Abstract
Background
Penicillium chrysogenum is a filamentous fungus that is employed as an industrial producer of β–lactams. The high β–lactam titers of current strains is the result of a classical strain improvement program (CSI) starting with a wild-type like strain more than six decades ago. This involved extensive mutagenesis and strain selection for improved β–lactam titers and growth characteristics. However, the impact of the CSI on the secondary metabolism in general remains unknown.
Results
To examine the impact of CSI on secondary metabolism, a comparative genomic analysis of β-lactam producing strains was carried out by genome sequencing of three P. chrysogenum strains that are part of a lineage of the CSI, i.e., strains NRRL1951, Wisconsin 54-1255, DS17690, and the derived penicillin biosynthesis cluster free strain DS68530. CSI has resulted in a wide spread of mutations, that statistically did not result in an over- or underrepresentation of specific gene classes. However, in this set of mutations, 8 out of 31 secondary metabolite genes (20 polyketide synthases and 11 non-ribosomal peptide synthetases) were targeted with a corresponding and progressive loss in the production of a range of secondary metabolites unrelated to β–lactam production. Additionally, key Velvet complex proteins (LeaA and VelA) involved in global regulation of secondary metabolism have been repeatedly targeted for mutagenesis during CSI. Using comparative metabolic profiling, the polyketide synthetase gene cluster was identified that is responsible for sorbicillinoid biosynthesis, a group of yellow-colored metabolites that are abundantly produced by early production strains of P. chrysogenum.
Conclusions
The classical industrial strain improvement of P. chrysogenum has had a broad mutagenic impact on metabolism and has resulted in silencing of specific secondary metabolite genes with the concomitant diversion of metabolism towards the production of β–lactams.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-2154-4) contains supplementary material, which is available to authorized users.
Keywords: Penicillium chrysogenum, Secondary metabolism, Sorbicillinoids, Polyketide synthases, Sequencing
Background
For the industrial production of β–lactams, the filamentous fungus Penicillium chrysogenum has been subjected to an intense classical strain improvement (CSI) program that consisted of repeating rounds of mutagenesis and selection. This program started in 1943 with a natural isolated strain NRRL1951 derived from an infected cantaloupe because of its high levels of β–lactam production under submerged fermentation conditions. The spontaneous variant NRRL1951 B25 showed almost three times the production levels of P. notatum strain NRRL 836 - the first industrial candidate adopted for the tank fermentation method of β–lactam production during the forties of last century. To achieve production of increasing levels of β–lactams and to ease the purification of the product, a comprehensive mutagenesis was applied to the NRRL1951 B25 strain [1]. This included harsh mutagenic techniques like UV irradiation, X-ray, and nitrogen mustard (methyl-bis(β-chloroethyl) amine) treatment as well as selection of the desired spontaneous variants for production and fermentation characteristics. The confidentiality of the industrial CSI programmes prohibits a comprehensive view on the exact mutagenesis steps during the generation of improved penicillin producing strains. Only few intermediate strains can be mapped on the CSI chart with the addressed method of treatment (Fig. 1). However, a major breakthrough in the CSI of NRRL1951 was the selection of the so-called Wisconsin series of improved strains that were derived from the X-1612 (implying X-ray treatment) mutant that was further evolved into the current international laboratory standard strain Wisconsin 54-1255 whose genome sequence became available in 2008 [2]. Currently, Wisconsin 54-1255 is the reference strain for genome, proteome and transcriptome comparative studies [3–7]. The genetic studies and genome sequencing resulted in an understanding of the mechanisms underlying the enhanced β-lactam production, which included the amplification of the penicillin biosynthesis gene cluster [8], an enhanced amino acid production and various aspects of cellular development [2].
Fig. 1.
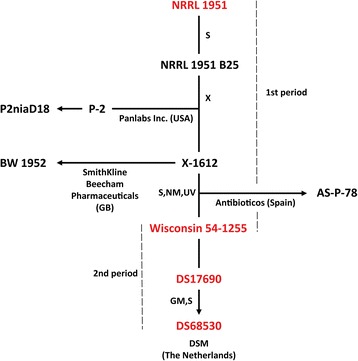
Schematic representation of main industrial β-lactam production improvement programs of P. chrysogenum. The main lineages of strains subjected to industrial β-lactam production classical strain improvement programs are shown. Strains NRRL1951 (4 mg/gDW), Wisconsin 54-1255 (20 mg/gDW), and DS17690 (95 mg/gDW) examined in this study are shown in red. In brackets the production levels of penicillin V are indicated according to reference [31]. The penicillin cluster free strain DS68530 that no longer produces penicillins is not included but it is derived from strain DS17690 by genetically removing the multiple penicillin biosynthetic gene clusters [28]. When reported, the techniques used for mutagenesis are indicated according to Newbert et al. [8]. Used abbreviations: S, selection of the spontaneous variant; X, X-Ray irradiation; UV, ultraviolet irradiation; NM, nitrogen mustard (methyl-bis(β-chloroethyl) amine) treatment; MG, other molecular genetic manipulations. Also indicated are the periods 1 and 2 followed in this study in order to identify major steps in the CSI
Unlike β-lactam biosynthesis, little is known about the consequences of CSI on the secondary metabolism in general. The genome sequence of Wisconsin 54-1255 revealed that in addition to β-lactams, the genome encodes a range of other secondary metabolite gene clusters that have been characterized only to a limited extent. More recently, advances in metabolome techniques systematically revealed a diversity of the secondary metabolites that can be produced by industrial variants of P. chrysogenum. For more than 30 of these compounds the genes involved in biosynthesis have been identified and partially characterized. This includes the non-ribosomal peptide synthetases involved in the production of toxins such as roquefortine and meleagrine [9–11], the pigment chrysogine (Ali, unpublished data), siderophores (Samol, unpublished data) and cyclic hydrophobic tetrapeptides [12]. However, this analysis also revealed that during CSI the diversity of the secondary metabolism by improved β-lactam producers has been significantly narrowed. For instance, elimination of the production of yellow pigments like sorbicillin and penitrinic acid, that were contaminants during antibiotic purification, was already achieved at the early stage of CSI [13]. However, the genetic origin of the production of these pigments and their bioactive properties has remained unstudied for long. In recent years many sorbicillin related bioactive compounds were re-identified in P. notatum [14], Trichoderma [15], and other fungal species isolated from different environments [16]. The broad spectra of activities of sorbicillins range from radical scavenging to cytotoxicity against L5178y leukemic cells, while the actual function and genetic basis for the production of these compounds in P. chrysogenum and other filamentous fungi are mostly unknown.
Here we performed a systematic genomic, expression and extracellular metabolome analysis to investigate the impact of CSI on secondary metabolism of P. chrysogenum. We demonstrate that CSI resulted in the silencing of many secondary metabolite pathways, whereas others remained unaffected. In addition, we identified the polyketide synthases responsible for the production of a vast group of pigments termed sorbicillinoids that were mutated and transcriptionally silenced at the early stages of the CSI programme.
Results
Mutational spread in the P. chrysogenum genome during classical strain improvement
To examine the mutational impact of the classical strain improvement (CSI) program on the genome of P. chrysogenum, we sequenced the genomes of four P. chrysogenum strains. These are the early β-lactam producing strain P. chrysogenum NRRL1951, which acts as a reference and two strains in the lineage that signify two different periods of mutagenesis. Here, we define period 1 as the early stage of CSI, using the Wisconsin 54-1255 strain as the hall mark. During this period CSI has had a major impact on the fermentation characteristics and pigment formation. This strain is the current international reference as its genome was sequenced. The second period entails strain DS17690 that has undergone major mutagenesis which amongst others has led to the amplification of the penicillin biosynthetic gene cluster. It is currently studied as a type strain. In addition, the penicillin biosynthesis cluster free strain DS68530 was sequenced that was derived from the latter strain. The absence of the multiple penicillin clusters facilitates the analysis of β-lactam unrelated secondary metabolites present in the extracellular metabolome (Fig. 1). The genomes of the four strains were sequenced using the Illumina HiSeq 2500 platform with near to complete coverage (99.70–99.91 %) (Table 1). Based on a comparison of the respective genomes, 455 SNPs were detected between the NRRL1951 and Wisconsin 54-1255 strain that accumulated during the first period of CSI. Within this set, 215 mutations occurred within coding regions, while 151 non-synonymus, 55 synonymus, 2 termination mutations, 6 frame shifts and one nonsense mutation were identified. During the second period of the CSI the multicopy strain DS17690 additionally obtained 2056 SNPs. This included 1187 non-coding, and 869 mutations within coding regions including 271 synonymus and 558 non-synonymus mutations, three SNPs at the terminal region, 13 frame shifts and 24 nonsense mutations (Table 2, Additional file 1: Table S1).
Table 1.
Quality statistics of the genome sequencing of P. chrysogenum strains within a CSI lineage
| Strain | Genome template size (bp) | Number of reads | Sample Yield (in MB) | Average Quality scores (Phred) | Insert size (bp) | Median coverage | Zero template coverage (bp) | Template coverage (%) | Mapped reads (%) |
|---|---|---|---|---|---|---|---|---|---|
| NRRL1951 | 32228535 | 38,686,435 | 11,682 | 35.20 | 285 | 350 | 28258 | 99,91 | 95,44 |
| Wisconsin 54-1255 | 32228535 | 34,422,618 | 10,394 | 35.20 | 288 | 313 | 27931 | 99,91 | 95,78 |
| DS17690 | 32228535 | 34,152,533 | 10,314 | 35.27 | 292 | 306 | 87244 | 99,73 | 95,61 |
| DS68530 | 32228535 | 49,691,784 | 15,006 | 35.19 | 302 | 340 | 95683 | 99,70 | 95,35 |
Table 2.
Mutational impact of CSI on a P. chrysogenum strain lineage
| Strain | Non-codinga | Codinga | ||||||
|---|---|---|---|---|---|---|---|---|
| Reference | Total | Synonymus | Non-synonymus | Termination mutation | Frameshift | Nonsense | ||
| Wisconsin 54-1244 | [2] | 455 | 240 | 55 | 151 | 2 | 6 | 1 |
| DS17690 | [28] | 2056 | 1187 | 271 | 558 | 3 | 13 | 24 |
| DS68530 | DSM | 18 | 3 | 7 | 8 | 0 | 0 | 0 |
aMutations mapped relative to strain NRRL1951
The use of the advanced next generation sequencing technique to the reference strain Wisconsin 54-1255 resulted in the identification of mismatches with the previously published sequence of this strain [2]. We identified 321 SNPs including 244 non-coding, 10 synonymus and 67 non-synonymus polymorphisms. This included 40 frame shifts, one termination and one nonsense mutation. These newly identified features are the result of the improved quality of the sequencing and correspondingly appeared in the genome assemblies of NRRL1951, DS17690 and DS68530 strain. To simplify the following comparative analysis these SNPs were subtracted from the mutational profiles of the subjected strains. An analysis using GASVPro [17] did not reveal any structural variation between the strains.
In order to assess the distribution of SNPs across genes involved in different cellular processes, we performed a functional classification of the entire gene set of the P. chrysogenum Wisconsin 54-1255 reference genome. In order to classify a maximal proportion of genes, we overlapped functional classifications from the COG, KOG and KEGG BRITE databases [18–20] into a single consensus prediction. The complete sets of identified SNPs between NRRL 1951 and Wisconsin 5412-55 and between Wisconsin 54-1255 and DS17690 were mapped to these functional categories. Subsequently, we performed a statistical comparison of the functional distribution of genes affected by the SNPs and the overall functional distribution of all genes in the genome, based on the hypergeometric distribution. Surprisingly, after Bonferroni multiple-testing correction, there was no statistically significant overrepresentation or underrepresentation of mutations in secondary metabolism, amino acid metabolism, transcriptional regulation, or any other functional category (Additional file 2: Table S2). Hence, strain selection seems to have had only very limited control over the mutations accumulated during the CSI, and it is likely that only a minority of the changes introduced by the mutagenesis procedure are responsible for the phenotypic improvements of the strains. The next key challenge was therefore to identify this important minority.
Distribution of mutations in secondary metabolism gene clusters
An inventory was made of mutations that were accumulated in key secondary metabolism genes during the CSI using strain NRRL1951 as a reference. The genome contains 11 NRPS and 20 PKS genes likely involved in secondary metabolite formation. In total, 9 putative secondary metabolite gene clusters accumulated mutational changes during both periods of the CSI (Fig. 2a). This affected 1 NRPS gene and 7 PKS genes. The mutations were all verified by DNA sequencing of the respective gene clusters.
Fig. 2.
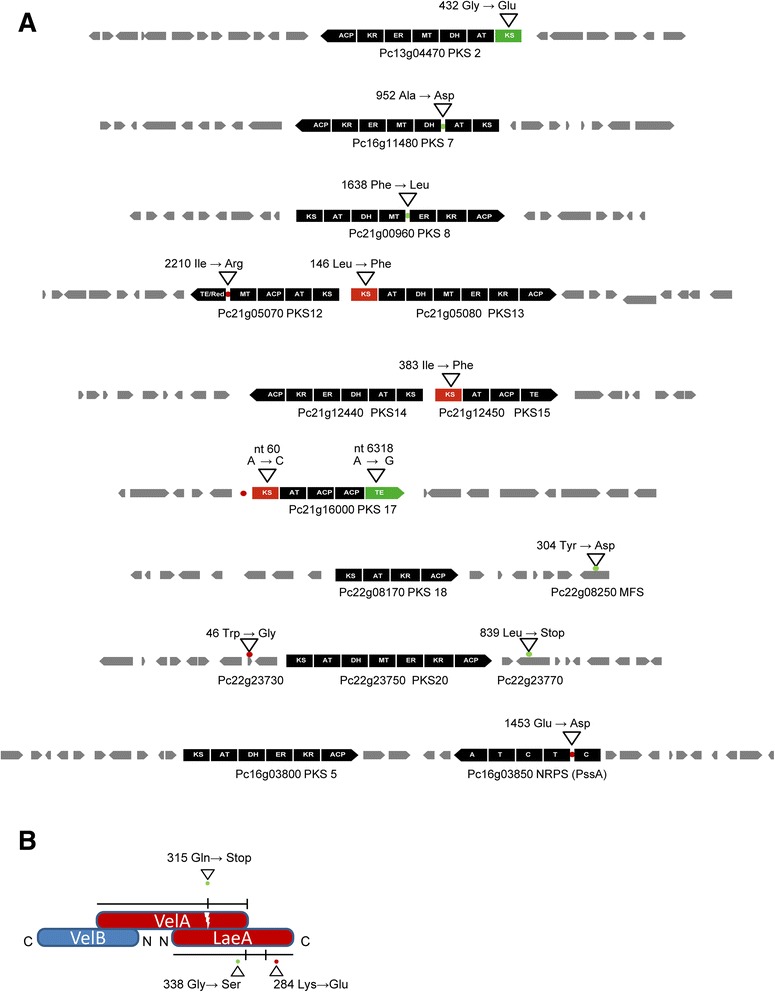
Mutational impact of the CSI program on secondary metabolite gene clusters of P. chrysogenum. a Mutations acquired by Wisconsin 54-1255 in period 1 of the CSI programme are colored in red. The mutagenic impact in period 2 is shown in green. Mutated PKS and NRPS domains are indicated. The grey arrows represent other genes of the putative gene clusters. Abbreviations used for PKS and NRPS domains: KS, keto-synthase; AT, acyltransferase; DH, dehydratase; MT, methyltransferase; ER, enoyl reductase; KR, ketoreductase; ACP, acyl carrier protein; TE, thioesterase; TE/Red, thioester reductase; A, adenylation; T, thiolation; C, condensation domain. b Schematic representation of the Velvet complex proteins VelA, VelB and LaeA with the mutations obtained during the CSI
The partially reducing (PR) PKS2 (Pc13g04470) gene cluster is part of predicted cluster of nineteen ORFs [21]. PKS2 shows 87 % identity to an unknown PKS of P. digitatum and strong similarity (46 % identity) to the lovastatin diketide synthase LovF of A. terreus. Two SNPs occurred at the second period of CSI and resulted in a glycine to glutamic acid substitution at position 432 within the ketosynthase domain of PKS2 that likely has inactivated the enzyme. The gene cluster of PR PKS7 (Pc16g11480) is predicted to consist of 18 ORFs [21]. A homologous cluster of five genes is found in the genome of A. fumigatus, but has no assigned function. PKS7 carries the SNP resulting in an alanine to aspartic acid substitution at position 952 located at the acyl transferase/dehydratase inter-domain region. This mutation was acquired in the second CSI period and presumably does not affect the function of the neighboring domains. The highly reducing (HR) PKS8 (Pc21g00960) is homologous (54 % identity) to an uncharacterized PKS enzyme of A. flavus and belongs to a putative cluster of 17 genes [21]. A single mutation resulted in a phenylalanine to leucine substitution at position 1638 in the C-(Met)/dehydratase inter-domain region and occurred in the second period of CSI. A further gene cluster contains two PKSs encoding genes HR PKS14 (Pc21g12440) and non-reducing (NR) PKS15 (Pc21g12450) and consists of 18 ORFs [21]. A mutation within the ketosynthase domain of PKS15 occurred during the first CSI period and resulted in isoleucine to phenylalanine substitution at position 383. A NR PKS encoded by Pc21g16000 (PKS17) shows similarity to the A. nidulans wA and A. fumigatus PksP naphthopyrone synthases. These enzymes are involved in the biosynthesis of a precursor for the formation of a conidial pigment [2]. A similar gene cluster of 11 ORFs is present in the genomes of A. niger and A. kawachii. Two nucleotide substitutions were detected within PKS17 causing no effect on the amino acid sequence. A third mutation is located within the terminator region of the PKS gene and is represented by an A to C nucleotide substitution. The actual impact of these mutations on pigment formation is unclear as the green coloring of the conidia was not affected during the CSI.
The gene cluster of PR PKS18 (Pc22g08170) shows 82 % identity to the 6-methylsalicylic acid synthase from P. griseofulvum and contains 16 ORFs. The P. griseofulvum homolog is involved in patulin biosynthesis and requires additional enzymatic conversions like decarboxylation, two hydroxylation and oxidation reactions [22]. In P. chrysogenum these enzymatic steps are presumably provided by the aminohydrolase and two cytochrome P450 enzymes encoded within the cluster. However, the essential gene encoding the isoepoxidon dehydrogenase cannot be identified in the genome of P. chrysogenum supporting the notion that this fungus does not produce patulin [2]. A single mutation was obtained during the second period of CSI and this affected a putative MFS transporter (Pc22g08250) encoding gene introducing a tyrosine to asparagine substitution at position 304. Finally, two neighboring, oppositely oriented NR PKS12 (Pc21g05070) and HR PKS13 (Pc21g05080) that belong to a gene cluster that potentially is related to sorbicillinoid biosynthesis, received SNPs in both coding sequences. PKS12 acquired an isoleucine to arginine substitution at C-(Met)/TE inter-domain region, while PKS13 obtained a mutation in the KS domain where leucine in position 146 is substituted by a phenylalanine. Both mutations were obtained at the first period of the CSI and thus were inherited already by strain Wisconsin 54-1255. The associated gene cluster is predicted to contain 18 ORFs [21].
The only NRPS gene that accumulated SNPs concerns PssA (Pc16g03850) that is involved in siderophore coprogen B biosynthesis under the iron depleted growth conditions. At the first CSI period the PssA encoding gene was mutated causing a glutamic acid for aspartic acid substitution at position 1453 within a non-conserved region. PssA is involved in the production of coprogen that still can be produced by the improved β-lactam producing strains (Samol, unpublished data). Therefore, this mutation apparently did not inactivate the enzyme.
Mutational changes within the Velvet complex
The Velvet complex is involved in the regulation of sexual development and secondary metabolism as demonstrated in various filamentous fungi including P. chrysogenum [3, 6, 7]. Since it is a multisubunit complex, we examined the impact of CSI on the core component of the Velvet complex. The P. chrysogenum orthologs LreA (Pc20g08380), LreB (Pc22g02540), FphA (Pc06g00040), KapA (Pc21g01970), VelB (Pc22g22320), and VosA (Pc22g06890) were all identical in the genomes of strains NRRL1951, Wisconsin 54-1255 and DS17690. However, mutational changes were found in Pc16g14010 and Pc13g13200 genes that encode LeaA and VelA, respectively. In the first period, the leaA gene acquired a nucleotide substitution at position 850 (T to C) which resulted in a substitution of lysine for glutamic acid at position of 284 of the protein. A second mutation occurred in period 2 and caused a nucleotide change at position 1012 (C to T) causing a glycine to serine substitution at position 338. In addition, a mutation was identified in the VelA encoding gene that occurred at the second CSI period. A C to T substitution at nucleotide position 943 caused a stop codon resulted in the formation of a truncation of the VelA protein from 562 to 315 amino acids long. These data indicate that the velvet complex has been a major target in the CSI, and this likely has impacted the expression of secondary metabolite encoding genes (Fig. 2b).
Expression of the secondary metabolism gene clusters
Since the velvet complex and many secondary metabolism genes acquired mutations during the CSI, the expression of the latter genes may have vastly changed along the lineage of strains. Therefore, we examined the expression of all 20 PKSs and 11 NRPS encoding genes in strains NRRL1951, Wisconsin 54-1255, DS17690 and the penicillin cluster free strain DS68530 (Fig. 3). Herein, the fungi were grown for 5 days and total RNA was isolated for qPCR analysis of the respective genes. The expression of γ-actin gene (Pc20g11630) was used as reference for normalization. The observed Ct values for the amplified γ-actin gene from the cDNA of NRRL1951, Wisconsin 54-1255, DS17690 and DS68530 were 19.9 ± 0.12; 20.11 ± 0.18; 20.48 ± 0.25 and 20.30 ± 0.18 respectively. This demonstrates nearly identical expression of the reference gene in the analyzed strains. The obtained expression profiles clearly indicates that the expression of the ACV synthetase, the key enzyme of β-lactam biosynthesis was dramatically enhanced upon CSI of Wisconsin 54-1255 as expected from the amplification of the β-lactam biosynthesis genes in strain DS17690 that contains 8 copies of this penicillin cluster. With the other NRPS and PKS genes, there were no marked expression changes except for three PKS genes. The two highly expressed PKS genes Pc21g05070 (PKS12) and Pc21g05080 (PKS13) were dramatically down regulated in Wisconsin 54-1255 and the derivatives DS17690 and DS68530. The NR PKS12 exhibits 43 % identity to citrinin polyketide synthase of Monascus purpureus while HR PKS13 is homologous to lovastatin diketide synthase LovF of A. terreus [2]. In addition, the cluster contains genes encoding a putative regulator (Pc21g05050), a monooxygenase (Pc21g05060), a transcriptional factor (Pc21g05090), a MFS transporter (Pc21g05100) and an oxidoreductase (Pc21g05110). All five genes of this putative cluster are also present in the genome of Trichoderma reesei and arranged in one locus. An identical gene cluster in the marine isolate of P. chrysogenum E01-10/3 has been previously associated with the biosynthesis of sorbicillactones A and B and thus by analogy, the aforementioned gene cluster may encode the biosynthesis pathway of yellow pigments sorbicillinoids in P. chrysogenum [23]. There is a significant decrease in expression of the seven genes of this cluster in the improved strains, whereas expression of flanking genes that are likely not part of the cluster was unaltered (Fig. 4). In addition, expression of PKS7 was increased substantially upon CSI of the NRLL1951 strain. Despite the significant overexpression of this PKS in the improved penicillin producing strain, the polyketide product has not yet been identified.
Fig. 3.
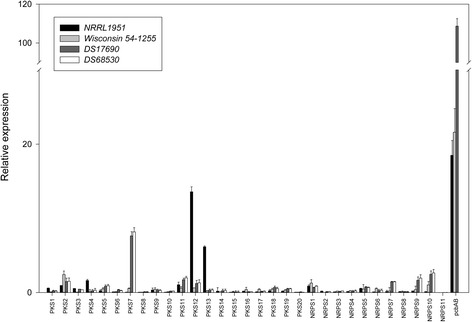
Comparison of secondary metabolite gene expression in strains NRRL1951, Wisconsin 54-1255, DS17690 and DS68530. Expression data was obtained with RNA isolated after 5 days of fungal growth. The error bars indicate the standard error of the mean of two biological samples including technical duplicates
Fig. 4.
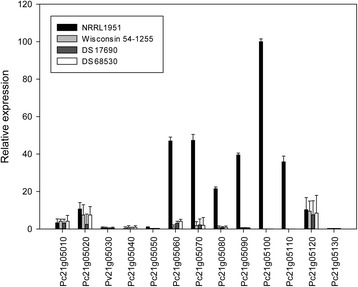
Relative expression of the putative gene cluster of sorbicillinoid biosynthesis in strains NRRL1951, Wisconsin 54-1255, DS17690 and DS68530. The transcriptional silencing of seven genes (Pc21g05050 – Pc21g05110) belonging to the putative sorbicillinoid gene cluster is shown. Expression data was obtained with RNA isolated after 5 days of fungal growth. The error bars indicate the standard error of the mean of two biological samples including technical duplicates
Comparative metabolomic profiling of P. chrysogenum strains
Next, we examined the extracellular metabolome focusing on secondary metabolites excreted into the spent culture medium. LC-MS and LC-MS/MS were used for identification. In this analysis, five β-lactams were detected in the growth media of all strains, except for the penicillin biosynthesis cluster-free strain DS68530 (Fig. 5). 6-APA (compound 1), penicillin G (2), V (3), isopenicillin N (4) and penicillin K (5) were identified by their expected molecular masses and on the basis of their calculated empirical formulas and by the use of standards, except for penicillin K. 6-APA and isopenicillin N were produced in the early stages of fermentation. As expected, the levels of these compounds were relatively low for the NRRL1951 strain, increased in strain Wisconsin 54-1255, and dramatically increased in DS17690, in particular during the late growth phase. 6-APA, isopenicillin N and the naturally produced penicillin K were the main products. Since the growth conditions did not involve any feed with the commercially used side chain precursors, penicillin G and V were presented very low amounts compared to other penicillins.
Fig. 5.
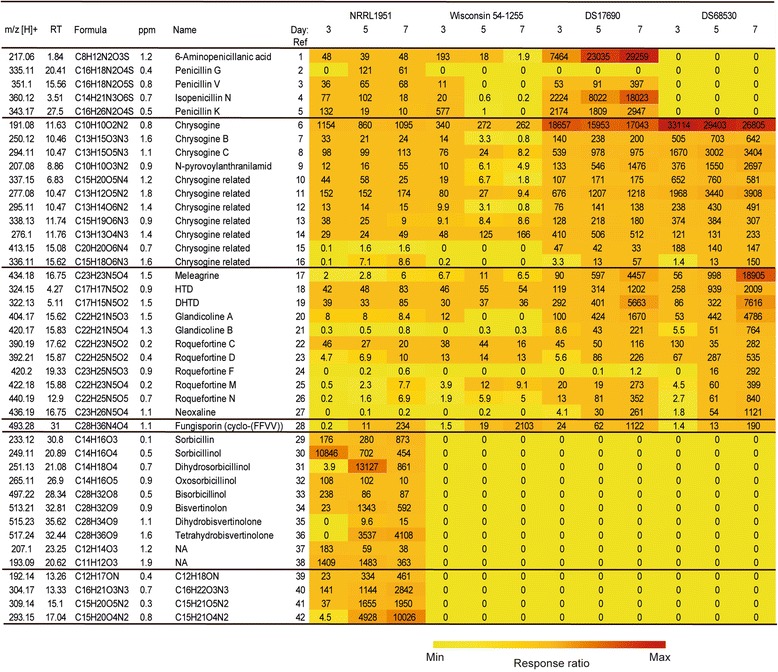
Response ratio of secondary metabolite concentrations present in the culture broth of strains NRRL1951, Wisconsin 54-1255, DS17690 and DS68530. Cell coloring corresponds to the internal standard corrected peak area of the detected metabolites in the culture broth after 3, 5 and 7 days of growth. The mass to charge ratio (m/z) of the protonated metabolites, retention time (RT) and empirical formulas are indicated. Abbreviations: HTD, histidyltryptophanyldiketopiperazine; DHTD, dehydrohistidyltryptophanyldi-ketopiperazine; NA, metabolites without structural data
Another group of compounds are the chrysogine related metabolites. This yellow pigment is produced in a biosynthetic pathway that has not yet been completely characterized, but at least four structurally resolved compounds can be identified including chrysogine (6), N-acetylalanylanthranilamide (chrysogine B) (7), N-malonylalanylanthranilamide (chrysogine C) (8) and N-pyrovoylanthranilamide (9), as well as 7 related but structurally unresolved molecules (10-16). These metabolites are already found in the culture broth after 3 days. Chrysogine was produced at high levels by the DS17690 strain, but in particular in the related penicillin cluster free DS68530 strain, production was at the highest levels.
A further class of compounds are the roquefortine and meleagrin related metabolites that are toxins [9–11]. The precursors for these toxins are (histidyltryptophanyldi-ketopiperazine (HTD) (18) and dehydrohistidyltryptophanyldi-ketopiperazine (DHTD) (19) that are only produced at low levels by strain NRRL 1951 and Wisconsin, but at substantial higher levels during the late growth phases by the DS17690 strain. Production further increased in the DS68530 strain. The same observation was made for the precursors of this pathway like glandicoline A-B (c20-21), roquefortine C, D, F, M, N (22-26) and neoxalin (27), as well as the final product meleagrin (17). It thus appears that like with the chrysogine, period 2 of the CSI has resulted in an enhanced production of roquefortines.
In the analysis, a unique group of 16 metabolites were identified in the culture broth of NRRL1951 that were absent in the other more evolved strains. Based on their exact masses and their MS spectra, empirical formulas could be assigned to these molecules with less than 2 ppm accuracy. Two groups of compounds could be discriminated (Fig. 5). The first group includes eleven molecules related to sorbicillins that are known to be produced by natural isolates of P. chrysogenum. Both, monomeric and dimeric sorbicillinoids were identified in the culture broth during seven days of culturing. The identified compounds fit to the proposed biosynthesis pathway [23, 24]. Major molecules are sorbicillin (29) and the derivative sorbicillinol (30) that already occur after 3 days of fermentation. Although dihydrosorbicillin was not detected, the accumulation of its oxidized derivative dihydrosorbicillinol (31) occurred after five days of cultivation. Oxosorbicillinol (32), and the oxidized compound bisvertinolon (34) (dimerization reaction with sorbicillinol) were identified. Further dimers were bisorbicillinol (33), dyhydrobisvertinulone (35) and tetrahydrobisvertinolone (36). In addition, two uncharacterized metabolites with a short (C11 and C12) carbon chain (37-38) were observed. The second group included four, presumably nitrogen containing compounds (39-42) (Fig. 6).
Fig. 6.
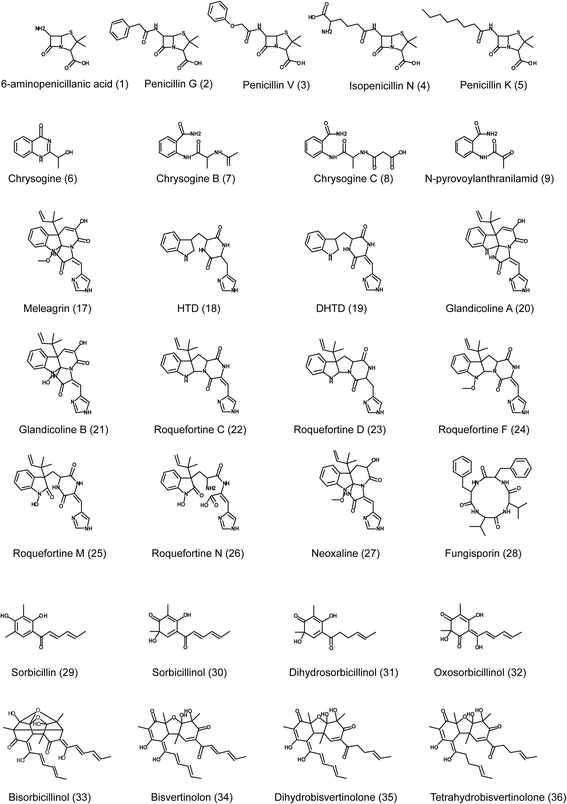
Metabolites characterized in this study. β-lactam related compounds (1-5), the intermediates of chrysogine pathway (6-9), roquefortine/meleagrin related compounds (17-27), the representative of the cyclic tetrapeptides - fungisporin (28) and the sorbicillinoids (29-36)
Discussion
Here we have examined the effect of the classical strain improvement (CSI) program on secondary metabolism by P. chrysogenum. Combined metabolome, transcriptome and genome profiling approaches were applied to a lineage of β-lactam producing strains. This concerned the natural isolate strain NRRL1951, the current laboratory reference Wisconsin 54-1255 and the high penicillin producer DS17690 which carries multiple copies of the penicillin biosynthesis cluster. In addition, a DS17690 derivative, which lacks the penicillin clusters, i.e., strain DS68530 was included in this analysis (Fig. 1). The genomes of the subjected strains were sequenced using the Illumina next generation technique that allowed us to determine the mutational impact on P. chrysogenum during CSI.
Of the 20 PKSs and 11 NRPS present in P. chrysogenum, the majority remains uncharacterized and has not been associated to the production of specific secondary metabolites. It is generally believed that many of these genes and gene clusters are transcriptionally silent or inactive. The introduction of the parental isolate NRRL1951 in our analysis revealed a number of mutations inherited by the current international laboratory standard strain Wisconsin 54-1244 during the first period of CSI. Further comparative analysis showed the accumulation of additional mutations during the evolvement of the industrial penicillin cluster multi-copy strain DS17690. Out of the 11 NRPS and 20 PKS associated gene clusters present in the genome, one NRPS and 7 PKS gene clusters accumulated mutational changes during both periods of the CSI. Three amino acid substitutions were located within the essential ketosynthase domains of PKS2, PKS13 and PKS15 that presumably led to a functional inactivation (Fig. 2a).
Significant changes of the secondary metabolite genes expression between the CSI derived mutants as well as the mutational impact on transcriptional factor encoding genes indicates that the regulation of secondary metabolism has been intensively altered during the CSI (Fig. 3). The three proteins VelA, VelB and LaeA of the velvet complex represent an important regulatory element in filamentous fungi. In A. nidulans, this heterotrimer has been associated with both cell development and secondary metabolism providing the first evidence on the regulatory cross-talk between these phenomena in filamentous fungi. The current structural model of the Velvet complex is derived from protein interaction studies performed in A. nidulans [25]. VelA is a scaffolding protein essential for recruiting VelB and LaeA. VelB interacts with the N-terminus of VelA while the C-terminus interacts with LaeA. It is important to emphasize that all recently published studies on the characterization of the Velvet complex proteins in P. chrysogenum were performed in strains derived from the CSI, i.e., the eight copy penicillin gene cluster strain DS17690 derived from Wisconsin 54-1255 and the two copy penicillin gene cluster strain P2niaD18 obtained during the alternative Panlabs strain improvement program [8]. Both overproducers originate from the parental NRRL1951 isolate but the P2 series diverged early at the CSI prior the generation of Wisconsin 54-1255. A pronounced phenotypic effect of the deletion of VelA and LaeA on β-lactam titers (up to 80 % decrease) in conjunction with a dramatic transcriptional deregulation of up to 10 % of genes present in the genome was demonstrated for the Panlabs derived P2niaD18 strain [3, 26]. In contrast, the penicillin gene expression and secondary metabolite production in DS17690 strain exhibited only moderate responses to the deletion of VelA and LaeA. The latter study, however, was performed under glucose-limited chemostat growth conditions, and it was suggested that the growth conditions weakened the phenotype [7]. However, since the Velvet complex has been repetitively targeted during the improvement of the DS17690 strain (Fig. 2b) this likely caused the weak phenotypes of the respective deletion strains. Indeed, the P2niaD18 strain [27] shows no alterations of the Velvet complex encoding genes and must therefore have remained functionally intact. Specifically, in LeaA, we identified two mutations resulting in amino acids changes while in VelA, in DS17690 strain a stop codon is introduced resulting in a C-terminal truncation by about 247 amino acids. VelA plays a primary role in the complex assembly where the C-terminus that was lost, is required for interaction with LaeA. Therefore, the velvet complex of the improved DS17690 is likely functionally impaired and this must have contributed to an improvement of the β-lactam production presumably as a collateral effect, increased or decreased expression of other secondary metabolite genes. In addition, pathway specific transcriptional regulation of secondary metabolism is crucial. We identified 19 mutated proteins specifying Zn2Cys6 binuclear cluster DNA-binding domains typical found in transcription regulators like GAL4 and six genes encoding proteins that are homologous to fungal specific transcription factors. Additionally, 29 transporter related genes were mutated during the CSI, eight ABC transporters and 15 transporters belonging to the major facilitator superfamily (MFS) (Supplementary XCL1). One of the identified MFS Pc22g08250 belongs to the putative secondary metabolite gene cluster harboring a PKS gene (Pc22g08170) that can be linked to the biosynthesis of patulin-like metabolites in P. chrysogenum [2].
The mutational and expression effects of CSI led to the significantly alterations in the secondary metabolite profile among the analyzed P. chrysogenum strains. Previously, genes and gene clusters have been identified for the production of roquefortine/meleagrine [11], cyclic tetrapeptides [12] and chrysogine (Ali, unpublished data) that all arise from the activity of NRPSs. By high-resolution mass spectrometry we generated a specific metabolite profile for each of the strains and demonstrate significant alterations in the pattern of the production of these secondary metabolites (Fig. 5). Importantly, during the CSI and with the onset of strain DS17690, the ability to produce toxins such as roquefortine/meleagrine and chrysogen increased. However, the transcriptional levels of the roqA and chyA genes was not significantly changed in the lineage of strains. Considering that the biosynthesis of these secondary metabolites is strongly dependent on nitrogen metabolism, it seems likely that the improvement in amino acid production [2] in the higher β-lactam producers is the indirect cause of this phenomenon. Moreover, the highest levels of these secondary metabolites where found in the DS68530 strain in which the multiple penicillin gene clusters have been removed [28]. This strain is genomically identical to the parental strain DS17690, also showing the same expression levels and timing of the expression of the corresponding NRPS encoding genes. Therefore, the observed increase in the aforementioned nonribosomal peptide products strongly supports our notion that their enhanced production is because of a re-diversion of nitrogen metabolism.
During the CSI, also the production of a suite of secondary metabolites was lost. Early P. chrysogenum isolates produced yellow pigments that were eliminated early on during CSI as they were found to be colored contaminants during antibiotic production. However, the genetic origin of pigment formation has remained obscure. Our secondary metabolite profiling indicates a group of structurally related compounds that are specifically produced by the NRRL1951 strain but are absent in the fermentation broth of Wisconsin 54-1255, DS17690 and DS68530. The empirical formulas generated based on mass accuracy (ppm < 2) correspond to known hexaketide yellow pigments termed sorbicillinoids. The production of sorbicillin, sorbicillinol, dihydrosorbicillinol, oxosorbicillinol, bisvertinolone, bisorbicillinol, and dihydrobisvertinolone was detected. We found that accumulation of the pathway precursors sorbicillin and sorbicillinol by a three days grown culture correlates with high levels of expression of a single PKS gene cluster in the NRRL1951 strain. qPCR analysis revealed the transcriptional silencing of the entire gene cluster of seven genes although expression of the flanking genes remained unchanged. Additionally, the genome sequencing analysis revealed two mutations accumulated in the two PKS encoding genes of the aforementioned cluster at the beginning of the CSI program. NR-PKS12 acquired an isoleucine to arginine substitution at the C-(Met)/DH inter-domain region. The neighboring oppositely oriented HR-PKS13 encoding gene obtained a mutation causing a leucine to a phenylalanine substitution in position 146 within the KS domain of the enzyme. The proposed model for sorbicillin biosynthesis, as the hexaketide precursor for all sorbicillinoid derivatives, foresees the function of a pair of NR-PKS and HR-PKS enzymes. The triacetate precursor is formed by HR-PKS following the non-reductive carbon chain extension by the second NR-PKS to form hexaketide. Taken together our results suggest this PKS gene cluster is responsible for the biosynthesis of sorbicillinoids in the early producer NRRL1951. The mutations found within the PKS gene cluster combined with the transcriptional silencing of the entire gene cluster shown for derivative strains Wisconsin 54-1255 and DS17690, demonstrates a multi-step complex eliminating effect of the CSI on the biosynthesis of these yellow pigments in P. chrysogenum. Importantly, the functional characterization of the aforementioned gene cluster through direct targeting of the PKS genes has not yet been achieved [23, 29], and will be subject of future work.
Conclusions
Our study concerned a combined genome, transcriptome and metabolome analysis of improved β-lactam producing strains of P. chrysogenum with a focus on secondary metabolism. Whereas several secondary metabolite genes were targeted by mutagenesis, overall there was not a single specific functional group of genes that was specifically mutagenized. Mutations appear to be randomly spread across the chromosomes and this is likely the result of the rather undirected approach used during CSI. The results suggest that the improved titers of the β-lactams achieved during the CSI are only partially reflected by the broad mutagenic impact of the CSI program. Whereas mutagenesis was random, the selection was focused on specific features such as the increased β-lactam titers; morphological aspects as well the fermentation characteristics. Thus, the strains collected many mutations that do not contribute to these features. The elevated production of nonribosomal peptide products such as chrysogine and roquefortine/meleagrin likely emerged from the enhanced nitrogen metabolic flux in improved penicillin producing strains. A group of yellow pigments including sorbicillinoids has been eliminated at the early stages of CSI. Production of sorbicillinoids is likely associated with a PKS gene cluster of seven genes that was transcriptionally silenced during the CSI and that acquired mutations at functionally essential PKS domains. These finding indicate a complex mechanism underlying the elimination of the production of sorbicillinoids from the secondary metabolism of P. chrysogenum and provide insight in the molecular basis of biosynthesis of this interesting group of bioactive compounds.
Methods
Strains and culture conditions
The parental P. chrysogenum isolate NRRL1951, Wisconsin 54-1255, the penicillin overproducer DS17690 (multi-copy penicillin gene cluster) and the penicillin free strain DS68530 (zero-copy penicillin gene cluster) used in this study were kindly provided by DSM Biotechnology Center. For secondary metabolite analysis and fungal RNA extraction, conidia were pre-grown in liquid YGG medium [30] for 24 h before transfer into minimal metabolite medium (SMP) (glucose, 0,5 g/l; lactose, 75 g/l; urea, 4.0 g/l; Na2SO4, 4.0 g/l; CH3COONH4, 5.0 g/l; K2HPO4, 2.12 g/l; KH2PO4, 5.1 g/l). All cultivations were performed at 25 °C in semi dark conditions using 100 ml shaking flasks with a volume 25 ml of SMP medium.
Genome analysis
Total genomic DNA (gDNA) was isolated from the mycelium after 4 days of cultivation in YGG liquid medium followed by enzymatic protoplasting with T. harzianum lytic enzyme using the adapted yeast genomic DNA isolation protocol (Promega). The next generation sequencing of the strain NRRL1951, Wisconsin 54-1255, DS17690 and DS68530 was performed using the Illumina HiSeq 2500 platform (BaseClear, Leiden, The Netherlands). FASTQ compressed files of the paired-end sequence reads were generated for each of the subjected strains using Illumina Casava pipeline version 1.8.3. Quality assessment was based on data passing the Illumina Chastity filtering. Subsequently, adapters were removed and the second quality assessment of the remaining reads was performed using the FASTQC quality control tool version 0.10.0. The resulted Average Quality scores (Phred > 35) was determined for each of the analyzed genomes (BaseClear, Leiden, The Netherlands). The alignment of the reads was generated with DNASTAR using the available sequence of Wisconsin 54-1255 as the reference. High quality reads of the genomes were obtained with near to complete coverage (99.70 – 99.91 % and median coverage 306-350) (Table 1). The resulted BAM files were used to obtain a list of single nucleotide polymorphisms (SNPs) with SeqManNGen11(DNASTAR) tool using 90 % of the SNP carrying reads as the cutoff parameter for selection. The structural variation analysis was performed with GASVPro [17].
LC-MS analysis
Metabolite analysis was performed using the Accella1250™ HPLC system coupled with the benchtop ES-MS Orbitrap Exactive™ (Thermo Fisher Scientific, San Jose, CA). A sample of 5 μL was injected onto a Shim-pack XR-ODS™ C18 column (3.0 x 75 mm, 2.2 μm) (Shimadzu, Japan) operating at 40 °C and flow rate of 300 μL/min. The linear gradient began with 90 % of solvent A (100 % water) and 5 % of solvent C (100 % acetonitrile) starting after 5 min of isocratic flow. The first linear gradient reached 60 % of C at 30 min, and the second 95 % of C at 35 min. A washing step for 10 min at 90 % of solvent C was followed by column equilibration for 15 min at the initial isocratic conditions. Solvent D (2 % formic acid) was continuously used to maintain a final 0.1 % of formic acid in the system. The column fluent was directed to the Exactive™ ES-MS Orbitrap operating at the scan range (m/z 80 – 1600 Da) and switching to positive/negative modes. Voltage parameters for the positive mode was 4.2 kV spray, 87.5 V capillary and 120 V of tube lens. Voltage parameters for the negative mode was 3 kV spray, -50 V capillary, -150 V tube lens. The capillary temperature was 325 °C, sheath gas flow 60 a.u., and auxiliary gas was off to maintain higher detection sensitivity for both positive and negative modes during analysis. The obtained raw files were processed using SIEVE software (Thermo Fisher Scientific, San Jose, CA). The resulting peak tables were used as the target list in which each feature was integrated in every individual sample. For more accurate integration, the discovered features were selected and transferred into Excalibur 2.1 (Thermo Fisher Scientific, San Jose, CA) processing tool. Auto-integration of the peaks was performed using base peak traces in mass range 10 ppm with a window of 60 s.
LC-MS sample preparation
For secondary metabolite analysis, samples of the culture broth were taken after 3, 5 and 7 days of growth in SMP medium. After centrifugation at 14,000 rpm for 10 min at 4 °C, 100 μL of the supernatant was supplemented with 16 μL of reserpine (600 μmol/mL) as the internal standard. After filtration using 0.2 μm polytetrafluorethylene (PTFE) syringe filters samples were quickly frozen in liquid nitrogen and stored at -80 °C.
qPCR analysis
Total RNA was isolated after 3 and 5 days of culturing of the fungal strains on SMP medium. The Trizol™ (Invitrogen) extraction method was used, with additional DNAse treatment using the Turbo DNA-free™kit (Ambion). The total RNA concentration was measured with a NanoDrop ND-1000™. For the synthesis of cDNA by iScript™ cDNA synthesis kit, 500 ng of RNA per reaction was used (Bio-Rad). Primers were designed around the introns in order to be able to discriminate between amplification on gDNA and cDNA and are listed in Table 3. For expression analysis, the γ-actin gene was used as a control for normalization. A negative reverse transcriptase (RT) control was used to determine the gDNA contamination in the isolated total RNA. The expression levels were analyzed for two biological samples that were split into technical duplicates. Measurements were done with a MiniOpticon™ system (Bio-Rad) using the Bio-Rad CFX™ manager software, with which the threshold cycle (CT) values were determined automatically by regression. The SensiMix™ SYBR Hi-ROX kit (Bioline) was used as a master mix for qPCR. The following thermocycler conditions were applied: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. Subsequently, a melting curve was generated to determine the specificity of the qPCRs.
Table 3.
Oligonucleotide primers used for qRT-PCR
| Primer name | Gene | Sequence 5′-3′ |
|---|---|---|
| PKS1F | Pc12g05590 | GCTACAGCCCTGACGCCATGG |
| PKS1R | Pc12g05590 | CTGCGCAGGTCTACATCGGTACC |
| PKS2F | Pc13g04470 | CCGAAGATGCCGGCGACGG |
| PKS2R | Pc13g04470 | CGCTGGTCTGCGATGTGGCC |
| PKS3F | Pc13g08690 | CGAGAGACCAGGATAAGGTTCTTGGC |
| PKS3R | Pc13g08690 | GGTGGTCTGTCACCACTCTTCCC |
| PKS4F | Pc16g00370 | CATGGTCAGCACCCTCAGTGCC |
| PKS4R | Pc16g00370 | CCAGGTCAGGCGTCGTACGC |
| PKS5F | Pc16g03800 | CGGGTGCTGCATAGATGTACTACGC |
| PKS5R | Pc16g03800 | GCTGGCCACGGAAGACAACGC |
| PKS6F | Pc16g04890 | CCTATTCGCGCCCTGATTATGGGC |
| PKS6R | Pc16g04890 | CGAGATTTGTCTTCACAGAACCCACC |
| PKS7F | Pc16g11480 | CACGATTTTAGCAAGTCAACCAGCGCG |
| PKS7R | Pc16g11480 | CTCGCTCTCCCAGAATGTCAAGGC |
| PKS8F | Pc21g00960 | GCCACACTCATCGGCACCACG |
| PKS8R | Pc21g00960 | GCTCCACAGAGCAACCAACCCG |
| PKS9F | Pc21g03930 | GACGTGGCCGGTGATGCCG |
| PKS9R | Pc21g03930 | GCGATGTTGCGGACGAGGCC |
| PKS10F | Pc21g03990 | CAGCGCCGAGTCCTACAGCC |
| PKS10R | Pc21g03990 | GTGGACCTTGGAGGATGTCTTGC |
| PKS11F | Pc21g04840 | CCTTGACGAATATCCGCACTCCG |
| PKS11R | Pc21g04840 | CAAGCCACAGCTGATGAAGCGC |
| PKS12F | Pc21g05070 | GTCGGAGGCAATTCGGGAAGGC |
| PKS12R | Pc21g05070 | GCAAAGTTCCACCACAATGCCGCG |
| PKS13F | Pc21g05080 | CCGAGGATCTCCGCCAGGC |
| PKS13R | Pc21g05080 | GGTTGTGCAGGTTCCAGGTGCC |
| PKS14F | Pc21g12440 | GCACCACCATCAGCCAAAGCATACC |
| PKS14R | Pc21g12440 | CCGAGGTCCATTGGAACTATGCGC |
| PKS15F | Pc21g12450 | CCAGTTGTCTGCAGCCGGCC |
| PKS15R | Pc21g12450 | GCCCAGATCACCGCCGTACG |
| PKS16F | Pc21g15160 | CAGCCGCGTAGTTTGCCTGGC |
| PKS16R | Pc21g15160 | GCACAGTGTGCTGAGGTTACGGC |
| PKS17F | Pc21g16000 | CTTGTCATCAGCAGCCCAGAGG |
| PKS17R | Pc21g16000 | CAATTTGCGGTGGCTGAGACGC |
| PKS18F | Pc22g08170 | GGTTGATACTCCTGGGACTGAATACAG |
| PKS18R | Pc22g08170 | GCTGCTGTGGATCCATCTGCTCG |
| PKS19F | Pc22g22850 | CGGTCAACCAGGGATCCAACTGC |
| PKS19R | Pc22g22850 | CTGAAGCGGTCTCTGTGTGGCC |
| PKS20F | Pc22g23750 | CGGTAATGTCCAGCTGGCACTCG |
| PKS20R | Pc22g23750 | CTTCAGGCACTTCTGTACCGGG |
| NRPS1F | Pc13g05250 | GCAGACCTGTATCCATCGCAA |
| NRPS1R | Pc13g05250 | GGAGGCAAGTGAAGGTGTGTT |
| NRPS2F | Pc13g14330 | GCGACAGCCGCCGGAGTAACTATGG |
| NRPS2R | Pc13g14330 | GAGAGACGGGGACACGCGTGATG |
| NRPS3F | Pc14g00080 | ACGTACGCTCGAGCTGGACT |
| NRPS3R | Pc14g00080 | GCCGTCGCGTTGATAATTGG |
| NRPS4F | Pc16g03850 | TGGTTGAAAGAGGGCAGTCTC |
| NRPS4R | Pc16g03850 | CGCGAACATACACAACACCAC |
| NRPS5F | Pc16g04690 | CTTTCCAGAACAGTTGGCTGGT |
| NRPS5R | Pc16g04690 | GCTGCATCTTACCCAGGTAATTG |
| NRPS6F | Pc16g13930 | CCACCCTTGTTCAGCCGCTGAATTCC |
| NRPS6R | Pc16g13930 | GGACGAGGCGAACAACATCGGAC |
| NRPS7F | Pc21g01710 | GCTATCTCGGTGGAGGATCTTCTGTCC |
| NRPS7R | Pc21g01710 | GTGCTGCTGAGAACACGGGATTGT |
| NRPS8F | Pc21g10790 | GTGAGGCAGCTTTGTTCAACACCATT |
| NRPS8R | Pc21g10790 | TTCTGCAGCAGGCTGTCGGCCTGAG |
| NRPS9F | Pc21g12630 | GAGCCAACTCTGTTGTCTACG |
| NRPS9R | Pc21g12630 | CAGGGCAATTTGCCTCATTCTG |
| NRPS10F | Pc21g15480 | CTTGGTGGATGCAGCGAAGG |
| NRPS10R | Pc21g15480 | CTGTGAGAGAGGCTCTTGAGTA |
| NRPS11F | Pc22g20400 | TTCGCGAACATCCGAAGAAGC |
| NRPS11R | Pc22g20400 | TCGGGCGAAGACACTGTTCA |
| pcbAB F | Pc21g21390 | CACTTGACGTTGCGCACCGGTC |
| pcbAB R | Pc21g21390 | CTGGTGGGTGAGAACCTGACAG |
| 12.10 F | Pc21g05010 | GATGCTTTCTTGAATAGCGGATCCACG |
| 12.10R | Pc21g05010 | GAACTTTGCGTTGATCGACTATTTCCACG |
| 12.20 F | Pc21g05020 | CTTAACATCTCCGATTCCCTACTCGG |
| 12.20R | Pc21g05020 | CAACCAATCCTGCGATCCATCCTCC |
| 12.30 F | Pc21g05030 | GGATAGCCAGAGAGTCAGAGAGACC |
| 12.30R | Pc21g05030 | GCGGACGGCAAGACTCAAGGCGCCTCC |
| 12.40 F | Pc21g05040 | GGAGAAAGCACGGCAATCGATAGTGG |
| 12.40R | Pc21g05040 | CCTATTCCTGTGCATGATTCTCGGCG |
| 12.50 F | Pc21g05050 | GAAGCGTGGATAGAGACCGAAGAGAAC |
| 12.50R | Pc21g05050 | GGAGCCAGACTCCGGAAAGGATACTG |
| 12.60 F | Pc21g05060 | GATAGTGAGTACAAATGCGCCTGGACC |
| 12.60R | Pc21g05060 | CGTTCAATACCGGAAATGGCTAGATTCG |
| 12.90 F | Pc21g05090 | CGTTAACTAATGACGCCACCTGTTGC |
| 12.90R | Pc21g05090 | GGAAAATAGTATCCCCAGCGATTGGC |
| 12.100 F | Pc21g05100 | CATCAGCACCGAGGTCTTCATTGTCG |
| 12.100R | Pc21g05100 | GCAACGCAATAGATGGTCAATGCCAG |
| 12.110 F | Pc21g05110 | CTGCAGCACTTCAGCATGGATGAAACC |
| 12.110R | Pc21g05110 | TCGTTGTGAGACTTGGATGCTCGGACG |
| 12.120 F | Pc21g05120 | CCTGCTTCTTAATCTTGCCCTGGC |
| 12.120R | Pc21g05120 | CCAAGCCGATGCCAAGAAGGAAGAG |
| 12.130 F | Pc21g05130 | CCTAAGTCCTTGATCAGAGCCTGG |
| 12.130R | Pc21g05130 | GTTGAGGTCGATACCCGTAAGTCATCC |
Acknowledgements
We acknowledge the financial support from the Netherlands Organization for Scientific Research (NWO) via the IBOS (Integration of Biosynthesis and Organic Synthesis) Program of Advanced Chemical Technologies for Sustainability (ACTS), the Kluyver Centre for Genomics and Industrial fermentation, which is part of the Netherlands Genomic initiative/Netherlands Organization for Scientific Research, and the Perspective Genbiotics program subsidized by Stichting toegepaste wetenschappen (STW). We thank Leonie Boenders-van Dijk (DSM Biotechnology Centre, Delft, NL) for help with the quality control analysis of genome data.
Abbreviations
- 6-APA
6-aminopenicillanic acid
- BLAST
Basic local alignment search tool
- CSI
Classical strain improvement
- COG
Clusters of orthologous groups
- GASVPro
Geometric Analysis of Structural Variants
- gDNA
Genomic DNA
- HR
Highly reductive
- KEGG
The Kyoto encyclopedia of genes and genomes database
- KOG
euKaryotic Ortholog Groups
- LC-MS
Liquid chromatography-Mass spectrometry
- NR
Non reductive
- NRPS
Nonribosomal peptide synthetase
- PKS
Polyketide synthase
- SNP
Single nucleotide polymorphism
- q-PCR
Real-time quantitative PCR
Additional files
The list of coding mutations acquired by the strain lineage of P. chrysogenum. (XLSX 52 kb)
The list of COG/KOG/BRITE functional categories and distribution of the mutated nucleotides. (XLSX 354 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
OS performed the experiments, analyzed the data and wrote the manuscript. MR performed the mass spectrometry analysis and analyzed the data. MM performed the bioinformatics analysis. PL performed the structural analysis. RV coordinated the mass spectrometry analysis. AD and RB designed and coordinated the project and revised the manuscript. All authors contributed to the writing of the manuscript and approved the final manuscript.
Contributor Information
Oleksandr V. Salo, Email: o.v.salo@rug.nl
Marco Ries, Email: marco.ries@gmx.de.
Marnix H. Medema, Email: marnix.medema@wur.nl
Peter P. Lankhorst, Email: Peter.Lankhorst@dsm.com
Rob J. Vreeken, Email: robvreeken@gmail.com
Roel A. L. Bovenberg, Email: Roel.Bovenberg@DSM.COM
Arnold J. M. Driessen, Email: a.j.m.driessen@rug.nl
References
- 1.Backus MP, Stauffer JF. The production and selection of a family of strains in Penicillium chrysogenum. Mycologia. 1955;47(4):429–463. doi: 10.2307/3755661. [DOI] [Google Scholar]
- 2.van den Berg MA, Albang R, Albermann K, Badger JH, Daran JM, Driessen AJ, et al. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat Biotechnol. 2008;26(10):1161–1168. doi: 10.1038/nbt.1498. [DOI] [PubMed] [Google Scholar]
- 3.Hoff B, Kamerewerd J, Sigl C, Mitterbauer R, Zadra I, Kurnsteiner H, et al. Two components of a velvet-like complex control hyphal morphogenesis, conidiophore development, and penicillin biosynthesis in Penicillium chrysogenum. Eukaryot Cell. 2010;9(8):1236–50. doi: 10.1128/EC.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jami MS, Barreiro C, Garcia-Estrada C, Martin JF. Proteome analysis of the penicillin producer Penicillium chrysogenum: characterization of protein changes during the industrial strain improvement. Mol Cell Proteomics. 2010;9(6):1182–1198. doi: 10.1074/mcp.M900327-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiel JA, van den Berg MA, Fusetti F, Poolman B, Bovenberg RA, Veenhuis M, et al. Matching the proteome to the genome: the microbody of penicillin-producing Penicillium chrysogenum cells. Funct Integr Genomics. 2009;9(2):167–184. doi: 10.1007/s10142-009-0110-6. [DOI] [PubMed] [Google Scholar]
- 6.Kopke K, Hoff B, Bloemendal S, Katschorowski A, Kamerewerd J, Kuck U. Members of the Penicillium chrysogenum velvet complex play functionally opposing roles in the regulation of penicillin biosynthesis and conidiation. Eukaryot Cell. 2013;12(2):299–310. doi: 10.1128/EC.00272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veiga T, Nijland JG, Driessen AJ, Bovenberg RA, Touw H, van den Berg MA, et al. Impact of velvet complex on transcriptome and penicillin G production in glucose-limited chemostat cultures of a beta-lactam high-producing Penicillium chrysogenum strain. Omics. 2012;16(6):320–333. doi: 10.1089/omi.2011.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newbert RW, Barton B, Greaves P, Harper J, Turner G. Analysis of a commercially improved Penicillium chrysogenum strain series: involvement of recombinogenic regions in amplification and deletion of the penicillin biosynthesis gene cluster. J Ind Microbiol Biotechnol. 1997;19(1):18–27. doi: 10.1038/sj.jim.2900411. [DOI] [PubMed] [Google Scholar]
- 9.Ries MI, Ali H, Lankhorst PP, Hankemeier T, Bovenberg RA, Driessen AJ, et al. Novel key metabolites reveal further branching of the roquefortine/meleagrin biosynthetic pathway. J Biol Chem. 2013;288(52):37289–37295. doi: 10.1074/jbc.M113.512665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Estrada C, Ullan RV, Albillos SM, Fernandez-Bodega MA, Durek P, von Dohren H, et al. A single cluster of coregulated genes encodes the biosynthesis of the mycotoxins roquefortine C and meleagrin in Penicillium chrysogenum. Chem Biol. 2011;18(11):1499–1512. doi: 10.1016/j.chembiol.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Ali H, Ries MI, Nijland JG, Lankhorst PP, Hankemeier T, Bovenberg RA, et al. A branched biosynthetic pathway is involved in production of roquefortine and related compounds in Penicillium chrysogenum. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0065328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ali H, Ries MI, Lankhorst PP, van der Hoeven RA, Schouten OL, Noga M, et al. A non-canonical NRPS is involved in the synthesis of fungisporin and related hydrophobic cyclic tetrapeptides in Penicillium chrysogenum. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0098212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cram DJ, Tishler M. Mold metabolites; isolation of several compounds from clinical penicillin. J Am Chem Soc. 1948;70(12):4238. doi: 10.1021/ja01192a076. [DOI] [PubMed] [Google Scholar]
- 14.Maskey RP, Grun-Wollny I, Laatsch H. Sorbicillin analogues and related dimeric compounds from Penicillium notatum. J Nat Prod. 2005;68(6):865–870. doi: 10.1021/np040137t. [DOI] [PubMed] [Google Scholar]
- 15.Abe N, Yamamoto K, Hirota A. Novel fungal metabolites, demethylsorbicillin and oxosorbicillinol, isolated from Trichoderma sp. USF-2690. Biosci Biotechnol Biochem. 2000;64(3):620–622. doi: 10.1271/bbb.64.620. [DOI] [PubMed] [Google Scholar]
- 16.Reategui RF, Wicklow DT, Gloer JB. Phaeofurans and sorbicillin analogues from a fungicolous Phaeoacremonium species (NRRL 32148) J Nat Prod. 2006;69(1):113–117. doi: 10.1021/np0504199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sindi SS, Onal S, Peng LC, Wu HT, Raphael BJ. An integrative probabilistic model for identification of structural variation in sequencing data. Genome Biol. 2012;13(3):R22. doi: 10.1186/gb-2012-13-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34(Database issue):D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4(1471-2105 (Electronic)):41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28(1):33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43(W1):W237–243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puel O, Galtier P, Oswald IP. Biosynthesis and toxicological effects of patulin. Toxins. 2010;2(4):613–631. doi: 10.3390/toxins2040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aa F, Abood A, Fisch KM, Osipow A, Davison J, Avramovic M, et al. Oxidative dearomatisation: the key step of sorbicillinoid biosynthesis. Chemical Science. 2014;5(2):523–527. doi: 10.1039/C3SC52911H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abe N, Arakawa T, Yamamoto K, Hirota A. Biosynthesis of bisorbicillinoid in Trichoderma sp. USF-2690; evidence for the biosynthetic pathway, via sorbicillinol, of sorbicillin, bisorbicillinol, bisorbibutenolide, and bisorbicillinolide. Biosci Biotechnol Biochem. 2002;66(10):2090–2099. doi: 10.1271/bbb.66.2090. [DOI] [PubMed] [Google Scholar]
- 25.Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320(5882):1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 26.Kosalkova K, Garcia-Estrada C, Ullan RV, Godio RP, Feltrer R, Teijeira F, et al. The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie. 2009;91(2):214–225. doi: 10.1016/j.biochi.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 27.Specht T, Dahlmann TA, Zadra I, Kurnsteiner H, Kuck U: Complete Sequencing and Chromosome-Scale Genome Assembly of the Industrial Progenitor Strain P2niaD18 from the Penicillin Producer Penicillium chrysogenum. Genome Announce 2014, 2(4). doi: 10.1128/genomeA.00577-14. [DOI] [PMC free article] [PubMed]
- 28.Harris DM, van der Krogt ZA, Klaassen P, Raamsdonk LM, Hage S, van den Berg MA, et al. Exploring and dissecting genome-wide gene expression responses of Penicillium chrysogenum to phenylacetic acid consumption and penicillinG production. BMC Genomics. 2009;10:75. doi: 10.1186/1471-2164-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bringmann G, Lang G, Gulder TAM, Tsuruta H, Muhlbacher J, Maksimenka K, et al. The first sorbicillinoid alkaloids, the antileukemic sorbicillactones A and B, from a sponge-derived Penicillium chrysogenum strain. Tetrahedron. 2005;61(30):7252–7265. doi: 10.1016/j.tet.2005.05.026. [DOI] [Google Scholar]
- 30.Kovalchuk A, Weber SS, Nijland JG, Bovenberg RA, Driessen AJ. Fungal ABC transporter deletion and localization analysis. Methods Mol Biol. 2012;835:1–16. doi: 10.1007/978-1-61779-501-5_1. [DOI] [PubMed] [Google Scholar]
- 31.Nijland JG, Ebbendorf B, Woszczynska M, Boer R, Bovenberg RA, Driessen AJ. Nonlinear biosynthetic gene cluster dose effect on penicillin production by Penicillium chrysogenum. Appl Environ Microbiol. 2010;76(21):7109–7115. doi: 10.1128/AEM.01702-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


