Version Changes
Revised. Amendments from Version 1
The first version of this paper was improved mainly by following the suggestions received from our reviewers. In this sense we updated the FastCentiScaPe web-tool, that is now working and that does not require the registration of the user's email anymore. The procedure simply requires a valid email to which the results will be sent as soon as they are ready. We also improved figure 1 by changing the layout. In the figure we highlighted three nodes, with a black arrow, that are used to show different topological characteristics. These three examples permit to describe the role of each node by adding few biological information and by proposing some directions for further investigations. The figure shows the results from both directed and undirected analysis and gives a comprehensive view of the data we obtained, related to the Stress centrality. Lastly we modified the description about the edge's weighting by adding a less concise and more clear explanation of its biological and topological meaning.
Abstract
The growing dimension and complexity of the available experimental data generating biological networks have increased the need for tools that help in categorizing nodes by their topological relevance. Here we present CentiScaPe, a Cytoscape app specifically designed to calculate centrality indexes used for the identification of the most important nodes in a network. CentiScaPe is a comprehensive suite of algorithms dedicated to network nodes centrality analysis, computing several centralities for undirected, directed and weighted networks. The results of the topological analysis can be integrated with data set from lab experiments, like expression or phosphorylation levels for each protein represented in the network. Our app opens new perspectives in the analysis of biological networks, since the integration of topological analysis with lab experimental data enhance the predictive power of the bioinformatics analysis.
Keywords: Cytoscape, centrality indexes, nodes, networks, topological analysis
Introduction
Biological processes can be abstracted as networks where the nodes represent biological entities and the edges represent interactions between them. Several kind of biological networks have been described: metabolic networks, gene networks, signal transduction networks and protein-protein interaction networks 1. Such networks are a static representation of the dynamics of complex biological processes, where molecular interactions give rise to cascades of reactions called pathways, determining the life processes of living organisms. Even if the scientific community is far from the capability of simulating the dynamical behaviour of such pathways, several important information can be extracted from the topological analysis of biological networks 2, 3 since the structure of a network can affect its function 4. In this context, different global parameters are commonly used to describe the properties of the whole network: centralities 5 are indexes that permit the identification of the most important actors by characterizing the nodes that, more than others, are good candidates as regulators of the underlying biological process.
In this paper we present CentiScaPe 2.1, a Cytoscape app 6, 7 for network centralities analysis. While the built-in NetworkAnalyzer tool is oriented in characterizing the global behaviour of the network, provided with several global network statistics, CentiScaPe is designed to identify the most relevant nodes and provides a more comprehensive set of centralities. The new version introduces the computation of centralities for directed and weighted networks, two features that are not available in any other Cytoscape app. As in the previous version it computes Average Distance, Diameter, Degree, Stress, Betweenness, Radiality, Closeness, Centroid Value and Eccentricity. New indexes as Eigenvector, Bridging centrality and Edge Betweenness have been added, making it the most complete app for network centrality analysis. Some hypothesis about the centralities interpretation and application in a real biological context are presented in the CentralitiesTutorial (see the Supplementary materials section).
A web version of CentiScaPe, i.e. FastCentiScaPe, is also available ( http://www.cbmc.it/fastcent/). It performs very fast computations for large networks by sending the network to a multiprocessor server. The centrality analysis results are sent to the user by e-mail in a Cytoscape readable, i.e. .xgmml, format.
The main aim of CentiScaPe is to produce results that could drive further lab experiments since the high score nodes identified by the computation should be considered as potential targets for drugs and could be the starting point for further investigations and for a deeper understanding of the underlying biological process.
Methods and implementation
The main usage of CentiScaPe is to rank the nodes of a network depending on their topological and experimental relevance. The numerical results are saved as node, edge or network attributes in the Cytoscape attributes browser, depending on the kind of parameters, so all the Cytoscape features for managing attributes are supported; after the computation the centralities are treated as normal Cytoscape attributes. CentiScaPe can be used in undirected networks 8, in directed networks and in weighted networks. Centralities for directed networks (see Supplementary materials: CentralitiesTutorial) are useful in the case of metabolic networks in which the direction describes the interaction between the substrates and reactants and the products of the chemical reactions and also in signal transduction networks, in which the direction depends on the flux of information. Considering the direction in the computation of centralities can lead to different results and interpretations than the undirected version.
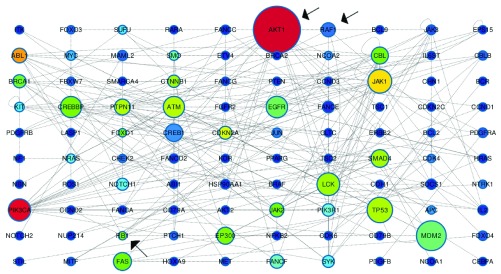
As an example, in Figure 1 we show the computation of the directed and undirected Stress applied to a network of Oncogenes (see Supplementary materials: Oncogenes.txt and Oncogenes_edge_directions.txt). Results of both the computations are shown and discussed.
Figure 1. Stress computed on oncogenes.
Size represents directed Stress, colour undirected Stress (blue=low, red=high).
The image, obtained using Cytoscape’s graphical tool, represents the different Stress values by using the colour and the size of the nodes. The size describes the value obtained by using the directed Stress: the bigger the node the higher the value; the colour describes the value obtained by using the undirected Stress: red is used for the highest value, blue for the lowest value. For example a large blue node requires particular attention because it is showing a node with a high centrality value if the network is considered as directed but with a very low value if the network is considered as undirected.
By analyzing the Oncogenes network we saw that the large red node, i.e. AKT1, shows how its Stress values are high using both algorithms. It plays a central role in different cell processes like metabolism, proliferation, cell survival, growth and angiogenesis. This role may highlight its high Stress value but, on the other hand, the high values suggest us to deeply investigate its characteristics; it is also involved in two different kind of cancer: breast and colorectal (see http://www.uniprot.org/uniprot/P31749). This evidence suggest that the node could be involved in cancer related processes but this assertion needs validation from several lab experiments.
Another interesting result is shown by the blue medium sized node RAF1. It shows how, using undirected Stress we obtained a low value, but by using the new algorithm we obtained an interesting Stress value. RAF1 was identified as a proto-oncogene with different and fundamental cellular functions (see http://www.omim.org/entry/164760). The results could be interpreted by saying that the directed network gives us a better understanding about how the gene, and its product, are involved in the development of cancer and could highlight that the use of the direction enhance our ability in describing a complex biological process.
The opposite situation is found in the third highlighted node, the small green node, RB1 in the right bottom corner. In this case the value computed with undirected Stress is not very high, but the value computed with the directed Stress is very low. RB1 is a gene involved in coding a protein involved in the retinoblastoma and other type of cancer like bladder cancer and osteogenic sarcoma. It was the first tumor suppressor gene found (see http://www.ncbi.nlm.nih.gov/gene/5925). As already said for RAF1, if the directed analysis is considered more reliable than the undirected one, then RB1 seems to be marginally involved in the Oncogenes network otherwise the undirected network is a better description. As already stated an experimental validation should be carried out in order to improve the results from the topological analysis and to better understand the role of each highlighted node.
All the results shown and described must be considered as a possible direction for further lab experiments. The main goal of this kind of analysis is to give us a comprehensive view that could be useful in describing the role of each node involved in a specific biological process and to drive future insights and investigations.
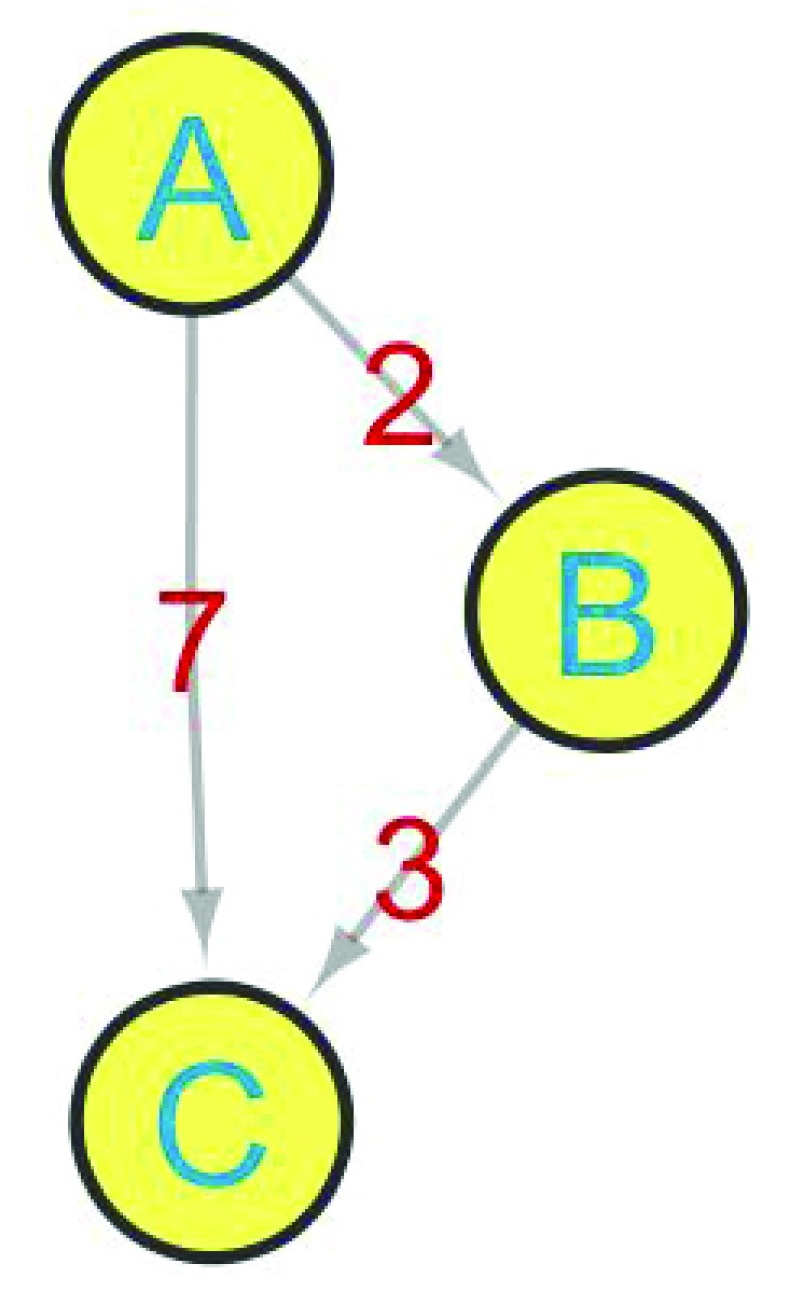
Second important features of the new version of CentiScaPe is the possibility of computing centralities for weighted networks, that are networks in which the edges are provided with an attribute that can be interpreted as a distance between the two connected nodes.
In the network in Figure 2 we have a distance ( dist) attribute for each edge. The values are dist( A, B) = 2, dist( B, C) = 3 and dist( A, C) = 7. Since A and C are connected by a single edge, in an unweighted computation, the distance from A to C is equal to one. But if the attributes of the edges are considered as distances, the shortest path between A and C is the one passing through B (= 2 + 3 = 5) since it is shorter, or lighter, than the one connecting A directly to C (= 7). The computation of weighted shortest paths will result in completely different values from the case wherein the weight is not considered. The user should consider that the weight is used in the sense that close nodes are more important than distant nodes. Therefore depending on the meaning of the attributes, one can use the real value or its reciprocal. For example if the attribute represents the speed of a reaction instead of a distance, the reciprocal should be used. This is because the higher the value of the speed, the nearer the nodes are: an increasing speed determines a decreasing reciprocal and the distance decrease by consequence.
Figure 2. A weighted network.
If the weight of the edge is not considered the shortest path from A to C is the directed edge, and the distance is one. If the weights are considered the shortest path from A to C is the one passing through B, and the distance is equals to 5.
An example of usage of weighted networks centralities analysis, can be found in Holly et al. 9 where an euclidean distance is given to each edge depending on the difference between the phosphorylation level of the proteins connected by that edge.
All the graphical features of the previous version of CentiScaPe, as the plot-by-node, the plot-by-centrality and the boolean-based result panel have been maintained in the new version. A complete guide can be found in Scardoni et al. 8 or in the CentiScaPe userguide available from the website.
Conclusions
CentiScaPe 2.1 have been enriched with new centrality parameters as Eigenvector, Bridging centrality and Edge Betweenness centrality and with the possibility to analyze directed and weighted networks. It allows integration centrality-based network analysis with experimental data by using nodes, edges or network attributes. The results of the computation can be used directly and are easily portable as Cytoscape attributes allowing the user to exploit all the other features of Cytoscape and its apps. Compared to the Cytoscape’s built-in tool, NetworkAnalyzer, CentiScaPe is an excellent integrative tool that allows the identification of potential target nodes from both the topological and the experimental point of view, and can be considered as an essential instrument for the characterizations of nodes in order to drive further experiments.
Software availability
Software available from the Cytoscape App Store
Latest source code
Source code as at the time of publication
https://bitbucket.org/F1000Research/centiscapepublic-archive
Archived source code as at the time of publication
License
Lesser GNU Public License 3.0: https://www.gnu.org/licenses/lgpl.html
Funding Statement
This work was supported by: Italian Association for Cancer Research (AIRC, IG 8690) (C.L.); Fondazione Cariverona; Nanomedicine project University of Verona and Fondazione Cariverona (C.L.). Directed centralities have been developed thanks to the GSoC2013 program.
[version 2; referees: 2 approved]
Supplementary materials
Supplementary files available from:
References
- 1. Caldarelli G: Scale-Free Networks: Complex Webs in Nature and Technology (Oxford Finance). Oxford University Press, USA.2007. 10.1093/acprof:oso/9780199211517.001.0001 [DOI] [Google Scholar]
- 2. Jeong H, Tombor B, Albert R, et al. : The large-scale organization of metabolic networks. Nature. 2000;407(6804):651–654. 10.1038/35036627 [DOI] [PubMed] [Google Scholar]
- 3. Barabási AL, Oltvai ZN: Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–113. 10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- 4. Strogatz SH: Exploring complex networks. Nature. 2001;410(6825):268–276. 10.1038/35065725 [DOI] [PubMed] [Google Scholar]
- 5. Koschützki D, Lehmann KA, Peeters L, et al. : Centrality indices. In Ulrik Brandes and Thomas Erlebach, editors, Network Analysis: Methodological Foundations, Springer.2005;16–61. 10.1007/978-3-540-31955-9_3 [DOI] [Google Scholar]
- 6. Cline MS, Smoot M, Cerami E, et al. : Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366–2382. 10.1038/nprot.2007.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saito R, Smoot ME, Ono K, et al. : A travel guide to Cytoscape plugins. Nat Methods. 2012;9(11):1069–76. 10.1038/nmeth.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scardoni G, Petterlini M, Laudanna C: Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25(21):2857–2859. 10.1093/bioinformatics/btp517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Currie HN, Vrana JA, Han AA, et al. : An approach to investigate intracellular protein network responses. Chem Res Toxicol. 2014;27(1):17–26. 10.1021/tx400247g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scardoni G, Tosadori G, Faizan M, et al. : F1000Research-centiscapepublic-archive. Zenodo. 2014. Data Source [Google Scholar]
- 11. Newman ME: Modularity and community structure in networks. Proc Natl Acad Sci U S A. 2006;103(23):8577–8582. 10.1073/pnas.0601602103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bhalla US, Iyengar R: Emergent properties of networks of biological signaling pathways. Science. 1999;283(5400):381–7. 10.1126/science.283.5400.381 [DOI] [PubMed] [Google Scholar]
- 13. Scardoni G, Laudanna C: Centralities based analysis of complex networks. In Yagang Zhang, editor, New Frontiers in Graph Theory InTech.2012. 10.5772/35846 [DOI] [Google Scholar]
- 14. Dijkstra EW: A note on two problems in connexion with graphs. Numerische Mathematik. 1959;1(1):269–271. 10.1007/BF01386390 [DOI] [Google Scholar]
- 15. Scardoni G, Montresor A, Tosadori G, et al. : Node interference and robustness: performing virtual knock-out experiments on biological networks: the case of leukocyte integrin activation network. PLoS One. 2014;9(2):e88938. 10.1371/journal.pone.0088938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gilbert D: JFreeChart. Reference Source [Google Scholar]
- 17. Liu YY, Slotine JJ, Barabási AL: Controllability of complex networks. Nature. 2011;473(7346):167–173. 10.1038/nature10011 [DOI] [PubMed] [Google Scholar]
- 18. Chuang HY, Hofree M, Ideker T: A decade of systems biology. Annu Rev Cell Dev Biol. 2010;26(1):721–744. 10.1146/annurev-cellbio-100109-104122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ivanciuc O, Balaban TS, Balaban AT: Design of topological indices. Part 4. Reciprocal distance matrix, related local vertex invariants and topological indices. J Math Chem. 1993;12(1):309–318. 10.1007/BF01164642 [DOI] [Google Scholar]