Abstract
An inflammatory component is present in the microenvironment of most neoplastic tissues, including those not causally related to an obvious inflammatory process. Several microRNAs, and especially miR-155, play an essential role in both the innate and adaptative immune response. Resveratrol (trans-3,4′,5-trihydroxystilbene) is a natural antioxidant with anti-inflammatory properties that is currently at the stage of preclinical studies for human cancer prevention. Here, we establish that, in human THP-1 monocytic cells as well as in human blood monocytes, resveratrol upregulates miR-663, a microRNA potentially targeting multiple genes implicated in the immune response. In THP-1 cells, miR-663 decreases endogenous activator protein-1 (AP-1) activity and impairs its upregulation by lipopolysaccharides (LPS), at least in part by directly targeting JunB and JunD transcripts. We further establish that the downregulation of AP-1 activity by resveratrol is miR-663 dependent and that the effects of resveratrol on both AP-1 activity and JunB levels are dose dependent. Finally, we show that resveratrol impairs the upregulation of miR-155 by LPS in a miR-663-dependent manner. Given the role of miR-155 in the innate immune response and the fact that it is upregulated in many cancers, our results suggest that manipulating miR-663 levels may help to optimize the use of resveratrol as both an anti-inflammatory and anticancer agent against malignancies associated with high levels of miR-155.
Introduction
Epidemiological studies suggest that as many as 25% of all cancers may be due to chronic inflammation (1–4). The connection between inflammation and cancer consists of an extrinsic pathway, driven by inflammatory conditions that increase cancer risk, and an intrinsic pathway, driven by genetic alterations that cause inflammation and neoplasia (2). Furthermore, inflammatory mediators released by cancer-related inflammation can induce genetic instability, leading to the accumulation of random genetic alterations in cancer cells (3). Beside resident cell types, such as fibroblasts and adipocytes, tumor microenvironment contains various inflammatory cell types infiltrating the tumor area in response to inflammatory stimuli, such as macrophages, neutrophils and mast cells (5–7). Tumor-associated macrophages are thought to play key roles in the production of various growth factors, angiogenic factors, proteinases, chemokines and cytokines, through cross talks with cancer cells and other tumor stromal cells (8–10). These factors in turn stimulate cell migration/motility, proliferation, survival, angiogenesis and metastasis, resulting in a dynamic environment that favors the progression of cancer, thus affecting the clinical outcome of malignant tumors (7). Tumor-associated macrophages have thus been described as ‘obligate partners for tumor cell migration, invasion and metastasis’ (11). Namely, in a genetic model of breast cancer in macrophage-deficient mice, the tumors developed normally but were unable to form pulmonary metastases in the absence of macrophages (12). As tumor metastasis is responsible for ∼90% of all cancer-related deaths, a better understanding of inflammation regulatory mechanisms may potentially allow to optimize the use of anticancer drugs that may target tumor-associated macrophages and lower tumor-specific inflammatory response (7).
MicroRNAs (miRNAs) are short non-coding RNAs that regulate the translation and/or degradation of target messenger RNAs (13), and whose molecular malfunctions are associated with major pathologies such as cancers (14) or autoimmune diseases (15). Several miRNAs have been implicated in the control of hematopoietic lineage differentiation (16–18). Among them, miR-155 has been implicated in the regulation of myelopoiesis and erythropoiesis, Type 1 helper T-cells differentiation, B cell maturation, IgG1 production, somatic hypermutations, gene conversion, class switch recombination, B- and T-cell homeostasis as well as in the regulation of the innate immune response [(19) and references therein]. Thus, miR-155 levels increase following lipopolysaccharides (LPS) treatment of THP-1 cells or Raw-264 macrophages, and miR-155 transgenic mice show enhanced sensibility to LPS-induced endotoxin shock (20–22). In contrast, miR-155-knockout mice are unable to mount a proper T-cell or B-cell immune responses (23). On the other hand, the immune response is limited by the upregulation of anti-inflammatory miRNAs such as miR-146a and miR-146b, which target interleukin-1 receptor-associated kinase 1 and tumor necrosis factor receptor-associated factor 6 (20,24). Importantly, several reports have recently established a direct link between elevated levels of miR-155 expression and the formation and development of tumors such as leukemias and breast, lung or gastric cancers (19). Elevated miR-155 levels are linked with enhanced cell proliferation (16,17), transgenic mice with B cells overexpressing miR-155 develop B-cell leukemia (25) and a sustained expression of miR-155 in hematopoietic stem cells causes a myeloproliferative disorder (26).
Resveratrol (trans-3,4′,5-trihydroxystilbene) is a dietary polyphenolic, non-flavonoid antioxidant derived from grapes, berries, peanuts and other plant sources. Resveratrol has various health benefits, such as cardiovascular and cancer preventive properties (27–29), and it is currently at the stage of preclinical studies for human cancer prevention (29,30). Some of the resveratrol protective activities are associated with the downregulation of the inflammatory response through inhibitory effects on transcription factors like nuclear factor-kappaB or activator protein-1 (AP-1) components c-Jun and c-Fos, including the inhibition of the release of pro-inflammatory mediators such as reactive oxygen, nitrogen species and prostaglandin E2 or of enzymes implicated in their synthesis (27,31,32). In adipocytes, resveratrol inhibits the production of the tumor necrosis factor-induced monocyte chemotactic protein-1, which has been implicated in chronic low-grade inflammation, a key feature of obesity type 2 diabetes (33).
To date, it is not known if the antitumor and anti-inflammatory effects of resveratrol may rely on the modulation of expression of either pro-inflammatory or anti-inflammatory miRNAs. To address this question, we pretreated human THP-1 monocytes with resveratrol before challenging them with LPS. We show that resveratrol impairs the upregulation of oncogenic pro-inflammatory miR-155 by LPS, at least in part through the upregulation of miR-663, a miRNA targeting JunB and JunD transcripts.
Materials and methods
Microarray analyses
RNAs extracted with TRIzol (Invitrogen, Carlsbad, CA) were subsequently subjected to DNase digestion (Turbo-DNase; Ambion, Austin, TX). Affymetrix and miRNA microarray analyses were done at the Ohio State University microarray facility.
Cell culture, treatments and transfection
THP-1 cells were electroporated using the AMAXA Kit (Lonza, Allendale, NJ). LPS stimulation was done with Salmonella enteritidis-derived LPS (100 ng/ml; Sigma-Aldrich, St Louis, MI). Whenever needed, cells were pretreated 14 h with either the vehicle (ethanol) or resveratrol 50 μM.
Isolation and treatment of human blood monocytes
Human peripheral blood monocytes were obtained from healthy donors. Monocytes were enriched using a Monocyte Isolation Kit with autoMACS Separator according to the manufacturer’s instructions (Miltenyi Biotec, Auburn, CA). They were subsequently grown for 24 h in RPMI 1640 medium with glutamax-I (Lonza) supplemented with 10% fetal bovine serum (Lonza) in standard conditions and exposed to 100 ng/ml colony stimulating factor-1. Cell viability was assessed using Trypan blue. Monocytes were then treated for 14 h with either the vehicle or resveratrol 30 μM before RNA extraction.
RNase-protection assays
Assays were performed using the mirVana miRNA Probe Construction Kit and the mirVana miRNA Detection Kit from Ambion. RNAs were incubated overnight at 46°C in the presence of 8 × 104 c.p.m. of antisense RNA probe and then digested for 40 min at 37°C using a 1/50 dilution of the provided RNAse A/RNase T1 solution.
Preparation of clones and mutagenesis
The 3′-untranslated regions (UTRs) of JunB and JunD were cloned by polymerase chain reaction (PCR) from HEK-293 cells genomic DNA and then inserted downstream of the Luciferase gene in the XbaI site of the pGL3-Control vector (Promega, Madison, WI). MiR-663 target sites were mutated using the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene; Agilent Technologies, Santa Clara, CA). Sequences of the mutated miR-663 target sites in JunB and JunD 3′-UTRs are given in supplementary Table II, available at Carcinogenesis Online.
Luciferase assays
Cells (1 × 106) in six-well plates were transfected with 0.4 μg of DNA (Promega pGL3-Control vector or derived constructs), pre-miR-663 or 663-I (50 nM final; Ambion) and 20 ng of Renilla Luciferase control vector (pRL-TK from Promega). Assays were performed 48 h after transfection using the Dual Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. The pre-miR Precursor Molecule-Negative Control #1 (Ambion) was used as a pre-miR control. AP-1 activity was measured using the Cignal AP-1 Luciferase Reporter Assay Kit (Super Array Bioscience Corporation; SABioscience, Qiagen, Valencia, CA).
Quantitative real-time PCR
MiR-155 quantitative real-time PCRs were performed using TaqMan MicroRNA Assays (Applied Biosystems, Foster City, CA). Values were normalized using either RNU-44 or RNU-48. Real-time PCRs were run in triplicates from three different complementary DNAs.
Western blots
Anti-JunB, anti-α-tubulin and anti-glyceraldehyde-3-phosphate dehydrogenase antibodies were from Cell Signaling Technology (Danvers, MA). Anti-JunD was from Abcam (Cambridge, MA). Anti-cMaf was from Santa Cruz Biotechnology (Santa Cruz, CA). The relative intensities of the bands of interest, prealably normalized to glyceraldehyde-3-phosphate dehydrogenase or α-tubulin, are given under each panel in percent of the respective control sample.
Results
Resveratrol upregulates miR-663
Affymetrix microarrays showed that resveratrol modulated the levels of a number of transcripts in THP-1 cells, expectedly upregulating those coding for anti-proliferation factors, whereas downregulating those coding for pro-proliferation factors (MIAME E-TABM-782). The most significant change was a roughly 20 times upregulation of LOC284801 transcripts, which contain the sequence of pre-miR-663 and thus represent miR-663 primary transcripts. In silico analysis using TargetScan (http://www.targetscan.org/) showed that miR-663 potentially targets transcripts of several genes implicated in the immune response (supplementary Table I is available at Carcinogenesis Online), and especially JunB, JunD and FosB, which encode AP-1 factors known to activate many cytokine genes in partnership with nuclear factor of activated T-cells factors (34).
Due to miR-663 base composition (20 CG/22 nt), no TaqMan probe is presently available for quantitative real-time PCR. However, miRNA microarrays also showed that resveratrol upregulates miR-663 by roughly 75% (MIAME E-TABM-780). This was further confirmed by RNase-protection assays (Figure 1). Furthermore, miRNA microarrays (MIAME E-TABM-1035) also showed that a lower dose (30 μM) of resveratrol still induced similar changes in miRNA populations from human blood monocytes, with the levels of miR-663 increasing roughly by 30%.
Fig. 1.
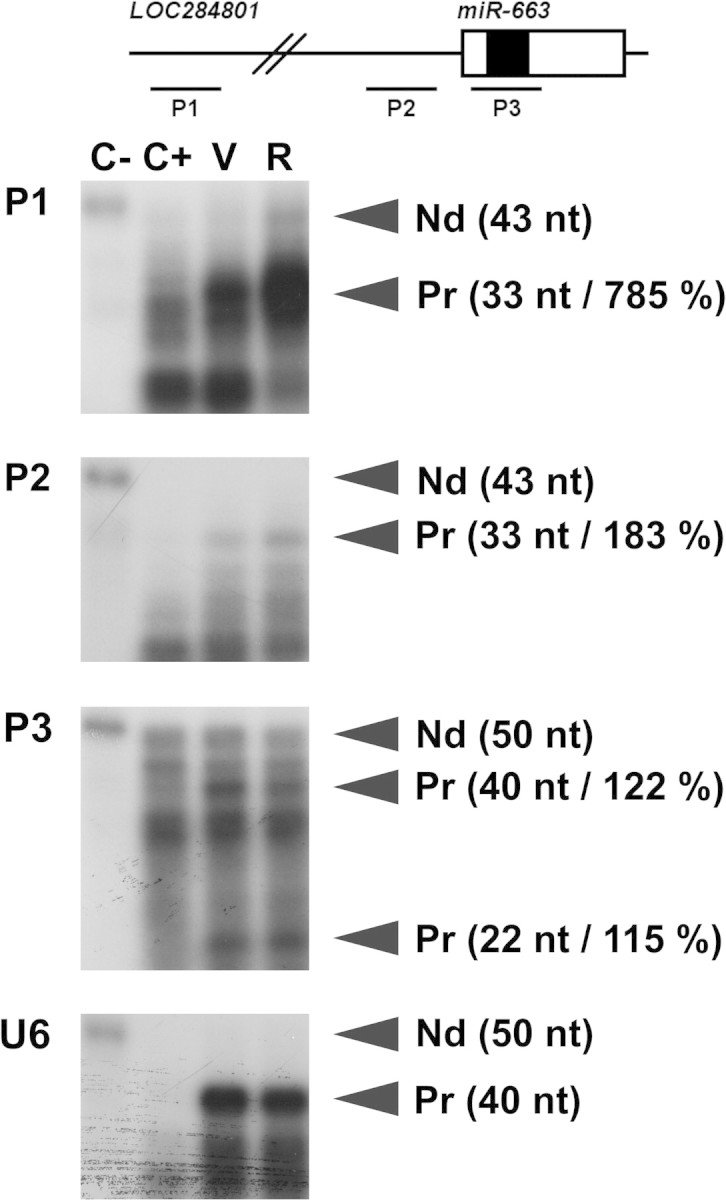
Resveratrol increases the levels of both miR-663 and LO284801 transcripts. Total RNAs (0.6 μg) extracted from THP-1 cells treated 14 h with the vehicle (V) or resveratrol (R) were hybridized with a radiolabelled RNA antisense probe (P1, P2, P3 or U6) in the presence of 4.4 μg of yeast transfer RNA. The schematic structure of the LO284801 locus, whose transcripts represent miR-663 primary transcripts, is presented on the unscaled top drawing. Open and filled boxes represent miR-663 precursor (pre-miR-663) and mature miR-663, respectively. P1 is located 1308 bp upstream of pre-miR-663 and corresponds to one of the Affymetrix probes. P2 is located 151 bp upstream of pre-miR-663. Pre-miR-663 and miR-663, respectively, generate 40 and 22 nt long protected fragments from P3, which corresponds to one of the miRNA microarrays probes. The relative intensity of the signal following resveratrol treatment is given between parentheses in percent of the vehicle sample, calculated using U6snRNA as an internal control. The efficiency of RNase digestion was assessed in parallel (two left lanes). C−, no RNase (15-fold dilution); C+, RNase; Nd, non-digested; Pr, protected fragment.
MiR-663 targets AP-1 activity as well as JunB and JunD transcripts
In the presence of LPS, both the surge of AP-1 activity and the upregulation of JunB, an AP-1 factor whose transcript levels peak within the first hour of LPS challenge (35) were impaired by miR-663 overexpression in THP-1 cells (Figure 2A and B, respectively). This indicates that transcripts encoding JunB and possibly other AP-1 factors upregulated during the mounting of the inflammatory response to LPS are targeted, directly or indirectly, by miR-663. In addition, a miR-663 inhibitory RNA (663-I) increased both AP-1 activity and JunB levels in non-challenged THP-1 cells, suggesting that miR-663 normally works to impair any sudden surge of AP-1 activity or JunB levels in resting cells (Figure 2A and B). Finally, luciferase assays using constructs containing JunB 3′-UTR with wild-type or mutated miR-663 target sites provided strong evidence that miR-663 targets JunB 3′-UTR directly (Figure 2C). Furthermore, the effects of both miR-663 and 663-I on JunD levels and those of miR-663 on the expression of a construct containing the 3′-UTR of JunD, a gene encoding another AP-1 factor, were quite similar (supplementary Figure 1A and B, respectively, is available at Carcinogenesis Online). Together, these results establish miR-663 as a new regulator of JunB and JunD levels and more generally of AP-1 activity.
Fig. 2.
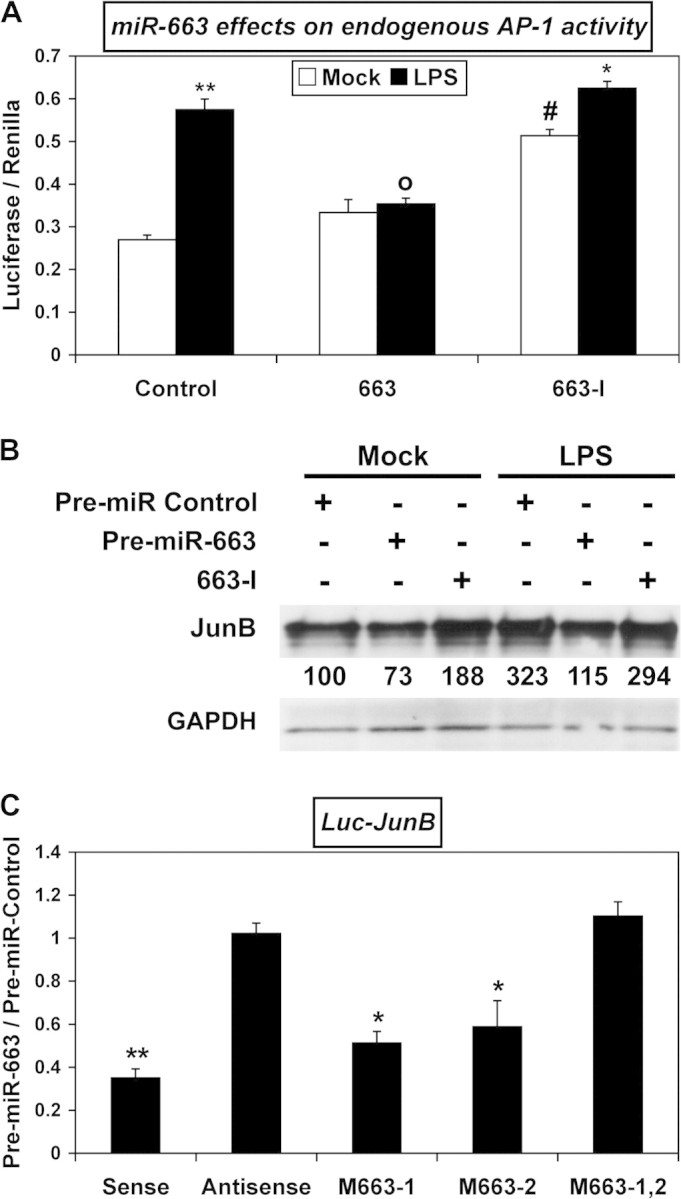
miR-663 decreases AP-1 activity and targets JunB. (A) AP-1 activity was measured in extracts of THP-1 cells transfected with the AP-1 luciferase reporter plasmid and with either a Control RNA, pre-miR-663 (663) or a miR-663 inhibitor RNA (663-I), with or without LPS challenge (6 h). Values represent the mean ± standard deviation (n = 3). * and **, significantly different from the corresponding mock-challenged sample, *P < 0.001 and **P < 0.0005. o, significantly different from LPS-challenged Control, P < 0.0008. #, significantly different from mock-challenged Control, P < 0.0001. (B) The levels of JunB in THP-1 cells transfected with either a pre-miR Control, pre-miR-663 or a miR-663 inhibitor RNA (663-I), with or without LPS challenge (6 h), were determined by western blotting. (C) Effects of pre-miR-663 on Luc-JunB constructs, both in sense and antisense orientation. Sequences of the two mutated miR-663 target sites in JunB sense constructs (M663) are given in supplementary Table II, available at Carcinogenesis Online. Values represent the mean ± standard deviation (n = 4). * and **, significantly different from Pre-miR-Control, *P < 0.001 and **P < 0.0001.
Of note, 663-I failed to further increase AP-1 activity or JunB levels following LPS challenge (Figure 2A and B, respectively). This suggested that either AP-1 activity and JunB already reached their maximum possible levels under LPS challenge in the presence of control RNA or that the innate immune response might reduce miR-663 endogenous levels. Indeed, the levels of both miR-663 and pre-miR-663 decreased roughly 25% within 30 min of LPS challenge (supplementary Figure 2 is available at Carcinogenesis Online). Of note, the levels of pri-miR-663 remained unchanged, suggesting that LPS signaling may decrease the processing of pri-miR-663 by Drosha without impacting the transcription of the miR-663 host gene.
Resveratrol decreases AP-1 activity through miR-663
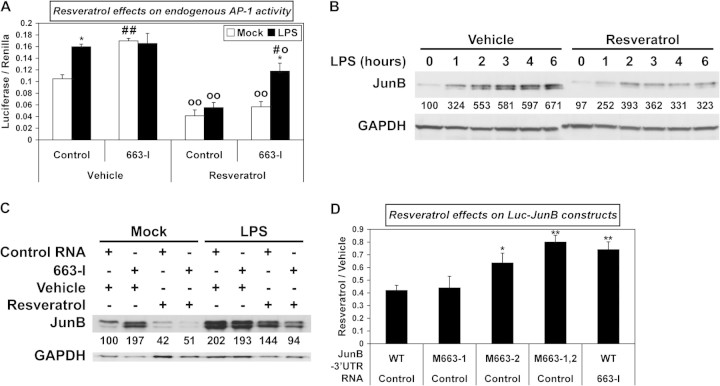
As resveratrol decreases the levels and binding activities of AP-1 factors c-Jun and c-Fos (32), whose transcripts do not contain any miR-663 target site, we then checked if it may also control AP-1 activity through the upregulation of miR-663. As expected, resveratrol decreased AP-1 activity in non-challenged THP-1 cells (Figure 3A). This effect was dose dependent (supplementary Figure 3A is available at Carcinogenesis Online). Resveratrol also proved capable to decrease JunB levels in unchallenged cells in a dose-dependent manner (supplementary Figure 3B is available at Carcinogenesis Online). Furthermore, resveratrol still decreased AP-1 activity following LPS challenge (Figure 3A). Also, in agreement with the results of Figure 2A, 663-I sharply increased AP-1 activity in untreated non-challenged cells, but not following LPS treatment (Figure 3A). Importantly, 663-I significantly impaired the downregulating effects of resveratrol on AP-1 activity following LPS challenge, indicating that JunB, JunD and/or possibly FosB levels decreased as a consequence of the upregulation of miR-663 by resveratrol. Accordingly, resveratrol impaired JunB upregulation following LPS-challenge (Figure 3B). 663-I, however, did not significantly impair the downregulation of AP-1 activity by resveratrol in non-challenged cells (Figure 3A), suggesting that the effects of resveratrol in resting cells are mainly directed on AP-1 components not targeted by miR-663, presumably c-Jun and c-Fos. Thus, the upregulation of miR-663 may allow resveratrol to change the composition, and most probably the activity, of AP-1 dimers following LPS challenge.
Fig. 3.
Resveratrol decreases AP-1 activity in a miR-663-dependent manner. (A) AP-1 activity was measured in extracts of THP-1 cells transfected with the AP-1 luciferase reporter plasmid and with either a Control RNA or a miR-663 inhibitor RNA (663-I), treated with either vehicle or resveratrol for 14 h and subsequently challenged or not with LPS for 6 h. Values represent the mean ± standard deviation (n = 3). *, significantly different from the corresponding mock-challenged sample, P < 0.005. o and oo, significantly different from the corresponding vehicle-treated sample, o, P < 0.03, oo, P < 0.001. #, significantly different from the LPS-challenged resveratrol-treated Control, P < 0.005. ##, significantly different from the mock-challenged Vehicle-treated Control, P < 0.0005. (B) The effects of a 14 h resveratrol pretreatment on the accumulation of JunB following LPS challenge were determined by western blotting. (C) The levels of JunB in THP-1 cells transfected with either a Control RNA or a miR-663 inhibitor RNA (663-I), treated with either vehicle or resveratrol for 14 h and subsequently challenged or not with LPS for 6 h, were determined by western blotting. (D) Effects of a 14 h resveratrol treatment on the expression of wild-type (WT) or mutated (M663) Luc-JunB constructs in THP-1 cells transfected with either a Control RNA or a miR-663 inhibitor RNA (663-I). Values represent the mean ± standard deviation (n = 4). * and **, significantly different from the corresponding WT/Control, *P < 0.007 and **P < 0.0009.
We then checked if the downregulation of JunB by resveratrol was miR-663-dependent. As shown previously for AP-1 activity (Figure 3A), 663-I increased sharply JunB levels in untreated non-challenged THP-1 cells but proved incapable of increasing JunB levels further following LPS challenge of untreated cells (Figure 3C). Of note, when cells had been prealably transfected, the upregulation of JunB by LPS was roughly 3-fold less (compare panels B and C of Figure 3). This suggests that transfection might by itself elicit some kind of response, with the result of decreasing the sensibility and thus the further response of THP-1 cells to LPS. Finally, 663-I seemingly increased slightly the capability of resveratrol to bring down JunB levels following LPS challenge. This counterintuitive result was not due to a lack of targeting of JunB transcripts by miR-663. Namely, as shown by luciferase assays, resveratrol as expected decreased the expression of Luc-JunB (Figure 3D) and Luc-JunD (supplementary Figure 1C is available at Carcinogenesis Online) constructs in a miR-663-dependent manner, thus giving a further evidence that it upregulates miR-663. It might be that 663-I could also impair the targeting of transcripts encoding some unidentified repressor of JunB by miR-663. Nevertheless, all together, these results indicate that resveratrol can downregulate AP-1 activity by both miR-663-dependent and miR-663-independent mechanisms.
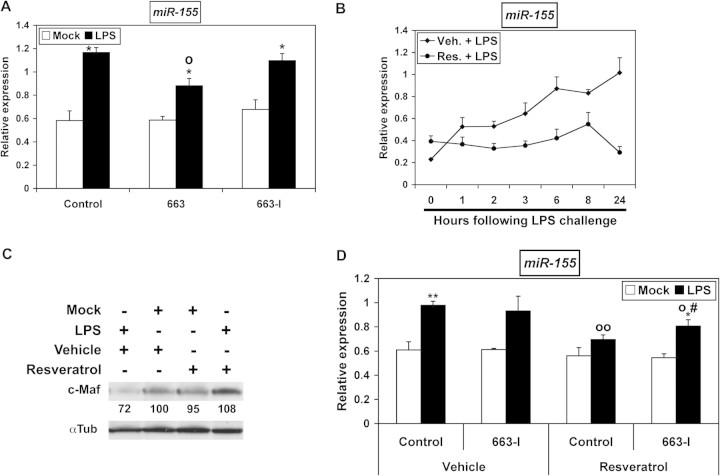
Resveratrol impairs the upregulation of oncogenic pro-inflammatory miR-155 by LPS at least partly through miR-663
The upregulation of miR-155 is the hallmark of inflammatory response following LPS treatment of THP-1 cells (21) or Raw-264 macrophages (22). In B cells, AP-1 factors such as JunB and FosB transcriptionally activate BIC, whose transcripts represent miR-155 primary RNAs (36). Given their effects on AP-1 activity, miR-663 and resveratrol would thus be expected to impair the upregulation of miR-155 by LPS. Indeed, transfecting THP-1 cells with pre-miR-663 limited miR-155 upregulation following LPS challenge (Figure 4A). In contrast, 663-I remained without obvious effects, which was probably due to the downregulation of miR-663 by LPS (supplementary Figure 2 is available at Carcinogenesis Online) and also possibly to the fact that the maximum possible levels of miR-155 were already reached in LPS-challenged control cells. Accordingly, resveratrol impaired the LPS-induced upregulation of miR-155 (Figure 4B) and consequently suppressed the downregulation of c-Maf, a validated target of miR-155 (23) (Figure 4C). In addition, 663-I slightly but significantly limited the capability of resveratrol to downregulate miR-155 (Figure 4D). Thus, some of the effects of resveratrol on oncogenic pro-inflammatory miR-155, and therefore also presumably some of its antitumor effects, are due to its upregulation of miR-663.
Fig. 4.
Resveratrol opposes the upregulation of miR-155 at least in part by upregulating miR-663. (A) The relative expression of miR-155 in THP-1 cells transfected with either a Control RNA, pre-miR-663 (663) or a miR-663 inhibitor RNA (663-I) and subsequently challenged or not with LPS for 6 h were determined by quantitative real-time PCR. Values represent the mean ± standard deviation (n = 3). *, significantly different from the corresponding mock-challenged sample, P < 0.06. o, significantly different from LPS-challenged Control, P < 0.005. (B) Time-course of miR-155 expression in LPS-challenged THP-1 cells prealably treated with vehicle or resveratrol for 14 hours, as determined by quantitative real-time PCR. Values represent the mean ± standard deviation (n = 3). (C) The levels of c-Maf in THP-1 cells pretreated with vehicle or resveratrol for 14 h and subsequently challenged or not with LPS for 6 h were determined by western blotting. (D) The relative expression of miR-155 in THP-1 cells transfected with either a Control RNA or a miR-663 inhibitor RNA (663-I) and then pretreated with vehicle or resveratrol for 14 h and subsequently challenged or not with LPS for 6 h were determined by quantitative real-time PCR. Values represent the mean ± standard deviation (n = 3). * and **, significantly different from the corresponding mock-challenged sample, *P < 0.014 and **P < 0.004. o and oo, significantly different from LPS-challenged vehicle-treated Control, oP < 0.02 and ooP < 0.0001. #, significantly different from LPS-challenged resveratrol-treated Control, P < 0.05.
Discussion
Our study provides the very first data concerning the effects of resveratrol on miRNAs and its ability to both upregulate miR-663 and downregulate oncogenic pro-inflammatory miR-155. It provides strong support that: (i) miR-663 targets JunB and JunD transcripts, consequently decreasing JunB and JunD levels as well as AP-1 activity; (ii) miR-663 also potentially targets transcripts encoding several other factors needed for the innate immune response; (iii) the downregulation of AP-1 activity by resveratrol is miR-663-dependent and (iv) resveratrol impairs the upregulation of miR-155 by LPS at least in part by upregulating the anti-inflammatory miR-663.
Transcripts from the LOC284801 locus on chromosome 20, which represent miR-663 primary RNAs, showed the highest statistically significant level change following resveratrol treatment of THP-1 cells, as deduced from Affymetrix microarray analysis. Although no probe is presently available to follow miR-663 accumulation by quantitative real-time PCR, due to its >92% CG content, miRNA microarrays, RNase protection and luciferase assays provided independent confirmations of the upregulation of mature miR-663 by resveratrol. As it is the case for many other miRNAs, miR-663 upregulation was not proportional to that of its primary transcripts, which probably reflects posttranscriptional regulations.
Our results confirm the downregulation of AP-1 activity by resveratrol. However, while previous reports mainly focused on resveratrol effects on c-Jun and c-Fos, our results show that resveratrol can specifically target JunB and JunD by increasing miR-663 levels and suggest that the same miRNA could also target FosB transcripts. This is an important result, for AP-1 factors include c-Jun, JunB, JunD, FosB, Fra-1 and Fra-2, as well as Jun dimerization partners JDP1 and JDP2 or the closely related ATF2, LRF1/ATF3 and B-ATF, so that potentially ∼18 different dimeric combinations may be formed. Thus, the capability of resveratrol to upregulate miR-663 to specifically target a subset of AP-1 dimers might have profound effects on the levels of expression of promoters to whom different AP-1 factors can compete to bind to. Due to the many roles of AP-1 factors both in inflammation (37) and cancer (38,39), the specific targeting of genes encoding a subset of AP-1 factors, by changing the composition of AP-1 dimers on key promoters, may possibly explain some of the resveratrol anti-inflammatory and anticancer properties. However, while 663-I increased luciferase activity produced from Luc-JunB and Luc-JunD constructs in the presence of resveratrol, it failed to impair the downregulation of JunB by resveratrol, indicating that miR-663-independent effects were also taking place. This was probably due to the fact that resveratrol downregulates the pro-inflammatory nuclear factor-kappaB activity (32), which is needed for JunB activation (40). Also, 663-I failed to increase JunB levels following LPS challenge. This is probably due in part to the fact that LPS challenge decreased miR-663 levels. It is also possible that JunB might have already reached its maximum possible levels under LPS challenge in control experiment, that LPS may limit miR-663 access to JunB transcripts or that miR-663 might target transcripts encoding an unidentified repressor of JunB.
Importantly, miR-663 also potentially targets transcripts encoding factors playing a role not only in toll-like receptor signaling and the innate immune response to LPS, but also in granulopoiesis, monopoiesis, interferon γ signaling, Th1 lymphocytes differentiation or TCR signaling. The functions of miR-663 are thus probably not restricted to the monocyte/macrophage lineage.
Our results also show that miR-663 impairs the upregulation of miR-155 by LPS. This effect may be due to the targeting of JunB or FosB, which have been shown to transcriptionally activate BIC, namely miR-155 host gene (36). Also, miR-663 potentially targets KSRP transcripts, which encode a RNA-binding protein implicated in the LPS-induced miR-155 maturation from its primary transcripts BIC (41). Given the facts (i) that AP-1 factors participate to the activation of many pro-inflammatory cytokine genes in partnership with nuclear factor of activated T-cells factors (37) and are implicated in the upregulation of miR-155 by LPS, (ii) that miR-155 upregulation is a hallmark of innate immune response (21,22) and (iii) that miR-663 levels decreased at the very beginning of LPS challenge (this manuscript), miR-663 is probably to work along with miR-146a and miR-146b to control the intensity of the innate immune response. Furthermore, beside its roles in myeloid differentiation and innate response, miR-155 has many other functions in the immune system, such as the regulation of myelopoiesis and erythropoiesis, Type 1 helper T-cells differentiation, B cell maturation, IgG1 production, somatic hypermutations, gene conversion, class switch recombination or B- and T-cell homeostasis (19). It will thus be very interesting to determine if miR-663 can also modulate miR-155 levels and functions in these different cell types.
Finally, the fact that resveratrol decreases both AP-1 activity and miR-155 levels at least in part through the upregulation of miR-663 could be critical in optimizing the use of resveratrol as an anti-inflammatory but also as an antitumor agent. Namely, several reports have recently established a direct link between elevated levels of miR-155 expression and the formation and development of tumors such as leukemias and breast, lung or gastric cancers (19). Elevated miR-155 levels are linked with enhanced cell proliferation (16,17), transgenic mice with B cells overexpressing miR-155 develop B-cell leukemia (25) and a sustained expression of miR-155 in hematopoietic stem cells causes a myeloproliferative disorder (26). These transgenic mice also showed that one of the many effects of miR-155 is the enhancement of cell proliferation. Of note, resveratrol impaired the upregulation of cyclins B1 and D3 by LPS (results not shown). This suggests that, beside opposing the proliferative effects of AP-1 factors, resveratrol may also oppose the proliferative activity of miR-155 through the upregulation of miR-663. Importantly, miR-663 was found to be downregulated in hormone refractory prostate cancer cells, along with miR-146a and miR-146b (42). Unfortunately, miR-663 is not present in mouse genome, thus impairing direct experimentation using knockout animals.
Nevertheless, while resveratrol anticarcinogenic potential has been linked with data primarily from human cell culture systems, evidence that resveratrol can inhibit carcinogenesis in several organ sites emerged from results of cancer prevention and therapy studies in laboratory animal models (29). However, previous results have shown that, at least when it comes to the immune system, the expression and thus potentially the function of miRNAs cannot be directly transposed from one species to another one. For example, the upregulation of anti-inflammatory miR-146a and miR-146b during the innate immune response has been observed only in human cell lines, such as THP-1 (20) or A549 cells (24), but not in mouse cell lines or in mouse spleen macrophage (22,43). As miR-663 is found only in primates, our results come as a warning that studies in animal may not always allow to predict accurately the molecular effects of resveratrol in human.
Supplementary material
Supplementary Figures 1–3 and Tables I and II can be found at http://carcin.oxfordjournals.org/
Funding
Comprehensive Cancer Center of the Ohio State University; Ligue Française contre le Cancer, BIVB; Conseil Régional de Bourgogne.
Supplementary Material
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AP-1
activator protein-1
- LPS
lipopolysaccharides
- miRNA
microRNA
- PCR
polymerase chain reaction
- UTR
untranslated region
References
- 1.Hussain SP, et al. Inflammation and cancer: an ancient link with novel potential. Int. J. Cancer. 2007;121:2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 2.Montovani A, et al. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 3.Colotta F, et al. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1082. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 4.Schetter AJ, et al. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2009;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balkwill F, et al. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 6.Coussens LM, et al. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ono M. Molecular links between tumor angiogenesis and inflammation: inflammatory stimuli of macrophages and cancer cells as targets for therapeutic strategy. Cancer Sci. 2008;99:1501–1506. doi: 10.1111/j.1349-7006.2008.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollard JW. Tumor-educated macrophages promote tumor progression and metastasis. Nat. Rev. Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 9.Yoshimura A. Signal transduction of inflammatory cytokines and tumor development. Cancer Sci. 2006;97:439–447. doi: 10.1111/j.1349-7006.2006.00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sica A, et al. Altered macrophage differentiation and immune dysfunction: tumor development. J. Clinic. Invest. 2007;117:1155–1165. doi: 10.1172/JCI31422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Condeelis J, et al. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Lin EY, et al. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal miRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, et al. MiRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 15.Tili E, et al. MicroRNAs, the immune system and rheumatic disease. Nat. Clin. Pract. Rheumatol. 2008;10:534–541. doi: 10.1038/ncprheum0885. [DOI] [PubMed] [Google Scholar]
- 16.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:243–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Tili E, et al. Expression and function of micro-RNAs in immune cells during normal or disease state. Int. J. Med. Sci. 2008;5:73–79. doi: 10.7150/ijms.5.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsitsiou E, et al. microRNAs and the immune response. Curr. Opin. Pharmacol. 2009;9:514–520. doi: 10.1016/j.coph.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tili E, et al. miR155: on the crosstalk between inflammation and cancer. Int. Rev. Immunol. 2009;28:264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 20.Taganov KD, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Connell RM, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl Acad. Sci. USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tili E, et al. Modulation of miR-155 and miR-125b levels following LPS/TNF-α stimulation, and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry MM, et al. Rapid changes in microRNA-146a expression negatively regulate the IL-1β-induced inflammatory response in human lung alveolar epithelium cells. J. Immunol. 2008;180:5689–5698. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl Acad. Sci. USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Connell RM, et al. Sustained expression of microRNA-155 in hematopoietic stem cell causes a myeloproliferative disorder. J. Exp. Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Csiszar A, et al. Resveratrol attenuates TNFα-induced activation of coronary arterial endothelial cells: role of NF-κB inhibition. Am. J. Physiol. Heart Circ. Physiol. 2006;291:H1694–H1699. doi: 10.1152/ajpheart.00340.2006. [DOI] [PubMed] [Google Scholar]
- 28.Shankar S, et al. Chemoprevention by resveratrol: molecular mechanisms and therapeutic potential. Front. Biosci. 2007;12:4839–4854. doi: 10.2741/2432. [DOI] [PubMed] [Google Scholar]
- 29.Bishayee A. Cancer prevention and treatment with resveratrol: from rodent studies to clinical trials. Cancer Prev. Res. 2009;2:409–418. doi: 10.1158/1940-6207.CAPR-08-0160. [DOI] [PubMed] [Google Scholar]
- 30.Athar M, et al. Resveratrol: a review of preclinical studies for human cancer prevention. Toxicol. Appl. Pharmacol. 2007;224:274–283. doi: 10.1016/j.taap.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leiro J, et al. Effects of cis-resveratrol on inflammatory murine macrophages: antioxidant activity and downregulation of inflammatory genes. J. Leukoc. Biol. 2004;75:1156–1165. doi: 10.1189/jlb.1103561. [DOI] [PubMed] [Google Scholar]
- 32.Das S, et al. Anti-inflammatory responses of resveratrol. Inflamm. Allergy Drug Targets. 2007;6:168–173. doi: 10.2174/187152807781696464. [DOI] [PubMed] [Google Scholar]
- 33.Zhu J, et al. Anti-inflammatory effect of resveratrol on TNF-α-induced MCP-1 expression in adipocytes. Bioch. Bioph. Res. Commun. 2008;369:471–477. doi: 10.1016/j.bbrc.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 34.Macian F, et al. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 35.Fujihra M, et al. Mechanism of lipopolysaccharide-triggered JunB activation in a mouse macrophage-like cell line (J774) J. Biol. Chem. 1993;268:14896–14905. [PubMed] [Google Scholar]
- 36.Yin Q, et al. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J. Biol. Chem. 2008;283:2654–2662. doi: 10.1074/jbc.M708218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blank V, et al. The Maf transcription factors: regulators of differentiation. Trends Biochem. Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 38.Ozanne BW, et al. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007;26:1–10. doi: 10.1038/sj.onc.1209759. [DOI] [PubMed] [Google Scholar]
- 39.Verde P, et al. Deciphering AP-1 function in tumorigenesis: Fra-ternizing on target promoters. Cell Cycle. 2007;6:2632–2639. doi: 10.4161/cc.6.21.4850. [DOI] [PubMed] [Google Scholar]
- 40.Krappmann D, et al. The IκB kinase complex and NF-κB act as master regulators of lipopolycaccharide-induced gene expression and control subordinate activation of AP-1. Mol. Cell. Biol. 2004;24:6488–6500. doi: 10.1128/MCB.24.14.6488-6500.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruggiero T, et al. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009;23:2898–2908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 42.Lin S-L, et al. Loss of miR-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417–424. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Androulidaki A, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.