Abstract
Background
sdw1/denso is one of the most important and useful semi-dwarf genes in barley breeding. At least four sdw1/denso alleles have been reported and HvGA20ox2 is considered as the candidate gene. Up to date, results of studies have not univocally proven the genetic relationship between sdw1/denso and HvGA20ox2.
Results
In the present study, a complete deletion of Morex_contig_40861 including both HvGA20ox2 and Mloc_56463 genes was identified at the sdw1 locus from a semi-dwarf mutant Riso no. 9265. Expression of the genes encoding gibberellin biosynthesis (HvGA20ox1 and HvGA3ox2) were increased in the mutant compared to the wild type Bomi, while the expression of GA catabolic gene HvGA2ox3 was decreased. Over-expression of HvGA20ox2 could rescue the semi-dwarf phenotype and increase GAs concentration.
Conclusions
We confirmed that a GA biosynthetic enzyme HvGA20ox2, acted as GA 20-oxidase, is the functional gene for the sdw1/denso semi-dwarfism. Lose of HvGA20ox2 is partially compensated by HvGA20ox1 and further feedback is regulated by gibberellin. We also deduced that the sdw1/denso allele itself affects later heading owing to its reduced endogenous GAs concentration.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-2116-x) contains supplementary material, which is available to authorized users.
Keywords: Barley, Deletion, Gene compensation, Gibberellin, Semi-dwarf
Background
Plant height is one of the most important agronomic traits in cereal crops that not only determines plant architecture, but also is closely associated with grain yield. A reduction in plant height usually leads to strong straw and high resistance to lodging, as happened in semi-dwarf rice and wheat cultivars. However, reduced plant height means lowering of canopy, which favors the epidemic spread of fungal diseases, resulting in an undesired increase of fungicide use, and a reduction of yield potential mainly due to smaller grain weight [1]. For optimizing the balance of plant height and yield, breeders have successfully utilized the semi-dwarf genes in cereal crop breeding since 1960s. The development and wide cultivation of semi-dwarf wheat and rice varieties led to a dramatic increase of cereal production worldwide [2], which is labeled as a ‘green revolution’.
The Green Revolution genes have been identified and isolated in the semi-dwarf varieties by reversed genetics. Among them, sd1 in rice is one of the most famous genes and has been widely used to produce semi-dwarf phenotype in both japonica and indica rice. It was reported that sd1 encoded one of the GA 20-oxidases and was involved in the last steps of gibberellin biosynthesis [3–6]. Semi-dwarf genes have also been extensively explored in barley breeding programs and more than 30 types of them have been described [7]. One of the most successfully used semi-dwarf genes in modern barley breeding is sdw1/denso, which was postulated as homologous to ga5 in Arabidopsis and sd1 in rice [8, 9]. The main phenotypic effect of the sdw1/denso gene is a 10-20 cm reduction of plant height, depending on environmental conditions [10]. Besides, there are some deleterious effects associated with sdw1/denso gene, such as late heading and maturity, decreased thousand-grain weight and high screening [10–15]. As for the relationship between grain yield and sdw1/denso, both positive and negative were reported depending on different genetic backgrounds [10, 11, 15]. However, with its suitable semi-dwarf phenotype and potentially increased yield, sdw1/denso has been introduced to numerous cultivars. For example, newly released more than 150 European cultivars carried the denso allele [16].
There are at least four independent alleles based on the allelism test done up to date [12, 17]. One spontaneous mutant was derived from a Danish variety Abed denso in 1946, as named accordingly [17]. Two alleles induced by X-ray with different parents were named as denso and sdw1, respectively. Interestingly, the denso mutant has been used to develop malting barley, while the sdw1 has been limited to feed barley [10, 12]. The fourth sdw1 allele was found in a M2 generation from the variety Bomi, treated by neutrons in the Stockholm reactor and named as Riso no. 9265. Although these mutants are characterized by semi-dwarf phenotype, all of them are sensitive to GA3 [18].
In 1993, sdw1/denso was first mapped on the 3HL as a phenotypic trait [19]. In our previous study, GA20ox2 homolog was identified using PCR method with the primers designed from the conserved domain of rice sd1 and considered as a candidate gene of sdw1/denso [8]. Meanwhile, it was found that the expression of HvGA20ox2 in denso mutant was reduced by 4-fold, but almost 60-fold in the sdw1 mutant, compared to the control [15]. However, there was no direct evidence to prove the HvGA20ox2 as the functional gene. In addition, the function of HvGA20ox2 was predicted based on its functional domain, being highly similar with those of sd1 (OsGA20ox2) and ga5 (AtGA20ox1). No evidence showed its function in vitro or in vivo. In the present work we investigated the sdw1 allele of Riso no. 9265 and identified a complete deletion contig, which includes two genes Mloc_56462 (HvGA20ox2) and Mloc_56463 (a putative methyltransferase PMT26). The function of the HvGA20ox2 gene was further analyzed using genetically-transformed Arabidopsis. Furthermore, real-time PCR assays revealed that transcript level of GA synthesis-related genes were significantly different between Bomi and Riso no. 9265. The current results showed that the semi-dwarf phenotype of Riso no. 9265 is attributed to the deletion of HvGA20ox2.
Results
Plant height, internode length and heading date of Bomi and Riso no. 9265
At maturity, plant height and length of spike, culm and internodes of Bomi and its semi-dwarf mutant Riso no. 9265 were measured (Table 1). Plant height of Bomi was 83.1 cm, and Riso no. 9265 was 63.3 cm, being only 76.2 % of its wild type parent, as reported previously [17]. No obvious difference was found in spike length between the two genotypes. Obviously, the reduction of plant height for the mutant is completely attributed to the shorter culm length. Culm length of Riso no. 9265 was 53.5 cm, being 19.6 cm shorter than that of Bomi. In fact, all internodes of the mutant were shorter than those of the wild type (Table 1). Moreover, the difference between the two genotypes was smaller for the length of the top two internodes and larger for the basal four internodes. Therefore, it may be assumed that the effect of the semi-dwarf gene in Riso no. 9265 is mainly on the basal internodes. In addition, Riso no. 9265 headed three days later than Bomi.
Table 1.
Internode length and plant height of Bomi and Riso no. 9265
| Traits | Bomi | % | Riso no.9265 | % |
|---|---|---|---|---|
| Length (cm) | Length (cm) | |||
| Plant height | 83.2 ± 1.1 | 63.5 ± 2.4** | ||
| Spike length | 10.1 ± 0.4 | 10.0 ± 0.5 | ||
| Culm height | 73.1 ± 0.9 | 100 | 53.5 ± 2.1** | 100 |
| First-internode length | 25.2 ± 1.0 | 34.5 | 20.1 ± 0.7* | 37.6 |
| Second-internode length | 14.9 ± 0.3 | 20.4 | 13.1 ± 0.7* | 24.5 |
| Third-internode length | 10.5 ± 0.4 | 14.4 | 7.6 ± 0.6** | 14.2 |
| Fourth-internode length | 10.8 ± 0.4 | 14.8 | 6.7 ± 0.4** | 12.5 |
| Fifth-internode length | 8.2 ± 0.6 | 11.2 | 4.2 ± 0.3** | 7.9 |
| Sixth-internode length | 3.5 ± 0.6 | 4.8 | 1.8 ± 0.2** | 3.4 |
Values are means ± SE, N = 20. *significant difference at 0.005 level; **significant difference at 0.001 level
HvGA20ox2 was deleted in Riso no. 9265
In this study, the complete genome sequence of HvGA20ox2 (Mloc_56462) was identified after blast against Gramene (http://www.gramene.org/) using the sequences of HvGA20ox2 from our previous study [8]. Two pairs of primers of HvGA20ox2 were designed to amplify its genomic sequence. In Bomi, both primer pairs produced single band with different sizes. One is 2974 bp and the other is 3165 bp. However, no PCR product was obtained in Riso no. 9265 (Fig. 1), indicating that HvGA20ox2 is lost in the mutant. Thus, the sequences of HvGA20ox2 were used to identify Contigs after blast against International Barley Genome Sequencing Consortium (http://webblast.ipk-gatersleben.de/barley/viroblast.php). One Morex_contig_40861, covering 21596 bp was identified, and it contains HvGA20ox2 and Mloc_56463 genes. Several primers covering Morex_contig_40861 were designed (Additional file 1: Table S1) and PCR amplification succeeded in Bomi, but failed in the mutant Riso no. 9265. Furthermore, we were able to amplify flanking genes (Mloc_3247, Mloc_45089, Mloc_51746 and Mloc_7311) of Morex_contig_40861, obtained using the latest barley assembly from Gramene and barley reference genome information from Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) (primers are listed in Additional file 1: Table S1). As a result, it was found that there is a large deletion spanned at least two genes HvGA20ox2 and Mloc_56463 in Riso no. 9265. A blast search for the Mloc_56463 protein sequence against NCBI identified a possible methyltransferase PMT26-like gene.
Fig. 1.
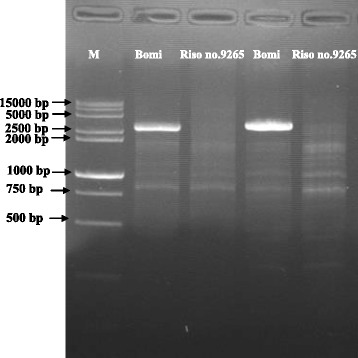
Dectection of the HvGA20ox 2 gene by PCR analysis. M, Marker; the right two lanes were produced by forward primer sdwF10 and reverse primer sdwR10; the left two lanes were produced by forward primer sdw9 and reverse primer sdwR13
Overexpression of HvGA20ox2 caused more gibberellin production in Arabidopsis
To evaluate if HvGA20ox2 is responsible for the semi-dwarf phenotype, we examined transgenic Arabidopsis plants that over-express HvGA20ox2 in wild type (Col-gl1) and a semi-dwarf mutant ga5-3. T1 transgenic plants were selected by BASTA analysis and PCR method. We obtained eleven and six transgenic lines in Col-gl1 and ga5-3 background, respectively. All of them displayed a higher growth rate. Homozygous T3 plants of transgenic Col-gl1 lines (OE-1 and OE-2) and transgenic ga5-3 lines (OE-3 and OE-4) were taken randomly for further analysis. The four transgenic plants differed greatly in the expression levels of HvGA20ox2. However, transcripts of barley GA oxidase-encoding gene were not found in both wild type and semi-dwarf mutant ga5-3 (Fig. 2).
Fig. 2.

Relative expression level of HvGA20ox 2 in transgenic Arabidopsis. nd, not detected. **means significant difference at 0.01 level. nd means not detected
The two independent transgenic lines (OE-1 and OE-2) showed GA-overdose phenotype although they differed in the expression level of HvGA20ox2. There was no obvious difference in root length of 7-day-old seedlings between wild type and HvGA20ox2 over-expression plants, but the latter had 50 % longer hypocotyl than the former (Table 2 and Fig. 3). In addition, the transgenic plants flowered relatively early.
Table 2.
Phenotypic parameters for wild type, ga5-3 and HvGA20ox 2 over-expressing transgenic Arabidopsis
| Traits | Col-gl1 | OE-1(Col-gl1) | OE-2(Col-gl1) | ga5-3 | OE-3(ga5-3) | OE-4(ga5-3) |
|---|---|---|---|---|---|---|
| Hypocotyl length (mm) | 2.05 ± 0.04 | 3.64 ± 0.10A | 4.04 ± 0.01A | 2.07 ± 0.04A | 3.64 ± 0.10AB | 3.75 ± 0.12AB |
| Root length (mm) | 36.1 ± 0.7 | 38.9 ± 1.8 | 40.1 ± 1.2 | 37.5 ± 0.67 | 40.3 ± 1.4 | 36.8 ± 1.7 |
| Flowering time (d) | 26.2 ± 0.35 | 24.6 ± 0.33A | 23.0 ± 0.25A | 26.7 ± 0.28 | 22.5 ± 0.23AB | 23.7 ± 0.24AB |
| Vegetative internode length (cm) | 1.9 ± 0.1 | 2.7 ± 0.1A | 2.9 ± 0.1A | 0.6 ± 0.1A | 3.0 ± 0.2AB | 2.7 ± 0.1AB |
| No.vegetative internodes | 3.4 ± 0.1 | 5.2 ± 0.2A | 5.0 ± 0.2A | 3.1 ± 0.1A | 4.8 ± 0.4AB | 4.9 ± 0.1AB |
| inflorescence internode length (cm) | 0.61 ± 0.01 | 0.68 ± 0.01A | 0.71 ± 0.02A | 0.38 ± 0.01A | 0.67 ± 0.02AB | 0.64 ± 0.02AB |
| No.inflorescence internodes | 34.4 ± 1.1 | 47.3 ± 1.2A | 43.9 ± 1.4A | 31.5 ± 1.1A | 43.7 ± 1.6AB | 47.3 ± 1.9AB |
| Final plant (cm) | 28.8 ± 0.5 | 46.8 ± 0.7A | 44.8 ± 1.0A | 14.6 ± 0.4A | 42.0 ± 1.7AB | 43.8 ± 1.1AB |
The values are the means ± SE
Capital letter A means significantly different from the wild type (Col-gl1); Capital letter B means significantly different from ga5-3 mutant
Fig. 3.

Overexpression of HvGA20ox 2 in Arabidopsis affects plants development. The pictures compared the growth of transgenic and untransgenic at 1-week (a) and 6-week (b)
To determine whether the over-expression constructs would affect the phonotype of ga5-3, we characterized the GA-affecting traits in ga5-3 transgenic lines (OE-3 and OE-4), including hypocotyl length, internodes length and number, flowering time, and final plant height. The results showed that ga5-3 transgenic lines (OE-3 and OE-4) also had the GA-overdose phenotype, with longer hypocotyls and internodes, more internode number, flowering earlier in comparison with those of wild type or ga5-3 mutant (Table 2 and Fig. 3). It can be seen from Fig. 4 that there was the dramatic difference in plant height among the four transgenic lines, wild type and ga5-3 throughout the whole growth stage.
Fig. 4.
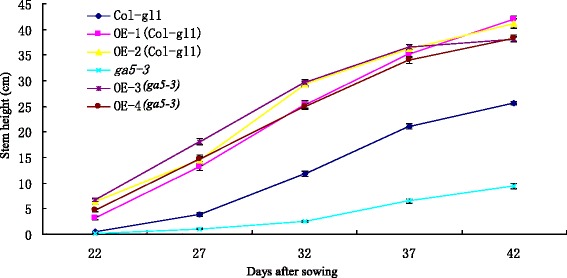
Stem heights of Arabidopsis over expressing HvGA20ox 2 gene
GA4+7 content were much higher than GA1+3 content in wild type. The transgenic plants had significantly higher GA1+3 content than both wild-type and ga5-3 plants, but the difference in GA4+7 content among them was much smaller. Moreover, GA1+3 content was lower in ga5-3 mutants than in wild-type plants, whereas both genotypes had the similar GA4+7 content (Fig. 5).
Fig. 5.

The content of active gibberellin in the 7-d-old seedlings of transgenic and control plants. Each column represents the mean of three repeats with ±SE bar. **means significant difference at 0.01 level
In order to determine whether the changes of phenotype and GA content observed in the transgenic lines are accompanied by the alteration in expression of GA biosynthesis and catabolism, three GA 20-oxidases, one GA 3-oxidase and one GA 2-oxidase, highly expressed in 7-day-old seedlings, were selected for further analysis. As shown in Fig. 6, the expression of AtGA20ox2 and AtGA3ox1 was greatly up-regulated in the AtGA20ox1 deficient mutant ga5-3, while the expression of AtGA2ox2 was significantly decreased in comparison with the wild type. No significant difference was found in the expression of AtGA20ox3 between ga5-3 and wild type. In the transgenic Arabidopsis with HvGA20ox2 over-expression, the expression of AtGA20ox1, AtGA20ox2 and AtGA3ox1 was distinctly decreased, however, AtGA2ox2 was dramatically increased relative to wild type and ga5-3 mutants.
Fig. 6.

The effect of over expression HvGA20ox 2 on expression of gibberellin biosythetic and catabolic genes. *indicades significant difference at 0.05 level; **means significant difference at 0.01 level.; nd means not detected
The expression of GA biosynthesis and catabolism genes were changed in Riso no. 9265 mutant
We determined the expression level of GA dioxygenase genes in stems of Bomi and Riso no. 9265 at the initial jointing stage. HvGA20ox1, HvGA3ox2 and HvGA2ox3 showed high transcript level. The relative mRNA expression of HvGA20ox1 and HvGA3ox2 were dramatically increased in the mutant Riso no. 9265 compared with those of Bomi. On the other hand, the mRNA expression of HvGA2ox3 was decreased in the Riso no. 9265 (Fig. 7).
Fig. 7.

Transcriptional changes of barley GA biosynthetic and catabolic genes in stem at the initial jointing stage. **means significant difference at 0.01 level
Discussion
Previously, we reported that barley HvGA20ox2 (Mloc_56462) was a candidate gene of sdw1/denso and different expression level of HvGA20ox2 was observed between denso and sdw1 (Jotun) alleles [8, 15]. Here, we reported that around 21 kb DNA fragment, including both HvGA20ox2 and Mloc_56463 genes was completely deleted in the mutant Riso no. 9265. Because of the incomplete barley reference genomes and repetitive nature of the barley genome [20], the boundary of deleted segments was not determined precisely in Riso no. 9265. Gene annotation showed that HvGA20ox2 has gibberellin 20-oxidase activity and may be involved in GA biosynthetic pathway. Moreover, the sdw1/denso mutant is GA sensitive and exogenous application of 10 ppm GA3 restored its plant height [18], which indicates that sdw1/denso mutants are GA deficiency. On the contrary, Mloc_56463 is a putative methyltransferase and has not been reported or predicted as a GA-related gene. Thus, HvGA20ox2 is considered as the only candidate responsible for the semi-dwarfism in Riso no. 9265.
The GA biosynthetic pathway has been extensively investigated and the enzymes involved have been well characterized in plants [21]. The early GA-biosynthetic steps are encoded by single-copy genes, while the final steps catalyzed by GA 20-oxidase, GA 3-oxidase and GA 2-oxidase, are encoded by multigene families. GA20ox activity removes carbon-20 in the formation of C19-GA skeleton [22, 23]. In Arabidopsis, GA20-oxidase genes are differentially expressed and involved in different developmental processes controlled by GA, but the plants that constitutively express each of three GA20oxs (AtGA20ox1, AtGA20ox2 and AtGA20ox3) behave as the control treated with GA [22, 23]. The GA-over-production phenotype was also found in HvGA20ox2 transgenic Arabidopsis (Table 2, Figs. 3 and 4). The phenotype was characterized by elongated hypocotyl and stems, early flowering, and higher growth rate. In addition, barley GA20ox2 could recover AtGA20ox1 lose-of-function mutant. These results suggest that HvGA20ox2 is the orthologous of AtGA20ox1 and acts as the oxidase at carbon-20 of GA biosynthetic pathway.
The bioactive GAs in plants are GA1, GA3, GA4 and GA7, but GA4 is a major active GA in Arabidopsis [24]. Both endogenous GA1+3 and GA4+7 in 7-day-old seedlings of the control and transgenic plants were quantified in this study. The results showed that transgenic plants had higher GA contents, especially for GA1+3 compared with the control. The increased GAs is associated with the changed phenotype (Figs. 3 and 4). Similar results have been reported that GA1 content was increased and GA4 content had little change in the 7-day-old Arabidopsis with AtGA20ox1 overexpressors [22]. In contrast, both GA1 and GA4 content had slight change in the shoot tip of Arabidopsis plant with over-expression AtGA20ox, while a 2- to 3-fold increase in GA4 content was observed in the rosette leaves of the transgenic lines [23]. Radi et al. [25] deduced that the dramatic difference in GAs content might be due to the variation in GA level among plant tissues and growth stage.
As indicated in Fig. 6, the transgenic lines overexpressing barley GA 20-oxidases showed a decrease in AtGA20ox1, AtGA20ox2, AtGA3ox1 and an increase in AtGA2ox2 expression, while ga5-3 was just opposite in the expression of these genes, which could be attributed to feedback from an increased bioactive GAs in HvGA20ox2 over-expressing transgenic Arabidopsis or decreased bioactive GAs in ga5-3, respectively. A similar regulatory role has been proposed for AtGA3ox1, whose transcript level is also feedback-regulated in transgenic Arabidopsis of over-expressed GA20ox [23, 26]. No considerable change was detected in AtGA20ox3 level of transgenic Arabidopsis and the control, suggesting that AtGA20ox3 gene might be less sensitive than AtGA20ox1 and AtGA20ox2 to the alteration of bioactive GAs.
However, not all over-expressing GA20ox plants displayed GA-overdose phenotype. In the case of CmGA20ox1, over-expression of Cucurbita maxima GA 20-oxidase in Arabidopsis resulted in accumulation of inactive GA25 and GA17, and reduction of GA4 content, which caused a slight decrease in stem elongation [25, 27]. Accordingly, GAs accumulation and changed expression levels of GA-regulated transcripts confirmed that HvGA20ox2 should be involved in regulation of GAs production. In barley semi-dwarf mutant Riso no. 9265, lose of HvGA20ox2 caused its GA deficiency and reduced plant height.
It was reported that GA promoted stem elongation and was found in the young tissues [22, 23]. HvGA20ox2 is highly expressed in stem and possiblely has effect on internode length [15]. Once jointing stage starts, the stems are young and internode region begins to elongate and grow rapidly. Therefore, the transcriptional levels of GA biosynthetic and catabolic genes were detected at the initial jointing stage. As a result, we found that GA biosynthesis genes (HvGA20ox1 and HvGA3ox2) were up-regulated and GA catabolic gene (HvGA2ox3) was down-regulated in Riso no. 9265 compared with Bomi, suggesting a type of feedback regulation from the low bioactive GAs because of the null mutation of HvGA20ox2. Feedback regulation of GA20ox and GA3ox and feed-forward regulation of GA2ox genes expression have been also shown in GA-deficient Arabidopsis mutants [27–29]. In rice sd1, the expression of OsGA20ox1 was increased in stem to compensate the sd1 effect [5, 30]. As a result, the increased expression of HvGA20ox1 could compensate the effect of HvGA20ox2 in the stem at least partially, and the feedback or feed-forward regulation of the GA dioxygenase helps maintaining a relative stable endogenous GA level. Consequently, Riso no. 9265 exhibited the observed plant height.
As mentioned above, we have characterized the barley semi-dwarf gene, sdw1/denso, and conclude that it encodes a GA biosynthetic enzyme, GA20ox, on the basis of the following results. Firstly, sdw1/denso mutant responds to exogenous GA3 [15, 18]. Secondly, the fourth sdw1 allele is a complete lose of HvGA20ox2 activity due to the deletion of HvGA20ox2 gene in Riso no. 9265 (Fig. 1). In contrast, denso allele and sdw1 of Jotun shows different expression of HvGA20ox2 [15]. Thirdly, the transgenic wild type Arabidopsis and semi-dwarf mutant ga5-3 by HvGA20ox2 gene showed GA over-dose phenotypes. Finally, GA-regulated transcripts were changed in Riso no. 9265 in the same way as those of some GA20oxs in rice and Arabidopsis [5, 23, 26, 30].
It was reported that sdw1/denso was associated with later heading [11, 31]. Upon first mapped denso to the long arm of chromosome 3H, Barua et al. [19] found a quantitative trait locus for heading date, which could not be genetically separated from the denso locus. Previously identified QTLs of heading date was also located on the region around denso in the Blenheim × E224/3 DH population [14]. Furthermore, it was found that sdw1 delayed maturity by around 3d based on eight populations [10]. We also found the QTL of development score was co-located with the HvGA20ox2 eQTL on 3HL [15]. However, it is still difficult to distinguish if it is pleiotropy of sdw1/denso or a tight linkage between the gene and one controlling later heading. In the present study, over-expression of HvGA20ox2 caused early flowering in Arabidopsis. The same is true for AtGA20oxs over-expressors [22, 23]. Both AtGA20ox1 and AtGA20ox2 act redundantly and affect many developmental processes, with AtGA20ox1 making the great contribution to internode and filament elongation, and AtGA20ox2 making the great contribution to flowering time and silique length in Arabidopsis [29]. It was also revealed that either AtGA20ox1 or AtGA20ox2 mutation delayed flowering under short-day condition,while only AtGA20ox2 delayed flowering slightly under long-day condition. Thus it can be assumed that the deficiency of AtGA20ox1 or AtGA20ox2 together with low GA affects flowering time, because GA acts a particularly important developmental switch between vegetative and reproductive development [32]. In the same way, it might be the deletion of HvGA20ox2 that causes later heading due to low GA concentration in Riso no. 9265. The hypothesis could be proved by the following facts. Firstly, GAs is involved in many developmental processes, including germination, stem extension, flowering and fruit set [22, 23, 29, 32]. Secondly, HvGA20ox2 and Mloc_56463 are absent in Riso no. 9265, unlike its wild type, while it is HvGA20ox2 that has gibberellin 20-oxidase activity and acts as a major determinant for GA production. Thirdly, GA deficiency was proved in sdw1/denso using GA sensitive experiment [15, 18]. Lastly, Riso no. 9265 mutant exhibited a late heading date. Similar results were found that semi-dwarf progenies with sdw1/denso matured generally later than their tall counterparts [10, 11, 31]. Considering all of these, it can be concluded that late heading in sdw1/denso could be the pleiotropy of HvGA20ox2.
Conclusions
The current study showed a complete deletion of over 21 kb DNA fragment including both HvGA20ox2 and Mloc_56463 genes in the mutant Riso no. 9265. HvGA20ox2 acts as GA 20-oxidase and is involved in gibberellin biosynthesis. The expression of the genes encoding GA biosynthesis (HvGA20ox1 and HvGA3ox2) are up-regulated and the expression of GA catabolic gene HvGA2ox3 is down-regulated in Riso no. 9265 in comparison with those in wild type Bomi, respectively. We conclude that sdw1/denso encodes one of the GA biosynthetic enzymes, GA 20-oxidase and the deletion of HvGA20ox2 as well as the compensatory of HvGA20ox1 and feedback regulation of gibberellin results in semi-dwarf phenotype of Riso no. 9265. We also deduced that sdw1/denso allele itself evoked later heading due to its reduced endogenous GAs concentration.
Methods
Plant materials and sampling
Semi-dwarf mutant Riso no. 9265, kindly provided by Dr. Chengdao Li of Murdoch University, Australia and its wild parent Bomi, kindly provided by Dr. Jing Zhang of Chinese Academy of Agricultural Sciences, China were used in this study. Both genotypes were planted in 2 × 6 m plots. The heading date was recorded as the number of days after sowed when 50 % of the spike emerged from the sheath. At maturity, 20 plants of each genotype were harvested at random for measurement of plant height, spike and internode length. Meanwhile, main stems of each genotype were taken at the initial jointing stage for measuring the expression levels of gibberellin biosynthetic and catabolic genes.
DNA isolation, PCR amplification and sequencing
Seedlings at tillering stage were sampled and DNA was extracted according to Murray and Thompson [33] with small modification. DNA samples were quantified using a Thermo Unicam UV300 and adjusted to a final concentration of 50 ng/μl for PCR analysis. PCR was performed on a Veriti 96 well thermocycler (Applied Biosystems). Sequencing of HvGA20ox2 isolated from barley was conducted using two-pair primers: (1) forward primer (sdwF10) 5’ CTAGCTCACACACCTCTCATCTCAT 3’, and reverse primer (sdwR10) 5’ GTTCCCGACAAAAATTCCGTGT 3’; (2) forward primer (sdwF9) 5’ CTCTCCCGCACACTCACTCGCAAC 3’, and reverse primer (sdwR13) 5’ GCGGTGAGGGGGCATGCATAT 3’. PCR reaction was comprised of 50 ng of template DNA, 0.3 μM of each primer, 1 × PCR buffer, 0.4 mm dNTP and 0.2 U PCR enzyme (KOD FX, TOYOBO) in a final volume of 10 μl. PCR cycling conditions consisted of an initial denaturation step of 94 °C for 3 min, followed by 30-35 cycles of 98 °C for 10 s, 60 °C for 30s, and 68 °C for 3 min.
Primers of Morex_contig_40861 and neighbor genes of HvGA20ox2 were listed in Additional file 1: Table S1. PCR reaction was comprised of 1 × Taq Mix (Bio life), 0.3 μM of each primer and 50 ng of template DNA. The following PCR amplification profile was used : denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 0.5-2 min depending on the size of amplilcons, and a final extension at 72 °C for 5 min. The amplification products were run in 1 % agrose gels and sequenced by Biosune Biotechnology Co. Ltd.
Plasmid construction and Arabidopsis transformation
For plasmid construction, full-length CDS of HvGA20ox2 was amplified from Bomi by PCR. The primers were designed as 5’ AGTACTCGAGCTCACACACCTCTCATCTCAT 3’ and 5’CTATGGATCCGAATCAGCCCGTGGAT 3’ with XhoI and BamH I sites, respectively. The amplified PCR product was confirmed by sequencing and cloned into T4 vector for further manipulation. Insert was confirmed by sequencing. The CDS paragraph of HvGA20ox2 digested with XhoI/ BamH I was cloned into the XhoI/ BamH Isite located between the 35S CaMV promoter and ocs terminator of the pFGC5941 binary vector, named as pFGC5941- HvGA20ox2. The binary vector was transformed into Agrobacterium tumefaciens by electroporation, and the floral dip method was used to transform Col-gl1 (Columbia ecotype, different from Col-o only in its glabrous leaf. Hereafter refer to wild type) and a semi-dwarf mutant ga5-3 (a T-DNA inserted mutant of AtGA20ox1 in Col-o, Salk016701) [34]. T1 transgenic plants were screened by spraying bialaphos solution twice at an interval of three days at cotyledon stage, and confirmed by PCR using following specific primers: Forward, 5’ GGAGCATCGTGGAAAAAGAAGA 3’ (from CaMV 35S promoter sequence) and Reverse, 5’ GGAGTCGCAGGGCTGGTGTCC 3 ’(from HvGA20ox2). The constructs were also verified by sequencing. For analysis of HvGA20ox2 gene expression level in transgenic plants, 7-day-old Col-gl1, ga5-3 and T3 transgenic seedlings were harvested for RNA extraction.
Growth conditions and phenotypic characterization of Arabidopsis
The seeds of Arabidopsis were stratified at 4 °C for 1-2 d and sown on soil at 24 °C under LDs (16 h of light). Plants for hypocotyl and root length measurements were grown vertically on one half times Murashige and Skoog media (Sigma, M6899) containing 1 % MES, 3 % sucrose and 0.46 % Gelrite with pH 5.8 under LDs, being measured after 7 d. Flowering time was scored as the days when buds could be detected with naked eyes. Other measurements were performed on the plants that had stopped flowering.
Real-time quantitative RT-PCR
RNA was extracted from the stems of Bomi and Riso no. 9265, as well as 7-day-old seedlings of T3 transgenic lines, Col-gl1 and ga5-3 using Spin Column Plant total RNA Purification Kit(Sanggon Biotech (Shanghai) Co., Ltd). cDNA was prepared from 1 μg RNA using AMV First Strand cDNA Synthesis Kit(Sanggon Biotech (Shanghai) Co., Ltd). qPCR reactions were performed using SYBR Green (SG Fast qPCR Master Mix(High Rox), BBI ) and the Applied Biosystems Stepone plus Real-time PCR System. The Real-time PCR assays were performed in triplicate for each cDNA sample. For determining transcription levels of barley GA20ox2 and genes encoding final biosynthesis of GA, HvACTIN and HvGAPDH for barley, and At1g13320 and At4g26410 for Arabidopsis were employed as reference genes [5, 35, 36]. Additional file 2: Table S2 listed the oligonuleotide sequences used for quantitative RT-PCR.
ELISA assay of gibberellin (GA1+3 and GA4+7)
Approximately 0.5 g fresh Arabidopsis tissue (7-day-old seedling) was homogenized in liquid nitrogen and extracted at 4 °C in cold 80 % (v/v) methanol containing 1 mM butylated hydroxytoluene for 4 h. The extracts were collected after centrifugation at 8000 g at 4 °C for 20 min. The residues were suspended in the same extraction solution and stored at 4 °C for 1 h, and then centrifuged again at 8000 g at 4 °C for 20 min. The two resulting supernatants were combined and passed through a C18 Sep-Pak cartridge (Waters, Milford, MA, USA). The efflux was collected and dried in N2. The residues was then dissolved in 0.01 M phosphate buffer solution (pH 7.5) and concentrations of GA1+3 and GA4+7 were determined in an indrect enzyme-linked immuno sorbent assay (ELISA) using GA3 and GA4 antibody, respectively. The ELISA KIT was obtained from Professor Baomin Wang (Chinese Agricultural University) and the methods were described in previous publications [37, 38].
Statistical analyses
Phenotypic differences of Bomi and Riso no. 9265 were tested by Student’s t test. In order to avoid heterogeneity of variance, a natural log transformation was applied to hypocotyl length,and a square transformation was applied to flowering time (days), plant height, root and internode length. Least significant difference (LSD) at 5 % probability was used to assess the difference between genotypes. For statistical analysis of qPCR data, cycle threshold (CT) values were used to determine Δ CT values (Δ CT = CTtarget –CTreference), and expression levels of target genes relative to reference gene were determined as 2-Δ CT. For comparison of GA concentration and the expression levels of GA-regulated transcripts between transgenic and wild type plants, ANOVA was used. Analysis of qRT-PCR efficiency showed that all amplicons of the genes used in this study were within the optimal range of 98.7-102.4 %.
Ethics Statement
In this study we only used plant material including barley and Arabidopsis. Based on the rule of BMC Genomics, no ethics statement was required for the collection of genetic material.
Acknowledgements
We thank Dr Xuelong Wu (Institute of Virology and Biotechnology, Zhejiang Academy of Agricultural Sciences, China) for technical assistance in plasmid constructs and Arabidopsis transformation. This work was supported by National Natural Science Foundation of China (Grant No. 31471495), Science and Technology Innovation Team Project of Zhejiang Province (2011R50026-9), China Agriculture Research System (CARS-05) and Young talent project of Zhejiang Academy of Agricultural Sciences.
Additional files
Table S1. Primers used to detect flanking genes or sequences of HvGA20ox 2. (XLS 18 kb)
Table S2. Oligonucleotide sequences used in qRT-PCR assays. (XLS 17 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
QJ, CL, JY and GZ made the experimental design. QJ and YS performed to identify the deleted sequences and qRT-PCR GA related genes. QJ and JZ were involved in phenotyping Bomi and Riso no. 9265. QJ and WH developed the transgenic Arabidopsis and characterized phenotype of Arabidopsis. QJ, JZ and JW conducted statistical analysis of the expression data and phenotypic data. QJ, CL and GZ wrote the manuscript, and all authors read and approved the final version.
Contributor Information
Jianming Yang, Email: jmyang@163.com.
Guoping Zhang, Phone: +86 571 88982115, Email: zhanggp@zju.edu.cn.
References
- 1.Zapata TC, Silva CP, Acevedo HE. Grain yield and assimilate partitioning in wheat isogenic plant height lines. Agric Tec. 2004;64(2):139–155. [Google Scholar]
- 2.Khush GS. Green revolution: preparing for the 21st century. Genome. 1999;42(4):646–655. doi: 10.1139/g99-044. [DOI] [PubMed] [Google Scholar]
- 3.Ashikari M, Sasaki A, Ueguchi-Tanaka M, Itoh H, Nishimura A, Datta S, et al. Loss-of-function of a rice gibberellin biosynthetic gene, GA20 oxidase (GA20ox-2), led to the rice ‘green revolution’. Breed Sci. 2002;52:143–150. doi: 10.1270/jsbbs.52.143. [DOI] [Google Scholar]
- 4.Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, et al. Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 2002;9:11–17. doi: 10.1093/dnares/9.1.11. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, et al. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 6.Spielmeyer W, Ellis MH, Chandler PM. Semidwarf (sd-1), green revolution rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci U S A. 2002;99:9043–9048. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuczyńska A, Surma M, Adamski T, Mikołajczak K, Krystkowiak K, Ogrodowicz P. Effects of the semi-dwarfing sdw1/denso gene in barley. J Appl Genetics. 2013;54:381–390. doi: 10.1007/s13353-013-0165-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia Q, ZHang J, Westcott S, Zhang X, Bellgard M, Lance R, et al. GA-20 oxidase as a candidate for the semidwarf gene sdw1/denso in barley. Funct Integr Genomics. 2009;9:255–262. doi: 10.1007/s10142-009-0120-4. [DOI] [PubMed] [Google Scholar]
- 9.Barboza L, Effgen S, Alonso-Blanco C, Kooke R, Keurentjes JJ, Koornneef M, et al. Arabidopsis semidwarfs evolved from independent mutations in GA20ox1, ortholog to green revolution dwarf alleles in rice and barley. Proc Natl Acad Sci U S A. 2013;110(39):15818–15823. doi: 10.1073/pnas.1314979110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellewell KB, Rasmusson DC, Gallo-Meagher M. Enhancing yield of semidwarf barley. Crop Sci. 2000;40:352–358. doi: 10.2135/cropsci2000.402352x. [DOI] [Google Scholar]
- 11.Thomas WTB, Powell W, Swanston JS. The effects of major genes on quantitatively varying characters in barley. 4. The GPert and denso loci and quality characters. Heredity. 1991;66:381–389. doi: 10.1038/hdy.1991.48. [DOI] [Google Scholar]
- 12.Mickelson HR, Rasmusson DC. Genes for short stature in barley. Crop Sci. 1994;34:1180–1183. doi: 10.2135/cropsci1994.0011183X003400050007x. [DOI] [Google Scholar]
- 13.Thomas WTB, Powell W, Waugh R, Chalmers KJ, Barua UM, Jack P, et al. Detection of quantitative trait loci for agronomic, yield, grain and disease characters in spring barley (Hordeum vulgare L.) Theor Appl Genet. 1995;91:1037–1047. doi: 10.1007/BF00223917. [DOI] [PubMed] [Google Scholar]
- 14.Powell W, Thomas WTB, Baird E, Lawrence P, Booth A, Harrower B, et al. Analysis of quantitative traits in barley by the use of amplified fragment length polymorphisms. Heredity. 1997;79:48–59. doi: 10.1038/hdy.1997.122. [DOI] [Google Scholar]
- 15.Jia Q, Zhang XQ, Westcott S, Broughton S. CakirM, Yang J, et al: Expression level of a gibberellin 20-oxidase gene is associated with multiple agronomic and quality traits in barley. Theor Appl Genet. 2011;122:1451–1460. doi: 10.1007/s00122-011-1544-5. [DOI] [PubMed] [Google Scholar]
- 16.Grausgruber H, Bointner H, Tumpold R, Ruckenbauer P. Genetic improvement of agronomic and qualitative traits of spring barley. Plant Breed. 2002;121:411–416. doi: 10.1046/j.1439-0523.2002.756385.x. [DOI] [Google Scholar]
- 17.Haahr V, von Wettstein D. Studies of induced, high yielding dwarf-mutant of spring barley. In: Gaul H, editor. Barley Genetics III.: Proceedings of the 3rd International Barley Genetics Symposium. München: Verlag Karl Thiemig; 1976. pp. 215–218. [Google Scholar]
- 18.Franckowiak JD, Pecio A. Coordinator’s report: a listing of genetic stocks. Barley Genet Newsl. 1992;21:116–126. [Google Scholar]
- 19.Barua UM, Chalmers KJ, Thomas WTB, Hackett CA, Lea V, Jack P, et al. Molecular mapping of genes determining height, time to heading, and growth habit in barley (Hordeum vulgare ) Genome. 1993;36:1080–1087. doi: 10.1139/g93-143. [DOI] [PubMed] [Google Scholar]
- 20.Mayer K, Waugh R, Brown J, Schulman A, Langridge P, Platzer M, et al. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- 21.Hedden P, Phillips AL. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/S1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- 22.Huang S, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown SM. Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol. 1998;118(3):773–781. doi: 10.1104/pp.118.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coles JP, Phillips AL, Croker SJ, García-Lepe R, Lewis MJ, Hedden P. Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J. 1999;17(5):547–556. doi: 10.1046/j.1365-313X.1999.00410.x. [DOI] [PubMed] [Google Scholar]
- 24.Talon M, Koornneef M, Zeevaart JAD. Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc Natl Acad Sci U S A. 1990;87:7983–7987. doi: 10.1073/pnas.87.20.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radi A, Lamge T, Niki T, Koshioka M, Lange MJP. Ectopic expression of pumpkin gibberellin oxidases alters gibberellin biosynthesis and development of transgenic Arabidopsis plants. Plant Physiol. 2006;140(2):528–536. doi: 10.1104/pp.105.073668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrera E, Bou J, García-Martínez JL, Prat S. Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant J. 2000;22(3):247–256. doi: 10.1046/j.1365-313x.2000.00736.x. [DOI] [PubMed] [Google Scholar]
- 27.Xu YL, Li L, Gage DA, Zeevaart JA. Feedback regulation of GA5 expression and metabolic engineering of gibberellin levels in Arabidopsis. Plant Cell. 1999;11(5):927–936. doi: 10.1105/tpc.11.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips AL, Ward DA, Uknes S, Appleford NE, Lange T, Huttly AK, et al. Isolation and expression of three gibberellin 20-Oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, Gong F, et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008;53:488–504. doi: 10.1111/j.1365-313X.2007.03356.x. [DOI] [PubMed] [Google Scholar]
- 30.Asano K, Takashi T, Miura K, Qian Q, Kitano H, Matsuoka M, et al. Genetic and molecular analysis of utility of sd1 alleles in rice breeding. Breed Sci. 2007;57:53–58. doi: 10.1270/jsbbs.57.53. [DOI] [Google Scholar]
- 31.Powell W, Caligari PDS, Thomas WTB, Jinks JL. The effects of major genes on quantitatively varying characters in barley. 2. The denso and daylength response loci. Heredity. 1985;54:349–352. doi: 10.1038/hdy.1985.47. [DOI] [Google Scholar]
- 32.Eriksson S, Bohlenius H, Moritz T, Nilsson O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell. 2006;18:2172–2181. doi: 10.1105/tpc.106.042317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucl Acids Res. 1980;8(19):4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clough JP, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 35.Boden SA, Weiss D, Ross JJ, Davies NW, Trevaskis B, Chandler PM, et al. EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell. 2014;26:1557–1569. doi: 10.1105/tpc.114.123794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan Y, Liu Y, Wu C, Chen S, Wang Z, Yang Z, et al. Water deficit affected flavonoid accumulation by regulating hormone metabolism in scutellaria baicalensis georgi roots. Plos One. 2012;7(10):e42946. doi: 10.1371/journal.pone.0042946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia YX, Tao FQ, Li WQ. Analysis of endogenous hormone levels to reveal the retardation by suppression of phospholipase Dα1 in Arabidopsis leaves during hormone-promoted senescence. Plant Diversity Res. 2014;36(4):485–496. [Google Scholar]


