Abstract
Several discoveries have paved the way to personalise cancer medicine and a tremendous gain of knowledge in genomics and molecular mechanisms of cancer progression cumulated over the last years. Big stories in biology commonly start in a simple model system. No wonder microRNAs have been identified as regulators of embryonic development in the nematode Caenorhabditis elegans. From the first identification in worms to the first-in-man microRNA-based clinical trial in humans, almost 20 years passed. In this review we follow the story of understanding microRNA alterations in cancer, describe recent developments in the microRNA field and critically discuss their potential as diagnostic, prognostic and therapeutics factors in cancer medicine. We will explain the rationale behind the use of microRNAs in cancer diagnosis and prognosis prediction, but also discuss the limitations and pitfalls associated with this. Novel developments of combined microRNA/siRNA pharmacological approaches will be discussed and most recently data about MXR34, the first-tested microRNA drug will be described.
Keywords: microRNAs, diagnosis, prognosis, treatment, worms, cancer
More than 20 years ago, two groups published their seminal work about the involvement of a small RNA sequence (lin-4) in diverse postembryonic developmental events in the nematode Caenorhabditis elegans (Lee et al, 1993; Wightman et al, 1993). Although at that time the term microRNA had not been created (in the first years after this discovery they had been referred to as small temporal RNAs), the group of Victor Ambros already proposed in their pioneering study fundamental principles of what microRNAs are and how this new class of noncoding RNA works. By determining the size of ∼22 nucleotides, and discovering the interaction of lin-4 with sequence complementary elements in a repeated sequence element in the 3′-untranslated region of lin-14 messenger RNA, they established at this time the basis for a new research direction in developmental biology, physiology and medicine. Another groundbreaking work was published by the group of Thomas Tuschl, where they clearly demonstrated a broader role for these small RNAs in biology and their existence in multiple organisms including vertebrates and humans (Lagos-Quintana et al, 2001). According to its relatively small size of about 20 nucleotides, this novel RNA species was termed microRNA. At this time the race to determine the biological function of microRNAs in human diseases was initiated. (Lagos-Quintana et al, 2001). Two years later, Calin et al published for the first time a direct link between microRNAs and human cancer. In their work, Calin et al. reported that miR-15 and miR-16 are located at chromosome 13q14, a region frequently deleted in B-cell chronic lymphocytic leukaemia. In more than two third of cases, these microRNA genes are deleted or their expression is downregulated by other events (Calin et al, 2002). Since then, many other important experimental and clinical discoveries have been reported by many different groups. For reasons of space restriction in this review, many of these excellent works could not be cited or discussed here. Summarising the main findings of the last 10 years, it clearly came out that microRNAs are differentially expressed between normal and cancer cells, that they are more or less reflecting tissue-specific expression signatures and that microRNAs can either promote (‘oncomiRs') or suppress tumour development and progression, thereby influencing all hallmarks of cancer (depending on the type of cell and tissue context, Figure 1; Calin and Croce, 2006). Besides their comprehensively and well-described intracellular functions, microRNAs have been found as circulating biomolecules in all body fluids (e.g., blood, urine, sputum or stool). Recently published studies propose that microRNAs are not only ‘passively' circulating byproducts, but also exert a role as intercellular messengers by exosome-mediated transfer between different cells in a ‘hormone-like' manner (Cortez et al, 2011). In this review, we are trying to discuss representative examples of the most recent and relevant developments of microRNA research in clinical oncology and their current status of applications in cancer patients. Overall, there are three major topics we are addressing: microRNAs in diagnosis, in prognosis and in therapy of cancer.
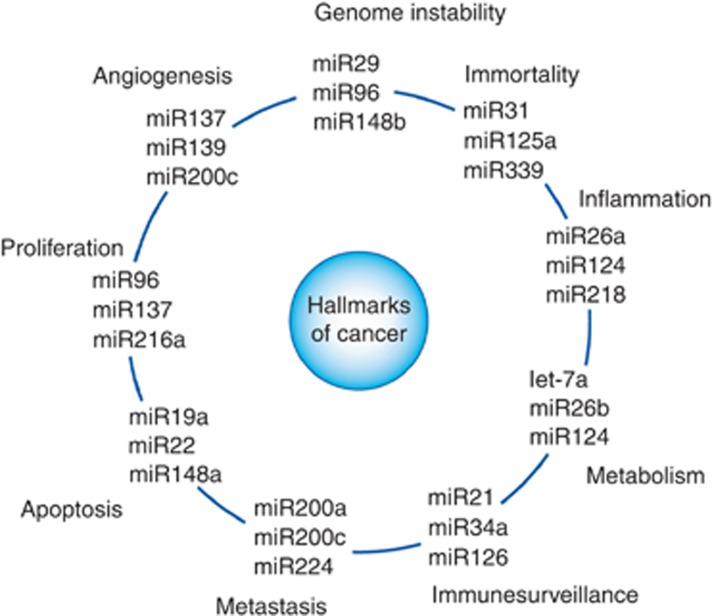
Figure 1.
Schematic illustration of the association between microRNAs and the hallmarks of cancer. Each hallmark shows three examples of microRNAs that influence the particular cellular function in certain types of cancer. Of note, some microRNAs influence more than one hallmark indicating to the multiple pathways regulated by them.
MicroRNAs in Cancer Diagnosis
In many if not all cancer patients, the tumour stage at diagnosis of the underlying malignant disease significantly influences risk of recurrence, progression and death. For many types of cancer, there is currently a lack of early detection methods or screening tests, making the issue of early cancer detection a promising field for microRNA-based diagnostics. On the basis of the above-mentioned features of (1) cancer cell and tissue-specific expression profiles and (2) circulation in body fluids, microRNAs exhibit some characteristics for ideal biomarkers. The relatively high chemical stability of microRNAs in fresh or even formalin-fixed tissues and body fluids is another advantage that increases their potential as diagnostic markers in comparison with longer messenger RNAs or long noncoding RNAs (Blondal et al, 2013). As microRNAs are released by healthy and cancerous cells, many attempts have been made to determine the meaning of the specific expression signatures as potential blood-, urine- or stool-based diagnostic markers. Especially in cancers where other early detection methods are not available, expensive or even harmful for patients, microRNA-based biomarkers might possess chances to get established in routine clinical practice. One important example is the potential of microRNAs in lung cancer early-stage detection. Previous studies have clearly indicated that by using low-dose computed tomography (CT)-based screening strategies in high-risk populations, higher detection rates of early-stage lung cancer, results in improved survival rates of patients (Aberle et al, 2011). However, owing to the associated relatively high costs and the risk of induction of secondary cancers by (even low) radiation exposure, the widespread clinical application of CT-based screening programs is controversially discussed. In this clinical setting, Montani and colleagues recently published data of a comprehensive large-scale validation study (n=1115) of a serum-based microRNA signature (‘miR-Test'). The authors tested this microRNA signature in high-risk individuals enrolled in the Continuous Observation of Smoking Subjects lung cancer screening programme. Of note, the overall accuracy, sensitivity and specificity of this microRNA assay were 74.9% (95% confidence interval (CI)=72.2–77.6%), 77.8% (95% CI=64.2–91.4%) and 74.8% (95% CI=72.1–77.5%), respectively. This study results are striking, as the authors used a well-designed cohort with high number of patients, both supporting meaningfully statistical conclusion (Montani et al, 2015). In context of lung cancer detection methods, another recently published study presented data about a sputum-based microRNA panel to identify lung cancer in indeterminate solitary lung nodules. In their study, Xing et al. used a training set to develop a biomarker panel of miR-21, -31 and -210, and validated this panel in larger independent sets of samples. Sensitivity and specificity ranged between 80 and 88% in all tested cohorts (Xing et al, 2015). In addition to the application of early cancer detection, another promising field of microRNA-based assays in cancer diagnosis is reasoned on the discovery that microRNA expression profiles are highly tissue and cell type specific, allowing the reliable classification of different types of cancer based on the microRNA profile (Rosenfeld et al, 2008).
This tissue specificity carries great potential for the diagnosis of cancer of unknown primary origin (CUP) or uncertain origin. Cancer of unknown primary origin alone constitutes 3–5% (50 000–70 000 cases) of all newly diagnosed cancers per year in the United States. Cancer of unknown primary origin presents a clinical challenge as the origin of tumour tissue is crucial for selecting proper treatment plan. Meiri and colleagues published the development and validation of a second-generation microRNA-based custom array that can assign CUP to 42 different types of cancer. In their study, the authors showed an impressive overall assay sensitivity of 85% in a large cohort of 509 CUP samples (Meiri et al, 2012). The sensitivity reached 90% for cases in which the assay reported a single answer (>80% of cases). Beyond the use of microRNAs for the classification of CUP, a series of other studies reported about the potential for discrimination of histological subtypes in certain organs. The rationale for developing microRNA assays for differentiating histological subtypes comes from limitations of current diagnostic standards: Low amounts of collected cancer cells by biopsy or only cytological smears make the microscopic diagnosis difficult in some cases and significantly increase the interobserver variability. For instance, Lebanony et al (2009) reported about a high-discrimination rate (sensitivity of 96% and specificity of 90%) for miR-205 to identify squamous cell carcinoma of the lung. Over the years, more advanced diagnostic microRNA assays have been developed. In one study, the authors propose a novel diagnostic microRNA-based assay (miRview lung, Rosetta Genomics Ltd.), which can differentiate between the four main types of lung cancer: squamous cell carcinoma of the lung, nonsquamous nonsmall cell lung cancer, carcinoid tumours and small cell carcinoma. On several hundred samples, this assay returned a result for >90% of the samples with overall accuracy of 94% (95% CI, 91–96%), with similar performance observed in pathologic and cytological samples (Gilad et al, 2012). Despite impressive results, the true value of these assays has to be interpreted by integrating considerations of the current practice in lung cancer management. On the basis of the approval of patient-tailored drugs (i.e., EGFR inhibitors and ALK inhibitors), the determination of the general mutational landscape in lung cancer tissue is getting more important than only the histological diagnosis, as the mutational spectrum directly and significantly influences the treatment plan. There is an ongoing discussion and also already initiated clinical trials (‘basket' and ‘umbrella' clinical trial design) that aim on histology-independent and aberration-specific clinical trials (Menis et al, 2014). The ability to classify histological subtypes by microRNA-based assays has also been successfully demonstrated for kidney cancer, pleura mesothelioma and other types of cancer (Benjamin et al, 2010; Spector et al, 2013).
Despite these promising findings, there are some general hurdles and limitations for these microRNA-based diagnostic tools. First, all of this data rely on retrospective cohorts and data collections, which are prone to error and selection bias. Therefore, the next logical step has to be a prospective validation and comparison (preferable in independent centralised review laboratories) to the diagnostic gold standard (i.e., the histopathological diagnosis) in a blinded manner. This comparison has to include the assay performance criteria-like specificity and sensitivity, as well as other parameters including cost effectiveness, duration of time from biopsy to diagnosis and applicability in routine diagnostic laboratories (including the necessity of special trained personal and devices). Other problems that are obviously to all gene expression assays (irrespective of whether quantitative PCR, array technology or RNA sequencing is used) are intratumoral and intercellular heterogeneity. Intratumoral heterogeneity is a major cause of misinterpretation of all molecular tests, as the molecular and microRNA profiles significantly differ between different areas of the tumour (Gerlinger et al, 2012). On the other hand, the expression profile of tumours is significantly influenced by bystander cells of the tumour stroma and contamination with stromal cells can lead to wrong conclusions and irreproducible microRNA expression results (Kent et al, 2014). Both intratumoral heterogeneity and cellular heterogeneity are essential points for the diagnostic process with microRNA-based assays. Alternative methods such as in situ hybridisation can offer a real localisation of microRNAs in tumour cells and might overcome some of these limitations, but owing to their rather semiquantitative nature, lack of standardisation and time-consuming procedure, have their own pitfalls.
MicroRNAs in Cancer Prognosis
The next level of information retrieved by the use of microRNA-based assays is the prediction of the individual risk of tumor progression and clinical endpoints. Traditionally, individual risk stratification and patient counselling mainly rely on clinical and pathological parameters. However, some of these parameters show a high interobserver variability (e.g., tumour grade or Ki-67 staining) and even predictive accuracy of the combination of such prognostic factors to prognostic scores is far from perfect (Pichler et al, 2011). Novel laboratory-based or molecular factors including microRNAs to these established prognostic factors and models can significantly increase the predictive ability (Szkandera et al, 2014). There are hundreds of published studies proposing the value of different microRNAs as prognostic biomarkers in every different type of cancer. As with many of such prognostic biomarker studies, many of them lack an independent validation and almost all of them are retrospective in their nature. The problems of aforementioned intratumoral and cellular heterogeneity apply also for prognostic biomarkers (Gerlinger et al, 2012; Kent et al, 2014). For these and other reasons, microRNA-based prognostic assays are far away from approval in clinical routine use and, similar to diagnostic microRNA-based biomarkers, large prospective studies are needed to evaluate their true value in a particular clinical scenario.
Nevertheless, there have been several interesting studies published and some of these microRNA-based prognosticators might warrant further clinical validation. For instance, a very recently published study measured the microRNA profile of colorectal cancer patients in primary tumors compared to metastasis (‘metastatic-signature') and identified 23 microRNAs as differentially expressed. Five of these microRNAs could be validated in a second cohort, in which four of them were downregulated (let-7i, miR-10b, miR-221, and miR-320a) and one was upregulated (miR-885-5p) in liver metastases compared with the primary tumour. Interestingly, low let-7i expression in primary tumour tissue predicted poor prognosis (HR=5.0, 95% CI=1.0–24.4, P=0.0479) as well as distant metastasis (OR=5.5, 95% CI=1.1–26.8, P=0.0334). High miR-10b expression in primary tumour tissue independently predicted distant metastasis (OR=4.9, 95% CI=1.2–19.7, P=0.0248). Furthermore, high serum miR-885-5p expression independently predicted prognosis (HR=2.9, 95% CI=1.1–7.5, P=0.0323), lymph node metastases (OR=3.0, 95% CI=1.3–7.2, P=0.0116) and distant metastases (OR=3.1, 95% CI=1.0–10.0, P=0.0456; Hur et al, 2015). The combination of prognostic studies in patient cohorts together with the consequently experimental proof and explanation of biological functions and molecular interactions of microRNAs can substantiate the prognostic significance of a given microRNA. In line with this, a very recently published study by Ling et al (2015) proposes that miR-224 is a negative prognostic factor in colorectal cancer patients. Multiple cohorts were used to demonstrate the prognostic value of miR-224 and by using in vitro and in vivo models, the authors experimentally confirmed miR-224 to promote tumour metastases. Similarly, miR-200a has been reported as prognostic relevant in a screening cohort of 110 colorectal cancer patients and has been validated in independent samples of the Cancer Genome Atlas. The authors of this study substantiate the prognostic value by showing experimental data about the involvement of miR-200a in epithelial–mesenchymal transition, a fundamental process for cancer metastases in colorectal cancer (Pichler et al, 2014). Recently, a large study in B-cell lymphoma patients demonstrated well-defined microRNA signatures for normal B cells as well as subsets of lymphoma cells. High expression levels of miR-155 were identified as significantly associated with rituximab plus cyclophosphamide, doxorubicin, vincristine and prednisone (R-CHOP) treatment failure (Iqbal et al, 2015). Studies like these generate data for potential biomarkers but might also be fundamental to discover druggable microRNAs for cancer therapy.
MicroRNAs in Cancer Therapy
Of all microRNA-based applications in cancer medicine, the therapeutic potential of microRNAs might be the most promising and challenging path. On the one hand, microRNAs interact with multiple targets including several mRNAs of the same signalling pathway, which might potentiate the efficacy of microRNA-based drugs. However, having several potential interactors will also carry risk of off target effects resulting in frequently occurring and severe adverse events in other organs. Numerous in vitro and in vivo studies have demonstrated efficacy for microRNAs to interfere with all hallmarks of cancer ultimately resulting in tumour regression and cancer cell death. The mode of action of microRNA-based drugs can either rely on restoring their loss of function (for tumour suppressive microRNAs) or inhibiting their gain of function (for oncomiRs). One innovative approach has been published by Nishimura and colleagues, where the authors presented data about a double targeting strategy by combining a microRNA together with a short interfering RNA (siRNA). The authors used a siRNA against the EphA2 oncogene in a preclinical model of ovarian cancer and boosted the antitumour effects by addition of miR-520-3d, which synergistically inhibited the EphA2 expression in cancer cells. Nishimura et al. used 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine nanoliposomes loaded with miR-520d-3p and EphA2 siRNA and clearly demonstrated a synergy of this combined treatment to shrink the tumours, which might have broad implications for innovative gene-silencing therapies in clinical trials (Nishimura et al, 2013). One of the most advanced microRNA-based therapeutic candidates currently evaluated in clinical trials is MRX34 (Mirna Therapeutics, TX, USA), a miR-34 mimetics that restores the function miR-34 in cancer cells. MiR-34 is frequently downregulated in human cancers and acts as a tumour suppressive microRNA. Most recent data of an ongoing multicenter phase I clinical trial protocol for patients with liver cancer and liver metastases of other cancers have been presented in April 2015 at the Annual Meeting of the American Association for Cancer Research (AACR). Interims safety data indicate a manageable profile of side effects and in white blood cells of patients the repression of expression of several potential miR-34 target oncogenes could be proven (Hong et al, oral presentation at AACR 2015). Another, though preclinical work, presented recently at the AACR includes data derived from an in vivo study in nonsquamous lung cancer demonstrating that miR-34 directly represses the checkpoint signalling molecule PD-L1 (programmed death ligand 1) and that MRX34 treatment leads to an increase in active tumour-infiltrating immune cells (CD8+) and a decrease in CD8+PD1+ tumour-infiltrating immune cells (Cortez et al, oral presentation at AACR 2015, abstract # 2875). Hopefully within the next several months we will receive more details about efficacy of this exciting first-in-class clinical trial. Besides the use of microRNAs as drug candidates themselves there is another emerging field related to microRNAs in cancer therapy. MicroRNA-based predictive biomarkers hold promise to inform about the probability of response rates of other (microRNA-unrelated) cancer drugs (Stiegelbauer et al, 2014). A representative example is the value of microRNAs for the prediction of epidermal growth factor receptor-directed therapies (e.g., cetuximab). In colorectal cancer patients, a single-nucleotide polymorphism in the let-7 binding site of the KRAS gene, has been proposed to predict the tumour responsiveness (a particular allele combination resulted in overall response rate of a 42% compared with a 9%) in cetuximab-treated patients (Zhang et al, 2011). Another example is miR-212, which has been involved in cetuximab-resistant cancer cells of head and neck carcinoma by directly regulating heparin-binding EGF-like growth factor (Hatakeyama et al, 2010). Table 1 summarises microRNAs with important roles in cancer diagnosis, prognosis or prediction of response to treatment. Taken together, all these directions are innovative and promising, but the proof of concept in preclinical models has to move forward and successfully pass confirmation in prospective clinical trials. The next up-coming years will verify whether these small molecules will help to substantially improve cancer diagnosis and treatment or just represent another small piece of the large puzzle.
Table 1. Examples of important studies that implicated microRNAs in cancer diagnosis, prognosis and prediction of response to cancer drugs.
| Purpose | MicroRNA(s) | Type of cancer | Sample | Authors |
|---|---|---|---|---|
| Early detection | 13 microRNA panel (‘miR-Test') | Lung cancer | Serum | Montani et al, 2015 |
| Diagnosis | 24 microRNA panel | Kidney cancer | Tissue | Spector et al, 2013 |
| Diagnosis | 3 microRNA panel (miR21, miR31 and miR210) | Lung cancer | Sputum | Xing et al, 2015 |
| Diagnosis | 3 microRNA panel (miR200c, -93-3p, -192) | Mesothelioma | Tissue | Benjamin et al, 2010 |
| Diagnosis | 8 microRNA panel (miR106a, -125a-5p, -129-3p, -205, -21, -29b, -375, -7) | Lung cancer | Tissue | Gilad et al, 2012 |
| Diagnosis | 64 microRNA panel | Cancer of unknown primary | Tissue | Meiri et al, 2012 |
| Diagnosis | 48 microRNA panel | Cancer of unknown primary | Tissue | Rosenfeld et al, 2008 |
| Prognosis | miR224 | Colorectal cancer | Tissue | Ling et al, 2015 |
| Prognosis | miR200a | Colorectal cancer | Tissue | Pichler et al, 2014 |
| Prognosis | let-7i, miR-10b, miR-885-5p | Colorectal cancer | Tissue | Hur et al, 2015 |
| Prognosis | miR155 | Lymphoma | Cells | Iqbal et al, 2015 |
| Prediction | miR212 | Head and neck cancer | Tissue | Hatakeyama et al, 2010 |
| Prediction | let7-binding site in KRAS gene | Colorectal cancer | Tissue or blood | Zhang et al, 2011 |
Acknowledgments
Dr Calin is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. He is supported also by the CLL Global Research Foundation. The work in Dr. Calin's laboratory is supported in part by the NIH/NCI grants 1UH2TR00943-01 and T32CA009599, Developmental Research Awards in Prostate Cancer, Multiple Myeloma, Leukemia (P50 CA100632) and Head and Neck (P50 CA097007) SPOREs, a SINF MDACC_DKFZ grant in CLL, a SINF grant in colon cancer, the Laura and John Arnold Foundation, the RGK Foundation and the Estate of C. G. Johnson, Jr. Dr. Pichler is supported by an Erwin-Schroedinger Scholarship of the Austrian Science Funds (project no. J3389-B23).
The authors declare no conflict of interest.
References
- Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 365(5): 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin H, Lebanony D, Rosenwald S, Cohen L, Gibori H, Barabash N, Ashkenazi K, Goren E, Meiri E, Morgenstern S, Perelman M, Barshack I, Goren Y, Edmonston TB, Chajut A, Aharonov R, Bentwich Z, Rosenfeld N, Cohen D (2010) A diagnostic assay based on microRNA expression accurately identifies malignant pleural mesothelioma. J Mol Diagn 12(6): 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, Dahlsveen IK (2013) Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 59(1): S1–S6. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6(11): 857–866. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99(24): 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA (2011) MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat Rev Clin Oncol 8(8): 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366(10): 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad S, Lithwick-Yanai G, Barshack I, Benjamin S, Krivitsky I, Edmonston TB, Bibbo M, Thurm C, Horowitz L, Huang Y, Feinmesser M, Hou JS St, Cyr B, Burnstein I, Gibori H, Dromi N, Sanden M, Kushnir M, Aharonov R (2012) Classification of the four main types of lung cancer using a microRNA-based diagnostic assay. J Mol Diagn 14(5): 510–517. [DOI] [PubMed] [Google Scholar]
- Hatakeyama H, Cheng H, Wirth P, Counsell A, Marcrom SR, Wood CB, Pohlmann PR, Gilbert J, Murphy B, Yarbrough WG, Wheeler DL, Harari PM, Guo Y, Shyr Y, Slebos RJ, Chung CH (2010) Regulation of heparin-binding EGF-like growth factor by miR-212 and acquired cetuximab-resistance in head and neck squamous cell carcinoma. PLoS One 5(9): e12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur K, Toiyama Y, Schetter AJ, Okugawa Y, Harris CC, Boland CR, Goel A (2015) Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J Natl Cancer Inst 107: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Shen Y, Huang X, Liu Y, Wake L, Liu C, Deffenbacher K, Lachel CM, Wang C, Rohr J, Guo S, Smith LM, Wright G, Bhagavathi S, Dybkaer K, Fu K, Greiner TC, Vose JM, Jaffe E, Rimsza L, Rosenwald A, Ott G, Delabie J, Campo E, Braziel RM, Cook JR, Tubbs RR, Armitage JO, Weisenburger DD, Staudt LM, Gascoyne RD, McKeithan TW, Chan WC (2015) Global microRNA expression profiling uncovers molecular markers for classification and prognosis in aggressive B-cell lymphoma. Blood 125(7): 1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent OA, McCall MN, Cornish TC, Halushka MK (2014) Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res 42(12): 7528–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294(5543): 853–858. [DOI] [PubMed] [Google Scholar]
- Lebanony D, Benjamin H, Gilad S, Ezagouri M, Dov A, Ashkenazi K, Gefen N, Izraeli S, Rechavi G, Pass H, Nonaka D, Li J, Spector Y, Rosenfeld N, Chajut A, Cohen D, Aharonov R, Mansukhani M (2009) Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol 27(12): 2030–2037. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5): 843–854. [DOI] [PubMed] [Google Scholar]
- Ling H, Pickard K, Ivan C, Isella C, Ikuo M, Mitter R, Spizzo R, Bullock MD, Braicu C, Pileczki V, Vincent K, Pichler M, Stiegelbauer V, Hoefler G, Almeida MI, Hsiao A, Zhang X, Primrose JN, Packham GK, Liu K, Bojja K, Gafa R, Xiao L, Rossi S, Song JH, Vannini I, Fanini F, Kopetz S, Zweidler-McKay P, Wang X, Ionescu C, Irimie A, Fabbri M, Lanza G, Hamilton SR, Berindan-Neagoe I, Medico E, Mirnezami AH, Calin GA, Nicoloso MS (2015) The clinical and biological significance of MIR-224 expression in colorectal cancer metastasis Gut; e-pub ahead of print 24 March 2015; doi:10.1136/gutjnl-2015-309372. [DOI] [PMC free article] [PubMed]
- Meiri E, Mueller WC, Rosenwald S, Zepeniuk M, Klinke E, Edmonston TB, Werner M, Lass U, Barshack I, Feinmesser M, Huszar M, Fogt F, Ashkenazi K, Sanden M, Goren E, Dromi N, Zion O, Burnstein I, Chajut A, Spector Y, Aharonov R (2012) A second-generation microRNA-based assay for diagnosing tumor tissue origin. Oncologist 17(6): 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menis J, Hasan B, Besse B (2014) New clinical research strategies in thoracic oncology: clinical trial design, adaptive, basket and umbrella trials, new end-points and new evaluations of response. Eur Respir Rev 23(133): 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montani F, Marzi MJ, Dezi F, Dama E, Carletti RM, Bonizzi G, Bertolotti R, Bellomi M, Rampinelli C, Maisonneuve P, Spaggiari L, Veronesi G, Nicassio F, Di Fiore PP, Bianchi F (2015) miR-test: a blood test for lung cancer early detection. J Natl Cancer Inst 107: 6. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Jung EJ, Shah MY, Lu C, Spizzo R, Shimizu M, Han HD, Ivan C, Rossi S, Zhang X, Nicoloso MS, Wu SY, Almeida MI, Bottsford-Miller J, Pecot CV, Zand B, Matsuo K, Shahzad MM, Jennings NB, Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK, Calin GA (2013) Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov 3(11): 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler M, Hutterer GC, Chromecki TF, Jesche J, Kampel-Kettner K, Rehak P, Pummer K, Zigeuner R (2011) External validation of the Leibovich prognosis score for nonmetastatic clear cell renal cell carcinoma at a single European center applying routine pathology. J Urol 186(5): 1773–1777. [DOI] [PubMed] [Google Scholar]
- Pichler M, Ress AL, Winter E, Stiegelbauer V, Karbiener M, Schwarzenbacher D, Scheideler M, Ivan C, Jahn SW, Kiesslich T, Gerger A, Bauernhofer T, Calin GA, Hoefler G (2014) MiR-200a regulates epithelial to mesenchymal transition-related gene expression and determines prognosis in colorectal cancer patients. Br J Cancer 110(6): 1614–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, Lebanony D, Goren Y, Silberschein E, Targan N, Ben-Ari A, Gilad S, Sion-Vardy N, Tobar A, Feinmesser M, Kharenko O, Nativ O, Nass D, Perelman M, Yosepovich A, Shalmon B, Polak-Charcon S, Fridman E, Avniel A, Bentwich I, Bentwich Z, Cohen D, Chajut A, Barshack I (2008) MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol 26(4): 462–469. [DOI] [PubMed] [Google Scholar]
- Spector Y, Fridman E, Rosenwald S, Zilber S, Huang Y, Barshack I, Zion O, Mitchell H, Sanden M, Meiri E (2013) Development and validation of a microRNA-based diagnostic assay for classification of renal cell carcinomas. Mol Oncol 7(3): 732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegelbauer V, Perakis S, Deutsch A, Ling H, Gerger A, Pichler M (2014) MicroRNAs as novel predictive biomarkers and therapeutic targets in colorectal cancer. World J Gastroenterol 20(33): 11727–11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkandera J, Gerger A, Liegl-Atzwanger B, Absenger G, Stotz M, Friesenbichler J, Trajanoski S, Stojakovic T, Eberhard K, Leithner A, Pichler M (2014) The lymphocyte/monocyte ratio predicts poor clinical outcome and improves the predictive accuracy in patients with soft tissue sarcomas. Int J Cancer 135(2): 362–370. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75(5): 855–862. [DOI] [PubMed] [Google Scholar]
- Xing L, Su J, Guarnera MA, Zhang H, Cai L, Zhou R, Stass SA, Jiang F (2015) Sputum microRNA biomarkers for identifying lung cancer in indeterminate solitary pulmonary nodules. Clin Cancer Res 21(2): 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Winder T, Ning Y, Pohl A, Yang D, Kahn M, Lurje G, Labonte MJ, Wilson PM, Gordon MA, Hu-Lieskovan S, Mauro DJ, Langer C, Rowinsky EK, Lenz HJ (2011) A let-7 microRNA-binding site polymorphism in 3'-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol 22(1): 104–109. [DOI] [PMC free article] [PubMed] [Google Scholar]