performed using MACS or FACS. (E,F) Line plots depicting the dynamics of genes involved in germline THY1+ SSC maintenance or self-renewal (E) or embryonic stem cell pluripotency (F). The X-axis is the chronological developmental time course, and the Y-axis is log2 (FPKM [fragments per kilobase per million mapped fragments] + 1). (G) Expression heat map summarizing signaling pathways involved in self-renewal or maintenance. Scale is log2 FPKM.
Transcriptional profiling involved strand-specific RNA sequencing (RNA-seq) of total RNA from biological replicates, whereas DNAme analyses involved whole-genome bisulfite sequencing (WGBS) using 101-base-pair (bp) paired-end reads and, typically, >20× genome coverage (for statistics and replicates, see Supplemental Table 1). To enable comparisons, we reprocessed published data from embryonic stem cells (ESCs) and PGCs (Stadler et al. 2011; Seisenberger et al. 2012). We also compared with our prior data sets of “AGSCs,” which are referred to here as “adult SSCs” to better align with the field nomenclature. Our profiling of 5hmC and histone modifications in germ cells used standard methods and were compared with existing ESC and/or PGC data sets (Mikkelsen et al. 2007; Ng et al. 2013).
Overall comparisons of postnatal spermatogonia
To compare overall profiles, we employed multidimensional scaling (MDS) (Fig. 1B) as well as pairwise comparisons (Fig. 1C). The MDS profiles reveal three groups: PGCs, postnatal spermatogonia/SSCs, and adult SSCs. The heterogeneous postnatal SSC stages form a relatively broad region in the MDS plot, requiring further analyses (correlation plots, clustering, and gene set analyses) to define their similarities and differences. As KIT+ cells show greatly reduced transplantation, we focused our comparisons on the highly transplantable SSC populations (THY1+, ID4+, and OCT4+); KIT+ comparisons are provided in the Supplemental Figures and Data Sets, with only the most notable features highlighted in the main text. Our data sets can be compared in multiple ways, and we first addressed changes in the majority population (THY1+) over the postnatal developmental time course and noted that high-ID4/OCT4 cells are also THY1+ (Fig. 1E) and therefore are included within the THY1+ population.
Transcriptional dynamics of THY1+-enriched SSCs during postnatal development
We first focused on genes with dynamic expression across the THY1+-enriched spermatogonia data sets (P0–P14) (Fig. 1D; Supplemental Fig. 2A) and performed gene ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses (Supplemental Table 2). Categories prominent at P0 include cell adhesion and morphogenesis (consistent with gonad colonization) and the piRNA system, which is active at this time (e.g., Piwil2 and Tdrd9) (Supplemental Table 2). P7 cells emphasize cell cycle/division, histone synthesis, RNA splicing, and translation, consistent with their proliferation and expansion within the seminiferous tubule. P12 cells experience changes in metabolic programs and signaling pathways. Thus, GO/KEGG analysis reflects the expected developmental transitions of postnatal SSCs, which are explored in pathway- and gene-specific detail below.
Developmentally regulated changes in transcription factors
We then examined developmental transcription dynamics in THY1+-enriched cells, choosing factors linked to processes of known function (or interest) in developing SSCs, and hereafter use the following FPKM (fragments per kilobase per million mapped fragments) scale: silent/low, <1; moderate, 2–5; high, 5–25; and very high, >25. We first examined whether factors important for PGC specification (e.g., Blimp1/Prdm1, AP2g/Tcfap2c, Wnt3, T, and Prdm14) remain present in SSCs. Interestingly, we observed silencing of these markers between P0 and P14 (Supplemental Fig. 2B), suggesting their lack of involvement in maintaining a germline stem cell state. For transcription factors of known importance in SSC development, we found Zbtb16/Plzf and Bcl6b low/silent in PGCs but highly activated from P0 to P14 (Fig. 1E), Lxh1 high from PGCs to P7 but silent by P12, Sox3 low to moderate in PGCs and at P0 but high or very high in SSCs, and Etv5 high or very high at all stages but noticeably lower in KIT+ cells.
Regarding pluripotency, certain key genes are expressed in early PGCs (e.g., Pou5f1/Oct4, Klf4, Nanog, Sox2, Lefty, and Prdm14), but a subset (e.g., Sox2 and Nanog) decline later in PGC development (Seisenberger et al. 2012; Lesch et al. 2013; Sachs et al. 2013). Accordingly, we found Nanog, Lefty, and Prdm14 silent at P0 and Sox2 silenced by P7 (Fig. 1F) Thus, SSCs lack many core pluripotency factors but express alternative adult stem cell factors, including noncoding RNAs (e.g., Lin28a) linked to pluripotency in postnatal SSCs (Supplemental Fig. 2C), prompting further study. Although HOX family genes are generally silent in SSCs, we found Hoxd8 and the HOX-related Rhox1, Rhox10, and Rhox13 genes expressed at low to moderate levels in SSCs, with Rhox13 peaking at P7 (Supplemental Fig. 2D). This aligns with recent work showing that Rhox13 is needed for progression from P3 to P7 (Song et al. 2012).
For proliferation, we found Myc and Mycn high in postnatal SSC stages but low in THY1+ adult SSCs. Also, Sox3 and Sox4 are silent in adult SSCs, whereas Sox5 and Sox30 are active, suggesting a possible handoff. Additional switches in transcription family members during development were observed for the TBX (e.g., Tbx2) and FOX (e.g., Foxj1) families, among others (Supplemental Table 2). Chromatin remodeling factors are often needed for major developmental transitions, and the germline also assembles and uses testis-specific histone proteins and linker histones. Notably, SSCs pass through developmental states that employ only the Brg1-containing BAF complex (neonate and adult) or only the Brm-containing BAF complex (at P7) (Supplemental Fig. 2E). Likewise for chromatin assembly factors, CAF1 complex members (Chaf1a/b), the testis-specific histone chaperones (Tspy1l/2), and other chromatin factors show clear developmental specificity (Supplemental Fig. 2E). Finally, we examined expression dynamics of the ZNF-KRAB family of proteins, which bind and repress retrotransposons in the germline (Supplemental Fig. 2F; Supplemental Table 2). We found the vast majority of ZNF-KRAB family genes expressed during this time course and cohorts with higher expression in PGCs, neonates, P7, or P14/adult (Supplemental Table 2); however, we did not observe clustering/colocation of ZNF-KRAB genes expressed at similar time points. Finally, we found transcripts encoding the adaptor/repressor protein TRIM28 (which interacts with ZNF-KRAB proteins and repressive chromatin factors) extremely high throughout postnatal development (FPKM ∼250), consistent with the high levels of ZNF-KRAB partners.
Signaling pathways impacting SSC self renewal and differentiation
We then examined signaling factors of known importance in SSC biology and/or culturing. LIF enhances SSC culturing and promotes STAT3 and KLF4 activity in ESCs (Hall et al. 2009). Accordingly, Stat3, Klf4, and LIF receptor (Lifr) are expressed at high levels throughout postnatal SSCs. Likewise, FGF2 is needed for SSC culturing (Ishii et al. 2012), and most FGF receptors (e.g., Fgfr1 and Fgfr3) are expressed in SSCs, but FGF2 is only expressed at P0. Regarding BMP signaling, BMP2, BMP4, and BMP6 are expressed at P0; reduced in prepubertal SSCs; and absent in adult SSCs, whereas many BMP receptors are expressed throughout SSC development (Fig. 1G). Thus, the results for BMP and FGF suggest transitions from autocrine to paracrine after P0.
Gfra1 is silent in PGCs but high in prepubertal SSCs (Fig. 1E; Supplemental Fig. 2G), and its ligand (Gdnf) is low (Supplemental Fig. 2G), consistent with known production by Sertoli cells (Meng et al. 2000; Hofmann et al. 2005). Curiously, both appeared low in THY1+ adult SSCs; however, immunostaining and Western blot analysis suggest translational control, as GFRa1 protein was still detected in Thy1+ SSCs (Supplemental Fig. 2H,I). Notably, Gfra2 and Gfra4 (which bind neurturin preferably to GDNF) were both high in THY1+ adult SSCs but not in postnatal stages, suggesting utilization of additional GFRA receptor subtypes in adult SSCs (Supplemental Fig. 2G–I). For the WNT pathway, canonical WNT ligands were absent in SSCs, whereas WNT receptors (Fzd and Lrp genes) and transducers were expressed in SSCs (Supplemental Fig. 2G), suggesting a paracrine mechanism. Notably, only neonates expressed noncanonical WNT receptors at moderate levels (e.g., Ror1/2), whereas all SSC stages expressed high Ryk. For RAS signaling, we found Hras, Rras, Rras2, and Nras all moderately to highly expressed in SSCs (Supplemental Fig. 2G).
Finally, regarding differences between THY1+ and KIT+ cells along this time course, we found THY1+ SSCs and KIT+ spermatogonia quite similar at P7 (r = 0.97) (Fig. 1B,C) but developing modest and increasing differences; by P14 (r = 0.94) (Fig. 1B,C), this modest difference is dominated by the activation of genes for meiosis and gametogenesis (Supplemental Table 2) and the lowering of certain SSC stem-like genes (e.g., Zbtb16/Plzf, Etv5) in KIT+-enriched cells, as these spermatogonia commit to the first wave of spermatogenesis.
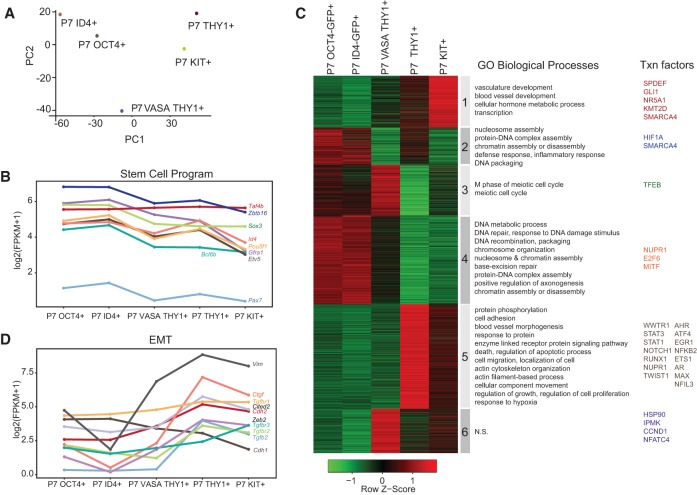
Features of SSC subpopulations at P7
As THY1+-enriched cells are heterogeneous, we sought to better understand similarities and differences among the subpopulations. First, correlation plots (Fig. 1C) and MDS plots (Figs. 1B, 2A) show the high similarity between high-ID4 and high-OCT4 cells at P7 (r = 0.98). Furthermore, high-OCT4 cells at P0 highly resembled high-OCT4 cells at P7 (r = 0.98), showing that high-OCT4 and high-ID4 cells differ only modestly in transcriptional profiles in these stages. However, as high-OCT4 cells (high GFP) are the minority at P0 and P7 (Supplemental Fig. 1B), we compared them with the larger population (THY1+ and/or VASA+), which revealed moderate differences (Fig. 2A), suggesting heterogeneity.
Figure 2.
Postnatal SSC subtypes can resemble stem-like or mesenchymal-like states. (A) MDS plot comparing transcriptional profiles of SSC populations at P7. (B–D) Line plots depicting the dynamics of genes involved in SSC maintenance self renewal (B) and epithelial–mesenchymal transition (D). The X-axis is the chronological developmental time course, and the Y-axis is log2 (FPKM + 1). (C) RNA-seq heat map of all transitioning THY1+ SSCs, with enriched GO terms at the right. Note that all cell purifications were performed using either FACS or MACS.
For comparison, we determined differentially expressed genes (FPKM > 1, greater than twofold change between any two cell types, yielding ∼4000 genes), which were subjected to clustering analyses. First, high-OCT4/ID4 cells express higher levels of many of the known stem cell markers for SSCs (e.g., Zbtb16/Plzf and Gfra1) compared with the THY1+ population (Fig. 2B). High-OCT4/ID4 cells also express higher levels of factors involved in DNA repair and chromatin (Fig. 2C, cluster 4). Interestingly, the THY1+-enriched population showed only modest reductions in stem-related genes but instead appeared more mesenchymal, with higher levels of many key mesenchymal markers (e.g., Zeb2 and Vimentin) (Fig. 2D), lower levels of key epithelial markers (e.g., Cdh1), and enrichment of GO categories such as cell adhesion, cell migration, and vasculature (Fig. 2C, clusters 1 and 5), categories that coenrich for mesenchymal genes. Finally, KIT+-enriched cells had lower levels of many SSC genes compared with the THY1+-enriched subtype.
SSCs display DNA hypomethylation and bivalency of developmental genes
Prior work revealed bivalent loci with underlying DNA hypomethylation at somatic developmental genes (and especially developmental transcription factors) in sperm and adult SSCs (Hammoud et al. 2014). To examine loci in postnatal SSCs, we performed chromatin immunoprecipitation (ChIP) experiments profiling the locations of H3K4me3 (typically correlated with activation) and H3K27me3 (typically correlated with silencing) in P7 THY1+-enriched SSCs. Here, we observed bivalent chromatin (coincident H3K4me3 and H3K27me3) and DNA hypomethylation at the promoters of many genes important for embryo development, including Hox, Sox, Fox, Tbx, and Gata family transcription factors ( e.g., HoxA locus) (Supplemental Fig. 3A,B), but not housekeeping genes. These properties are shared with PGCs, ESCs, adult SSCs, and sperm (Seisenberger et al. 2012; Lesch et al. 2013; Sachs et al. 2013; Hammoud et al. 2014), reinforcing the emerging notion that this bivalent/DNA hypomethylation status of developmental genes might be generally present throughout the entire germline cycle. We note that genes shown to be bivalent in THY1+-enriched SSCs were likewise silent in high-OCT4/ID4 cells but were not directly tested for bivalency here. Finally, this bivalent/DNA hypomethylated state was also observed at P7 at the promoters or enhancers of the silent/poised Nanog, Sox2, Lefty, and Prdm14 genes, as noted previously in adult SSCs (Hammoud et al. 2014).
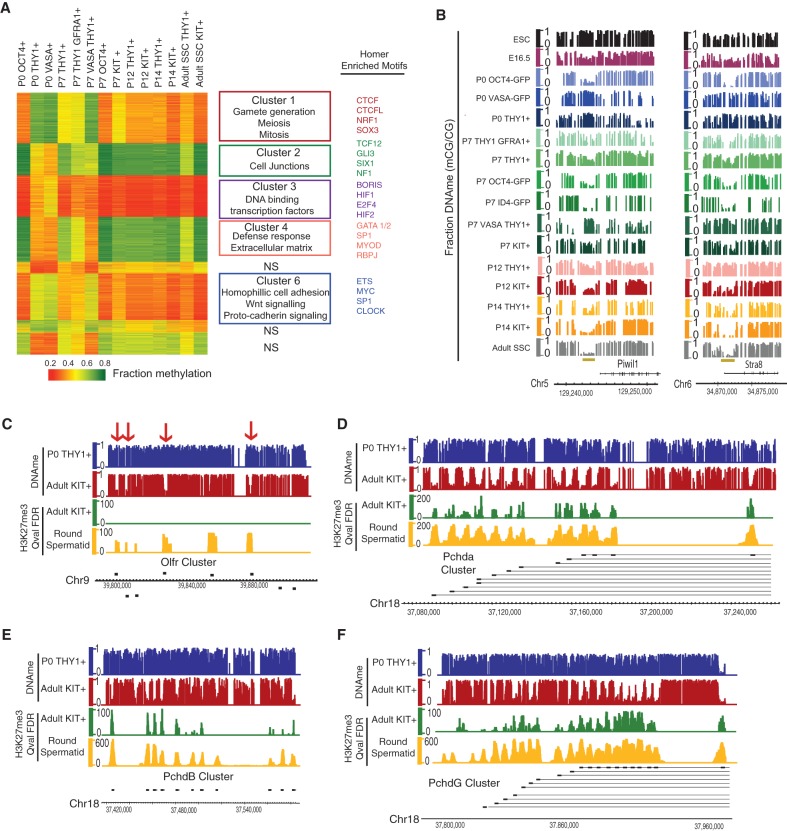
DNAme reprogramming in THY1-enriched SSCs of genes for gametogenesis, olfactory receptors, and protocadherins (PCs)
To examine DNAme dynamics across germline development, we set thresholds for changes in CG methylation (>30% change) at either repetitive elements or gene promoters (2 kb, promoters centered on the transcriptional start site [TSS], yielding ∼3000 dynamic promoters) and performed clustering, de novo transcription factor-binding motif analyses, and GO/KEGG analyses (Fig. 3A). We note that high-ID4 profiles were omitted from Figure 3A due to low to moderate sequencing depth, but where coverage met thresholds, the methylation status was virtually identical to high OCT4 (e.g., Fig. 4D; data not shown).
Figure 3.
DNAme and chromatin dynamics in SSC subtypes. (A) K-means clustering (n = 6) of DNAme (mean fraction CG methylation) at TSS regions (±1 kb) of promoters with ≥30% change in methylation. Pairwise comparisons of all germ cell stages (summed) yielded differentially methylated promoters (DMRs; criteria: three or more CpGs, eight or more reads per C, ≥30% change in fraction CG methylation). Enriched GO terms are in the middle column. At the right, HOMER motif analysis reveals distinctive transcription factors for clusters 1–8. P-value < 1/100. Due to low to moderate sequencing coverage, the high-ID4 data set was removed from the differential analysis but is included in subsequent snapshots. (B) DNA hypomethylation of meiotic and spermatogenic genes is completed by P14. Piwil1 (left) and Stra8 (right) genomic snapshots (mouse ESC and E16.5 methylation data were obtained from Stadler et al. 2011; Seisenberger et al. 2012, respectively). (C–F) Genes with known neural monoallelic expression (e.g., Olfr and Pchd) lose methylation during germ cell development and acquire H3K27me3 in round spermatids. Genomic snapshots of Olfr, PchdA, PchdB, and PchdG clusters (adult SSC and round spermatid data are from prior work [Hammoud et al. 2014]).
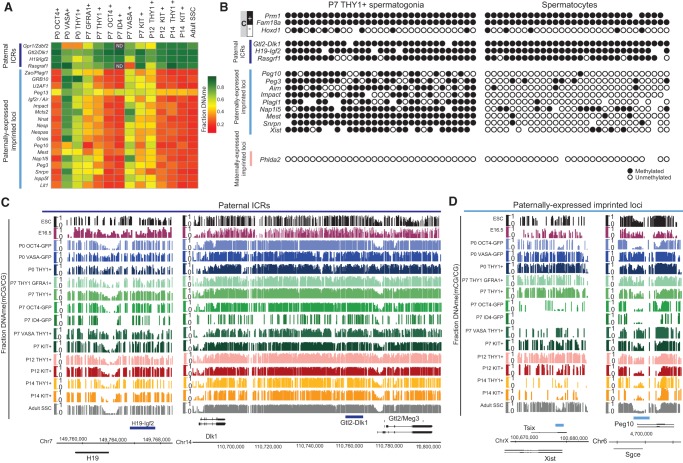
Figure 4.
THY1+-enriched SSCs have improper imprinting (high DNAme) at most paternally expressed imprinted loci. (A) Heat map summarizing the fraction DNAme of the DMR at all known paternal imprinting control regions (ICRs) and paternally expressed imprinted loci. Grey boxes with ND (not determined) within are regions with insufficient sequencing coverage in high-ID4 data sets. (B) Single-cell DNAme validation of 16 loci in P7 THY1+-enriched SSCs and in spermatocytes using the Fluidigm Biomark system. Genomic loci analyzed include known methylated (M) and unmethylated (U) control loci, paternally imprinted ICRs (highlighted in dark blue), paternally expressed imprinted loci (highlighted in light blue), and maternally expressed imprinted loci (highlighted in pink). (C) DNAme genomic snapshots of paternally imprinted ICRs (e.g., H19/Igf2 and Dlk1/Gtl2). The dark-blue bar depicts the ICR. (Y-axis) Fraction CG DNAme. (D) DNAme genomic snapshots of paternally expressed imprinted loci (e.g., Xist [left] and Peg10 [right]). (Y-axis) Fraction CG DNAme in ESCs, PGCs, and prepubertal and adult SSCs. The blue bar depicts previously defined imprinted loci.
Examination of changes in promoter DNAme (via clustering) revealed considerable differences between the epithelial-like and stem-like high-OCT4 population and the alternative mesenchymal-like THY1+- and/or VASA-enriched cell types. For example, within clusters 1 and 6 (Fig. 3A), promoters from THY1+- or VASA+-enriched cells are methylated at P0 and then progressively lose methylation over the developmental time course. In contrast, these promoters in high-OCT4 cells are already hypomethylated at P0 and remain so throughout development. Cluster 1 is highly enriched for categories of sexual reproduction, meiosis, and gametogenesis and includes Piwil1, Sohlh2, Mael, Ctcfl, Stra8, Rad51, Sycp, and Syce (Fig. 3B). Recent studies established that meiotic genes are DNA-methylated in PGCs but fully DNA-hypomethylated in adult SSCs (and “poised” by low/moderate H3K4me3) (Hammoud et al. 2014) but had not addressed when DNA hypomethylation occurs. Our results suggest that these genes are differentially DNA-methylated in these two populations. Furthermore, we observed a bimodal allelic distribution of DNAme in THY1+- or VASA+-enriched cells between P0 and P7, suggesting that alleles gradually (but asynchronously) convert from largely methylated to largely unmethylated rather than synchronous partial/diminishing methylation (Supplemental Fig. 3C). Thus, in the mesenchymal-like THY1+- or VASA+-enriched cells, meiotic gene promoters lose DNAme in advance of their future expression in spermatocytes. Curiously, we found low/moderate H3K4me3 at these meiotic promoters in P7 THY1+-enriched SSCs, far prior to their expression in spermatocytes (data not shown), and high levels of Tet2 and Tet3 transcripts (Supplemental Fig. 4C). However, whether the presence of H3K4me3 alone deters DNMTs or whether active DNA demethylation machinery is used remains to be determined.
Interestingly, two prominent gene families are also present in Figure 3A clusters 1 and 6: PCs and olfactory receptors. Olfactory receptor genes are located either in gene clusters (e.g., on chromosomes 11, 13 and 15) or as individual genes. Here, with THY1+- or VASA+-enriched cells, we observed ∼50 olfactory receptor genes (scattered throughout the clusters) undergoing DNA demethylation, primarily between P7 and P12. Specifically, both the promoter and the entire gene undergo pronounced DNA demethylation (Fig. 3C; Supplemental Fig. 3D,E) and further acquire H3K27me3 during the round spermatid stage (Fig. 3C). Most PCs reside in one of three linked PC clusters on chromosome 18, termed the α, β, and γ classes (Fig. 3D–F). At these PC clusters, we observed striking DNA demethylation focally focused at each of the separate promoters for each variable exon (58 of 58 PC genes) (Fig. 3D–F) but generally not at the dispersed PC δ class (only one of 20 genes). Regarding mechanism, we did not observe 5hmC at these loci (data not shown). Taken together, in THY1+- or VASA+-enriched cells, a large fraction of olfactory receptor genes and especially PCs undergo extensive DNAme/chromatin reprogramming during SSC development; notably, these two gene families share neuronal utilization, combinatorial regulation, and known monoallelic expression. Finally, beyond PC and olfactory receptor genes, we observed a large number of promoters (∼500) that likewise undergo pronounced focal DNA demethylation during SSC development, including melanocortin receptors, cytokines, and interleukins.
Finally, we observed clear DNAme addition at the vast majority of L1 and IAP elements occurring between E16.5 PGCs and P0 prospermatogonia (Supplemental Fig. 3F,G), consistent with different rates of DNAme acquisition as PGCs develop into P0 prospermatogonia. Here, the very small fraction that avoids acquiring full DNAme at P0 (or remains unmethylated throughout germline development) is found in intergenic regions that are not linked to known gene promoters.
A subpopulation of postnatal SSCs bears high DNAme at most paternally expressed imprinted loci
Previous studies in the male germline have suggested that the establishment of all paternal/maternal imprints and imprinting control regions (ICRs) begin in utero and are completed by birth. This expectation was met when examining high-OCT4 SSCs and was also met for all other SSC subpopulations tested when solely considering the paternally imprinted ICRs (Igf2/H19, Dlk1/Gtl2, and Rsgrf1), which were generally fully methylated (>0.8 fraction methylation) at birth (Fig. 4A–C). Interestingly, in THY1+- or VASA+-enriched SSCs at P0 or P7 (but not high-OCT4 SSCs), we found ∼70% (24 of 37 genes) of all known paternally expressed imprinted (those normally DNA hypomethylated) genes to be DNA methylated (Fig. 4A,B,D). As additional confirmation, THY1+ and GFRa1+ SSCs at P7 also showed DNAme at the same genes. As a further test, single-cell DNAme assays were performed using Fluidigm Biomark arrays on P7 THY1+ SSCs separately isolated from an alternative mouse colony. This analysis, which relies on a methylation-sensitive enzymatic digestion (Lorthongpanich et al. 2013), likewise revealed clear DNAme of promoters of the paternally expressed imprinted loci tested (Airn, Igf2, Impact, Mest, Nap1l5, Peg10, Peg3, Plagl1, Snrpn, and Xist) (Fig. 4B,D; Supplemental Fig. 4A,B; data not shown). Moreover, these results confirmed the normal/expected high DNAme at known methylated paternal ICRs (H19-Igf2, Gtl2-Dlk1, and Rasgrf1) (Fig. 4B,C). Furthermore, whereas the promoters of most maternally expressed imprinted genes showed the expected promoter DNA hypomethylation in all subpopulations tested, three genes (Meg3, Cdkn1c, and Gnas) deviated and bore DNAme at P0 in THY1+- or VASA+-enriched SSCs but not in high-OCT4 SSCs (which display normal imprinting). Curiously, many of the genes that displayed improper imprinting displayed improper expression as well (Supplemental Fig. 4A,B). However, all paternal and maternal imprints and gene expression patterns resolve to the normal/expected pattern in late juveniles (P14) (Supplemental Fig. 4A,B). Taken together, whereas high-OCT4 cells (and high-ID4 cells, where threshold coverage is available) displayed expected imprinting patterns, other subpopulations (P0 THY1+, P0 VASA-cre [GFP-sorted], P7 THY1+ GFRa1+, P7 THY1+, and single-cell formats) displayed unexpected patterns (Fig. 4C,D); with these, the majority of paternally expressed imprinted genes and three maternally expressed imprinted genes lacked full imprinting at P0/P7 but resolved to the expected imprinting patterns by P12/P14, prior to the onset of puberty and adult gametogenesis.
Discussion
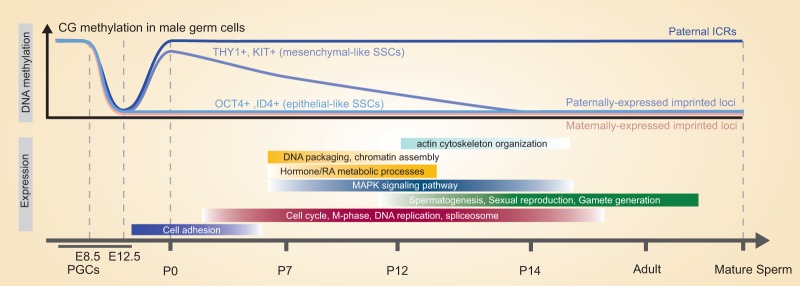
Germline stem cells are specified at approximately E5.5 as PGCs and soon after undergo remarkable phases of genome-wide DNA demethylation—coupled to imprint erasure—followed by the re-establishment of parental imprints prior to gametogenesis. Beyond imprinting, these cells pass through multiple developmental stages from PGCs to adult SSCs that involve complex migration and proliferation phases and culminate in adult SSCs that balance self-renewal and differentiation through communication with niche cells. Although prior genetic and molecular work provided significant insights into involved genes and physiology, much remained unknown regarding the transcription, chromatin, imprinting, and signaling programs that drive or accompany these developmental phases and also imprinting regulation. Here, we conducted extensive genomic profiling of several postnatal subtypes to reveal the transcription networks, chromatin programs, and signaling systems (inferred by transcription) that drive and/or accompany these developmental stages, providing a foundation for functional studies and revealing several unexpected features (Fig. 5).
Figure 5.
Summary schematic depicting changes in DNAme and transcription during germ cell development. (Top panel) Whereas postnatal SSCs with high OCT4/ID4 (epithelial-like SSCs) display normal imprinting patterns, THY1+-enriched SSCs (mesenchymal-like SSCs) at P0–P7 display imprinting defects (high DNAme) at paternally expressed imprinted genes and certain monoallelically expressed genes but resolve to normal/expected patterns before puberty. THY1+-enriched SSCs transcriptomes enrich for particular GO categories during development, aligned with needed processes and the germ cell–niche codevelopment.
To aid in the interpretation of these data, we first presented challenges and limitations. One clear challenge was the known heterogeneity of postnatal SSCs, requiring isolation and comparison of multiple subtypes during development. Here, we profiled multiple subtypes and focused on markers best correlated with transplantation (and comparisons with poorly transplanting KIT+ spermatogonia). However, these profilings were not exhaustive, so informative subtypes likely remain untested. Second, SSC subtypes were isolated by cell surface markers (e.g., THY1 or KIT) or GFP sorting (e.g., for high OCT4 and ID4) using MACS or FACS, respectively. Both methods are enrichment rather than purification procedures, and genes expressed at high levels in rare contaminating cells can impact RNA-seq profile interpretations. Third, our GFP sortings involved transgenic animals of nonidentical genetic background, which could impact profiles. Fourth, our interpretations assumed that changes in transcription impact protein levels, which remain untested. Nevertheless, these data sets provide high-resolution genomic profiling of multiple SSC/spermatogonial subpopulations spanning from birth to puberty, providing foundational data sets for comparisons and analyses.
Transcription and chromatin programs of germline stem cells
Here, we profiled the majority THY1+ SSC population from P0 to P14 and compared it with prior data sets from PGCs and adult SSCs, revealing many dynamic changes. First, we found PGC specification factors declining in P0 prospermatogonia and low/absent in SSCs, strongly suggesting that PGC specification factors are not required for the maintenance of SSC identity. In addition, several transcription factors linked to pluripotency and self-renewal in ESCs are absent in SSCs (e.g., SOX2, PRDM14, NANOG, and LEFTY), while others that contribute to self-renewal in ESCs and other stem cells remain present, prompting future work on their involvement in self-renewal. Regarding chromatin, we extend to P7 SSCs our earlier observations in adult SSCs that bivalency and DNA hypomethylation reside at the promoters or enhancers of the silent/poised Nanog, Sox2, Lefty, and Prdm14 genes (Hammoud et al. 2014), which we speculate may underlie their ability to be activated following normal fertilization (or during their conversion to pluripotent embryonic germ cells in vitro).
Regarding DNAme changes in the germline, DNAme is low in early PGCs (reflecting recent genome-wide DNA demethylation). However, we found that bulk DNAme levels are largely restored by P0. In keeping with this, L1 and LTR elements are largely highly methylated by P0. Furthermore, we found the piRNA system highly expressed in P0 prospermatogonia (e.g., Piwil2 and Tdrd9), consistent with their function in DNAme maintenance and retrotransposon silencing at this stage. Moreover, we observed stage-specific expression of many ZNF-KRAB family proteins. Thus, we reveal the developmental transcriptional timing of many factors involved in innate immune defense against transposons.
Signaling pathway dynamics in developing germline stem cells
Our examination of signaling pathways (inferred by transcription) reveals changes in signaling pathway components during SSC development. For FGF and BMP pathways, our work supports a shift from autocrine to largely paracrine signaling (using the niche) as postnatal SSCs develop into adult SSCs. In contrast, our profiles support WNT signaling through a paracrine system, as canonical WNT ligands are generally silent in SSCs, but the receptors (Fzd and Lrp genes) are expressed. Notably, we also found noncanonical WNT receptors expressed but only in neonates. We also observed particular transitions in GDNF signaling components during development. For example, very high levels of the receptor (Gfra1) and partnered signaling factors (e.g., Ret) were observed in the juvenile, in contrast to low expression levels in adult SSCs. However, we found GFRA1 protein still clearly present in adult SSCs although at lower levels than in differentiating spermatocytes. Instead, adult SSCs express high levels of GFRA2, which preferably binds neurturin to GDNF. Together, these results provide new information for designing more advanced cell culturing systems for SSCs and for genetic investigation.
SSC subtypes differ transcriptionally, revealing epithelial-like or mesenchymal-like properties
Transcriptional profiles of high-OCT4 and high-ID4 cells proved highly similar (r = 0.98), and prior work reveals them both as highly transplantable subtypes. In accordance with their stem-like potential, they express factors known to promote SSC maintenance (e.g., Zbtb16/Plzf and Gfra1) at levels moderately higher than THY1+-enriched cells (which also transplant well). Notably, KIT+-enriched cells had even lower levels of these SSC markers compared with the THY1+-enriched subtype, which may result in poor transplantation. GO categories enriched include DNA repair and chromatin organization, which may help ensure genome integrity. Perhaps the most striking difference was the higher levels of many key mesenchymal markers (e.g., Zeb2 and Vimentin) (Fig. 2D) in the THY1+-enriched subtypes, along with lower levels of key epithelial markers (e.g., Cdh1), and enrichment of GO categories such as cell adhesion and migration (Fig. 2C, clusters 1 and 5), categories that coenrich for mesenchymal genes. Here, we speculate that this heterogeneous population of SSCs may transition between more epithelial-like states and more mesenchymal-like states, which helps enable proliferation, migration, and attachment of these SSCs to the basement membrane of the seminiferous tubule during these postnatal stages.
Dynamics of monoallelic genes in THY1+-enriched SSCs
Interestingly, THY1+-enriched SSCs, but not high-OCT4/ID4 SSCs, display DNAme/chromatin changes at a large proportion of olfactory receptor and PC loci during postnatal SSC development. However, all SSC subtypes arrive at the same DNAme status for these genes by P14: hypomethylated. Notably, these two gene families share neuronal utilization, combinatorial regulation, and known monoallelic expression (Singh et al. 2003; Esumi et al. 2005; Chess 2013). Finally, beyond PC and olfactory receptor genes, we observed a large number of promoters (∼500) that likewise undergo pronounced focal DNA demethylation during development in the THY1+-enriched subtype, including melanocortin receptors, cytokines, and interleukins. These gene promoters are generally DNA hypomethylated in both mature sperm (Hammoud et al. 2014) and oocytes (Smith et al. 2012). Here, future studies are needed to determine which transcription and chromatin factors conduct this phase of reprogramming and their impact on SSC biology.
Dynamics of imprinting in developing germline stem cells
Interestingly, we found that THY1+ and KIT+ cells from neonates and P7 mice display normal imprinting of paternal and maternal ICRs but surprisingly lack full imprinting of most paternally expressed imprinted genes and three specific maternally expressed imprinted genes (Fig. 5). However, these genes/loci attain their full/expected imprinting prior to puberty. In contrast, the high-OCT4/ID4 subtypes displayed expected paternal imprints throughout postnatal development, supporting recent work in high-OCT4 neonatal SSCs (Kubo et al. 2015). Here we note that the subset of THY1+ and KIT+ spermatogonia that shows both imprinting defects and mesenchymal-like features may contribute to the pool of spermatogonia that participates in the first wave of gametogenesis (Yoshida et al. 2006). Future work will examine targeting proteins and the mechanism of demethylation (passive vs. active, including TET family proteins) as well as whether the imprinting/transcription status of these imprinted genes (and the monoallelic genes described above) affects proliferation or the more mesenchymal-like properties of these SSC subtypes.
The extensive changes in DNAme/chromatin occurring during the postnatal phase of mouse SSC development are striking and focused on regulatory regions rather than the repetitive regions observed in PGCs. Whether these same phenomena extend to humans remains unknown; however, epidemiological studies in humans and animals suggest that caloric restriction or overeating during the prepubertal period impacts risk for cardiovascular disease, obesity, and diabetes in the next generation (Kaati et al. 2002; Skorupa et al. 2008; Ng et al. 2010; Ost et al. 2014; Rechavi et al. 2014). Therefore, these epidemiological findings underscore a phase of prepubertal germline plasticity where heritable perturbations to the epigenome may occur.
Materials and methods
Mouse husbandry and germ cell isolation
All mice were maintained on a normal 12-h/12-h light/dark cycle. Isolation of either the THY1+ or c-KIT+ stem cell fraction was carried out with a MACS separator (Miltenyi Biotec) using anti-CD117 antibody (KIT) or anti-CD90.2 (Thy1) (Miltenyi Biotec). Quantitative PCR was used to confirm stem cell purity. RNA and DNA was harvested from SSCs as biological replicates as described in the detailed Supplemental Material and are available for all RNA-seq data sets. FACS analysis involved isolations from P0 Vasa-GFP+ (The Jackson Laboratory, 006954), P0 and P7 Oct4-GFP+ (The Jackson Laboratory, 008214), and P7 Id4-GFP+ (J. Oatley's laboratory). Cells were sorted using FACSCalibur (BD Biosciences). The percentage of live cells was >95%, by exclusion of propidium iodide.
ChIP combined with deep sequencing (ChIP-seq)
ChIP was performed as described previously (Hammoud et al. 2014). Prior to library preparation, ChIP samples were amplified using SEQX (SEQX-50RXN, Sigma Aldrich) due to very low immunoprecipitation yield. After amplification and primer removal, libraries were prepared using standard Illumina pipeline. Libraries were sequenced using 50-bp single-end reads on an Illumina HiSeq 2000 or 2500. The antibodies used were anti-H3K27ac (Active Motif, 39135), H3K4me3 (Active Motif, 39159), and H3K27me3 (Upstate Biotechnology, 07-449).
Single-cell DNAme
Single-cell DNAme analysis was carried out based on prior methods (Lorthongpanich et al. 2013). Spermatocytes and spermatogonia were single-cell FACS-sorted into 96-well plates. DNAme-sensitive restriction digest was performed using Haiti (New England Biolabs). Long and short primers were designed for each analyzed site (Supplemental Table 3). Preamplification was then performed by initial denaturation for 10 min at 95°C followed by 22 cycles of 30 sec of denaturation at 95°C and 4 min of annealing/extension at 60°C. Site-specific real-time amplification was performed on 48.48 dynamic arrays using the Biomark System (Fluidigm).
Immunostaining analysis
Mouse testes were fixed in 4% PFA overnight at 4°C, cut, and analyzed. Immunostaining was performed using the primary antibodies anti-GFRa1 (ab8026), anti-Cd90 (ab3105), anti-Oct4 (ab196585), and anti-GFP (Fischer, PIMA515256) followed by Alexa fluor secondary antibodies 488, 594, and 647 (Invitrogen). Nuclear counterstaining was performed using DAPI (Invitrogen). Fluorescent images were acquired using a Leica Sp5 or an Olympus FluoView FV1000 BX2.
Mouse RNA extraction and library preparation
RNA extractions were performed following Ambion standard protocol (Ambion Life Technologies). Total RNA was DNase-treated (Ambion, AM1907). Long directional RNA-seq libraries (Ribozero-treated) were constructed according to Illumina's protocol and sequenced using a 50-bp single-end format on an Illumina HiSeq 2000 or 2500.
Mouse BisSeq and library preparation
Extracted genomic DNA (50 ng–1 µg) was spiked with 1% unmethylated λ DNA (Promega), and the library was constructed using the EpiGnome Methyl-Seq sample prep kit (Epicenter, Inc.) and sequenced using a 101-bp paired-end format on an Illumina HiSeq 2000 or 2500.
Bioinformatics analysis
Bioinformatics analysis was performed as previously described (Hammoud et al. 2014). Briefly, Fastq files from BisSeq libraries were aligned to the mm9 mouse genome assembly using Novoalign (Novocraft, Inc.) and analyzed using the USEQ package (http://useq.sourceforge.net). ChIP-seq libraries were aligned using Bowtie (http://bowtie-bio.sourceforge.net). RNA-seq alignments were done using TopHat version 2.0.9 (http://tophat.cbcb.umd.edu). ChIP-seq, RNA-seq, and DNAme BisSeq downstream analysis were done using the USEQ package, Cufflinks suite, and cummeRbund R package.
Data access
All data described in this study may be downloaded from Gene Expression Omnibus under the accession project GSE62355. This includes raw Fastq files and processed files for BisSeq, ChIP-seq, RNA-seq, and 5hmC enrichment experiments.
Supplementary Material
Acknowledgments
We thank B. Dalley for sequencing expertise; Ken Boucher for statistical analysis; David Nix, Tim Mosbruger, Brett Milash, Darren Ames, and Tim Parnell for bioinformatics assistance; and Candice Wike for microscopy assistance. We thank Susanna Dolci for germ cell purification protocols, and Marco Bezzi and Shun Xie Teo for help with mouse husbandry. We acknowledge the technical expertise provided by the Advanced Molecular Pathology Laboratory at Institute of Molecular and Cell Biology. Financial support includes the Biomedical Research Council of A*STAR (Agency for Science, Technology, and Research, Singapore), and Joint Council A*STAR grant 1234c00017 (to D.H.P.L., C.L.L., and E.G.), the Department of Urology (genomics), the Howard Hughes Medical Institute (genomics and biologicals), and CA24014 for the Huntsman Cancer Institute core facilities. S.S.H. is funded by the Helen Hay Whitney Foundation. B.R.C. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.261925.115.
References
- Chan F, Oatley MJ, Kaucher AV, Yang QE, Bieberich CJ, Shashikant CS, Oatley JM. 2014. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev 28: 1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A. 2013. Random and non-random monoallelic expression. Neuropsychopharmacology 38: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culty M. 2009. Gonocytes, the forgotten cells of the germ cell lineage. Birth Defects Res C Embryo Today 87: 1–26. [DOI] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, et al. 2011. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9: 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R. 2007. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J 26: 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T. 2005. Monoallelic yet combinatorial expression of variable exons of the protocadherin-α gene cluster in single neurons. Nat Genet 37: 171–176. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Zylicz JJ, Surani MA. 2012. Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet 28: 164–174. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. 2013. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339: 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, Morfopoulou S, Humphreys P, Mansfield W, Walker R, et al. 2009. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell 5: 597–609. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Low DH, Yi C, Carrell DT, Guccione E, Cairns BR. 2014. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15: 239–253. [DOI] [PubMed] [Google Scholar]
- Hess RA, Renato de Franca L. 2008. Spermatogenesis and cycle of the seminiferous epithelium. Adv Exp Med Biol 636: 1–15. [DOI] [PubMed] [Google Scholar]
- Hofmann MC, Braydich-Stolle L, Dym M. 2005. Isolation of male germ-line stem cells; influence of GDNF. Dev Biol 279: 114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Kanatsu-Shinohara M, Toyokuni S, Shinohara T. 2012. FGF2 mediates mouse spermatogonial stem cell self-renewal via upregulation of Etv5 and Bcl6b through MAP2K1 activation. Development 139: 1734–1743. [DOI] [PubMed] [Google Scholar]
- Kaati G, Bygren LO, Edvinsson S. 2002. Cardiovascular and diabetes mortality determined by nutrition during parents’ and grandparents’ slow growth period. Eur J Hum Genet 10: 682–688. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Boyar FZ, Driscoll DJ. 2004. Dynamic histone modifications mark sex chromosome inactivation and reactivation during mammalian spermatogenesis. Proc Natl Acad Sci 101: 16583–16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluin PM, de Rooij DG. 1981. A comparison between the morphology and cell kinetics of gonocytes and adult type undifferentiated spermatogonia in the mouse. Int J Androl 4: 475–493. [DOI] [PubMed] [Google Scholar]
- Kubo N, Toh H, Shirane K, Shirakawa T, Kobayashi H, Sato T, Sone H, Sato Y, Tomizawa S, Tsurusaki Y, et al. 2015. DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis. BMC Genomics 16: 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. 2004. Culture conditions and single growth factors affect fate determination of mouse spermatogonial stem cells. Biol Reprod 71: 722–731. [DOI] [PubMed] [Google Scholar]
- Lesch BJ, Dokshin GA, Young RA, McCarrey JR, Page DC. 2013. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc Natl Acad Sci 110: 16061–16066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorthongpanich C, Cheow LF, Balu S, Quake SR, Knowles BB, Burkholder WF, Solter D, Messerschmidt DM. 2013. Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science 341: 1110–1112. [DOI] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, et al. 2000. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287: 1489–1493. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. 2010. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature 467: 963–966. [DOI] [PubMed] [Google Scholar]
- Ng JH, Kumar V, Muratani M, Kraus P, Yeo JC, Yaw LP, Xue K, Lufkin T, Prabhakar S, Ng HH. 2013. In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev Cell 24: 324–333. [DOI] [PubMed] [Google Scholar]
- Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM. 2007. Developmental acquisition of genome-wide DNA methylation occurs prior to meiosis in male germ cells. Dev Biol 307: 368–379. [DOI] [PubMed] [Google Scholar]
- Oatley JM, Oatley MJ, Avarbock MR, Tobias JW, Brinster RL. 2009. Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development 136: 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M. 2009. A signaling principle for the specification of the germ cell lineage in mice. Cell 137: 571–584. [DOI] [PubMed] [Google Scholar]
- Ohno R, Nakayama M, Naruse C, Okashita N, Takano O, Tachibana M, Asano M, Saitou M, Seki Y. 2013. A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development 140: 2892–2903. [DOI] [PubMed] [Google Scholar]
- Ost A, Lempradl A, Casas E, Weigert M, Tiko T, Deniz M, Pantano L, Boenisch U, Itskov PM, Stoeckius M, et al. 2014. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell 159: 1352–1364. [DOI] [PubMed] [Google Scholar]
- Rechavi O, Houri-Ze'evi L, Anava S, Goh WS, Kerk SY, Hannon GJ, Hobert O. 2014. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 158: 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs M, Onodera C, Blaschke K, Ebata KT, Song JS, Ramalho-Santos M. 2013. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep 3: 1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. 2012. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 48: 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. 2005. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 278: 440–458. [DOI] [PubMed] [Google Scholar]
- Singh N, Ebrahimi FA, Gimelbrant AA, Ensminger AW, Tackett MR, Qi P, Gribnau J, Chess A. 2003. Coordination of the random asynchronous replication of autosomal loci. Nat Genet 33: 339–341. [DOI] [PubMed] [Google Scholar]
- Skorupa DA, Dervisefendic A, Zwiener J, Pletcher SD. 2008. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging cell 7: 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484: 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HW, Dann CT, McCarrey JR, Meistrich ML, Cornwall GA, Wilkinson MF. 2012. Dynamic expression pattern and subcellular localization of the Rhox10 homeobox transcription factor during early germ cell development. Reproduction 143: 611–624. [DOI] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthes P, Kokkinaki M, Nef S, Gnirke A, et al. 2013. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep 3: 2179–2190. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495. [DOI] [PubMed] [Google Scholar]
- Turner JM. 2007. Meiotic sex chromosome inactivation. Development 134: 1823–1831. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. 2006. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development 133: 1495–1505. [DOI] [PubMed] [Google Scholar]
- Yoshimizu T, Sugiyama N, De Felice M, Yeom YI, Ohbo K, Masuko K, Obinata M, Abe K, Scholer HR, Matsui Y. 1999. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev Growth Differ 41: 675–684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.