Abstract
Background:
Objective identification of key miRNAs from transcriptomic data is difficult owing to the inherent inconsistencies within miRNA target-prediction algorithms and the promiscuous nature of miRNA-mRNA target relationship.
Methods:
An integrated database of miRNAs and their ‘relevant' mRNA targets was generated from validated miRNA and mRNA microarray data sets generated from patient-derived prostate epithelial normal and cancer stem-like cells (SCs) and committed basal (CB) cells. The effect of miR-542-5p inhibition was studied to provide proof-of-principle for database utility.
Results:
Integration of miRNA-mRNA databases showed that signalling pathways and processes can be regulated by a single or relatively few miRNAs, for example, DNA repair/Notch pathway by miR-542-5p, P=0.008. Inhibition of miR-542-5p in CB cells (thereby achieving miR-542-5p expression levels similar to SCs) promoted efficient DNA repair and activated expression of Notch reporters, HES1 and Survivin, without inducing dedifferentiation into SCs.
Conclusions:
Our novel framework impartially identifies therapeutically relevant miRNA candidates from transcriptomic data sets.
Keywords: miRNA-mRNA integration, prostate cancer, cancer stem cell, Notch pathway, DNA repair, epithelial hierarchy
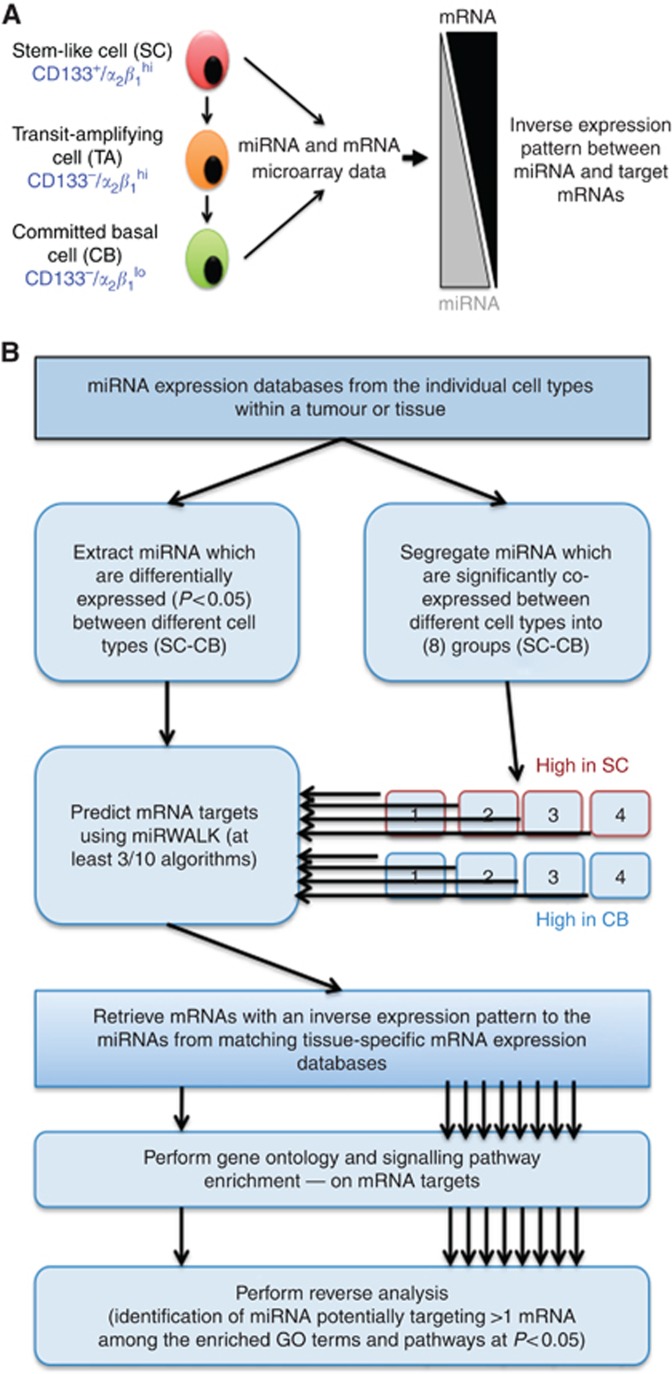
There are <2000 known human miRNAs, which influence the expression of at least 60% of cell proteins in a tissue-specific manner (Friedman et al, 2009), achieving precise orchestration of cell fate decisions. Sequencing of whole genomes, fractionated hierarchical sub-populations, and utilisation of sophisticated computational algorithms have revealed that miRNAs could be exploitable as both biomarkers and therapeutic targets for a number of diseases, particularly cancers (Catto et al, 2011; Iorio and Croce, 2012). However, objective identification and functional validation of candidate miRNAs remain difficult, owing to inherent inconsistencies with miRNA target-prediction algorithms and the promiscuous nature of miRNA-mRNA target relationship. To identify cell type-specific biomarker/therapeutically relevant miRNAs in prostate cancer, we have integrated miRNA (Rane et al, 2015) and mRNA (Birnie et al, 2008) microarray data sets. These data sets profiled miRNA and mRNA expression from patient-derived, human prostate stem-like cells (SC-CD133+/α2β1hi) and their differentiated progeny: committed basal (CB-CD133−/α2β1lo) cells (Figure 1A).
Figure 1.
Generation of integrated miRNA-mRNA data sets using expression data from prostate SC and CB cells. (A) A schematic of human prostate epithelial hierarchy (left) showing a stem-like cell (SC) with a basal phenotype differentiating into committed basal (CB) cell via transit-amplifying (TA) cell. The nature of inverse expression relationship between miRNA and its target mRNAs is depicted on the right side. For the purpose of data integration, only the expression profiles from SC and CB cells were used. (B) Algorithm used to generate integrated miRNA-mRNA data set. Two approaches were taken: one without co-expression analysis on miRNA data and second with co-expression analysis. The eight sub-groups in co-expression analysis were kept separate at each subsequent step in the analysis.
Materials and Methods
Sample procurement
Patient prostate tissue samples were obtained after written consent and full ethical approval (LREC 07/H1304/121). Tissue collection and epithelial cell culture were performed as described before (Collins et al, 2005; Rane et al, 2014).
miRNA-mRNA microarray data integration
The details of samples, methods and the platform used for miRNA (accession code: GSE59156) and mRNA (accession code: E-MEXP-993) microarray expression database generation were described before (Birnie et al, 2008; Rane et al, 2015). The co-expression analysis was performed using Pearson correlation as the distance metric and Ward's linkage method. miRNA target prediction was performed using eight different prediction algorithms on the miRWalk website (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/; Lewis et al, 2005; Dweep et al, 2011). Target mRNAs not inversely expressed in the appropriate homologous cell type were removed. The pathway enrichment scores were computed on the inversely expressed mRNAs using standard hypergeometric tests. For more details see Supplementary File S1.
Expression analysis
miRVana kit (Life Technology, Paisley, UK) was used to extract RNA. miRNA expression was assessed with miScript primer assays (Qiagen GmbH, Hilden, Germany). mRNA expression was assessed by qRT-PCR and normalised first to RPLP0 and then with control expression using respective TaqMan probes (Life Technology). Immunofluorescence studies were performed as described before (Frame et al, 2013). For details of antibodies used, see Supplementary Table S1. Nuclear γ-H2AX foci and BRCA1/RAD51 positive nuclei were counted manually.
Transfection of miRNA inhibitor
CB cells were transfected with 100 nM miScript inhibitor for miR-542-5p (miR-542-5p-i) or miScript inhibitor negative control (Qiagen GmbH) using viromer BLUE (Lipocalyx GmbH, Halle (Saale), Germany) for 72 h according to the manufacturer's protocol.
Irradiation of cells and colony-forming efficiency
Irradiation of cells, and colony-forming efficiency were described previously (Rane et al, 2015).
Statistical analysis
All experiments were performed on 3 BPH and 3 PCa samples from individual patients. Errors are the standard deviation of mean. The significance was determined using Student's two-tailed t-test, *P<0.05, **P<0.01, ***P<0.001.
Results
miRNA-mRNA microarray database integration
To integrate the miRNA and mRNA microarray data sets, we firstly leveraged the observation that expressed levels of functionally relevant miRNAs and their target mRNAs should be inversely correlated, as mammalian miRNAs are known to suppress not only protein expression, but also the transcript levels of their targets (Guo et al, 2010). Having identified differentially expressed miRNAs in SC vs CB cells (P<0.05), the targets for each miRNA were selected based on positive target-prediction call by at least three prediction algorithms and on an inverse expression pattern (Figure 1A and B). The analysis implied that signalling pathways/processes (e.g. DNA repair associated with miR-542-5p/99a/100, P<0.01) were regulated by a single or relatively few miRNAs, each of which affected multiple genes associated with the same pathway (Supplementary File S2). To investigate whether diverse miRNAs can have similar pathway-regulating functions, we employed a second approach, where differentially expressed miRNAs (SC vs CB) were K-means clustered into eight distinct sets of co-expressed miRNAs prior to re-running the previous analysis for each cluster separately (Figure 1B and Supplementary File S3). Such co-expressed clusters were expected to have coordinated functions, which should be evident through their targets and their seed-sequence similarity (Lee et al, 2004; Supplementary File S4). The first evidence in support of this hypothesis was the observation that miRNAs with known functional associations (e.g., let-7 miRNAs and miRNAs of miR-17-92 family clustered within the same sub-group; Supplementary File S3). Notch signalling has been shown to have a significant role in prostate cancer cell fate (Carvalho et al, 2014; Supplementary Figure S1) Therefore it was of interest to further investigate miR-542-5p (identified by both in silico analyses), which can regulate Notch pathway-associated protein MAML1 (Supplementary File S4).
Proof-of-principle experiments to validate miRNA-mRNA database predictions
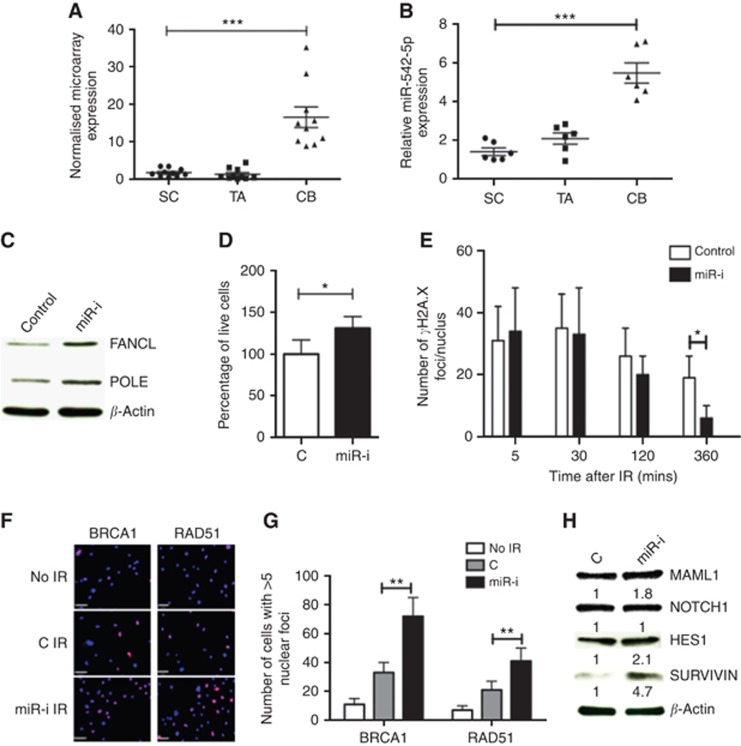
To test the validity of in silico predictions from the above two approaches, we measured the functional impact of miR-542-5p inhibition on prostate epithelial fate through FANCL, POLE1 and MAML1. Little is known about the role of miR-542-5p in prostate cancers, but its predicted targets, FANCL (Rajendra et al, 2014) and POLE1 (Palles et al, 2013), are vital mediators of DNA repair, whereas MAML1 is a critical transcriptional co-activator of Notch receptors that stimulates transcription of Hes1 (Wu et al, 2000). We have previously shown that the SCs from normal prostate and malignant tissues are relatively radio-resistant (Frame et al, 2013); however, they can be sensitised to radiation by concurrent inhibition of Notch signalling by the gamma-secretase inhibitor RO4929097 (Tolcher et al, 2012; Supplementary Figure S1). As miR-542-5p is suppressed in SC (compared with CB; Figure 2A and B), we hypothesised that this lower expression of miR-542-5p in SCs contributes to radioresistance. Inhibition of miR-542-5p expression in CB cells (Supplementary Figure S2) resulted in elevated expression of POLE1 and FANCL (Figure 2C). We also observed better cellular recovery (Figure 2D), quicker γ-H2A.X recovery (Figure 2E) and more efficient BRCA1 and RAD51 foci formation (Figure 2F and G) after exposure of the CB cells to 5 Gy radiation. The efficient DNA repair was also accompanied by activation of Notch signalling as demonstrated by upregulation of MAML1 protein (1.8-fold) and Notch targets HES1 (2.1-fold) and Survivin (4.7-fold; Figure 2H). A similar trend was also observed at the mRNA level, especially for Notch effector HES1 and Survivin (Supplementary Figure S3).
Figure 2.
Proof-of-principle experiments to validate miRNA-mRNA database predictions using miR-542-5p. (A) miR-542-5p expression in miRNA microarray data set and (B) qRT-PCR validation on independent samples. CB cells were then transfected with 100 nM miR-542-5p-inhibitor (‘miR-I') or negative control (‘C') for 3 days. The expression of FANCL and POLE proteins was assessed by western blot (C). These cells were irradiated with 5 Gy IR before counting live cell count after 48 h of IR (D), γ-H2AX immunofluorescence foci/nucleus recovery (E), BRCA1 and RAD51 nuclear positivity 2 h after IR (F) Blue: DAPI and lilac: BRCA1/RAD51, and quantification is shown in G. Protein expression changes in MAML1 and Notch-associated effector proteins HES1 and Survivin (H). Error bars=60 μm. Each experiment represents mean of 3 BPH and 3 PCa and plotted as mean±s.d. *P<0.05, **P<0.01, ***P<0.001 (Student's t-test). Abbreviations: SC=stem-like cells; TA=transit-amplifying cells; CB=committed basal cells; BPH=benign prostatic hyperplasia; PCa=treatment naive Gl-7 prostate cancer; miR-I=miR-542-5p inhibitor; C=inhibitor negative control.
Absence of a stem-like phenotype in miR-542-5p inhibited CB cells
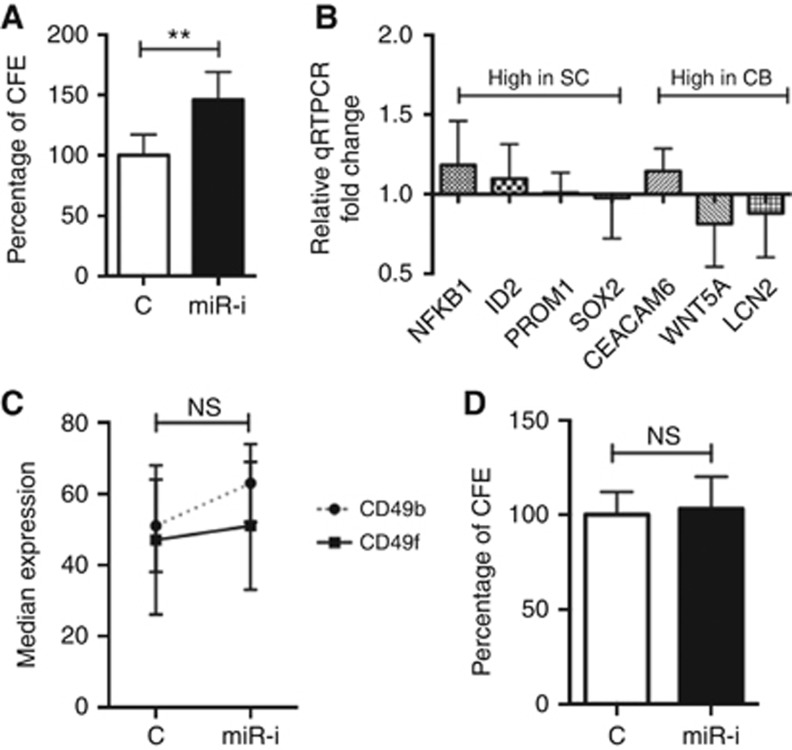
A substantial (50%) increase in colony-forming ability (Figure 3A) was also noted in miR-542-5p inhibited CB cells after radiation treatment (5 Gy). As we have shown that SCs are more radio-resistant than CB cells (Frame et al, 2013), the observed efficient DNA repair after miR-542-5p inhibition in CB cells could either be due to the direct effects of POLE1, FANCL and MAML1 overexpression, or the result of dedifferentiation of CB into SCs (or both). However, inhibition of miR-542-5p did not result in any detectable dedifferentiation of CB cells (Figure 3B–D). Thus, miR-542-5p inhibition in CB cells resulted solely in efficient DNA repair and Notch activation, as predicted by our miRNA-mRNA data integration data sets.
Figure 3.
Effect of miR-542-5p inhibition on differentiation state of prostate epithelial cells. (A) Colony-forming efficiency (CFE) of miR-542-5p inhibitor (or negative control)-transfected CB cells after exposure to 5 Gy radiation (n=3 BPH and PCa each). Before radiation, CB cells were transfected with miR-542-5p inhibitor for 72 h. CB cells transfected with miR-542-5p inhibitor (or negative control) for 72 h and subjected to qRT-PCR analysis for known SC/CB associated genes (B), FACS analysis for CD49b/f (C) and colony-forming efficiency (D). Each experiment represents the mean of 3 BPH and 3 PCa and plotted as mean±s.d. (except C). **P<0.01 (Student's t-test). Abbreviations: SC=stem-like cells; CB=committed basal cells; BPH=benign prostatic hyperplasia; PCa=treatment naive Gl-7 prostate cancer; miR-I=miR-542-5p inhibitor; C=inhibitor negative control; NS=not significant.
Discussion
The central theme of this work was to devise a means for the identification of functionally relevant miRNAs and their target mRNAs. Our approach selects only tissue-specific mRNA targets in the specific cellular context, such as SC fate determination in present study. As the reduction in target mRNA expression may not always be evident (as we have assumed), our investigation may have underestimated the overall influence of miRNAs in prostate carcinogenesis. However, the causal relationship between some of the identified miRNA and their target pathways/processes, for example, miR-99a/100 regulation of DNA repair (Mueller et al, 2013; Rane et al, 2015), clearly demonstrates the validity of our approach. Furthermore, the importance of studying miRNAs in a specific cellular phenotype has recently been underscored by the discovery that the expression changes in miR-143/145 in purified mouse colon stromal cells, rather than epithelial components (as previously assumed from unfractionated tissue/tumour biopsies) has functional relevance in regenerating mouse intestine (Chivukula et al, 2014). This pathway/process-based approach also provides an initial framework to address the long-standing issue of co-operative or functionally inter-related nature of miRNA targets in a practical setting. The database integration methods we have described can be readily adapted for any other tissue types to generate functionally relevant and therapeutically viable candidates, as exemplified here by miR-542-5p. In combination with alternative approaches such as large-scale miRNA functional screens, in an appropriate cellular environment (as we demonstrate), our approach should bring miRNA-driven therapeutics closer to direct clinical application.
Acknowledgments
We thank all the patients and urology surgeons L Coombes, G Cooksey and J Hetherington (Castle Hill Hospital, Cottingham, UK). We also thank Dr Davide Pellacani for useful discussions. The work was funded by PRO-NEST Marie-Curie Grant (to JKR), The Finnish Funding Agency for Technology and Innovation Finland Distinguished Professor programme, and Academy of Finland: project no. 132877 (to AY and MN), project support from Cancer Research Technology (to RA), and Yorkshire Cancer Research (to ATC and NJM).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Birnie R, Bryce SD, Roome C, Dussupt V, Droop A, Lang SH, Berry PA, Hyde CF, Lewis JL, Stower MJ, Maitland NJ, Collins AT (2008) Gene expression profiling of human prostate cancer stem cells reveals a pro-inflammatory phenotype and the importance of extracellular matrix interactions. Genome Biol 9(5): R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L, Schlomm T, Visakorpi T (2011) MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol 59(5): 671–681. [DOI] [PubMed] [Google Scholar]
- Carvalho FL, Simons BW, Eberhart CG, Berman DM (2014) Notch signaling in prostate cancer: a moving target. Prostate 74(9): 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivukula RR, Shi G, Acharya A, Mills EW, Zeitels LR, Anandam JL, Abdelnaby AA, Balch GC, Mansour JC, Yopp AC, Maitra A, Mendell JT (2014) An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell 157(5): 1104–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ (2005) Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 65(23): 10946–10951. [DOI] [PubMed] [Google Scholar]
- Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk–database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J Biomed Inform 44(5): 839–847. [DOI] [PubMed] [Google Scholar]
- Frame FM, Pellacani D, Collins AT, Simms MS, Mann VM, Jones GD, Meuth M, Bristow RG, Maitland NJ (2013) HDAC inhibitor confers radiosensitivity to prostate stem-like cells. Br J Cancer 109(12): 3023–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1): 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466(7308): 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Croce CM (2012) MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4(3): 143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P (2004) Coexpression analysis of human genes across many microarray data sets. Genome Res 14(6): 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1): 15–20. [DOI] [PubMed] [Google Scholar]
- Mueller AC, Sun D, Dutta A (2013) The miR-99 family regulates the DNA damage response through its target SNF2H. Oncogene 32(9): 1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I, Sherborne A, Chubb D, Carvajal-Carmona LG, Ma Y, Kaur K, Dobbins S, Barclay E, Gorman M, Martin L, Kovac MB, Humphray S Consortium C, Consortium WGS Lucassen A, Holmes CC, Bentley D, Donnelly P, Taylor J, Petridis C, Roylance R, Sawyer EJ, Kerr DJ, Clark S, Grimes J, Kearsey SE, Thomas HJ, McVean G, Houlston RS, Tomlinson I (2013) Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 45(2): 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendra E, Oestergaard VH, Langevin F, Wang M, Dornan GL, Patel KJ, Passmore LA (2014) The genetic and biochemical basis of FANCD2 monoubiquitination. Mol Cell 54(5): 858–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane JK, Droop AP, Pellacani D, Polson ES, Simms MS, Collins AT, Caves LS, Maitland NJ (2014) Conserved two-step regulatory mechanism of human epithelial differentiation. Stem Cell Rep 2(2): 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane JK, Scaravilli M, Ylipaa A, Pellacani D, Mann VM, Simms MS, Nykter M, Collins AT, Visakorpi T, Maitland NJ (2015) MicroRNA expression profile of primary prostate cancer stem cells as a source of biomarkers and therapeutic targets. Eur Urol 67(1): 7–10. [DOI] [PubMed] [Google Scholar]
- Tolcher AW, Messersmith WA, Mikulski SM, Papadopoulos KP, Kwak EL, Gibbon DG, Patnaik A, Falchook GS, Dasari A, Shapiro GI, Boylan JF, Xu ZX, Wang K, Koehler A, Song J, Middleton SA, Deutsch J, Demario M, Kurzrock R, Wheler JJ (2012) Phase I study of RO4929097, a gamma secretase inhibitor of Notch signaling, in patients with refractory metastatic or locally advanced solid tumors. J Clin Oncol 30(19): 2348–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD (2000) MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet 26(4): 484–489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.