Abstract
Background
Dinoflagellates are unicellular marine and freshwater eukaryotes. They possess large nuclear genomes (1.5–245 gigabases) and produce structurally unique and biologically active polyketide secondary metabolites. Although polyketide biosynthesis is well studied in terrestrial and freshwater organisms, only recently have dinoflagellate polyketides been investigated. Transcriptomic analyses have characterized dinoflagellate polyketide synthase genes having single domains. The Genus Symbiodinium, with a comparatively small genome, is a group of major coral symbionts, and the S. minutum nuclear genome has been decoded.
Results
The present survey investigated the assembled S. minutum genome and identified 25 candidate polyketide synthase (PKS) genes that encode proteins with mono- and multifunctional domains. Predicted proteins retain functionally important amino acids in the catalytic ketosynthase (KS) domain. Molecular phylogenetic analyses of KS domains form a clade in which S. minutum domains cluster within the protist Type I PKS clade with those of other dinoflagellates and other eukaryotes. Single-domain PKS genes are likely expanded in dinoflagellate lineage. Two PKS genes of bacterial origin are found in the S. minutum genome. Interestingly, the largest enzyme is likely expressed as a hybrid non-ribosomal peptide synthetase-polyketide synthase (NRPS-PKS) assembly of 10,601 amino acids, containing NRPS and PKS modules and a thioesterase (TE) domain. We also found intron-rich genes with the minimal set of catalytic domains needed to produce polyketides. Ketosynthase (KS), acyltransferase (AT), and acyl carrier protein (ACP) along with other optional domains are present. Mapping of transcripts to the genome with the dinoflagellate-specific spliced leader sequence, supports expression of multifunctional PKS genes. Metabolite profiling of cultured S. minutum confirmed production of zooxanthellamide D, a polyhydroxy amide polyketide and other unknown polyketide secondary metabolites.
Conclusion
This genomic survey demonstrates that S. minutum contains genes with the minimal set of catalytic domains needed to produce polyketides and provides evidence of the modular nature of Type I PKS, unlike monofunctional Type I PKS from other dinoflagellates. In addition, our study suggests that diversification of dinoflagellate PKS genes comprises dinoflagellate-specific PKS genes with single domains, multifunctional PKS genes with KS domains orthologous to those of other protists, and PKS genes of bacterial origin.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-2195-8) contains supplementary material, which is available to authorized users.
Keywords: Gene diversification, Horizontal gene transfer, Spliced-leader trans-splicing, Polyketide synthase, Bacterial PKS, NRPS, Zooxanthellamide D, Symbiodinium minutum, Dinoflagellates, Genome-wide survey
Background
Dinoflagellates are unicellular eukaryotes found in both marine and freshwater environments. Some are crucial symbionts of reef-building corals, and others sometimes cause toxic algal blooms [1]. Dinoflagellates are rich sources of structurally unique and bioactive secondary metabolites and are of interest to natural product chemists, biologists, and ecologists. These metabolites are unique in size, structure, and potency, and many are of polyketide origin [2–4].
Dinoflagellate toxins have been classified into three main categories: (i) polycyclic polyethers, (ii) macrolides, and (iii) linear polyethers [2, 4]. The majority of these compounds display remarkable biological activities, including ion channel modulation, phosphatase inhibition, hemolysis, mycotoxicity, and cytotoxicity [5–7]. One possible explanation for their high potency is to compensate for high dilution when they are released into the water [8]. Much is known regarding the biosynthesis of polyketides from terrestrial and freshwater organisms; however, only in the last decade have dinoflagellate polyketides been investigated.
Polyketides are synthesized by specific enzymes called polyketide synthases, through a series of condensation and reduction reactions involving at least three protein domains. These include ketosynthase (KS), acyl transferase (AT), and acyl carrier protein (ACP) (PP-binding) domains. In addition, polyketide synthesis may involve three optional domains: ketoreductase (KR), dehydratase (DH), and enoylreductase (ER) [9]. In 2008, full-length transcripts of Type I-like, modular PKS were sequenced from Karenia brevis, with seven out of eight transcripts containing single PKS domains, a feature typical of Type II PKS [10]. Eichholz et al. [11] characterized five transcripts for Type I-like, PKS-encoding KS proteins that are expressed as monofunctional units, from the dinoflagellates, Alexandrium ostenfeldii and Heterocapsa triquetra. Transcriptomic analysis of the non-toxic Heterocapsa circularisquama, revealed 61 polyketide synthase-encoding expressed sequence tags (EST) contigs, including one contig with two domains (KS-KR) [12]. Similar analysis revealed Type I-like polyketide synthases in the toxic dinoflagellate, Gambierdiscus polynesiensis, the main producer of ciguatoxins [13]. Meyer et al. [14] reported finding all genes essential for polyketide toxin synthesis in Azadinium spinosum, known to produce azaspiracid toxins. Recently, Kohli et al. catalogued 162 unique transcripts encoding complete KS domains in two species of Gambierdiscus, which are putatively involved in polyketide biosynthesis [15].
Among marine dinoflagellates, the Genus Symbiodinium includes major coral symbionts that are also associated with other invertebrate taxa (Porifera, Mollusca, and Platyhelminthes) [16, 17]. The draft genome of S. minutum, encoding ~42,000 protein-coding genes, has provided an opportunity for better understanding of its PKS system [18]. Snyder et al. [19] reported Type I PKSs in several dinoflagellates, including Symbiodinium sp.; however, there has been no detailed survey of genes involved in polyketide synthesis in S. minutum. We probed the S. minutum genome with respect to enzymes involved in polyketide synthesis and phylogenetically analyzed the KS domains of PKSs. We found a non-ribosomal peptide synthetase-polyketide synthase (NRPS-PKS) hybrid and confirmed that PKSs of S. minutum belong to the protistan Type I PKS group, along with some unexpected sequences associated with a bacterial clade.
Results
Diversification of KS domain-containing genes in the S. minutum genome
In total, 65 genes with ketoacyl synthase domains (Pfam IDs: PF00109) were screened from the predicted 41,925 genes in the S. minutum genome (http://marinegenomics.oist.jp/genomes/gallery). Using BLASTP searches, we also checked S. minutum genes similar to reported PKSs and confirmed the aligned sequences manually. After removing sequences for partial domains, 25 genes that encoded full KS domains in S. minutum were selected for sequence characterization (Table 1; see Additional file 1: Table S1) and molecular phylogenetic analysis. Sequence comparisons with KS domains showed that the most similar genes are those reported from other dinoflagellates, although several genes were unexpectedly most similar to bacterial (Bacillus) genes. Eleven KS domain-containing genes likely encode multifunctional proteins with other domains related to PKS synthesis (AT, ACP, KR, DH, and ER) (Table 1). Careful examination of the S. minutum genome identified 25 intron-rich genes for KS sequences (Table 1) that are expressed under standard culture conditions (see Additional file 1: Figure S1). Only one KS gene (symbB1.v1.2.039083.1) is likely to be more highly expressed than genes [18] for RNA polymerase (data not shown). Quantitative expression analysis under different conditions will be useful for functional predictions. An interesting feature was the presence of tandemly aligned KS genes (symbB1.v1.2.015790.t1, symbB1.v1.2.015788.t2 and symbB1.v1.2.015789.t1) on scaffold 1186.1, in addition to two KS genes on scaffold 514.1 (Table 1). Since domain combinations are not conserved completely, duplication and/or splitting are hypothesized as the mechanisms for these expansions (see Fig. 1). This hypothesis is not unreasonable, considering a recent report of dinoflagellates possessing por (protochlorophyllide oxidoreductase) gene duplicates [20]. Gene duplications can have metabolic advantages and can eventually become fixed in a population.
Table 1.
KS domain-containing genes in Symbiodinium minutum
| Gene ID | Total AA | domaina | BLASTP best hit of KS domain in NCBI database (Accession #) Organism | E-value | Identity/ Similarities (% AA) | Scaffold # of S.minutum genome V.1.2 | Assembled transcriptome ID |
|---|---|---|---|---|---|---|---|
| symbB1.v1.2.000535.t1 | 4838 | KS | GAE32993.1 Bacillus hemicellulosilyticus JCM 9152 | 2.00E-47 | 45/60 | 31.1 | symbB1.EST_k37c20_20326 |
| symbB1.v1.2.001307.t1 | 797 | KS | AIW63287.1 Azadinium spinosum | 5.00E-128 | 35/51 | 57.1 | symbB1.EST_k37c20_6869 |
| symbB1.v1.2.001928.t1 | 1164 | KS | ABQ85796.1 Karenia brevis | 3.00E-174 | 35/50 | 55.1 | symbB1.comp17616_c0_seq1 |
| symbB1.v1.2.002919.t1 | 1105 | KS | ABQ85802.1 Karenia brevis | 4.00E-123 | 35/49 | 160.1 | symbB1.EST_k37c20_17447 |
| symbB1.v1.2.008781.t1 | 2107 | DH-KR-PP-PP-KS-DH | KFG59574.1 Toxoplasma gondii RUB | 0 | 34/49 | 514.1 | symbB1.comp69166_c0_seq1 |
| symbB1.v1.2.008782.t1 | 1848 | KR-PP-KS-KR-PP-KS | WP_044601528.1 Candidatus magnetoglobus multicellularis | 1.00E-161 | 34/54 | 514.1 | symbB1.comp53648_c0_seq1 |
| symbB1.v1.2.012436.t1 | 10601 | See Fig. 2 | CAD29793.1 Planktothrix agardhii NIVA-CYA | 4.00E-60 | 39/56 | 860.1 | symbB1.comp70898_c0_seq1 |
| symbB1.v1.2.013880.t1 | 1071 | KS | ABQ85796.1 Karenia brevis | 2.00E-95 | 28/47 | 991.1 | symbB1.comp24939_c0_seq1 |
| symbB1.v1.2.015788.t2 | 920 | KR-PP-KS-AT | CDJ53564.1 Eimeria brunetti | 6.00E-171 | 40/56 | 1186.1 | symbB1.comp40305_c0_seq1 |
| symbB1.v1.2.015789.t1 | 1216 | KR-PP-KS | CDI87737.1 Eimeria praecox | 2.00E-153 | 37/53 | 1186.1 | symbB1.EST_k37c20_822 |
| symbB1.v1.2.015790.t1 | 427 | KS | WP_035347688.1 Bacillus hemicellulosilyticus | 5.00E-97 | 47/64 | 1186.1 | symbB1.comp52059_c0_seq1 |
| symbB1.v1.2.015913.t1 | 1671 | KS | ABQ85796.1 Karenia brevis | 0 | 37/55 | 1171.1 | symbB1.comp20639_c0_seq1 |
| symbB1.v1.2.017689.t1 | 2261 | KS | AIW63289.1 Azadinium spinosum | 3.00E-155 | 48/62 | 1368.1 | symbB1.EST_k37c20_9081 |
| symbB1.v1.2.019160.t1 | 1068 | KS | AIW63288.1 Azadinium spinosum | 0 | 37/55 | 1555.1 | symbB1.EST_k37c20_8679 |
| symbB1.v1.2.020241.t1 | 656 | KS | ABQ85802.1 Karenia brevis | 1.00E-156 | 44/59 | 1693.1 | symbB1.EST_k37c20_3634 |
| symbB1.v1.2.022565.t1 | 1547 | KS | ABQ85802.1 Karenia brevis | 7.00E-136 | 35/48 | 2011.1 | symbB1.EST_k37c20_6838 |
| symbB1.v1.2.027279.t1 | 615 | KS | XP_005823341.1 Guillardia theta | 0 | 72/82 | 2789.1 | symbB1.comp12619_c0_seq1 |
| symbB1.v1.2.027671.t1 | 957 | AM-PP-KS-AT | KGC29420.1 Burkholderia pseudomallei | 0 | 40/54 | 2857.1 | symbB1.EST_k37c20_5813 |
| symbB1.v1.2.028834.t1 | 2431 | KS | AFW98413.1 Alexandrium ostenfeldii | 1.00E-147 | 37/53 | 3094.1 | symbB1.EST_k37c20_17396 |
| symbB1.v1.2.030435.t1 | 481 | KS-AT | XP_008886813.1 Hammondia hammondi | 1.00E-107 | 42/59 | 3430.1 | symbB1.comp58270_c0_seq1 |
| symbB1.v1.2.036002.t1 | 2987 | AM-PP-KS-KR-DH | AAR87760.2 Bacillus cereus | 2.00E-110 | 31/46 | 4981.1 | symbB1.EST_k37c20_11234 |
| symbB1.v1.2.036410.t1 | 3519 | AM-KS-KR-PP-KS-DH-KR-PP-KS | AFE09917.1 Corallococcus coralloides DSM 2259 | 0 | 33/48 | 5132.1 | symbB1.comp56297_c0_seq6 |
| symbB1.v1.2.037839.t1 | 604 | KS | AIW63289.1 Azadinium spinosum | 0 | 61/72 | 5703.1 | symbB1.comp5164_c0_seq1 |
| symbB1.v1.2.039083.t1 | 980 | AT-KS-PP | XP_005785854.1 Emiliania huxleyi CCMP1516 | 5.00E-58 | 45/55 | 6338.1 | symbB1.EST_k37c20_7312 |
| symbB1.v1.2.040026.t1 | 582 | KS | AFW98411.1 Alexandrium ostenfeldii | 0 | 68/81 | 6945.1 | symbB1.comp4031_c0_seq1 |
a AM AMP-binding, AT Acyltransferase, DH Dehydratase, ER Enoylreductase, KR Ketoreductase, KS Ketosynthase, PP PP-binding
Fig. 1.
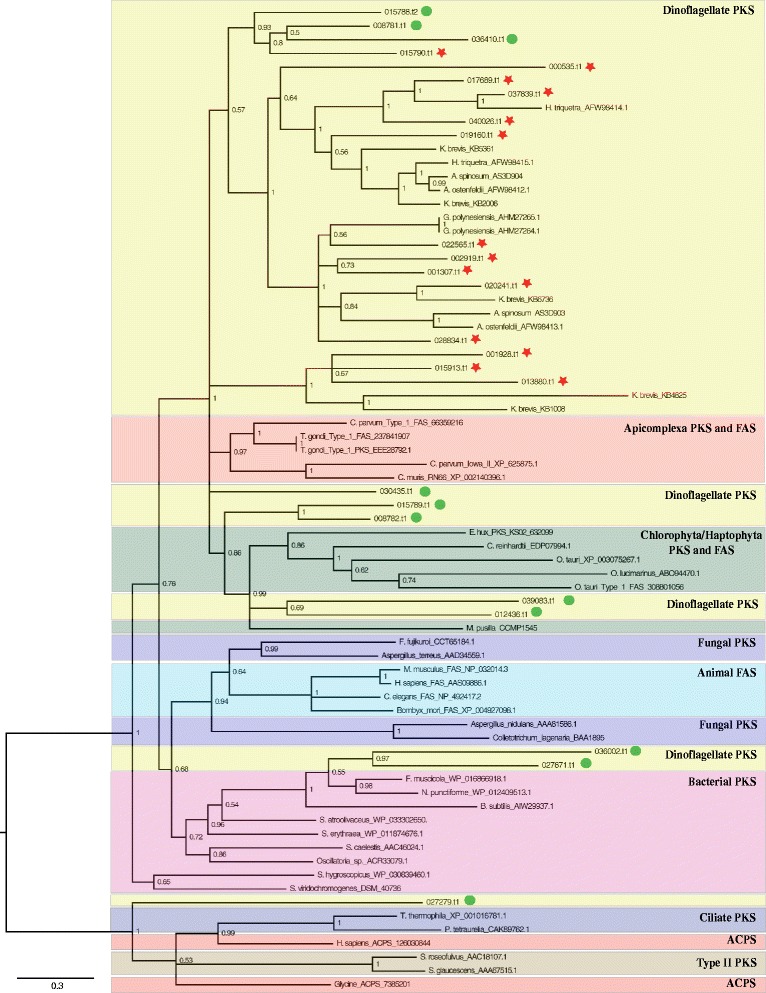
A molecular phylogenetic tree of Type I KS domains from prokaryotic and eukaryotic PKS and FAS, analyzed by Bayesian inference, reveals the diversification of the KS domain gene family. Symbiodinium minutum possesses genes belonging to three major groups within this gene family. Type II PKS and acyl carrier protein synthases (ACPS) were used as outgroups. Numbers at nodes indicate posterior probabilities. Details regarding S. minutum sequences are provided in Table 1. Red stars indicate S. minutum proteins with single PKS-related domains. Green circles indicate S. minutum proteins with multiple PKS-related domains. Dinoflagellate KSs (yellow) are classified as a well-supported group within the protistan Type I PKS clade.
Bayesian inference and maximum likelihood analysis of the 25 KS sequences were carried out with acyl carrier protein synthase (ACPS) and Type II PKS sequences as outgroups to understand relationships of KS sequences from S. minutum compared with those of other dinoflagellates. After alignment and trimming, a sequence of 235 amino acids (aa) was used for analysis. Molecular phylogenetic analyses significantly placed most sequences with those of other dinoflagellate Type I PKSs (Fig. 1). Bayesian inference clearly demonstrated that 22 S. minutum proteins and other protistan proteins were included in that clade (Fig. 1). There was strong Bayesian support (posterior probability: 1.00) for a ‘protistan’ clade of Type I PKS sequences comprising of apicomplexan, dinoflagellate, chlorophyte, and haptophyte sequences (Fig. 1). These major groups formed two separate sub-groups within the protistan clade. One sub-group with all existing sequences from A. ostenfeldii, A. spinosum, G. polynesiensis, and H. triquetra, included 10 proteins of S. minutum with one PKS-related domain (KS). The second sub-group, with chlorophyte/haptophyte proteins, contained four S. minutum sequences with multiple PKS-related domains. The third sub-group, with bacterial proteins, contained two S. minutum proteins (symbB1.v1.2.027671 and symbB1.v1.2.036002). Interestingly, the sequences were more closely related to cyanobacterial KS sequences than to other eukaryote sequences. A similar pattern has been reported in K. brevis, in which some sequences grouped with cyanobacterial proteins [21]. Dinoflagellate proteins were not found in clades of animal fatty acid synthase (FAS) and fungal PKS. It is worth mentioning that NRPS-PKS hybrid (symbB1.v1.2.012436.t1) was in a clade with PKS and FAS of chlorophyta/haptophyta (Emiliania huxleyi, Ostreococcus tauri, Chlamydomonas reinhardii, Ostreococcus lucimarinus and Micromonas pusilla), which encode the largest PKS protein in S. minutum. Maximum likelihood analysis provided additional support for a “protistan clade” containing Type I PKS sequences (Additional file 1: Figure S2).
To explore the possibility of spliced leader trans-splicing from a large transcriptome to single-domain, protein-coding transcriptomes, we mapped transcriptome data from the TSS (transcription start site). The mapping of SL (spliced leader)-removed TSS (red lines in Figure S1) showed that each of three gene models (symbB1.v1.2.000535 on scaffold 31, symbB1.v1.2.020241 on scaffold 1693, symbB1.v1.2.028834 on scaffold 3094) predicts two transcripts by SL trans-splicing (red arrows in Figure S1). They are genes with single PKS-related domains (Table 1; Fig. 1). Therefore, proteins with multiple PKS-related domains are likely to be expressed in cultured S. minutum.
Presence of a hybrid NRPS-PKS gene in S. minutum
One gene model of 10,601 amino acids was identified as a hybrid NRPS-PKS, based on PFAM domain analysis and confirmed by anti-SMASH (antibiotics & Secondary Metabolite Analysis Shell) [22]. It was composed of eight modules, three NRPSs, and five PKSs (Fig. 2a). The first NRPS is followed by three PKS modules, which contain at least a KS domain with different domain combinations. A second NRPS with condensation (C) and adenylation (A) is also present, followed by an additional NRPS-like assembly (C-HxxPF-A). Thioesterase (TE) domains are usually located in the final NRPS module where they catalyze product release [23]. Substrates of adenylation (A) domains in the NRPS module can be predicted based on residues in the binding pocket [24]. The reported A domains in NRPS proteins are responsible for recruiting amino acids into the final product. In this case, the first A domain has the sequence DLFNLSLI while the second A domain has DVWxFSLI. In silico methods were used to infer a hypothetical metabolite produced by the hybrid NRPS-PKS gene. Based on antiSMASH server prediction, the hybrid cluster could result in a natural product consisting of a core scaffold made from two amino acids (cysteine and serine) (Fig. 2b). I-TASSER [25] 3D protein structure prediction showed that the AT domain at the start of the cluster resembled an acyl-carrier-protein malonyltransferase (data not shown) and may provide malonyl groups for polyketide biosynthesis.
Fig. 2.
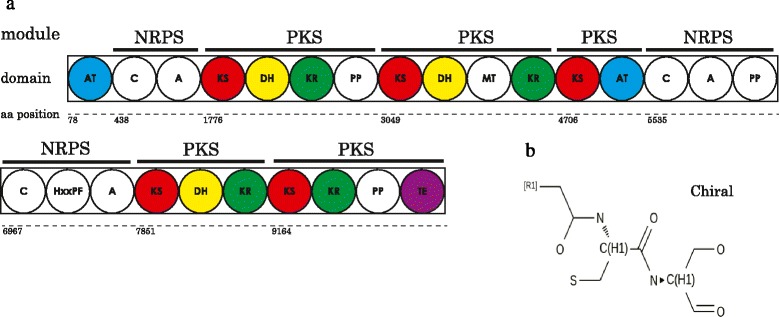
An unusual, very large NRPS-PKS protein is present in the Symbiodinium minutum genome. a Consensus schematic representation of the NRPS-PKS hybrid from S. minutum based on the PFAM database and anti-SMASH. Acyl carrier protein (ACP) is represented as phosphopantetheine (PP). The third A domain didn’t yield any prediction for a specific substrate using anti-SMASH. Amino acid (aa) positions show starting points for major domains. AT: acyl tansferase, C: condensation, A: adenylation, KS: ketosynthase, DH: dehydratase, KR: ketoreductase, MT: methyltransferase, TE: thioesterase b core scaffold of the hypothetical molecule, based on assumed PKS-NRPS co-linearity
Surveys of other PKS domains and conserved N-terminal sequences of S. minutum KS
BLAST screening of the S. minutum genome and comparisons of domain structure resulted in identification of candidates for all domains involved in polyketide synthesis: KS, AT, ACP (PP), KR, DH, and ER. First, we examined whether functionally important amino acid residues required for enzymatic activity (cysteine, histidine, and lysine) are present near the DTACSS-motif of S. minutum KS enzymes. We found that most sequences (21/25) contain these residues (see Additional file 2: Figure S3). Other domains can also be identified by comparing sequences and signature motifs: HxxxGxxxx with two active residues (histidine and proline) in DH [26], LxHxxxGGVG in ER with important residues (leucine, histidine, valine, and glycine) [26], GxGxxGxxxA in KR with three glycine and one alanine as essential residues [27] and the signature motif, GHSLG, in AT domains (see Additional file 2: Figure S4). The phylogenetic relationship of the PKS N-terminal region among dinoflagellates is shown in a maximum likelihood phylogeny (see Additional file 2: Figure S5). The tree reflects the same dinoflagellate resolution found in the KS-based phylogeny (Fig. 1) with one main sub-clade consisting of A. spinosum, A. ostenfeldii, H. triqueta, and K. brevis, one sub-clade comprising only one S. minutum and two K. brevis sequences, and a third clade consisting of three S. minutum sequences. Alignment of N-terminal regions revealed several conserved amino acid positions, including the highly conserved ExExGYLG in most dinoflagellates (see Additional file 2: Figure S5). In Symbiodinium, only one of the 25 sequences contained the signature GYLG and three other variants, DYLG, EYLG, and GYMG. Many variations were found in other sequences at the same position showing the diversified nature of the N-terminus in S. minutum (see Additional file 2: Figure S5).
Identification of ZAD-D in cultured S. minutum by NanoLC-MS
ZAD (zooxanthellamide D) was identified based on high-resolution mass data (Table 2). A NanoLC-MS (positive ion) profile of the methanol extract of S. minutum showed ions at m/z 1072.60 (10.6 min) and 1050.57 for the [M + Na]+ and [M + H]+, respectively (Additional file 3: Figure S6). The ammonium adduct [M + NH4]+ at m/z 1067.59 (10.6 min) was also observed. It should be noted that other polyhydroxy molecules were also observed in the crude methanol extract, but none of them corresponded to other reported zooxanthella polyhydroxy molecules (Table 3).
Table 2.
High-resolution MS of the target molecule, ZAD-D45
| Ions | Obs. Mass (m/z) | Theo. Mass | Delta (mmu) | Formula |
|---|---|---|---|---|
| [M + H]+ | 1050.5657 | 1050.5632 | 2.46 | C54H84O19N |
| [M + NH4]+ | 1067.5922 | 1067.5898 | 2.41 | C54H87O19N2 |
| [M + Na]+ | 1072.5476 | 1072.5452 | 2.46 | C54H83O19NNa |
Table 3.
Compounds isolated from Symbiodinium sp. and their biological activities
| Molecules | Reported m/z | Biological activity | References |
|---|---|---|---|
| Neosymbioimine | 404.154 | ─ | [54] |
| Norcarothenoids | 630.3557 | Growth-inhibitory activity against cancer cells | [55] |
| Symbiodinolide | 2859 | Activates N-type Ca 2+ channel | [56] |
| Symbioimine | 378.1368 | Anti-resorptive and Anti-inflammatory | [54] |
| Symbioramide | 582 | Ca2+-ATPase activator | [57] |
| Symbiospirols | 1229 | Inhibitory effect on protein kinase C | [58] |
| Zooxanthellatoxin A (ZT-A) | 2872 | Vasoconstrictive substance | [59] |
| Zooxanthellatoxin B (ZT-B) | 2829.9 | Vasoconstrictive substance | [60] |
| Zooxanthellamide A | 2715.4 | ─ | [61] |
| Zooxanthellamide B | 2697.4 | ─ | [62] |
| Zooxanthellamide C (ZAD-C) | 2697.4016 | Vasoconstrictive activity | [63] |
| Zooxanthellamide D (ZAD-D) | 1072.6 | Cytotoxicity against human carcinoma cell lines | [45], This study |
| Zooxanthellamine (ZA) | 498.321 | ─ | [64] |
| Zooxanthellactone (ZL) | 327.2317 | ─ | [65] |
Discussion
The KS domain is the most conserved domain of Type I PKS proteins and has divergent homologs, which permit comparative phylogenetic analysis of PKSs [28]. Our phylogenetic trees resolved previously reported clades [11, 14, 29]. Addition of novel sequences from S. minutum to the KS phylogenetic dataset provided evidence for three groups of dinoflagellate KS during PKS gene evolution. Other protist groups that diverged earlier also retain their KS evolutionary signatures and remain within well-supported clades, as shown by our analysis. A smaller clade comprising only S. minutum and K. brevis sequences showed alterations in their active sites (see Additional File 2: Figure S5). This topology may indicate a history of early gene duplication within the dinoflagellate clade.
As in PKSs, NRPSs also produce diverse secondary metabolites and have modular organizations with each module assuming specific functions. Formation of hybrid systems or clusters has been reported in bacteria [30, 31]. Fungal and bacterial NRPS and PKS have gained attention in recent years, mainly due to their complex evolutionary histories [32–34]. Lawrence et al. [35] provided evidence for horizontal gene transfer (HGT) of the hybrid NRPS-PKS gene from a putative bacterial donor in the Burkholderiales and suggested a HGT early in the history of the fungal Phylum Ascomycota. Bushley and Turgeon [34] identified NPS genes encoding NRPS and NRPS-like proteins in fungal genomes and suggested mechanisms for this modular architecture. In Aspergillus spp., genes involved in secondary metabolite biosynthesis tend to be located in subtelomeric regions, which may contribute to their rapid evolution [36, 37]. Gene transfer from cyanobacteria to dinoflagellates has been suggested by López-Legentil et al. [21] and this could explain the grouping of two S. minutum sequences with cyanobacterial sequences. The hybrid gene symbB1.v1.2.012436.t1 shares features with chlorophyte/haptophyte sequences used in our analysis. A data mining study found a surprisingly high number of hybrid NRPS-PKS gene clusters across three domains of life and this might be a consequence of long-term convergence between NRPS and PKS [38]. Our survey showed that the S. minutum genome encodes only one NRPS domain-containing gene. Hybrid NRPS-PKSs have been reported in other dinoflagellates (K. brevis [19] and H. circularisquama [12]). Dinoflagellate genomes are punctuated with a high number of simple and complex repeats and well known for frequent recombination events [39, 40]. Additionally, these genomes contain genes in high copy numbers, an indication of frequent gene duplication events during dinoflagellate evolution [41]. Shoguchi et al. [18] predicted that a total of 17,703 genes of S. minutum might have originated by gene duplication. It will be interesting to determine what types of natural products are synthesized by this NRPS-PKS gene and what role they do play in S. minutum.
Eichholz et al. [11] speculated that the N-terminus is related to the monofunctional nature of KS domains and may play a role in structural rearrangements, substrate docking, or protein-protein interactions. The N-termini of PKS multi-enzymes contain regularities in amino acid sequences. Recent studies have highlighted the potential role of these regions as “linkers” and their interactions with linker regions at the C-termini of PKS multi-enzymes [42, 43]. A low degree of conservation was noted within the N-terminal ExExGYLG signature sequence of Symbiodinium KS sequences. The GYLG conserved sequence has also been reported in G. polynesiensis, along with several variants (DYLG, HYLG, YYLG, GLLG and ALLG) [13]. Alteration of this signature has also been reported in A. spinosum (AFLG) [14]. Eichholz et al. [11] found that PKS domains are expressed as monofunctional units and that this feature may be unique to dinoflagellates. However, a transcript containing more than one domain has been reported in which one EST contig encoding two Type I PKS domains was found in the transcriptome of H. circularisquama, raising the possibility that there may be multi-modular PKS genes in dinoflagellates [12]. One feature suggesting this possibility is the presence of the ACP (PP arm), upstream of the KS domain. Such a group could serve as a swinging arm to present substrates to catalytic sites on PKS. Modular Type I PKS proteins have been reported in a closely related apicomplexan (Cryptosporidium parvum) and in a haptophyte (Emiliana huxleyi) with several different enzymatic domains arranged in distinct modules [29, 44]. Given that there is partial or complete absence of the conserved GYLG sequence in the N-terminus and that ACP precedes the KS domains, our genome-wide survey provides evidence for multifunctional PKS genes in the Symbiodinium genome along with monofunctional units.
ZAD-D is a linear polyhydroxylated polyketide and has been reported from Symbiodinium strain JCUCS-1 [45]. It is related to amphidinols isolated from the dinoflagellate, Amphidinium sp. [45]. This molecule is a polyhydroxy amide consisting of a C22-acid moiety and a C32-amine moiety; it furnishes three tetrahydropyran rings and six isolated butadiene chromophores. Apart from ZAD-D, other unknown polyhydroxy molecules were found in the methanol extract, and characterization of these unknown compounds could be interesting (see Additional File 3 Figure S6). Other natural products have been isolated from Symbiodinium sp. that displayed significant biological activities (Table 3). Hybrid NRPS-PKS systems are capable of incorporating both amino acids and short carboxylic acids into final products, eventually leading to greater chemical structural diversity. It is not yet known what type of natural products are synthesized by the hybrid NRPS-PKS reported here; further work is needed to characterize its end products as well as products of other PKSs in order to determine their role in S. minutum.
Conclusions
We demonstrate that three structural types of enzymes for polyketide synthesis, single-domain PKS, multi-domain PKS, and NRPS-PKS hybrid, are present in the dinoflagellate, S. minutum. Based on the ketosynthase domain, dinoflagellate PKSs can be evolutionarily classified into three groups. It is not yet clear why S. minutum possesses a polyketide biosynthetic pathway and how these multifunctional PKS proteins have evolved. Genomic characterization of dinoflagellate PKS genes will likely provide insights for combinatorial biosynthesis of polyketides with wide range of applications. Due to large and complex dinoflagellate genomes, it is more difficult to perform comprehensive analyses; however, cultured S. minutum, which has a comparatively small genome, might provide further insights into these phenomena.
Methods
Culture
Symbiodinium minutum, originally provided by Dr. Mary Alice Coffroth, of the University of New York, Buffalo, was cultured in autoclaved, artificial seawater containing 1X Guillard’s (F/2) marine-water enrichment solution (Sigma-Aldrich: G0154), plus three antibiotics, ampicillin (100 μg/mL), kanamycin (50 μg/mL), and streptomycin (50 μg/mL) [18]. Mass culturing was performed in a 2 L final volume by inoculating, and sampling at stationary phase one month later. A 12 h light/dark regime at 25 °C was maintained with a TOMY incubation chamber CLE-303.
Transcriptome mapping
Transcriptome sequences of S. minutum were obtained in our previous study [18]. RNAseq reads have been deposited at DDBJ Sequence Read Archive (http://trace.ddbj.nig.ac.jp/dra/index_e.html) [DRR003865-DRR003871]. Reads from the Transcription start site (TSS) library have been also deposited at the DDBJ Sequence Read Archive [DRR023220-DRR023221]. Spliced leader (SL) sequences (DCCGTAGCCATTTTGGCTCAAG ) (D = T, A, or G) were removed from single TSS reads (99-bp). SL sequences were deleted to yield 77-bp reads that were then used for mapping onto the genome [18]. Reads were mapped onto S. minutum genome version 1 [DDBJ/EMBL/GenBank: DF239013-DF260911 (scaffolds)], using TopHat with default parameters [46]. SAMtools software was used for visualization of read coverage [47].
Phylogenetic analysis
Amino acid sequences of KS domains were obtained from NCBI Genbank with additional sequences from Eichholz et al [11]. Type I and II PKS and FAS sequences, representing 39 different taxa, were used for Bayesian inference and maximum likelihood analysis. These represent major clades from prokaryote, fungal, animal, apicomplexan, haptophyte, and chlorophyte PKSs. Data included KS sequences from other dinoflagellates (K. brevis, A. ostenfeldii, H. triqueta, G. polynesiensis, and A. spinosum). The genome browser, MarinegenomicsDB (http://marinegenomics.oist.jp/genomes/gallery) [48] was accessed in order to retrieve PKS sequences. Multiple amino acid alignment was performed with the MUSCLE algorithm [49] included in MEGA 6 [50]. A maximum likelihood phylogenetic tree was generated with MEGA 6 using a Le-Gasquel amino acid replacement matrix with 1000 bootstraps. Bayesian inference was conducted with MrBayes v.3.2 [51] using the same replacement model and run for four million generations and four chains until the posterior probability approached 0.01. Statistics and trees were summarized using a burn-in of 25 % of the data [10]. Trees were edited using Figtree (http://tree.bio.ed.ac.uk/software/figtree/).
Active sites of KS, AT, DH, ER, and KR domains and the N-terminus of the KS domain
In order to investigate whether active sites of major enzyme motifs were conserved, dinoflagellate sequences from NCBI were aligned with reference sequences from A. ostenfeldii, A. spinosum, H. triquetra, and K. brevis. For the N-termini of dinoflagellate KS sequences, other regions were separated from the KS domain and searched against the NCBI and PFAM databases [52]. Multiple alignments and phylogenies of the truncated sequences were calculated as described above for the KS domains.
NRPS-PKS gene cluster
The antiSMASH server [22] was used to identify non-ribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) domains. FASTA format protein sequences were used as input. Results were compared and further annotation was conducted using PFAM database [52]. NRPSpredictor2 was used to identify binding specificity of the A domains in the NRPS modules [53]. I-TASSER was used to identify the AT domain associated with the PKS module [25].
Polyol extraction from S. minutum culture
Cultured cells were collected by centrifugation (9,000 g and 14,000 g, 10 min, 10 °C). After discarding the supernatant, the cell pellet was extracted with methanol (three times) at room temperature. Methanol (100 μL) was added to the biomass (37 mg, wet weight) followed by vortexing (1 min), sonication (10 min), and centrifugation (14,000 g, 10 min, 10 °C) to yield a methanol extract. The resulting clear solution was transferred to a new tube. By adding methanol (100 μL) to the residue, a second methanol extraction was carried out in the same fashion. The clear second methanol extract was combined with the first and stored at –30 °C. Additional methanol (100 μL) was added to the residue, vortexed (1 min), and kept overnight at room temperature. After centrifugation, the third methanol extract was pooled with the previous extracts (total 300 μL), and designated as the crude extract. To remove lipophilic materials, an aliquot (50 μL) of the crude extract was suspended in 50 μL water-methanol (90:10) containing 0.5 % formic acid. The suspension was vortexed (30 sec) and centrifuged (14,000 g, 10 min, 10 °C) to give a clean solution. The clean solution was transferred into a new tube (stock solution) and the insoluble part was discarded. The stock solution was kept at –30 °C before NanoLC-MS analysis or immediately analyzed after dilution.
NanoLC-MS analysis of the Symbiodinium methanol extract
A Thermo Scientific hydride (LTQ Orbitrap) mass spectrometer was used for MS data collection. The mass spectrometer was equipped with an HPLC (Paradigm MS4, Michrom Bioresources Inc.), an auto-sampler (HTC PAL, CTC Analytics) and a nanoelectrospray ion source (NSI). High-resolution MS spectra were acquired at 60,000 resolution in FTMS mode (Orbitrap), full mass range m/z 400–2,000 Da with 200 °C capillary temperature, 1.9 kV spray voltage in positive ion mode. The lipid-depleted crude extract (stock solution) was diluted 1:50 by adding water-methanol (50:50) containing 0.25 % formic acid and separated on a capillary ODS column (50 × 0.18 mm, 3 μm, C18, Supelco). A 20-min gradient was used for polyol separation (10 % B for 0.0–2.0 min, 10–100 % B for 2.0–10.0 min, hold 100 % B for 10.0–15.0 min, equilibration 10 % B for 15.1–20.0 min; where solvent A is water:acetonitrile 98:2 and solvent B is water:acetonitrile 2:98, both containing 0.1 % formic acid; flow rate 2.0 μL/min, injection 2.0 μL loop).
Availability of supporting data
The genome and transcriptome data of Symbiodinium minutum are available at http://marinegenomics.oist.jp/genomes/downloads?project_id=21.
Acknowledgments
We thank Steven D. Aird for editing the manuscript. We also thank Sutada Mungpakdee for the useful comments from the preliminary surveys for PKS genes. The authors are grateful to Dr. Mary Alice Coffroth for providing the S. minutum isolate. The authors acknowledge Drs. Yutaka Suzuki and Sumio Sugano for TSS sequencing and the OIST sequencing section for RNAseq sequencing. This work was supported in part by Grants-in-Aids from MEXT (No. 25440182, 221S0002) and by generous support of Okinawa Institute of Science and Technology Graduate University to the Marine Genomics Unit.
Abbreviations
- ACP
Acyl carrier protein
- ACPS
Acyl carrier protein synthase
- antiSMASH
Antibiotics and Secondary Metabolite Analysis Shell
- AT
Acyl transferase
- DH
Dehydratase
- ER
Enoylreductase
- EST
Expressed sequence tags
- FAS
Fatty acid synthase
- HPLC
High-performance liquid chromatography
- I-TASSER
Iterative Threading ASSEmbly Refinement
- KR
Ketoreductase
- KS
Ketosynthase
- MT
Methyltransferase
- NRPS-PKS
Non-ribosomal peptide synthetase-polyketide synthase
- PKS
Polyketide synthase
- TE
thioesterase
Additional files
Table S1. Predicted domains from transcriptome contigs Figure S1. Expression of KS domain-containing genes on scaffolds of S. minutum. Read coverages of RNAseq (gray line) on KS domain-containing genes (surrounded by green) show expression in our standard cultured conditions. In addition, the SL sequence containing reads (red line) from transcription start site (TSS) library suggest large multifunctional genes are expressed as a transcript that is not trans-spliced. Red arrows show trans-spliced sites, located internally in KS domain-containing genes. Figure S2. Molecular phylogenetic tree of Type I and Type II KS domains from prokaryotic and eukaryotic PKS and FAS, analyzed by maximum likelihood. Type II KS and acyl carrier protein synthases (ACPS) were used as outgroups. Bootstrap values ≥ 50 % are marked at appropriate nodes. Details regarding S. minutum sequences are provided in Table 1. (PDF 9386 kb)
Figure S3. (A) Alignment of KS domains with those of other dinoflagellates and animal PKSs and FASs. Asterisks indicate conserved amino acids required for catalytic activity. (B) Additional Symbiodinium and K. brevis sequences that are consistent with previous reports. Figure S4. Alignments of motifs within AT, ACP, KR, ER, and DH domains. Active sites within the motifs are boxed with dashed lines. Figure S5. (A) Multiple alignment of the truncated, conserved, N-termini of dinoflagellate KSs (B) Additional Symbiodinium sequences with the divergent signatures. Lower panel: Maximum likelihood tree of the N-termini computed with 1000 bootstrap replicates. Bootstrap values ≥ 50 % are marked at appropriate nodes. (PDF 2705 kb)
Figure S6. (A) NanoLC-MS (positive ion) profile of the methanol extract of Symbiodinium minutum. Top Chromatogram, Center Top: Extract ion (m/z 1072.60, 10.6 min), Center bottom: (m/z 1050.57, 10.6 min), Bottom: MS spectrum. (B) MS spectrum (positive ion) of the methanol extract (expanded). (PDF 145 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Author’s contributions
GB, ES, and MR wrote the manuscript. GB, KH, and ES analyzed the genomic data. MR performed mass sample preparation, mass data acquisition, and data interpretation. NS and ES designed and led the research program. All authors read and approved the manuscript.
Contributor Information
Girish Beedessee, Phone: +81-98-966-8653, Email: girish.beedessee@oist.jp.
Kanako Hisata, Email: kanako@oist.jp.
Michael C. Roy, Email: mcroy@oist.jp
Noriyuki Satoh, Email: norisky@oist.jp.
Eiichi Shoguchi, Email: eiichi@oist.jp.
References
- 1.Wang DZ. Neurotoxins from marine dinoflagellates: a brief review. Mar Drugs. 2008;6:349–71. doi: 10.3390/md6020349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rein KS, Borrone J. Polyketides from dinoflagellates: Origins, pharmacology and biosynthesis. Comp Biochem Physiol B Biochem Mol Biol. 1999;124:117–31. doi: 10.1016/S0305-0491(99)00107-8. [DOI] [PubMed] [Google Scholar]
- 3.Rein KS, Snyder RV. The biosynthesis of polyketide metabolites by dinoflagellates. Adv Appl Microbiol. 2006;59:93–125. doi: 10.1016/S0065-2164(06)59004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kellmann R, Stüken A, Orr RJ, Svendsen HM, Jakobsen KS. Biosynthesis and molecular genetics of polyketides in marine dinoflagellates. Mar Drugs. 2010;8:1011–48. doi: 10.3390/md8041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craik DJ, Scanlon MJ. Marine Toxins as a starting point for drugs. In: Botana LM, editor. Seafood and Freshwater Toxins: Pharmacology, Physiology, and Detection. New York: Marcel Dekker Inc; 2000. pp. 715–40. [Google Scholar]
- 6.Botana LM. Phycotoxins: Chemistry and Biochemistry. Oxford: Blackwell Publishing; 2007. [Google Scholar]
- 7.Camacho FG, Rodriguez JG, Miron AS, Garcia MCC, Belarbi EH, Chisti Y, et al. Biotechnological significance of toxic marine dinoflagellates. Biotechnol Adv. 2007;25:176–94. doi: 10.1016/j.biotechadv.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Newman DJ, Cragg GM. Marine natural products and related compounds in clinical and advanced preclinical trials. J Nat Prod. 2004;67:1216–38. doi: 10.1021/np040031y. [DOI] [PubMed] [Google Scholar]
- 9.Gokhale RS, Tuteja D. Biochemistry of polyketide synthases. In: Rehm H-J, Reed G, editors. Biotechnology set, second edition. Weinheim: Wiley-VCH Verlag GmbH; 2001. Part X: p. 341-372.
- 10.Monroe EA, Van Dolah FM. The toxic dinoflagellate Karenia brevis encodes novel type I-like polyketide synthases containing discrete catalytic domains. Protist. 2008;159:471–82. doi: 10.1016/j.protis.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Eichholz K, Beszteri B, John U. Putative monofunctional type I polyketide synthase units: A dinoflagellate-specific feature? PLoS One. 2012;7 doi: 10.1371/journal.pone.0048624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salcedo T, Upadhyay RJ, Nagasaki K, Bhattacharya D. Dozens of toxin-related genes are expressed in a nontoxic strain of the dinoflagellate Heterocapsa circularisquama. Mol Biol Evol. 2012;29:1503–6. doi: 10.1093/molbev/mss007. [DOI] [PubMed] [Google Scholar]
- 13.Pawlowiez R, Morey JS, Darius HT, Chinain M, Van Dolah FM. Transcriptome sequencing reveals single domain Type I-like polyketide synthases in the toxic dinoflagellate Gambierdiscus polynesiensis. Harmful Algae. 2014;36:29–37. doi: 10.1016/j.hal.2014.04.013. [DOI] [Google Scholar]
- 14.Meyer JM, Rödelsperger C, Eichholz K, Tillmann U, Cembella A, McGaughran A, et al. Transcriptomic characterisation and genomic glimps into the toxigenic dinoflagellate Azadinium spinosum, with emphasis on polyketide synthase genes. BMC Genomics. 2015;16:27. doi: 10.1186/s12864-014-1205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohli G, John U, Figueroa RI, Rhodes L, Harwood DT, Groth M, Bolch JS, Murray SA. Polyketide synthesis genes associated with toxin production in two species of Gambierdiscus (Dinophyceae) BMC Genomics. 2015;16:410. doi: 10.1186/s12864-015-1625-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trench RK. Cellular and molecular interactions in symbioses between dinoflagellates and marine invertebrates. Pure Appl Chem. 1981;53:819–35. doi: 10.1351/pac198153040819. [DOI] [Google Scholar]
- 17.Blank RJ, Trench RK. Speciation and symbiotic dinoflagellates. Science. 1985;229:656–8. doi: 10.1126/science.229.4714.656. [DOI] [PubMed] [Google Scholar]
- 18.Shoguchi E, Shinzato C, Kawashima T, Gyoja F, Mungpakdee S, Koyanagi R, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 2013;23:1399–408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 19.Snyder RV, Gibbs PDL, Palacios A, Abiy L, Dickey R, Lopez JV, et al. Polyketide synthase genes from marine dinoflagellates. Mar Biotechnol. 2003;5:1–12. doi: 10.1007/s10126-002-0077-y. [DOI] [PubMed] [Google Scholar]
- 20.Hunsperger HM, Randhawa T, Cattolico RA. Extensive horizontal gene transfer, duplication, and loss of chlorophyll synthesis genes in the algae. BMC Evol Biol. 2015;15:16. doi: 10.1186/s12862-015-0286-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López-Legentil S, Song B, DeTure M, Baden D. Characterization and localization of a hybrid non-ribosomal peptide synthetase and polyketide synthase gene from the toxic dinoflagellate Karenia brevis. Mar Biotechnol. 2010;12:32–41. doi: 10.1007/s10126-009-9197-y. [DOI] [PubMed] [Google Scholar]
- 22.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, et al. antiSMASH 2.0--a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–12. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strieker M, Tanović A, Marahiel MA. Nonribosomal peptide synthetases: structures and dynamics. Curr Opin Struct Biol. 2010;20:234–40. doi: 10.1016/j.sbi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donadio S, Katz L. Organization of the enzymatic domains in the multifunctional polyketide synthase involved in erythromycin formation in Saccharopolyspora erythraea. Gene. 1992;111:51–60. doi: 10.1016/0378-1119(92)90602-L. [DOI] [PubMed] [Google Scholar]
- 27.Aparicio JF, Molnar I, Schwecke T, Konig A, Haydock SF, Khaw LE, et al. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene. 1996;169:9–16. doi: 10.1016/0378-1119(95)00800-4. [DOI] [PubMed] [Google Scholar]
- 28.Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Phylogenomic analysis of Type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci U S A. 2003;100:15670–5. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.John U, Beszteri B, Derelle E, Van de Peer Y, Read B, Moreau H, et al. Novel insights into evolution of protistan polyketide synthases through phylogenomic analysis. Protist. 2008;159:21–30. doi: 10.1016/j.protis.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Dittmann E, Neilan BA, Börner T. Molecular biology of peptide and polyketide biosynthesis in cyanobacteria. Appl Microbiol Biotechnol. 2001;57:467–73. doi: 10.1007/s002530100810. [DOI] [PubMed] [Google Scholar]
- 31.Chang ZX, Flatt P, Gerwick WH, Nguyen VA, Willis CL, Sherman DH. The barbamide biosynthetic gene cluster: a novel marine cyanobacterial system of mixed polyketide synthase (PKS)-non-ribosomal peptide synthetase (NRPS) origin involving an unusual trichloroleucyl starter unit. Gene. 2002;296:235–47. doi: 10.1016/S0378-1119(02)00860-0. [DOI] [PubMed] [Google Scholar]
- 32.Khaldi N, Collemare J, Lebrun M, Wolfe KH. Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol. 2008;9:R18. doi: 10.1186/gb-2008-9-1-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt I, Lumbsch HT. Ancient horizontal gene transfer from bacteria enhances biosynthetic capabilities of fungi. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bushley KE, Turgeon BG. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol Biol. 2010;10:26. doi: 10.1186/1471-2148-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrence DP, Kroken S, Pryor BM, Arnold AE. Interkingdom gene transfer of a hybrid NPS/PKS from bacteria to filamentous ascomycota. PLoS One. 2011;6 doi: 10.1371/journal.pone.0028231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, Anderson MJ, et al. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang PK, Horn BW, Dorner JW. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet Biol. 2005;42:914–23. doi: 10.1016/j.fgb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci U S A. 2014;111:9259–64. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaeckisch N, Yang I, Wohlrab S, Glöckner G, Kroymann J, Vogel H, et al. Comparative genomic and transcriptomic characterization of the toxigenic marine dinoflagellate Alexandrium ostenfeldii. PLoS One. 2011;6 doi: 10.1371/journal.pone.0028012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McEwan M, Humayun R, Slamovits CH, Keeling PJ. Nuclear genome sequence survey of the Dinoflagellate Heterocapsa triquetra. J Eukaryot Microbiol. 2008;55:530–5. doi: 10.1111/j.1550-7408.2008.00357.x. [DOI] [PubMed] [Google Scholar]
- 41.Hou Y, Lin S. Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: gene content estimation for dinoflagellate genomes. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broadhurst RW, Nietlispach D, Wheatcroft MP, Leadlay PF, Weissman KJ. The structure of docking domains in modular polyketide synthases. Chem Biol. 2003;10:723–31. doi: 10.1016/S1074-5521(03)00156-X. [DOI] [PubMed] [Google Scholar]
- 43.Richter CD, Nietlispach D, Broadhurst RW, Weissman KJ. Multienzyme docking in hybrid megasynthetases. Nat Chem Biol. 2008;4:75–81. doi: 10.1038/nchembio.2007.61. [DOI] [PubMed] [Google Scholar]
- 44.Zhu G, LaGier MJ, Stejskal F, Millership JJ, Cai X, Keithly JS. Cryptosporidium parvum: the first protist known to encode a putative polyketide synthase. Gene. 2002;298:79–89. doi: 10.1016/S0378-1119(02)00931-9. [DOI] [PubMed] [Google Scholar]
- 45.Fukatsu T, Onodera K, Ohta Y, Oba Y, Nakamura H, Shintani T, et al. Zooxanthellamide D, a polyhydroxy polyene amide from a marine dinoflagellate, and chemotaxonomic perspective of the Symbiodinium Polyols. J Nat Prod. 2007;70:407–11. doi: 10.1021/np060596p. [DOI] [PubMed] [Google Scholar]
- 46.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koyanagi R, Takeuchi T, Hisata K, Gyoja F, Shoguchi E, Satoh N, et al. MarinegenomicsDB: An integrated genome viewer for community-based annotation of genomes. Zool Sci. 2013;30:797–800. doi: 10.2108/zsj.30.797. [DOI] [PubMed] [Google Scholar]
- 49.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Röttig M, Medema MH, Blin K, Weber T, Rausch C, Kohlbacher O. NRPSpredictor2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011;39:W362–7. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kita M, Ohishi N, Washida K, Kondo M, Koyama T, Yamada K, et al. Symbioimine and neosymbioimine, amphoteric iminium metabolites from the symbiotic marine dinoflagellate Symbiodinium sp. Bioorg Med Chem. 2005;13:5253–8. doi: 10.1016/j.bmc.2005.05.064. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki M, Watanabe K, Fujiwara S, Kurasawa T, Wakabayashi T, Tsuzuki M, Iguchi K, Yamori T. Isolation of peridinin-related norcarotenoids with cell growth inhibitory activity from the cultured dinoflagellate of Symbiodinium sp., a symbiont of the Okinawan softcoral Clavularia viridis, and analysis of fatty acids of the dinoflagellate. Chem Pharm Bull. 2003;51:724–7. doi: 10.1248/cpb.51.724. [DOI] [PubMed] [Google Scholar]
- 56.Kita M, Ohishi N, Konishi K, Kondo M, Koyama T, Kitamura M, et al. Symbiodinolide, a novel polyol macrolide that activates N-type Ca2+ channel, from the symbiotic marine dinoflagellate Symbiodinium sp. Tetrahedron. 2007;63:6241–51. doi: 10.1016/j.tet.2007.02.093. [DOI] [Google Scholar]
- 57.Kobayashi J, Ishibashi M, Nakamura H, Hirata Y, Yamasu T, Sasaki T, Ohizumi Y. Symbioramide, a novel Ca2 + -ATPase activator from the cultured dinoflagellate Symbiodinium sp. Experientia. 1988;15:800–2. doi: 10.1007/BF01959173. [DOI] [PubMed] [Google Scholar]
- 58.Tsunematsu Y, Ohno O, Konishi K, Yamada K, Suganuma M, Uemura D. Symbiospirols: novel long carbon-chain compounds isolated from symbiotic marine dinoflagellate Symbiodinium sp. Org Lett. 2009;11:2153–6. doi: 10.1021/ol900299x. [DOI] [PubMed] [Google Scholar]
- 59.Nakamura H, Asari T, Murai A, Kan Y, Kondo T, Yoshida K, et al. Zooxanthellatoxin-A, a potent vasoconstrictive 62-membered lactone from a symbiotic Dinoflagellate. J Am Chem Soc. 1995;117:550–1. doi: 10.1021/ja00106a071. [DOI] [Google Scholar]
- 60.Nakamura H, Asari T, Fujimaki K, Maruyama K, Murai A. Zooxanthellatoxin-B, vasoconstrictive congener of zooxanthellatoxin-A from a symbiotic dinoflagellate Symbiodinium sp. Tet Lett. 1995;36:7255–8. doi: 10.1016/0040-4039(95)01503-A. [DOI] [Google Scholar]
- 61.Onodera K, Nakamura H, Oba Y, Ojika M. Zooxanthellamide A, a novel polyhydroxy metabolite from a marine dinoflagellate of Symbiodinium sp. Tetrahedron. 2003;59:1067–71. doi: 10.1016/S0040-4020(02)01630-7. [DOI] [Google Scholar]
- 62.Onodera K, Nakamura H, Oba Y, Ojika M. Zooxanthellamide B, a novel large polyhydroxy metabolite from a marine dinoflagellate of Symbiodinium sp. Biosci Biotechnol Biochem. 2003;68:955–8. doi: 10.1271/bbb.68.955. [DOI] [PubMed] [Google Scholar]
- 63.Onodera K, Nakamura H, Oba Y, Ohizumi Y, Ojika M. Zooxanthellamide Cs: vasoconstrictive polyhydroxylated macrolides with the largest lactone ring size from a marine dinoflagellate of Symbiodinium sp. J Am Chem Soc. 2005;127:10406–11. doi: 10.1021/ja050810g. [DOI] [PubMed] [Google Scholar]
- 64.Nakamura H, Kawase Y, Maruyama K, Muria A. Studies on Polyketide Metabolites of a Symbiotic Dinoflagellate, Symbiodinium sp.: A New C30 Marine Alkaloid, Zooxanthellamine, a Plausible Precursor for Zoanthid Alkaloids. Bull Chem. Soc. Jpn. 1998;71:781–7. [Google Scholar]
- 65.Onodera K, Fukatsu T, Kawai N, Yoshioka Y, Okamoto T, Nakamura H, Ojika M. Zooxanthellactone, a Novel-Lactone-type Oxylipine from Dinoflagellates of Symbiodinium sp.: Structure, Distribution, and Biological Activity. Biosci Biotechnol Biochem. 2004;68:848–52. doi: 10.1271/bbb.68.848. [DOI] [PubMed] [Google Scholar]


