Summary
The heritability of B cell chronic lymphocytic leukemia (CLL) is relatively high; however, no predisposing mutation has been convincingly identified. We show that loss or reduced expression of death-associated protein kinase 1 (DAPK1) underlies cases of heritable predisposition to CLL and the majority of sporadic CLL. Epigenetic silencing of DAPK1 by promoter methylation occurs in almost all sporadic CLL cases. Furthermore, we defined a disease haplotype, which segregates with the CLL phenotype in a large family. DAPK1 expression of the CLL allele is downregulated by 75% in germline cells due to increased HOXB7 binding. In the blood cells from affected family members, promoter methylation results in additional loss of DAPK1 expression. Thus, reduced expression of DAPK1 can result from germline predisposition, as well as epigenetic or somatic events causing or contributing to the CLL phenotype.
Keywords: HUMDISEASE
Introduction
Chronic lymphocytic leukemia (CLL) is one of the most common types of adult leukemias and is currently incurable. There were 8190 new cases of CLL in the United States in 2004 (Kasper and Harrison, 2005). Familial occurrence of CLL has been noted in up to 10% of cases (Yuille et al., 2000); however, large pedigrees with many affected individuals are exceedingly rare. Most familial agglomerations consist of two or three affected first- or second-degree relatives (Sellick et al., 2005). Large case-control studies concluded that the risk ratio (RR), a measurement of the disease frequency in first-degree relatives of probands, was higher for CLL than for most other cancers (Goldgar et al., 1994). While the average RR for all cancers in a US study was approximately 2.1, CLL showed a RR of 5.0, the fourth highest of all cancers (Goldgar et al., 1994, Risch, 2001). For CLL a RR of 7.5 was recently calculated in Sweden (Goldin et al., 2004). In a study of the entire Icelandic population, lymphoid leukemia had the second highest RR of all malignancies studied (Amundadottir et al., 2004).
This evidence of high heritability has prompted investigators to conduct genome-wide searches for linkage in samples from CLL families. The findings, in particular from one US and one European-led consortium (Goldin et al., 2003, Sellick et al., 2005), have been limited due to weak evidence for linkage at several loci, most of which were different between the two studies. Loci in chromosome bands 11p11 and 13q21, respectively, were backed by statistically significant evidence, but no gene has been implicated to date.
Studies uncovering epigenetic aberrations in CLL have accelerated the search for CLL-related genes. A genome-wide DNA methylation analysis of CLL samples identified almost 200 novel epigenetically silenced genes in CLL. The study concluded that on average 4.8% of all CpG islands in a CLL genome could be targeted for aberrant DNA methylation and associated gene silencing (Rush et al., 2004). The role of aberrant methylation in CLL was highlighted by the finding of preferential promoter methylation-directed gene silencing of ZAP-70 and TWIST2 in subgroups of CLL defined by IgVH mutational status (Corcoran et al., 2005, Raval et al., 2005). The emerging concept from these findings is that DNA methylation contributes significantly to global expression changes that have been described for CLL (Rosenwald et al., 2003).
Here, we combined epigenetic and genetic data to identify a novel putative tumor suppressor in CLL. We present evidence that epigenetic silencing and/or germline mutations in death-associated protein kinase 1 (DAPK1), a positive mediator of apoptosis, contribute to familial CLL and that this gene is silenced in virtually all cases of sporadic CLL.
Results
Frequent Epigenetic Inactivation of DAPK1 in CLL
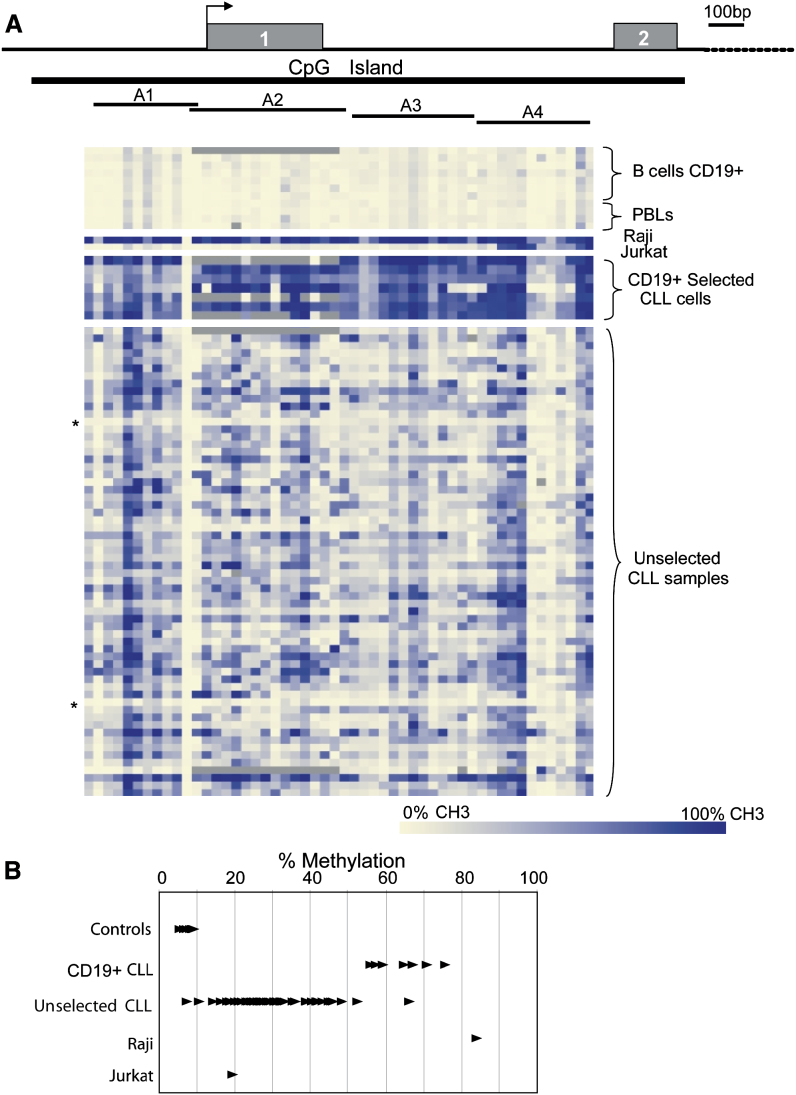
DAPK1 was initially isolated as a positive mediator of apoptosis induced by interferon-γ (INF-γ) (Deiss et al., 1995). Previous studies, using methylation-specific PCR (MSP), demonstrated low frequencies of aberrant DNA methylation of the DAPK1 promoter in CLL (Chim et al., 2006, Katzenellenbogen et al., 1999). We expanded the analysis of DNA methylation events in the DAPK1 CpG island and performed quantitative high-throughput analysis of DNA methylation in four amplicons using the MassARRAY system (Figure 1A). The number of CpGs studied in amplicons A1–A4 was 17, 22, 20, and 29, respectively. The average DNA methylation frequency in four samples of normal peripheral blood mononuclear cells (PBMCs) isolated from healthy volunteers and seven samples of normal CD19+ B cells was 6.3% and 7.6% (range 5.1%–8.9%), respectively. In contrast, we saw an average of 64% (range 55.9%–83.9%) DNA methylation in seven CD19+ selected CLL samples (Figure 1B). Next, we evaluated the PBMCs from 62 sporadic CLL patient samples. These CLL samples showed varying levels of DAPK1 promoter methylation, ranging from 7.2% to 66.1%. Only 2 out of 62 CLL samples (marked as ∗ in Figure 1A) showed methylation levels in the normal range (<11%), whereas all other samples showed levels >11%. The distribution of DNA methylation levels in CLL samples is significantly different from the one seen in normal cells (p < 0.0001). Raji cells were methylated (83.9%), whereas Jurkat cells were unmethylated in regions A1–A3 but methylated in A4. In addition, we tested the DNA methylation status in several leukemia/lymphoma cell lines—Ramos (Burkitt lymphoma), BJAB (atypical Burkitt lymphoma [EBV−]), MEC-1 (CLL), MEC-2 (CLL), Wac3CD5 (CLL), Daudi (Burkitt lymphoma), 697 (ALL), RS11846 (Non-Hodgkin lymphoma), and RS 4;11 (ALL) —using COBRA for the two most densely methylated regions in the DAPK1 promoter. With the exception of Wac3CD5, all cell lines demonstrated aberrant DNA methylation as compared to CD19+ selected B cells (Figures S1A and S1B). Chromatin immunoprecipitation (ChIP) assay, using antiacetylated histone H3 and H4 antibodies, showed that in Raji cells (methylated), the DAPK1 promoter was not associated with either ac-H3 or ac-H4 histones, while in WaC3CD5 and Jurkat cell lines (unmethylated) acetylated histones were found within this region (Figures S1C–S1F).
Figure 1.
DAPK1 Promoter Methylation in CLL Samples
(A) Schematic representation of the DAPK1 gene showing the location of the CpG island (black bar) and the four bisulfite reaction amplicons (A1–A4). The arrow indicates the predicted transcription start site. The lower panel shows a graphical display of quantitative DNA methylation data for the DAPK1 promoter region. Each square represents a single CpG or a group of two or three CpGs analyzed, and each row represents a sample. Methylation frequencies extend from light yellow (0%) to dark blue (100%). Gray indicates unavailable data. Samples included seven CD19+ selected control B cells, four control normal PBMCs, seven CD19+ selected CLL cells, and PBMCs from 62 sporadic CLL samples, Raji and Jurkat cells. Asterisks indicate CLL samples that showed less than 11% methylation.
(B) Plot of average percentage methylation in regions A1–A4, in controls (seven CD19+ selected B cells and four PBL samples; total = 11), CD19+ selected CLL cells (n = 7), unselected CLL cells (n = 62), and Raji and Jurkat cell lines.
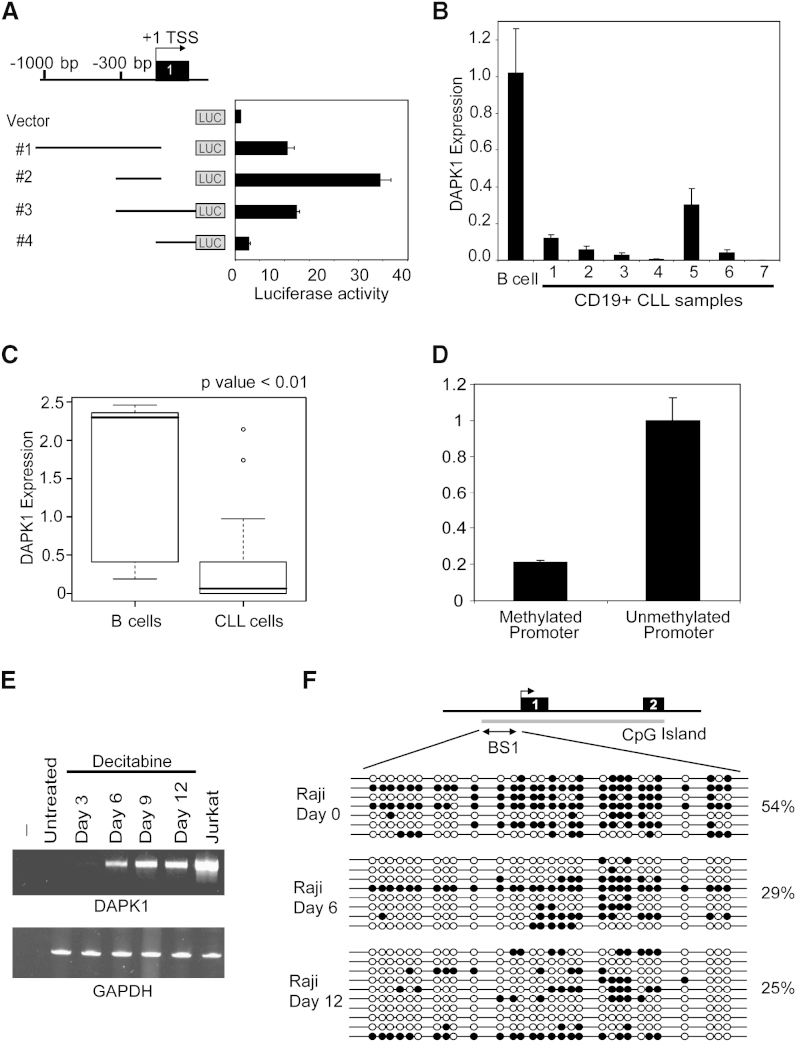
To study the relevance of promoter methylation for DAPK1 expression, we developed luciferase reporter constructs to test DAPK1 promoter activity in Jurkat cells. DAPK1 promoter construct no. 2 (c.1-1545–c.1-1151 bp), covering bisulfite region A1 showed a 30-fold increase in the reporter activity relative to the vector control (Figure 2A). Semiquantitative RT-PCR analysis of CD19+ CLL samples used in Figure 1A showed reduced DAPK1 expression in all seven samples (Figure 2B). Furthermore, DAPK1 expression in 50 unselected CLL cells also showed statistically significant (p < 0.01) reduction as compared to normal CD19+ B cells (Figure 2C). The functional relevance of DAPK1 promoter methylation was tested in a reporter assay in which the promoter sequence of construct no. 2 (Figure 2A) was ligated either following SssI treatment or untreated. Luciferase expression was 5-fold reduced in the methylated construct, further augmenting the finding that DAPK1 promoter methylation results in gene silencing (Figure 2D).
Figure 2.
Epigenetic Silencing of DAPK1 in CLL
(A) pGL3 luciferase constructs ligated with different DAPK1 5′ upstream inserts (nos. 1–4) were transfected into Jurkat cells, and reporter activity was studied 48 hr after transfection. pGL3 vector without insert was used as a negative control. Error bars indicate standard deviation (SD).
(B) Quantitative DAPK1 expression in normal B cells and seven CD19+ selected CLL samples. The expression in CLL samples is shown relative to the expression in B cells (defined as 1.0). Error bars indicate SD.
(C) Box plot of DAPK1 expression in 50 CLL samples as measured by semiquantitative RT-PCR and compared to its expression in six normal B cell samples. The distribution is significantly different (p < 0.01).
(D) Luciferase assays in 293T cells with either methylated or unmethylated DAPK1 construct no. 2. Error bars indicate SD.
(E) DAPK1 expression in Raji cells treated with 0.5 μM decitabine. RT-PCR was performed for DAPK1 and GAPDH on untreated and treated cell lines. Jurkat cells were used as a positive control.
(F) Bisulfite sequences in BS1 region (c.1-1509–c.1-1262) for untreated, 6 and 12 days decitabine-treated (0.5 μM) Raji cells. Each row represents a clone. The open circles indicate unmethylated CpG, and closed circles indicate methylated CpG dinucleotides. The overall methylation frequency is given in percentage. Error bars indicate SD.
Next, we treated Raji cells with 0.5 μM Decitabine for 3, 6, 9, and 12 days. RT-PCR for DAPK1 indicated that Decitabine treatment resulted in gradual upregulation of DAPK1 expression, while untreated Raji cells did not show any detectable expression (Figure 2E). Bisulfite sequencing of the BS1 region showed that 116 of the tested 210 CpG dinucleotides (55%) were methylated in untreated Raji cells. Treatment with Decitabine resulted in significantly (p < 0.001) reduced DNA methylation with 29% (70/240) on day 6 and 25% (68/270) on day 12 (Figure 2F).
DAPK1 Regulates Apoptosis in a Lymphoid Cell Line
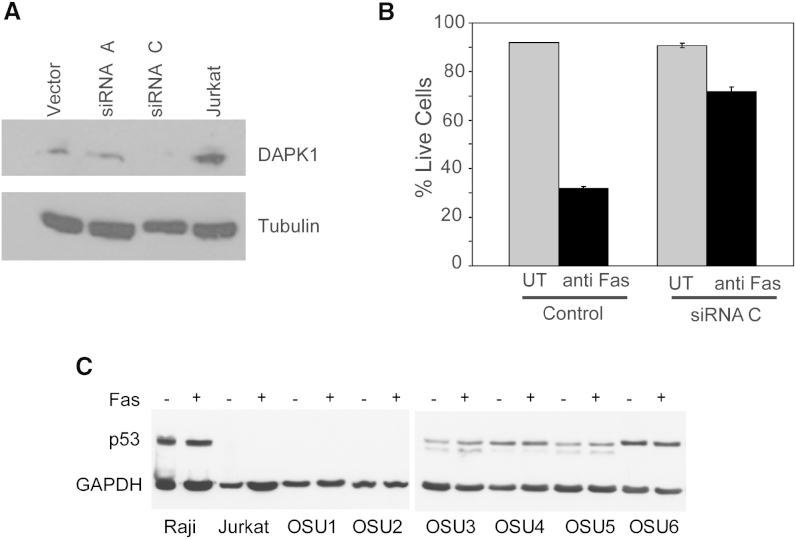
We stably transfected Jurkat cells with DAPK1 siRNA that resulted in its downregulation (Figure 3A). Cells transfected with vector alone or DAPK1 siRNA were treated with activating anti-Fas antibody, and apoptosis was examined using annexin-V-FITC/Propidium Iodide (annexin/PI) flow cytometry. Jurkat cells with inhibited DAPK1 expression showed a statistically significant increase in resistance to apoptosis compared to cells transfected with vector alone (p < 0.001; Figure 3B). These data demonstrated that DAPK1 is involved in Fas-induced extrinsic apoptosis in lymphoid cells. Next, we incubated 5 × 107 cells from nine CLL patients with or without Fas-activating antibody (50 or 100 ng/ml) for 24 hr and examined apoptosis by annexin/PI flow cytometry (Table S1). No significant effect on the percentage of nonapoptotic cells relative to the untreated control was seen (data not shown). Immunoblot analysis to assess potential changes in p53 expression following treatment showed no detectable differences (Figure 3C). In conclusion, these experiments are in line with our current knowledge that DAPK1 is a mediator of Fas-induced apoptotic signaling and loss of DAPK1 in CLL cells renders resistance to apoptosis (Cohen et al., 1999, Raveh et al., 2001).
Figure 3.
DAPK1 Regulates Apoptosis in Lymphoid Cells
(A) DAPK1 expression in Jurkat cells stably transfected with either vector alone, DAPK1 siRNA-A, or DAPK1 siRNA-C as measured by western blot. Tubulin expression served as a control.
(B) Percent live cells were measured in Jurkat cells stably transfected with vector alone or DAPK1 siRNA-C, treated with activating-Fas antibody (100 ng/ml). After 16 hr, cells were harvested and suspended in binding buffer with annexin V-FITC and propidium iodide, followed by flow cytometry to assess cell death. Error bars indicate SD.
(C) p53 expression in cells treated with either no, 50 ng/ml, or 100 ng/ml Fas-activating antibody.
A CLL Family with Linkage to Chromosome 9
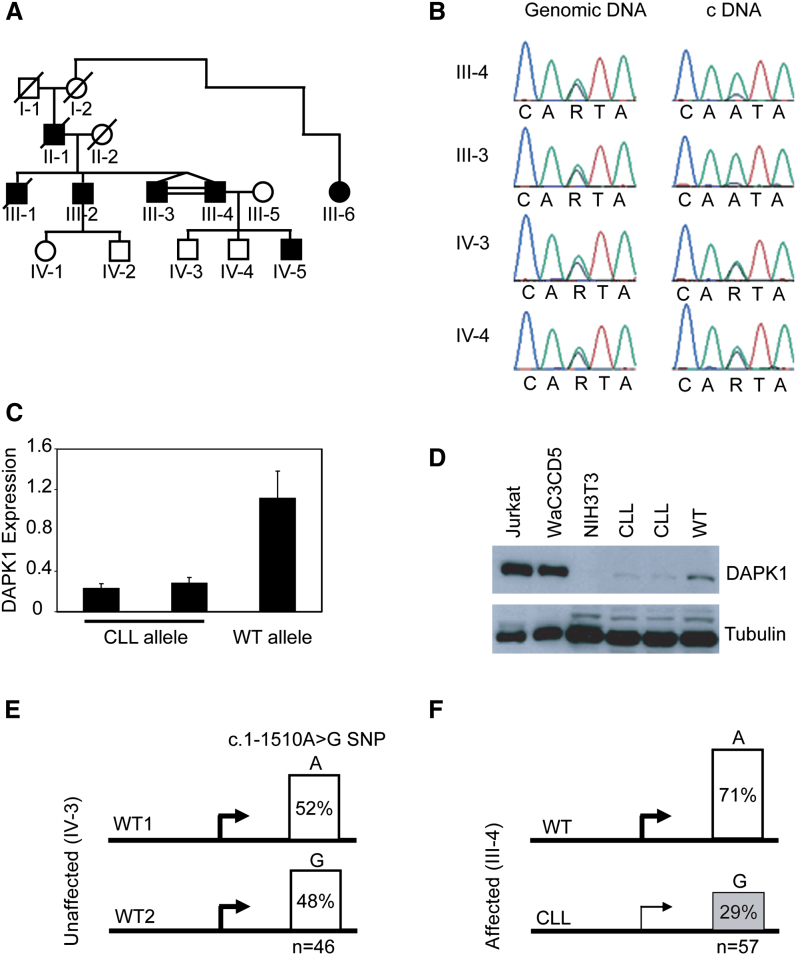
Lynch and colleagues previously documented an extended family in which a father and four sons were diagnosed with CLL (Lynch et al., 2002). We have since identified additional family members, both affected and unaffected (Figure 4A). Genome-wide linkage analysis with a panel of 400 microsatellite markers was performed using samples from individuals III-2, III-3, III-4, IV-3, IV-4, and IV-5. This identified a region on chromosome 9 between markers D9S175–D9S1776 with the highest nonparametric linkage (NPL) score of 0.96. The next highest scores were less than 0.42. High-resolution genotyping identified a common haplotype of 707 kb in all affected family members for whom samples were available. The segment of the presumed CLL haplotype includes three known genes (DAPK1, CTSL, and CCRK) and 11 predicted genes. Based on the epigenetic data indicating frequent loss of DAPK1 expression, we hypothesized that DAPK1 might be the predisposing gene mutated in this family. All exons and splice sites of DAPK1 were sequenced from DNA of the skin fibroblasts of four affected (III-2, III-3, III-4, and IV-5) as well as two unaffected family members (IV-3 and IV-4) and compared to the genomic sequence (NM_004938). Although five DNA variants were detected (one in exon 3, two in exon 16, and two in exon 26), none was unique for the CLL haplotype. These variants were useful in RT-PCR analysis of DAPK1 and showed that gene expression was highly reduced in the CLL allele of patients III-4 and III-3, whereas both wild-type (WT) alleles were equally expressed in unaffected individuals IV-3 and IV-4 (Figure 4B).
Figure 4.
DAPK1 Expression in CLL Family 4532
(A) Pedigree of CLL family no. 4532. Open circle and square represent unaffected female and male, respectively, while closed circle and square represent affected female and male.
(B) Sequencing of SNP c.114A > G from genomic DNA and cDNA of two unaffected (IV-3 and IV-4) and two affected (III-3 and III-4) individuals.
(C) RT-PCR on RNA isolated from monochromosomal hybrid clones with either WT or the CLL chromosome 9 from fibroblast cells of individual III-4 is shown. RPL4 expression was used as an internal control. Error bars indicate SD.
(D) DAPK1 protein expression from WT and CLL alleles in monochromosomal hybrids. Jurkat and WaC3CD5 cells were used as positive controls, and NIH3T3 cells were used as the negative control to show specificity to human DAPK1.
(E and F) The RT-PCR product amplifying SNP c.1510A > G in unaffected (IV-3; E) and affected (III-4; F) fibroblast cell lines was cloned, and individual clones were genotyped. Shown is the percentage of A and G clones in IV-3 and III-4, and n is the number of clones studied. The difference in allelic expression of DAPK1 in III-4 and IV-3 was statistically significant (p < 0.01).
Allelic Expression Imbalance in DAPK1 in Affected Family Members
As a next step we developed monochromosomal mouse-human hybrid clones containing either the WT (one clone) or the CLL (two clones) chromosome 9 from patient III-4. Semiquantitative RT-PCR (Figure 4C) and immunoblotting (Figure 4D) showed reduced DAPK1 expression from the two clones containing the CLL chromosome (25% or less) when compared to the clones containing the WT chromosome (100%). Reduced DAPK1 expression from the CLL allele was further confirmed in diploid cells. We cloned RT-PCR products containing exon 16 comprising the informative single nucleotide polymorphism (SNP) c.1510A > G from an unaffected (IV-3) and an affected (III-4) family member. Individual clones were genotyped by PCR using allele-specific primers. In individual IV-3, the ratio of A:G clones was close to the expected ratio of 1:1 (Figure 4E), while in individual III-4, the number of A-clones (WT allele) was approximately four times that of the G clones (CLL allele; Figure 4F), and this difference was statistically significant (p < 0.01).
Detection of a Rare Germline Mutation by DAPK1 Genomic Sequencing
In search of a germline mutation in DAPK1 regulatory sequences, we extended our genomic sequencing efforts. BAC clones derived from both the affected and the unaffected alleles of family member III-3 were generated, and two overlapping BAC clones for the CLL (CR956620 and CR956432) and the WT allele (CT009543 and CR974482) were chosen for shotgun sequencing. Approximately 400 kbp were sequenced for each allele, extending from 45 kb upstream to 100 kb downstream of DAPK1, covering the entire gene. No major rearrangements were detected in the sequence. In total, 281 single nucleotide differences were observed between the two alleles, out of which 162 were reported SNPs and 87 were located within repeat elements. We further eliminated 28 of the remaining 32 SNPs as candidate mutations, since they occurred in additional controls tested. One SNP seen in the CLL allele, c.1-6531A > G, was not found among 383 control samples from the US (n = 281) and from Northern Europe (n = 102). Screening of 263 CLL cases from the US (n = 129) and from Northern Europe (n = 134) identified one additional CLL sample from Scandinavia with SNP c.1-6531A > G. This Scandinavian sample (no. 102) showed the presence of this SNP in purified CLL cells and in T cells.
We further investigated samples from 75 CLL patients with one affected relative in order to determine if DAPK1 is a target for genetic mutations. We amplified and sequenced all exonic sequences (including splice sites) and the c.1-6531 suppressor region. As control samples we included DNAs from Wac3CD5 and two healthy donors. We identified numerous SNPs in exons 1 (noncoding), 3, 4, 16, 18, 19, and 26; some of them had already been reported as common polymorphisms (Table S2). We were unable to identify a missense mutation, a splice-site mutation, or an additional case with c.1-6531A > G.
HOXB7 Represses DAPK1 Transcription
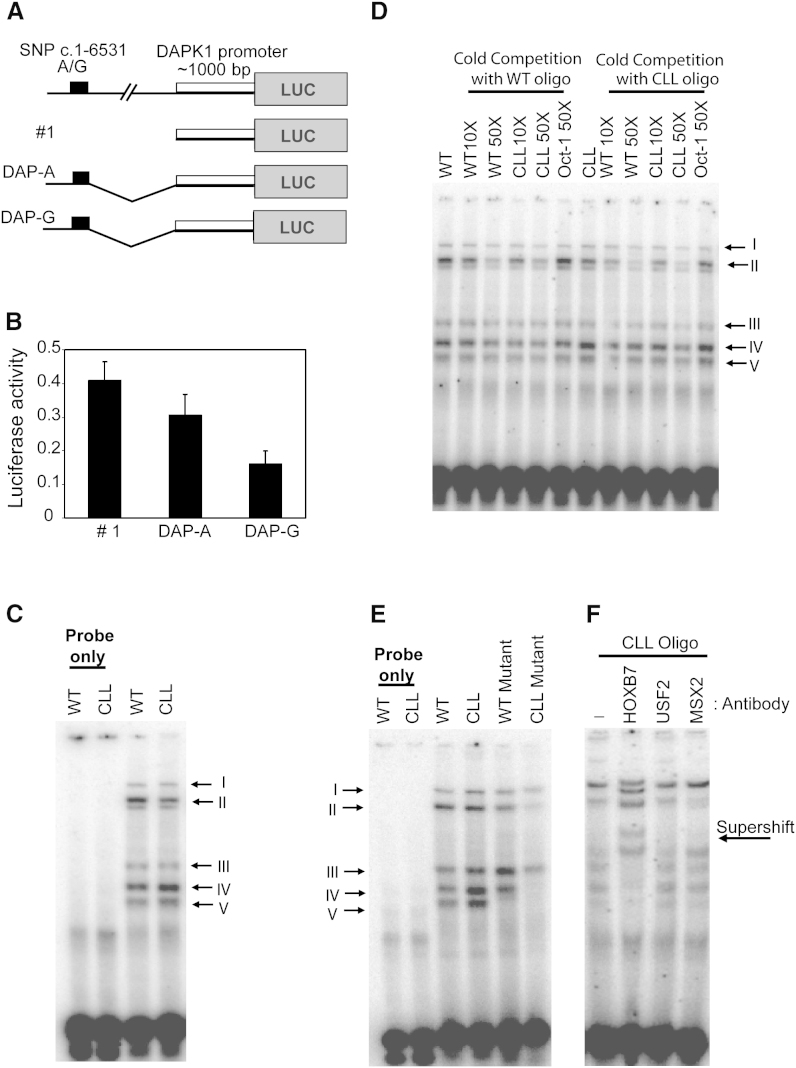
To study the effect of c.1-6531A > G on DAPK1 transcription, corresponding CLL and WT luciferase reporter constructs were designed (Figure 5A). A 357 bp fragment including either c.1-6531A (DAP-A) or c.1-6531G (DAP-G) was ligated upstream into luciferase construct no. 1 containing the DAPK1 promoter. Transcription from the DAP-A and DAP-G construct was suppressed by 39% and 70%, respectively, compared to control construct no. 1 (Figure 5B). This suggested that both sequences contain a suppressor element; however, the CLL-derived sequence, (DAP-G), displayed stronger effects.
Figure 5.
An A to G Change at c.1-6531 bp Regulates DAPK1 Expression in CLL Family
(A) A 357 bp PCR product with SNP c.1-6531A > G was ligated upstream to DAPK1 promoter (c.1-2215–c.1-1151) luciferase construct with either A (DAP-A) or G (DAP-G) as SNP.
(B) The DAPK1 promoter alone (no. 1), DAP-A, or DAP-G constructs were transfected into Jurkat cells, and the luciferase activity was measured. Error bars indicate SD.
(C) Nuclear extracts from Jurkat cells were analyzed by EMSA assay using WT or CLL oligo. Five specific bands (I–V) are marked.
(D) Ten- or fifty-fold molar excess concentrations of cold oligos were used for competition assays. An unlabeled Oct-1 oligo was used as a negative control.
(E) EMSA assays were performed using WT, WT mutant, CLL, and CLL mutant oligos. In mutant oligos, the adjacent bases to the c.1-6531 SNP were mutated.
(F) For the supershift assay, antibodies against HOXB7, USF2, and MSX2 were added to the Jurkat nuclear extract, and gel shift was studied using the CLL oligo.
To identify the potential suppressor molecule that binds at c.1-6531, an electrophoretic mobility shift assay (EMSA) was performed using oligonucleotides with either the WT oligo (A) or the CLL oligo (G). Using nuclear extracts from Jurkat cells, multiple bands were observed with both oligos (I–V; Figure 5C). Competition assays with unlabeled oligo confirmed specificity (Figure 5D). The specificity of binding was further illustrated by mutating one base on either side of the A or G within the respective oligos. Figure 5E shows that while the mutation in the WT oligo resulted in elimination of band V, the mutation in the CLL oligo resulted in elimination of bands IV and V. This result showed that protein binding at band IV is affected by the c.1-6531A > G mutation. Although EMSA is not a quantitative assay, we observed in multiple independent experiments that the intensity of band IV with the CLL oligo was stronger than with the WT oligo, hinting at a differential binding strength (Figures 5C–5E). Transcription factor binding-site prediction suggested that HOX family proteins might have differential affinity to A or G at the c.1-6531 SNP. In supershift assays using two HOX family proteins (HOXB7 and MSX2), only the HOXB7 antibody induced a clear shift, suggesting that HOXB7, or another closely related HOX protein, interacts with this site (Figure 5F).
Next, we tested DAPK1 as well as HOXB7 expression in B cells, T cells, and granulocytes of three healthy donors and selected B cells from CLL patients. DAPK1 expression was highest in normal B cells, and about one-fifth or one-third of the expression is seen in T cells and in granulocytes, respectively (Figure S2). As shown above, expression of DAPK1 was further reduced (or not detectable) in selected CLL cells. HOXB7 was expressed equally in B and T cells, however much reduced in granulocytes. Interestingly, HOXB7 was variably expressed in selected CD19+ CLL cells with several samples showing increased expression (Figure S2).
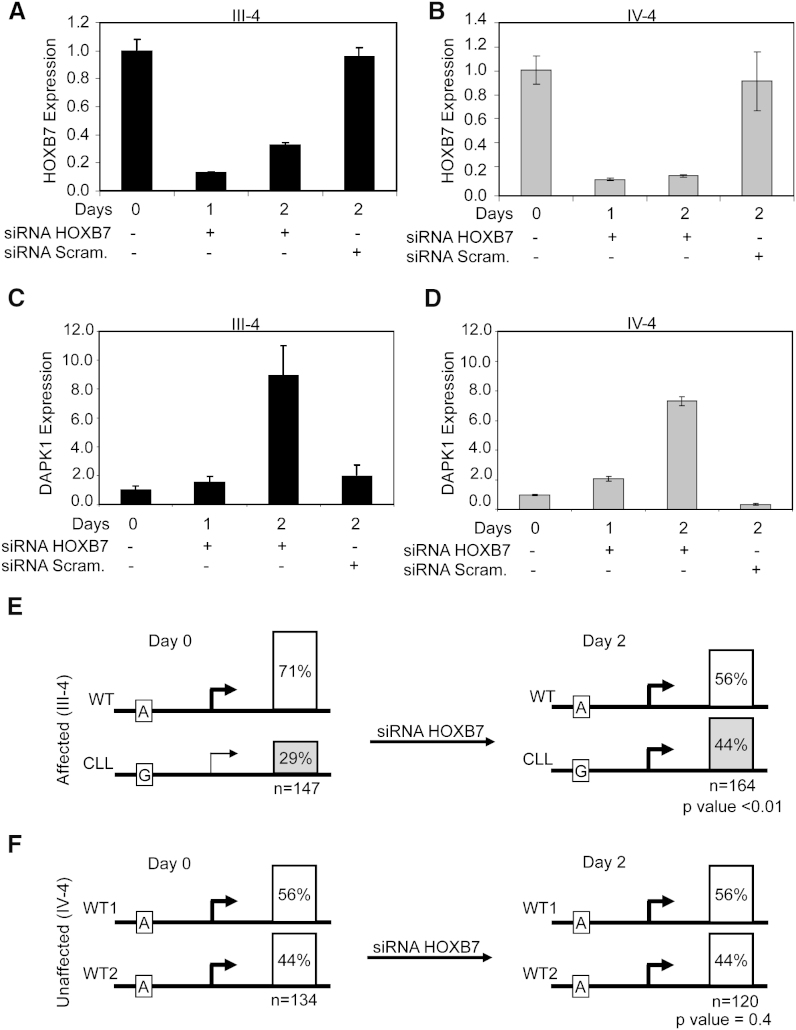
To explore whether HOXB7 affects DAPK1 expression, we transfected skin fibroblasts from an affected (III-4) and an unaffected family member (IV-4) with HOXB7 siRNA. Quantitative RT-PCR showed successful HOXB7 downregulation in both samples on day 1 following transfection, while scrambled siRNA did not show this downregulation (Figures 6A and 6B). DAPK1 upregulation in HOXB7 siRNA-transfected fibroblasts was observed in both III-4 and IV-4, showing that HOXB7 is a repressor of DAPK1 expression (Figures 6C and 6D). In addition, allele-specific expression analysis was studied in skin fibroblast cells before and after siRNA HOXB7 transfection in family members III-4 and IV-4. RT-PCR products (days 0 and 2) comprising informative SNP c.1510A > G were cloned, and individual clones were genotyped. In affected family member III-4, the increase in DAPK1 expression from the CLL allele was significantly higher than in the WT allele (p < 0.01), while the relative ratio between the two DAPK1 alleles in unaffected family member IV-4 remained unchanged (p = 0.4) (Figures 6E and F6). These results show that DAPK1 expression is downregulated by HOXB7 and suggest that c.1-6531A > G increases the affinity of HOXB7 binding, resulting in stronger repression of DAPK1 from the CLL allele.
Figure 6.
Downregulation of DAPK1 Expression by HOXB7
(A and B) Fibroblast cell lines from affected (III-4) and unaffected (IV-4) family members were transfected with 60 nM of HOXB7 siRNA or scrambled siRNA, and HOXB7 expression was studied at different time points by semiquantitative SYBR green RT-PCR. HOXB7 expression in untreated cells was set as 1.0. Error bars indicate SD. (C and D) DAPK1 expression was studied in the fibroblast cell lines transfected with HOXB7 siRNA or scrambled siRNA for different time points by quantitative SYBR green RT-PCR. DAPK1 expression in untreated cells was set as 1.0. Error bars indicate SD.
(E and F) Allele-specific expression of DAPK1 in an affected (E) and unaffected (F) family member was studied in fibroblasts from III-4 and IV-4, before and after transfection of HOXB7 siRNA. RT-PCR products including SNP c.1510A>G were cloned and genotyped. Shown is the percentage of the number of clones from the two alleles, and (n) indicates number of clones studied.
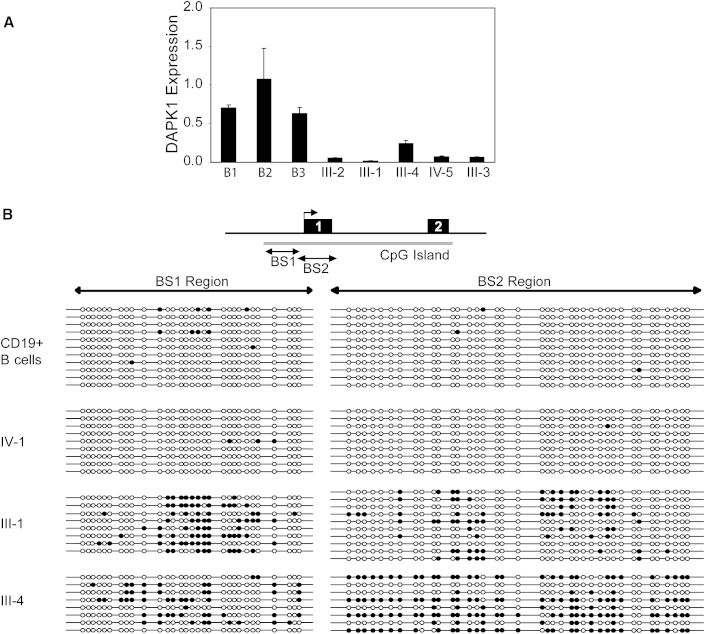
DAPK1 Promoter Methylation in CLL Cells of Affected Family Members
Our results indicated that expression of the CLL allele was reduced to 25% compared to the WT allele in affected family members. Thus, total DAPK1 expression was reduced to approximately 62.5% of that of normal levels. We hypothesized that additional DAPK1 repression occurred due to promoter methylation. Indeed, DAPK1 expression in PBMCs of five affected family members was remarkably downregulated when compared to three normal CD19+ cell controls (Figure 7A). Selected CD19+ B cells from blood of unaffected family members were not available as a control. Bisulfite sequencing of PBMC DNA from affected (III-1 and III-4) and unaffected family members (IV-1) for the BS1 and BS2 regions showed that both regions were highly methylated in the two affected family members (Figure 7B). Altogether, these data suggest that DAPK1 expression in the affected family members is significantly reduced by a combination of epigenetic and genetic aberrations.
Figure 7.
DAPK1 Expression and Promoter Methylation in CLL Cells of Family No. 4532
(A) RT-PCR for DAPK1 with GAPDH as an internal control on RNA extracted from blood cells of no. 4532 family members III-1, III-2, III-3, III-4 and IV-5 and from selected CD19+ normal B-cells from 3 healthy volunteers. Error bars indicate SD.
(B) DNA methylation analysis in CLL cells from CD19+ selected control B-cells, total PBMCs from one unaffected family member (IV-1) and two affected individuals (III-1 and III-4). Bisulfite treated DNA was amplified for the BS1 and BS2 regions. Each row represents a clone. The open circles indicate unmethylated and closed circles indicate methylated CpGs.
Discussion
We identified downregulaion of DAPK1 expression as a mechanism predisposing to CLL in a large family with seven individuals. A single nucleotide change enhances the binding affinity of transcription factor HOXB7 and results in downregulation of DAPK1 transcription. Importantly, DAPK1 is not only a target in familial CLL but is also inactivated in the majority of sporadic cases of CLL by epigenetic mechanisms. Interestingly, it has long been known that some high-penetrance genes (e.g., MLH1, BRCA1, p16) identified in familial cancers are also frequently silenced by epigenetic mechanisms in sporadic cancers (Esteller, 2002).
DAPK1 is an actin-filament-associated, calcium calmodulin-dependent, serine/threonine kinase that promotes apoptosis in response to various stimuli, including Fas, INF-γ, and TNF-α (Bialik and Kimchi, 2006). Increased DAPK1 expression leads to death-associated cellular alterations and cell morphology changes (Cohen et al., 1997). The DAPK1 promoter contains TGF-β response elements as well as p53-binding sites (Martoriati et al., 2005). DAPK1 overexpression results in upregulation of p53, suggesting a signaling feedback loop in which DAPK1 and p53 regulate each other's expression. DAPK1 suppresses cMYC- and E2F-induced cell transformation by activating p19ARF/p53-dependent apoptosis and also blocks tumor metastasis in vivo (Inbal et al., 1997, Raveh et al., 2001). DAPK1 inhibits extracellular signal-regulated kinase (ERK) activity, counteracts its survival signal (Chen et al., 2005), and promotes cytochrome c release from the mitochondria in response to TGF-β-induced apoptosis (Jang et al., 2002).
The observed frequent silencing of DAPK1 by promoter methylation in CLL could be one of the events required by the leukemic cells to escape cell death mediated by either the intrinsic or extrinsic pathways of apoptosis. Downregulation of DAPK1 by transfecting siRNA in Jurkat cells, an acute T cell leukemia cell line, resulted in reduced susceptibility to Fas-induced apoptosis. This suggests involvement of DAPK1 in the extrinsic apoptosis pathway in lymphocytes. Thus, it is reasonable to speculate that DAPK1 silencing may be an additional mechanism contributing to malignant progression by conferring resistance to apoptosis.
Past efforts have identified numerous affected genes in CLL, most in regions of somatic genetic alterations, mainly deletions, which are frequently observed in sporadic CLLs (Stilgenbauer and Dohner, 2005). For example, deletion of 13q14 is commonly seen in up to 50% of sporadic CLL (Dohner et al., 2000). Several candidate tumor-suppressor genes from this region have been proposed, including miR15 and miR16, two microRNAs targeting the oncogene BCL2 (Bullrich et al., 2001, Cimmino et al., 2005, Mertens et al., 2002).
Mapping efforts in CLL families have been relatively unsuccessful in identifying genes predisposing to CLL. Several candidate genes and predisposing polymorphisms have been proposed, including ARLTS1, a gene that resides on chromosome 13q14 (Calin et al., 2005) and P2RX7, located on chromosome 12q24 (Wiley et al., 2002). However, this predisposition was not supported in follow-up studies (Dao-Ung et al., 2004, Sellick et al., 2006, Thunberg et al., 2002).
How can these previous linkage results be reconciled with our findings regarding the role of DAPK1? Heritable germline mutations of DAPK1 itself may not underlie the disease in many of the larger CLL families that contributed to the previous linkage studies. The existence of multiple linkage peaks, none or few of which are statistically significant, suggests considerable locus heterogeneity or the existence of multiple mutated genes as recently proposed in an association study using nonsynonymous SNPs in candidate genes (Caporaso, 2006, Rudd et al., 2006). Since our results implicate downregulation of DAPK1 not only as the primary susceptibility factor in a large family, but also as a major event in sporadic CLL, it is quite possible that trans-acting factors, such as RNA genes (Calin and Croce, 2006, Esquela-Kerscher and Slack, 2006), play roles in the deregulation of DAPK1, and other genes, in CLL.
The occurrence of reduced DAPK1 expression in combination with frequent promoter methylation in tumor cells of familial CLL is intriguing. It is possible that reduced expression itself becomes a trigger for DAPK1 promoter methylation, which is initiated by transcriptional silencing (or reduced expression), followed by chromatin condensation and histone tail modifications, and finally by DNA methylation. This sequence of gene silencing events has been shown for target genes in the estrogen receptor signaling pathway in breast cancer cells (Leu et al., 2004) and for glutathione S-transferase in prostate cancer cells (Stirzaker et al., 2004). To explain the frequent occurrence of promoter methylation in sporadic CLL, one may postulate DAPK1 silencing due to modulation of upstream signals. One of these signals, HOXB7, was identified in this work. HOXB7 is a homeobox-containing transcription factor mediating a variety of developmental processes, including hematopoietic differentiation and lymphoid development (Lill et al., 1995, Shen et al., 1989). A scenario that may explain frequent targeting of the DAPK1 promoter by aberrant DNA methylation is the aberrant recruitment of DNA methyltransferase activity to the DAPK1 promoter by oncogenic proteins (Brenner et al., 2005, Di Croce et al., 2002).
In conclusion, we have identified DAPK1 as a novel putative tumor suppressor gene in CLL. The combination of linkage analysis and epigenetic studies demonstrated that downregulation of DAPK1 is common in CLL cells. The frequent silencing in sporadic CLL suggests that aberrant silencing of DAPK1 is an early and required event in leukemogenesis. Screening of familial CLL cases for DAPK1 promoter methylation could help in early diagnosis of CLL, which is a late onset disease. In addition, as DNA methylation events are reversible, our finding provides hope for the development of novel treatment regimens in CLL involving epigenetic therapies for gene reactivation.
Experimental Procedures
Patient Selection, Sample Collection, and Cell Lines
Blood and fibroblast cell lines were obtained from patients with B cell CLL through the CLL Research Consortium (CRC), from the Ohio State University (OSU), Creighton University (CU), and Uppsala University Hospital tissue banks. CLL patients and unaffected individuals from OSU, CU, and CRC provided written informed consent using Institutional Review Board-approved protocols. Written consent for Uppsala samples was provided according to the declaration of Helsinki. Seven CD19-selected sporadic CLL samples were obtained by positive selection using magnetic beads conjugated to anti-CD19 (MACS, Miltenyi Biotec, Auburn, CA). The resulting cells are at least 95% CD19 positive. All patients had immunophenotypically defined CLL as outlined by the modified NCI criteria (Cheson et al., 1996) and for Uppsala samples, according to the WHO classification. Cell lines WaC3CD5, Jurkat, Raji, lymphoblastoid cell lines generated from III-4, and skin fibroblasts from family 4532 were maintained in culture.
Quantitative DNA Methylation
Quantitative high-throughput DNA methylation analysis was done by MassARRAY system as described elsewhere (Ehrich et al., 2005). Four bisulfite reactions (A1–A4) were designed, which covered 17, 20, 22, and 29 CpGs, respectively, and extend from c.1-1573 to c.1-239 bp. The primer sequences are available upon request. For follow-up methylation studies on selected samples, bisulfite sequencing was performed as described earlier (Rush et al., 2004). Two bisulfite reactions covering regions c.1-1509–c.1-1262 (BS1) and c.1-1281–c.1-903 (BS2) were amplified.
Semiquantitative Reverse Transcriptase PCR (RT-PCR)
RT-PCR was performed and analyzed as described previously (Raval et al., 2005) using SUPERSCRIPT First-Strand Synthesis kit (Invitrogen). SYBRgreen PCR was done in triplicates with IQ SYBR Green Supermix (Bio-Rad, Hercules, CA).
Development of Monochromosomal Mouse-Human Hybrid Clones
Lymphoblastoid cells from samples III-4 was used to generate monoallelic clones as described previously (Papadopoulos et al., 1995). Genotyping confirmed one clone with WT and two clones with CLL chromosome 9.
Expression Vectors and Luciferase Reporter Assay
A modified pGL3 basic vector (Yu et al., 2004) was used to ligate the PCR-amplified DAPK1 promoter constructs. The SV40 promoter reporter construct served as positive control. Monochromosomal mouse-human hybrid cells were used to amplify SNP c.1-6531A > G in a 357 bp fragment with either the A or the G allele. The PCR products were ligated into the pGL3-promoter vector at the XhoI site, and the SV40 promoter was replaced by DAPK1 promoter region (c.1-2215–c.1-1151 bp) at the BglII and HindIII sites to create DAP-G or DAP-A constructs. Constructs were confirmed by sequencing. For transfection into Jurkat cells, a density of 1.5 × 105 cells/well was used in a 24-well plate with no serum for 2 hr. After 2 hr 1 μg plasmid pGL3 vector and 20 ng of pRL-TK internal control vector (Promega) were cotransfected into cells using polyethylenimine (Polysciences, Warrington, PA). After 4 hr, RPMI medium with 10% FCS was added to each well to make up the volume to 1 ml. The cells were further incubated for 48 hr, and the luciferase assay was performed according to manufacturer's instructions (Promega). Luciferase activity was normalized using pRL-TK activity. Each experiment was performed in triplicate. The in vitro methylation assay was performed as recently described (Yu et al., 2005).
siRNA Transfection
For stable transfection of DAPK1 siRNA into Jurkat cells, the pRS vector with different DAPK1 siRNA inserts (siRNA A and C) from OriGene, (Rockville, MD) were used. Ten micrograms of pRS-DAPK1 siRNA A, siRNA C, or pRS vector alone were transfected into the amphotropic Pheonix packaging cell line (60% confluent) using Superfect (Qiagen). Virus-containing medium was collected from the Pheonix cells after 48 hr, and cell debris was removed by centrifugation. One milliliter of fresh medium plus 1 ml of infectious medium, containing either pRS-DAPK siRNA A, siRNA C, or pRS vector alone were added to 2.5 × 105 Jurkat cells/well, and the 6-well plate was centrifuged at 2300 rpm for 90 min at room temperature in a Beckman centrifuge GPH, Rotor 3.7. Following the centrifugation step, 2 ml of fresh medium was added, and, 48 hr after incubation in 5% CO2, cells were selected for puromycin resistance using medium supplemented with 2 μg/ml Puromycin (Sigma, St Louis, MO). For transient transfection of HOXB7 siRNA (Ambion, Austin, TX) into fibroblast cells, 2 × 105 cells were plated, and 24 hr later cells were transfected using polyethylenimine (Polysciences, Warrington, PA), as described before.
Apoptosis and Flow Cytometric Studies
Jurkat cells stably transfected with pRS vector control or pRS-DAPK siRNA C were treated with 100 ng/ml anti-Fas (human activating, clone CH11) antibody (Upstate Biotechnology, Lake Placid, NY) for 16 hr and resuspended in binding buffer containing annexin V-fluorescein isothiocyanate (FITC) and propidium iodide according to the supplier's instuctions (BD Biosciences, San Diego, CA), and assessed by flow cytometry using a Beckman-Coulter model EPICS XL cytometer (Beckman-Coulter, Miami, FL). Each sample was run in triplicate.
Immunoblot Analysis
Immunoblotting was performed by transferring proteins to a nitrocellulose membrane (Hybond-ECL, Amersham Biosciences, Germany). Following the DAPK1 (Sigma-Aldrich, St. Louis, MO) and α-tubulin antibody (Oncogene, Boston, MA) staining, the proteins were detected with chemiluminescent substrate (SuperSignal, Pierce, Rockford, IL).
SSCP Analysis and Genotyping of Alleles by Colony PCR
PCR-amplified fragments were analyzed by single-strand conformation polymorphism (SSCP) as previously described (Liechti-Gallati et al., 1999). Variant bands were reamplified and used for direct sequencing on ABI Prism 3730 DNA analyzer (Applied Biosystems). For genotyping of the alleles for c.1-1510A > G SNP, cDNA was amplified and cloned. Colony PCR was performed in each samples by using primers specific to either “A” or the “G” SNP.
Electrophoretic Mobility Shift Assay (EMSA)
The oligonucleotides used for EMSA were WT oligo 5′-cttgccttggtcgtgattacctacagatgcctgaat-3′, WT oligo mutant 5′-cttgccttggtcgtaactacctacagatgcctgaat-3′, CLL oligo 5′-cttgccttggtcgtggttacctacagatgcctgaat-3′, and CLL oligo mutant 5′-cttgccttggtcgtagctacctacagatgcctgaat-3′. The double-stranded oligonucleotides were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase enzyme (NEB). The free probe was removed by purification in G50 Sephadex spin columns. The binding reaction was conducted at room temperature for 20 min with 5 μg of nuclear extract, 40,000 dpm (0.08–0.4 ng) of radiolabeled oligonucleotide probe, in 1x Ficoll buffer (10 mM Tris (pH 7.5), 1 mM DTT, 1 mM EDTA, and 4% Ficoll), 250 ng of poly(deoxyinosinic-deoxycytidylic acid) in 75 mM KCl, and double-distilled H2O to make the volume to 15 μl. For supershift assay, 1 μg each of HOXB7, MSX2 (Cemines, Golden, CO), and USF2 (Santa Cruz Biotechnologies, Santa Cruz, CA) was added, and the mixture was incubated at room temperature for an additional 30 min. DNA-protein complexes were fractionated by electrophoresis in 6% nondenaturating polyacrylamide gel, and radioactivity in the gels was visualized by autoradiography analyzed using the STORM860 image analyzer (Amersham Biosciences, NJ). Nuclear extracts from Jurkat cells were made as described previously (Frissora et al., 2003).
Statistics
Wilcoxon rank sum test was performed to compare the percent DAPK1 promoter methylation between CLL samples and normal controls and also to compare DAPK1 expression in 50 unselected CLL samples with that of CD19+ normal B cells. To obtain proportions for the number of “A” or “G” clones in unaffected and affected family members and for the methylated and unmethylated CpGs within DAPK1 promoter in Raji cells before and after decitabine treatment, the values were compared using the Z test (http://faculty.vassar.edu/lowry/VassarStats.html).
Acknowledgments
The authors wish to thank all members of the Plass and Byrd labs for critical discussions; Mattias Jansson from Uppsala University: and the Sequencing Division at the Wellcome Trust Sanger Institute for excellent technical assistance: Ramana Davuluri, Sandya Liyanarachchi, and Greg Singer for statistical support; Nickolas Papadopoulos for the conversion to haploidy; and Paivi Lahermo of the Finnish Genome Center for genotyping and linkage analyses. A.R. is supported by a T32 CA106196 fellowship in Cancer Genetics. B.H. is supported by a grant from the Dr. Mildred Scheel Foundation for Cancer Research, Germany. This publication was supported by National Cancer Institute grants CA110496 (J.C.B., C.P., and A.R.), CA101956 (C.P. and J.C.B.) CA81534 to the CLL Research Consortium (J.C.B., M.G., and T.J.K.), P30 CA16058 (A.d.l.C., C.P., and J.C.B.), The Leukemia and Lymphoma Society of America (J.C.B. and C.P.), and the D. Warren Brown Foundation (J.C.B. and C.P.). Further support came from revenue from Nebraska cigarette taxes awarded to Creighton University by the Nebraska Department of Health and Human Services (NDHHS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the state of Nebraska or NDHHS. Support was also received by NIH grant 5U01 CA86389 and the Swedish Cancer Society. C.P. is a Leukemia and Lymphoma Society scholar, and J.C.B. is a Leukemia and Lymphoma Society clinical scholar. P.C. and S.B. were supported by the Wellcome Trust.
Published: May 31, 2007
Footnotes
Supplemental Data include two figures and one table and can be found with this article online at http://www.cell.com/cgi/content/full/129/5/879/DC1/.
Contributor Information
Albert de la Chapelle, Email: albert.delachapelle@osumc.edu.
Christoph Plass, Email: christoph.plass@osumc.edu.
Supplemental Data
References
- Amundadottir L.T., Thorvaldsson S., Gudbjartsson D.F., Sulem P., Kristjansson K., Arnason S., Gulcher J.R., Bjornsson J., Kong A., Thorsteinsdottir U., Stefansson K. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 2004;1:e65. doi: 10.1371/journal.pmed.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialik S., Kimchi A. The Death-Associated Protein Kinases: Structure, Function, and Beyond. Annu. Rev. Biochem. 2006;75:189–200. doi: 10.1146/annurev.biochem.75.103004.142615. [DOI] [PubMed] [Google Scholar]
- Brenner C., Deplus R., Didelot C., Loriot A., Vire E., De Smet C., Gutierrez A., Danovi D., Bernard D., Boon T. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullrich F., Fujii H., Calin G., Mabuchi H., Negrini M., Pekarsky Y., Rassenti L., Alder H., Reed J.C., Keating M.J. Characterization of the 13q14 tumor suppressor locus in CLL: identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res. 2001;61:6640–6648. [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin. Oncol. 2006;33:167–173. doi: 10.1053/j.seminoncol.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Trapasso F., Shimizu M., Dumitru C.D., Yendamuri S., Godwin A.K., Ferracin M., Bernardi G., Chatterjee D., Baldassarre G. Familial cancer associated with a polymorphism in ARLTS1. N. Engl. J. Med. 2005;352:1667–1676. doi: 10.1056/NEJMoa042280. [DOI] [PubMed] [Google Scholar]
- Caporaso N. Chips, candidate genes, and CLL. Blood. 2006;108:415–416. [Google Scholar]
- Chen C.H., Wang W.J., Kuo J.C., Tsai H.C., Lin J.R., Chang Z.F., Chen R.H. Bidirectional signals transduced by DAPK-ERK interaction promote the apoptotic effect of DAPK. EMBO J. 2005;24:294–304. doi: 10.1038/sj.emboj.7600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheson B.D., Bennett J.M., Grever M., Kay N., Keating M.J., O'Brien S., Rai K.R. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996;87:4990–4997. [PubMed] [Google Scholar]
- Chim C.S., Fung T.K., Wong K.F., Lau J.S., Liang R. Frequent DAP kinase but not p14 or Apaf-1 hypermethylation in B-cell chronic lymphocytic leukemia. J. Hum. Genet. 2006;51:832–838. doi: 10.1007/s10038-006-0029-x. [DOI] [PubMed] [Google Scholar]
- Cimmino A., Calin G.A., Fabbri M., Iorio M.V., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen O., Feinstein E., Kimchi A. DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity. EMBO J. 1997;16:998–1008. doi: 10.1093/emboj/16.5.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen O., Inbal B., Kissil J.L., Raveh T., Berissi H., Spivak-Kroizaman T., Feinstein E., Kimchi A. DAP-kinase participates in TNF-alpha- and Fas-induced apoptosis and its function requires the death domain. J. Cell Biol. 1999;146:141–148. doi: 10.1083/jcb.146.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran M., Parker A., Orchard J., Davis Z., Wirtz M., Schmitz O.J., Oscier D. ZAP-70 methylation status is associated with ZAP-70 expression status in chronic lymphocytic leukemia. Haematologica. 2005;90:1078–1088. [PubMed] [Google Scholar]
- Dao-Ung L.P., Fuller S.J., Sluyter R., Skarratt K.K., Thunberg U., Tobin G., Byth K., Ban M., Rosenquist R., Stewart G.J., Wiley J.S. Association of the 1513C polymorphism in the P2X7 gene with familial forms of chronic lymphocytic leukaemia. Br. J. Haematol. 2004;125:815–817. doi: 10.1111/j.1365-2141.2004.04976.x. [DOI] [PubMed] [Google Scholar]
- Deiss L.P., Feinstein E., Berissi H., Cohen O., Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995;9:15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- Di Croce L., Raker V.A., Corsaro M., Fazi F., Fanelli M., Faretta M., Fuks F., Lo Coco F., Kouzarides T., Nervi C. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- Dohner H., Stilgenbauer S., Benner A., Leupolt E., Krober A., Bullinger L., Dohner K., Bentz M., Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- Ehrich M., Nelson M.R., Stanssens P., Zabeau M., Liloglou T., Xinarianos G., Cantor C.R., Field J.K., van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Slack F.J. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- Frissora F., Chen H.C., Durbin J., Bondada S., Muthusamy N. IFN-gamma-mediated inhibition of antigen receptor-induced B cell proliferation and CREB-1 binding activity requires STAT-1 transcription factor. Eur. J. Immunol. 2003;33:907–912. doi: 10.1002/eji.200323657. [DOI] [PubMed] [Google Scholar]
- Goldgar D.E., Easton D.F., Cannon-Albright L.A., Skolnick M.H. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J. Natl. Cancer Inst. 1994;86:1600–1608. doi: 10.1093/jnci/86.21.1600. [DOI] [PubMed] [Google Scholar]
- Goldin L.R., Ishibe N., Sgambati M., Marti G.E., Fontaine L., Lee M.P., Kelley J.M., Scherpbier T., Buetow K.H., Caporaso N.E. A genome scan of 18 families with chronic lymphocytic leukaemia. Br. J. Haematol. 2003;121:866–873. doi: 10.1046/j.1365-2141.2003.04372.x. [DOI] [PubMed] [Google Scholar]
- Goldin L.R., Pfeiffer R.M., Li X., Hemminki K. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood. 2004;104:1850–1854. doi: 10.1182/blood-2004-01-0341. [DOI] [PubMed] [Google Scholar]
- Inbal B., Cohen O., Polak-Charcon S., Kopolovic J., Vadai E., Eisenbach L., Kimchi A. DAP kinase links the control of apoptosis to metastasis. Nature. 1997;390:180–184. doi: 10.1038/36599. [DOI] [PubMed] [Google Scholar]
- Jang C.W., Chen C.H., Chen C.C., Chen J.Y., Su Y.H., Chen R.H. TGF-beta induces apoptosis through Smad-mediated expression of DAP-kinase. Nat. Cell Biol. 2002;4:51–58. doi: 10.1038/ncb731. [DOI] [PubMed] [Google Scholar]
- Kasper D.L., Harrison T.R. 16th edn. McGraw-Hill, Medical Pub. Division; New York: 2005. Harrison's principles of internal medicine. [Google Scholar]
- Katzenellenbogen R.A., Baylin S.B., Herman J.G. Hypermethylation of the DAP-kinase CpG island is a common alteration in B-cell malignancies. Blood. 1999;93:4347–4353. [PubMed] [Google Scholar]
- Leu Y.W., Yan P.S., Fan M., Jin V.X., Liu J.C., Curran E.M., Welshons W.V., Wei S.H., Davuluri R.V., Plass C. Loss of estrogen receptor signaling triggers epigenetic silencing of downstream targets in breast cancer. Cancer Res. 2004;64:8184–8192. doi: 10.1158/0008-5472.CAN-04-2045. [DOI] [PubMed] [Google Scholar]
- Liechti-Gallati S., Schneider V., Neeser D., Kraemer R. Two buffer PAGE system-based SSCP/HD analysis: a general protocol for rapid and sensitive mutation screening in cystic fibrosis and any other human genetic disease. Eur. J. Hum. Genet. 1999;7:590–598. doi: 10.1038/sj.ejhg.5200338. [DOI] [PubMed] [Google Scholar]
- Lill M.C., Fuller J.F., Herzig R., Crooks G.M., Gasson J.C. The role of the homeobox gene, HOX B7, in human myelomonocytic differentiation. Blood. 1995;85:692–697. [PubMed] [Google Scholar]
- Lynch H.T., Weisenburger D.D., Quinn-Laquer B., Watson P., Lynch J.F., Sanger W.G. Hereditary chronic lymphocytic leukemia: an extended family study and literature review. Am. J. Med. Genet. 2002;115:113–117. doi: 10.1002/ajmg.10686. [DOI] [PubMed] [Google Scholar]
- Martoriati A., Doumont G., Alcalay M., Bellefroid E., Pelicci P.G., Marine J.C. dapk1, encoding an activator of a p19ARF-p53-mediated apoptotic checkpoint, is a transcription target of p53. Oncogene. 2005;24:1461–1466. doi: 10.1038/sj.onc.1208256. [DOI] [PubMed] [Google Scholar]
- Mertens D., Wolf S., Schroeter P., Schaffner C., Dohner H., Stilgenbauer S., Lichter P. Down-regulation of candidate tumor suppressor genes within chromosome band 13q14.3 is independent of the DNA methylation pattern in B-cell chronic lymphocytic leukemia. Blood. 2002;99:4116–4121. doi: 10.1182/blood.v99.11.4116. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N., Leach F.S., Kinzler K.W., Vogelstein B. Monoallelic mutation analysis (MAMA) for identifying germline mutations. Nat. Genet. 1995;11:99–102. doi: 10.1038/ng0995-99. [DOI] [PubMed] [Google Scholar]
- Raval A., Lucas D.M., Matkovic J.J., Bennett K.L., Liyanarachchi S., Young D.C., Rassenti L., Kipps T.J., Grever M.R., Byrd J.C., Plass C. TWIST2 Demonstrates Differential Methylation in Immunoglobulin Variable Heavy Chain Mutated and Unmutated Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2005;23:3877–3885. doi: 10.1200/JCO.2005.02.196. [DOI] [PubMed] [Google Scholar]
- Raveh T., Droguett G., Horwitz M.S., DePinho R.A., Kimchi A. DAP kinase activates a p19ARF/p53-mediated apoptotic checkpoint to suppress oncogenic transformation. Nat. Cell Biol. 2001;3:1–7. doi: 10.1038/35050500. [DOI] [PubMed] [Google Scholar]
- Risch N. The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol. Biomarkers Prev. 2001;10:733–741. [PubMed] [Google Scholar]
- Rosenwald A., Wright G., Leroy K., Yu X., Gaulard P., Gascoyne R.D., Chan W.C., Zhao T., Haioun C., Greiner T.C. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J. Exp. Med. 2003;198:851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd M.F., Sellick G.S., Webb E.L., Catovsky D., Houlston R.S. Variants in the ATM-BRCA2-CHEK2 axis predispose to chronic lymphocytic leukemia. Blood. 2006;108:638–644. doi: 10.1182/blood-2005-12-5022. [DOI] [PubMed] [Google Scholar]
- Rush L.J., Raval A., Funchain P., Johnson A.J., Smith L., Lucas D.M., Bembea M., Liu T.H., Heerema N.A., Rassenti L. Epigenetic profiling in chronic lymphocytic leukemia reveals novel methylation targets. Cancer Res. 2004;64:2424–2433. doi: 10.1158/0008-5472.can-03-2870. [DOI] [PubMed] [Google Scholar]
- Sellick G.S., Webb E.L., Allinson R., Matutes E., Dyer M.J., Jonsson V., Langerak A.W., Mauro F.R., Fuller S., Wiley J. A high-density SNP genomewide linkage scan for chronic lymphocytic leukemia-susceptibility loci. Am. J. Hum. Genet. 2005;77:420–429. doi: 10.1086/444472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick G.S., Catovsky D., Houlston R.S. Familial chronic lymphocytic leukemia. Semin. Oncol. 2006;33:195–201. doi: 10.1053/j.seminoncol.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Shen W.F., Largman C., Lowney P., Hack F.M., Lawrence H.J. Expression of homeobox genes in human erythroleukemia cells. Adv. Exp. Med. Biol. 1989;271:211–219. doi: 10.1007/978-1-4613-0623-8_22. [DOI] [PubMed] [Google Scholar]
- Stilgenbauer S., Dohner H. Genotypic prognostic markers. Curr. Top. Microbiol. Immunol. 2005;294:147–164. doi: 10.1007/3-540-29933-5_9. [DOI] [PubMed] [Google Scholar]
- Stirzaker C., Song J.Z., Davidson B., Clark S.J. Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res. 2004;64:3871–3877. doi: 10.1158/0008-5472.CAN-03-3690. [DOI] [PubMed] [Google Scholar]
- Thunberg U., Tobin G., Johnson A., Soderberg O., Padyukov L., Hultdin M., Klareskog L., Enblad G., Sundstrom C., Roos G., Rosenquist R. Polymorphism in the P2X7 receptor gene and survival in chronic lymphocytic leukaemia. Lancet. 2002;360:1935–1939. doi: 10.1016/S0140-6736(02)11917-9. [DOI] [PubMed] [Google Scholar]
- Wiley J.S., Dao-Ung L.P., Gu B.J., Sluyter R., Shemon A.N., Li C., Taper J., Gallo J., Manoharan A. A loss-of-function polymorphic mutation in the cytolytic P2X7 receptor gene and chronic lymphocytic leukaemia: a molecular study. Lancet. 2002;359:1114–1119. doi: 10.1016/S0140-6736(02)08156-4. [DOI] [PubMed] [Google Scholar]
- Yu L., Liu C., Bennett K., Wu Y.Z., Dai Z., Vandeusen J., Opavsky R., Raval A., Trikha P., Rodriguez B. A NotI-EcoRV promoter library for studies of genetic and epigenetic alterations in mouse models of human malignancies. Genomics. 2004;84:647–660. doi: 10.1016/j.ygeno.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Yu L., Liu C., Vandeusen J., Becknell B., Dai Z., Wu Y.Z., Raval A., Liu T.H., Ding W., Mao C. Global assessment of promoter methylation in a mouse model of cancer identifies ID4 as a putative tumor-suppressor gene in human leukemia. Nat. Genet. 2005;37:265–274. doi: 10.1038/ng1521. [DOI] [PubMed] [Google Scholar]
- Yuille M.R., Matutes E., Marossy A., Hilditch B., Catovsky D., Houlston R.S. Familial chronic lymphocytic leukaemia: a survey and review of published studies. Br. J. Haematol. 2000;109:794–799. doi: 10.1046/j.1365-2141.2000.02111.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.