Abstract
Complete sets of cloned protein-encoding open reading frames (ORFs), or ORFeomes, are essential tools for large-scale proteomics and systems biology studies. Here we describe human ORFeome version 3.1 (hORFeome v3.1), currently the largest publicly available resource of full-length human ORFs (available at www.openbiosystems.com). Generated by Gateway recombinational cloning, this collection contains 12,212 ORFs, representing 10,214 human genes, and corresponds to a 51% expansion of the original hORFeome v1.1. An online human ORFeome database, hORFDB, was built and serves as the central repository for all cloned human ORFs (http://horfdb.dfci.harvard.edu). This expansion of the original ORFeome resource greatly increases the potential experimental search space for large-scale proteomics studies, which will lead to the generation of more comprehensive datasets.
Keywords: Human ORFeome, Gateway system, Clone resource, MGC collection, Nucleotide substitution rate, OMIM, GO slim, visant, hORFDB, High-throughput cloning
With the availability of complete genome sequences for many organisms [1], [2], [3], [4], [5], [6], [7], it is now possible to begin systematically to identify all functional genomic elements. Of particular interest are the elements of the genes that encode proteins, called open reading frames (ORFs). Full-length cDNA collections, which contain 5′ and/or 3′ UTRs in addition to the ORF, have been generated for several organisms, including Arabidopsis thaliana [8], Drosophila melanogaster [9], and Homo sapiens [10], [11]. While these collections are of immense value, they do not serve directly as ORF resources, but rather as collections of potential ORFs that must first be subcloned without UTRs before subsequent analysis of the encoded proteins can be performed [12]. One such example is the Mammalian Gene Collection (MGC) [10], [13]. This extensive collection of cDNAs was cloned into a vector that is not immediately useful for downstream functional experimentation. Ideally, clones should be archived in a convenient vector that would allow for high-throughput transfer of ORFs into a variety of different expression vectors, such as Gateway [12], [14] or any other recombinational cloning system [15], [16], [17].
In an effort to generate usable ORF collections, large-scale cloning projects, with the goal of cloning all predicted ORFs into flexible, recombinational vectors, have been described for a few model organisms including Brucella melitensis [18], Saccharomyces cerevisiae [19], and Caenorhabditis elegans [20], [21], [22]. These ORFeome resources [12] represent essential tools for large-scale protein characterization and therefore serve as a necessary bridge between genome annotation and systems biology.
Previously, we have described human ORFeome v1.1 [23], in which we used cDNAs from the MGC as templates to clone more than 8000 full-length ORFs. The utility of the resource was exemplified by its use in the generation of a large-scale human protein–protein interaction or “interactome” map, in which 6.4 × 107 (8000 × (8000 + 1)) possible pair-wise combinations were tested for yeast two-hybrid (Y2H) interactions, resulting in the identification of 2754 Y2H interactions between the products of 1549 ORFs [24]. Since linear increases in the number of ORFs in the ORFeome collection result in quadratic expansions in the biological search space that can be tested, the expansion of the human ORFeome will play an essential role in enhancing this interactome mapping effort as well as other systematic ORF studies. For example, using a matrix-based Y2H approach (testing all pair-wise combinations), an increase of 4000 ORFs (from 8000 to 12,000) would allow for the testing of 14.4 × 107 combinations, corresponding to an additional 8 × 107 pair-wise combinations and a 125% increase in the search space. Likewise, a more complete ORFeome resource will yield more comprehensive datasets for all systematic studies of ORF function from protein arrays [25] to high-content screening [26].
One of the main strategies of systems biology, the integration of genome-wide data generated by multiple orthogonal proteomic techniques [27], has been hampered by incomplete datasets. As the complete human ORFeome becomes one of the standard sets of clones used in reverse proteomic studies, the number of analyzed proteins in large-scale experiments should gradually improve, facilitating the integration of these data and ultimately leading to a better understanding of the properties of biological systems.
Here, we describe human ORFeome version 3.1 (hORFeome v3.1), a resource of 12,212 distinct ORFs, and introduce an improved human ORFeome database.
Results
Defining hORFeome v3.1
We define an ORF as the protein coding sequence of a gene from its start to its stop codon and excluding the 5′ and 3′ UTRs. A major milestone of the human ORFeome project will be the generation of the complete ORFeome, defined as the collection of protein-encoding ORFs representing at least one splicing isoform for every gene predicted in the human genome. Subsequently this resource will include splice variants and polymorphic variants for each gene.
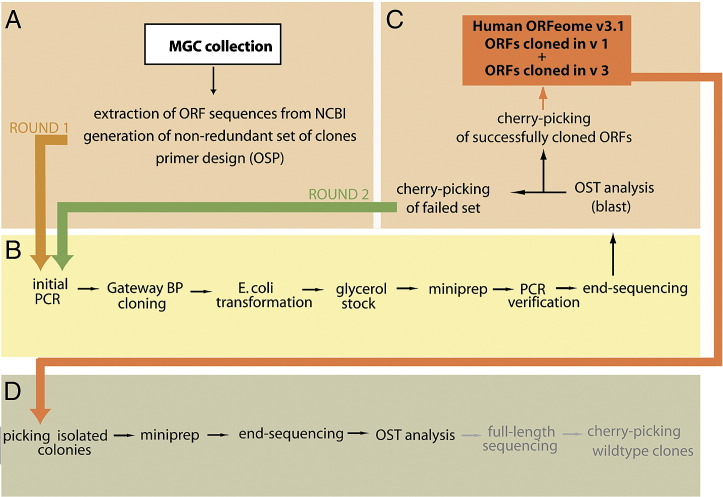
In the first human ORFeome project (hORFeome v1.1) we used directed PCR on the available set of cDNAs from MGC successfully to clone 8107 ORFs into the Gateway entry vector. In our second iteration of the human ORFeome effort, referred to here as the human ORFeome 3 project, we have attempted to clone ORFs from an additional 6027 cDNAs that are not part of v1.1. These cDNAs can be divided into two classes: 4806 clones correspond to newly available MGC clones mostly obtained by random cDNA library screening. The second class of cDNAs corresponds to 1221 MGC clones that failed to clone during the first human ORFeome project. All ORFs were passed through a semiautomated pipeline (Fig. 1) that allowed for efficient cloning and data analysis. First, clones shorter than 100 nucleotides (a threshold three times smaller than the convention of 300 nucleotides), and clones for which no complete coding sequence was available from NCBI, were eliminated from further analysis. Isoforms or polymorphic clones of the same gene were processed individually and treated as separate ORFs. ORFs that we failed to clone in the first round were attempted a second time and if successfully cloned were consolidated with the ORFs successfully cloned in hORFeome v1.1. This consolidated ORFeome collection is called human ORFeome version 3.1 (Fig. 1). Even-numbered version names have been reserved for ORFeome collections that contain single isolated wild-type clones for each ORF [21].
Fig. 1.
Automated human ORFeome pipeline. (A) A filter computationally removed ORFs, extracted from MGC cDNAs, that were not full-length; short ORFs (< 100 nucleotides); and redundantly cloned ORFs. Isoforms and SNP variants of each gene were retained and treated as individual clones. (B) Clones were PCR amplified, Gateway cloned, and sequenced at the 5′ end using universal primers. (C) The resulting ORF sequence tags (OSTs) were aligned to the ORFeome database containing all attempted ORF sequences. Clone attempts that produced a PCR band but whose 5′ OST did not correspond to the expected cDNA underwent a second round of cloning. Successfully cloned ORFs from hORFeome v1 and v3 were combined to form hORFeome v3.1. (D) To investigate the quality of this resource, we picked isolated colonies for 564 ORFs and sequenced them at their 5′ and 3′ ends. In the upcoming ORFeome version 4 project, clones without mutations in their end sequences will undergo full-length sequencing to generate a resource of wild-type clones for each ORF in the hORFeome v3.1.
ORF sequence tag (OST) analysis
Following BP recombinational cloning and transformation, ORFs were sequenced from the 5′ end to confirm their identity. Sequencing reads were truncated after the first 400 nucleotides (or fewer if the sequence read was short or of low quality before the 400th nucleotide) and used as queries for BLAST alignment [22] against an internal database containing sequences for all of the ORFs we attempted to clone. ORFs whose 5′ OST aligned to the predicted sequence and contained the predicted start codon were scored as successfully cloned.
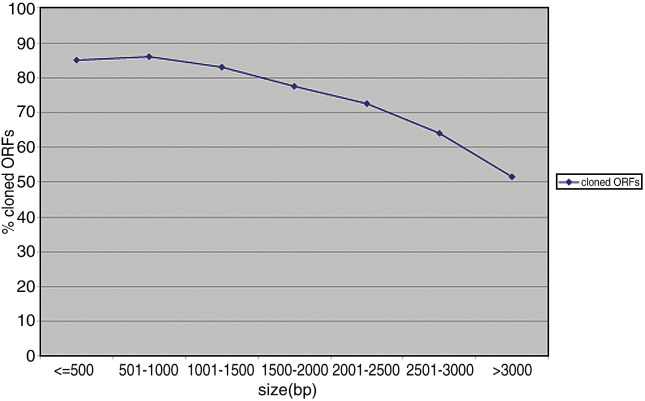
Following two rounds of cloning, we successfully isolated 4111 ORFs. Of these, 659 corresponded to ORFs that we failed to clone in the first version of the human ORFeome project [23], representing a 54% recovery (659/1220). Since the primers used here were identical to those used in our first attempt [23], the initial cloning failures were likely due to technical errors. As previously observed, the success rate correlated with the size of the ORFs, with small ORFs showing a higher success rate than larger ORFs (see Supplementary Fig. 1) [22].
In total, hORFeome v3.1 contains 12,212 ORFs, corresponding to 10,214 genes, representing a 51% expansion of the original human ORFeome resource. The ORFs range in size from 102 to 5499 bp and include 650 polymorphic ORFs and 1160 ORFs that correspond to multiple splice forms.
Quality assessment of hORFeome v3.1
hORFeome v3.1 is a collection of clones that were generated by PCR from unique, individual cDNA templates. Among the PCR products from individual templates are clones with mutations that originate during primer synthesis and clones that acquired mutations during PCR amplification. Following recombination, clones can also contain empty Gateway donor vector in which the toxic ccdB gene, which normally prevents growth of the empty vector, is no longer functional due to mutation [12], [14]. Since our cloning strategy generates minipools rather than individual isolated clones for each ORF, we did extensive sequence analysis on a set of individual isolated clones to assess the overall quality of hORFeome v3.1.
A thorough investigation of the quality of hORFeome v3.1 was carried out by isolating single colonies from a large number of minipools and end-sequencing them from the 5′ and 3′ ends. Five hundred sixty-four ORFs (six plates) were chosen at random from hORFeome v3.1 (three plates previously generated during the human ORFeome project 1 and three plates of newly cloned ORFs) and six single isolated colonies were picked from each well. These 3384 clones (6 plates × 94 wells × 6 colonies) were end-sequenced using two different pairs of sequencing primers, corresponding to two forward and two reverse oligonucleotides that anneal to distinct vector sequences. In total 13,536 sequence reads were generated (3384 clones × 2 pairs of primers × 2 reads) and only high-quality sequence reads (at least 100 nucleotides with a PHRED score of >19) were retained for further analysis. We expected to see mutations that arise from two sources: mutations in the primer sequence likely originated during primer synthesis, while those that were found in the ORF were most likely due to PCR-induced errors. If this were the case we should find different rates of mutation depending on the source of mutation.
We identified mutations in 9.8% of the primers from ORFeome project 1 and in 2.6% of the primers from ORFeome project 3. This difference in primer quality is most likely due to a less error-prone primer synthesis protocol used for ORFeome project 3. The analysis of 4,068,518 nt of ORF sequence (excluding primer sequence) revealed 316 mutations that were distributed among 275 sequences (Table 1). The resulting misincorporation rate using KOD polymerase (Novagen) [28] amounts to one nucleotide substitution every 12,875 bp. This mutation rate is higher than previously reported in hORFeome v1.1 (one mutation every ∼ 35,000 bp) using the same polymerase, but that analysis was limited to only 70,000 nt [23]. Nevertheless, this rate is substantially lower than the mutation rate observed in the C. elegans ORFeome (1/1500 bp), which was generated using a high-fidelity Taq DNA polymerase [21]. Considering the much larger dataset analyzed here (4 × 106 in v3.1 vs 7 × 104 in v1.1), this study provides the most extensive quality assessment of any large-scale ORFeome cloning project to date.
Table 1.
Summary of the analysis of the nucleotide substitution rate in ORF and primer sequences in human ORFeome v3.1
| No. of analyzed nucleotides | No. of mutations | 1 mutation every x nucleotides | No. of analyzed sequences | No. of mutated sequences | Percentage of mutated sequences | |
|---|---|---|---|---|---|---|
| ORF sequences | 4 × 106 | 316 | 12,875 | 9400 | 275 | 2.0 |
| Primer sequences | 17 × 104 | 588 | 293 | 9118 | 557 | 6.1 |
hORFeome v3.1 properties
Distribution of ORFs on chromosomes
Most MGC clones were generated by screening a diverse set of cDNA libraries for full-length cDNAs [29], [30]. The probability of finding a particular clone is dependent on its representation in the library; therefore, it may be difficult to identify cDNAs that are expressed under restricted conditions or in small subsets of cells. Given this expression bias, are our cloned ORFs distributed equally throughout the genome, or are there regions that are relatively under- or overrepresented with respect to cloned ORFs? For example, in C. elegans, there is a marked underrepresentation of cloned ORFs on chromosome 5, in a region containing a large cluster of G-protein-coupled receptors [21].
We used BLAT to align the cloned ORFs to the human genome using UCSC's human genome build Golden Path hg35 [31], [32]. The number of ORFs associated with each chromosome was then compared to the number of RefSeq models [33], defined as the most comprehensive nonredundant set of full-length cDNAs. On 22 chromosomes ORF cloning was uniformly successful, with a cloning success rate ranging between ∼ 42 and ∼ 53%. In contrast, cloned ORFs on chromosome 21 were slightly underrepresented (Table 2).
Table 2.
Summary of successfully cloned ORFs compared to RefSeq annotations on each chromosome
| Chromosome | No. of RefSeqs | No. of ORFs | Percentage of success |
|---|---|---|---|
| 1 | 2396 | 1207 | 50.3 |
| 2 | 1499 | 775 | 51.7 |
| 3 | 1294 | 676 | 52.2 |
| 4 | 838 | 416 | 49.6 |
| 5 | 1030 | 514 | 49.9 |
| 6 | 1227 | 620 | 50.5 |
| 7 | 1077 | 565 | 52.4 |
| 8 | 780 | 397 | 50.8 |
| 9 | 904 | 439 | 48.5 |
| 10 | 942 | 435 | 46.2 |
| 11 | 1474 | 675 | 45.8 |
| 12 | 1219 | 604 | 49.5 |
| 13 | 367 | 189 | 51.5 |
| 14 | 748 | 395 | 52.8 |
| 15 | 695 | 346 | 49.8 |
| 16 | 972 | 511 | 52.6 |
| 17 | 1342 | 667 | 49.7 |
| 18 | 321 | 156 | 48.6 |
| 19 | 1539 | 773 | 50.2 |
| 20 | 762 | 321 | 42.1 |
| 21 | 372 | 116 | 31.2 |
| 22 | 62 | 30 | 48.3 |
| X | 573 | 303 | 52.9 |
| Y | 963 | 408 | 42.4 |
| All | 23,396 | 11,538 | 49.3 |
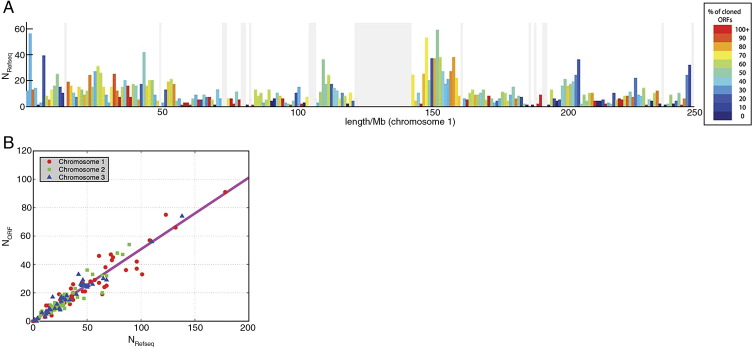
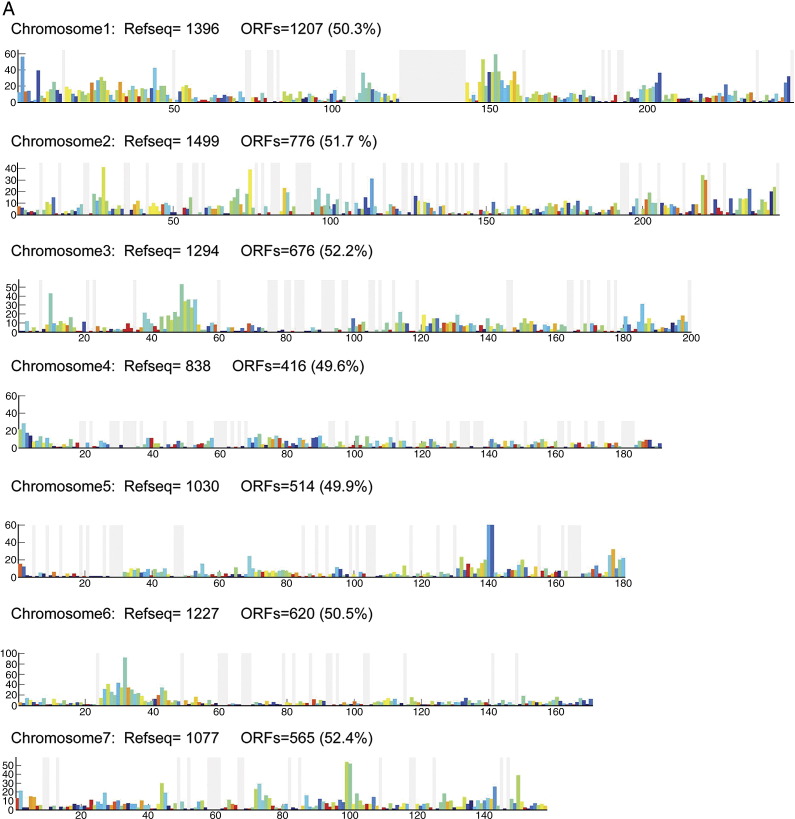
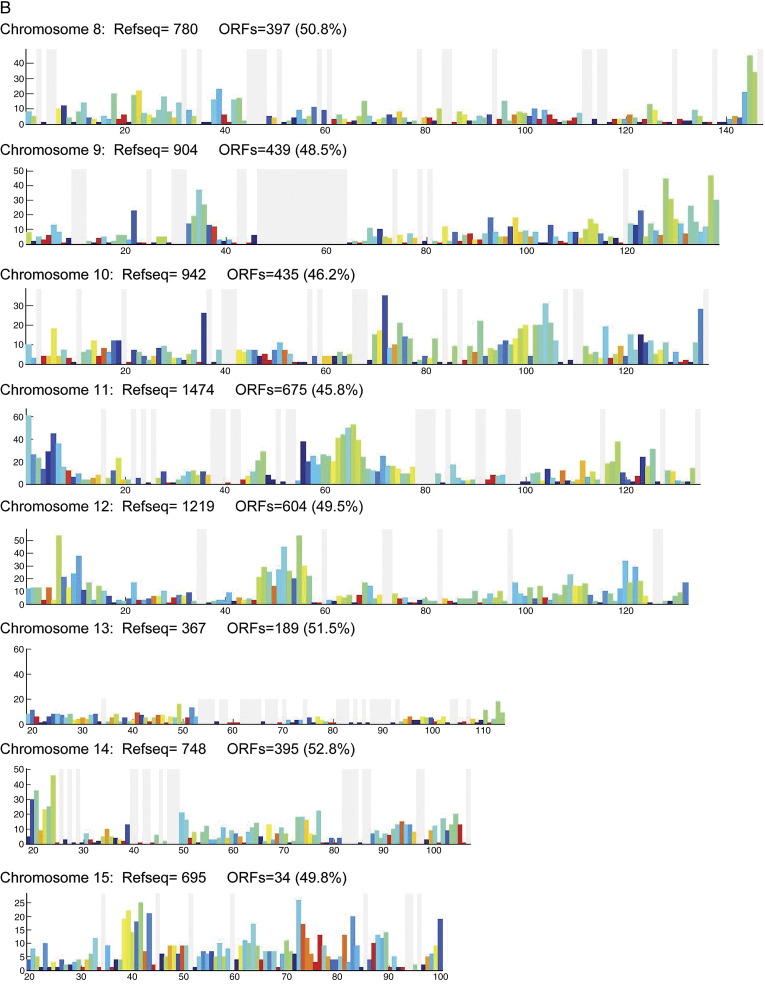
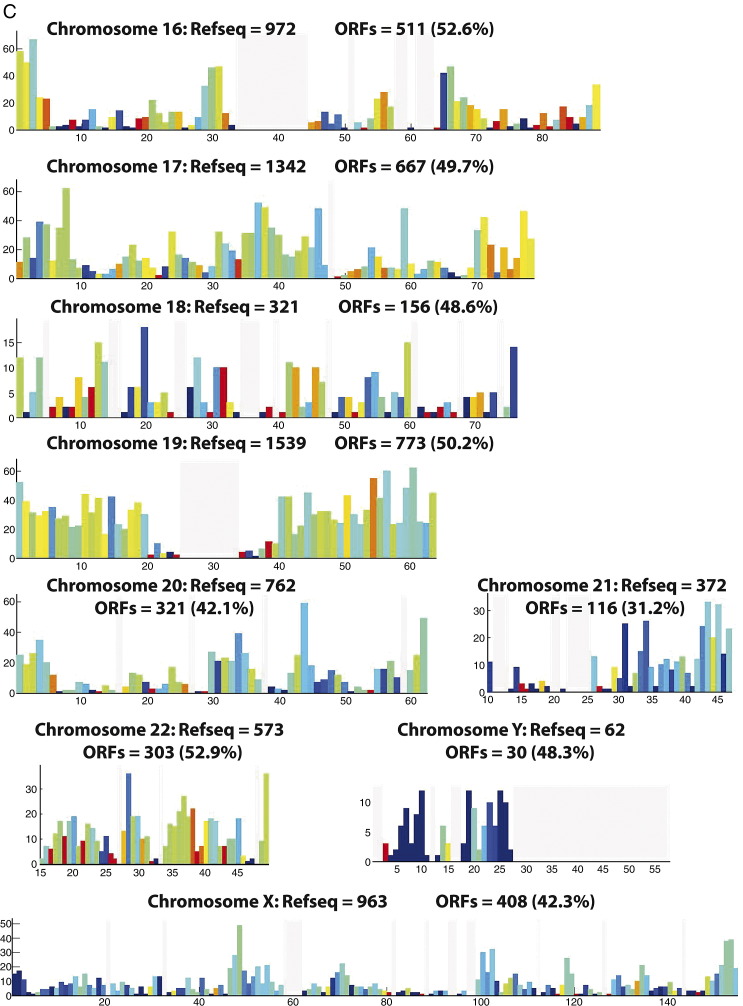
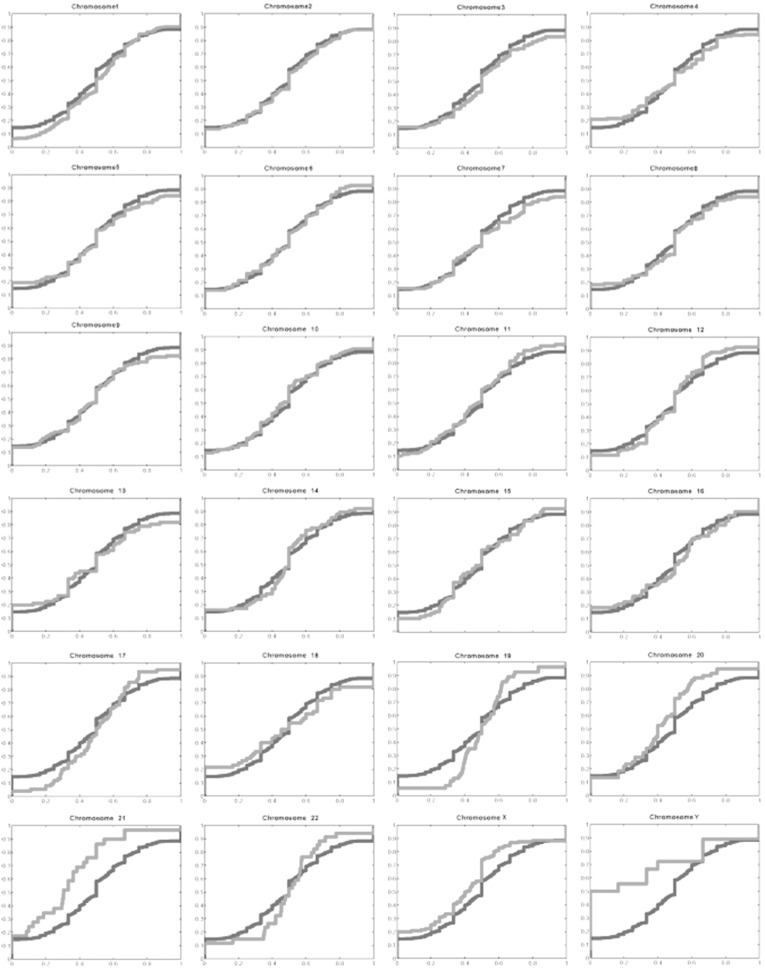
To investigate ORF distribution along each chromosome, we divided each chromosome into 1-Mb bins and counted the number of ORFs in each bin. We calculated the cloning success rate in each bin as the ratio of the number of cloned ORFs to RefSeq sequences (Fig. 2A). To check quantitatively whether there is a bias toward sparse or dense RefSeq regions in the cloning success rate, we plotted the number of cloned ORFs versus the number of RefSeq models for each bin for three chosen chromosomes (Fig. 2B). We find that the ORF density is linearly proportional to the RefSeq density and that the overall cloning success rate is ∼ 49% for every bin of chromosomes, showing that the cloned ORFs are equally distributed within chromosomes and that there are no regions of obvious over- or underrepresentation. We then compared the distribution of the local success rates among chromosomes and noticed a significantly different local success rate distribution on chromosomes 19, 20, 21, X, and Y (Supplementary Fig. 3). On chromosomes 20, 21, and X, this shift could be explained by the lower overall cloning success rate. On chromosomes 19 and Y, for which the cloning success rate was high, this shift might be due to erroneous gene annotation or related to the fact that these two chromosomes are among the shortest of chromosomes.
Fig. 2.
Distribution of cloned ORFs within each chromosome. (A) To determine whether chromosomes contain regions that are under- or overrepresented in the ORFeome, we divided each chromosome into 1-Mb bins and counted the number of cloned ORFs and the number of RefSeq sequences in each bin. The x axis represents the length (Mb) of chromosome I and the y axis the number of RefSeq sequences in each bin. The colors of the bars reflect the percentage of RefSeqs in each bin that were cloned in the ORFeome, as indicated by the color key. If the cloning success rate was uniformly independent of the position on the chromosome, every bar should be colored the same. Gray lines correspond to bins without RefSeq models and the wide gray vertical region in the middle of the chromosome corresponds to the centromere (Supplementary Fig. 2 shows graphs of the remaining chromosomes). (B) The number of cloned ORFs in bins 1 Mb in length, NORF, shown as a function of the number of predictions in the same respective bins, NRefSeq. Three chromosomes were taken as examples in this graph (chromosomes 1, 2, and 3). The straight line represents the linear regression to the data points. While only three of the chromosomes have been shown for clarity, the fitting yields NORF = (0.49 ± 0.006)NRefSeq + (0.42 ± 0.32) if all chromosomes are taken into account, predicting an overall cloning success rate of about 49% for every chromosomal bin.
GO Slim terms
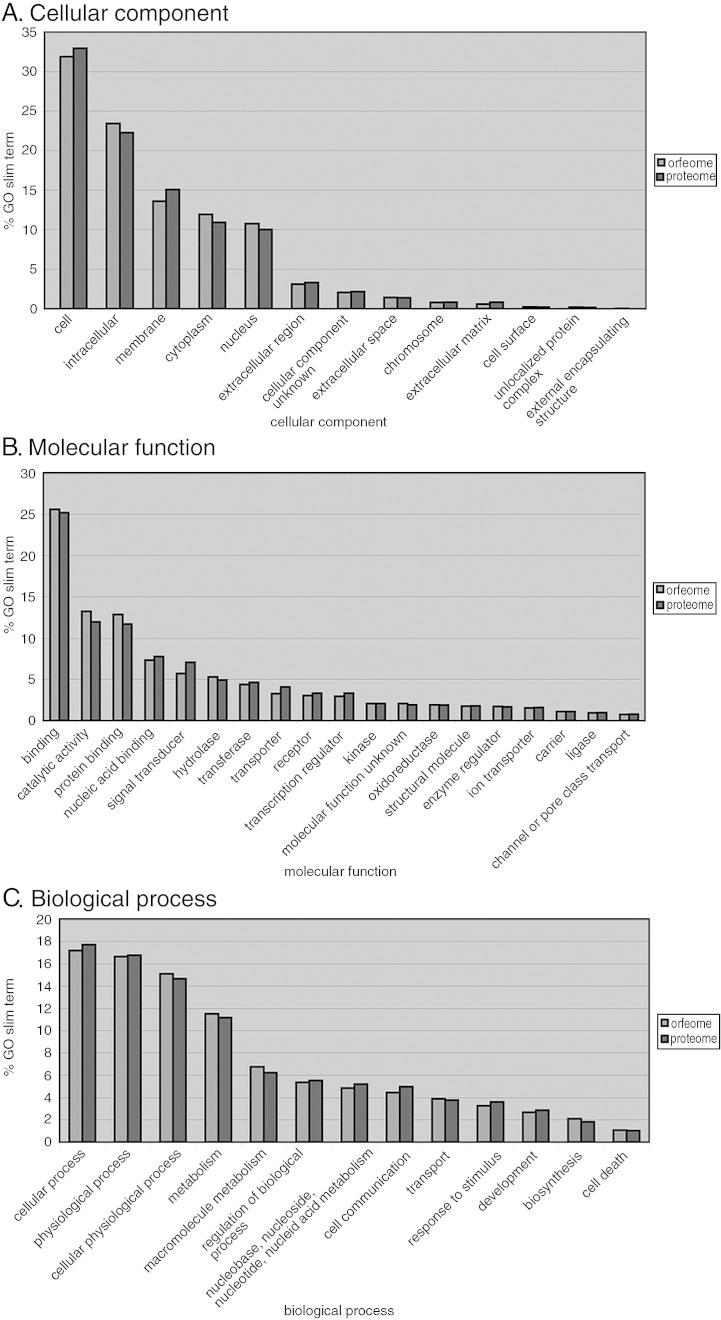
We turned to Gene Ontology (GO) annotations [34], [35] to assess whether specific functional categories were over- or underrepresented in human ORFeome version 3.1. Instead of the full GO hierarchy, we used the broader GO Slim terms of each GO branch (cellular component, biological process, and molecular function). We compared the fraction of each GO term found in clones in the ORFeome to the fraction found in the entire proteome (Fig. 3). We find that the ORFeome has a very similar profile of functional categories compared to the complete human proteome, with no obvious over- or underenriched categories.
Fig. 3.
Classification of cloned ORFs by GO Slim terms. To identify over- or underrepresented functional categories of proteins in the ORFeome, we classified ORFs by GO Slim terms within their three GO branches, (A) cellular component, (B) molecular function, and (C) biological process, and compared the fraction of each GO Slim term found in the ORFeome to that of the entire proteome. No GO Slim term in any of the three branches is over- or underrepresented in the ORFeome.
Disease genes
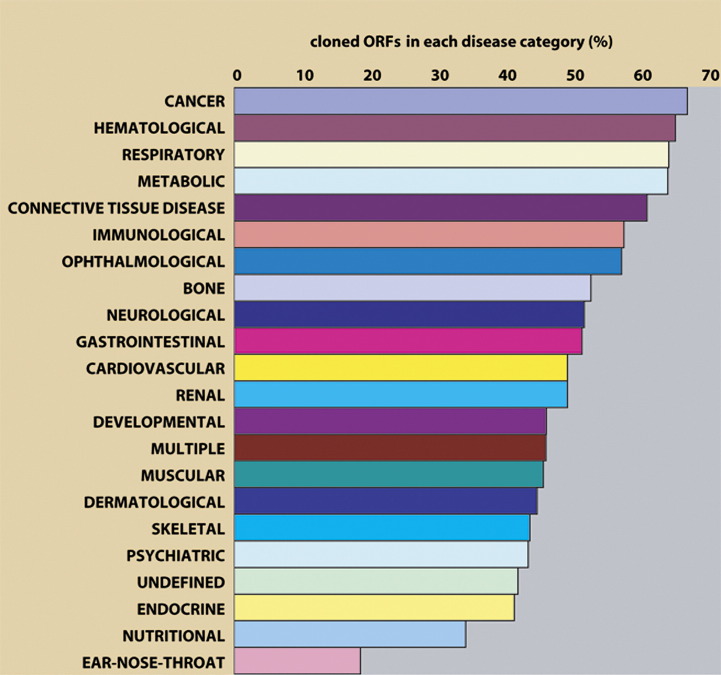
Disease-associated genes are obviously of great interest to the research community. The OMIM (Online Mendelian Inheritance in Man) database [35] represents the central repository for information about inherited disease-related genes. OMIM currently contains information for about 2801 genes that are associated with 1585 different diseases. hORFeome v3.1 contains 956 disease genes associated with 828 distinct diseases described in OMIM (Fig. 4). We classified all OMIM diseases into 22 categories (containing between 6 and 239 different diseases) based on the physiological system affected. We then determined how many diseases in each disease category were represented by at least one ORF in hORFeome v3.1. We could identify ORFs associated with 40–60% of the diseases within a given category, except a few slightly over- (cancer, hematological diseases) or underrepresented (ear–nose–throat-related diseases) categories. For example, v3.1 contains ORFs for 86 of 132 diseases that belong to the cancer category. Despite the good representation of OMIM genes in the ORFeome, only 9.7% of all cloned ORFs have been associated with an inherited disease. The generation of large ORF collections, such as hORFeome v3.1, will be crucial for the identification and characterization of additional disease associations.
Fig. 4.
Representation of disease genes in hORFeome v3.1. The list of inherited diseases and their associated genes was retrieved from the OMIM database, and the diseases were grouped into 22 disease categories based on the physiological system affected. The length of each bar represents the percentage of diseases in each disease category for which we cloned at least one associated ORF.
hORFDB 3.1 Web site
A new Web site (http://horfdb.dfci.harvard.edu) that improves both the user interface and the back end has been developed. Searches on the hORFDB 3.1 Web site can be performed for single or multiple clones using different queries, including MGC name, GI, GenBank accession number, EntrezGene ID, OST accession number, symbol, or plate position. The database can also be searched by description or keyword for ORFs involved in specific biological functions or diseases. The result page of a successfully cloned ORF provides information about the location of the ORF in the ORFeome resource, primer and GenBank sequences, and alternative IDs and descriptions for the ORF.
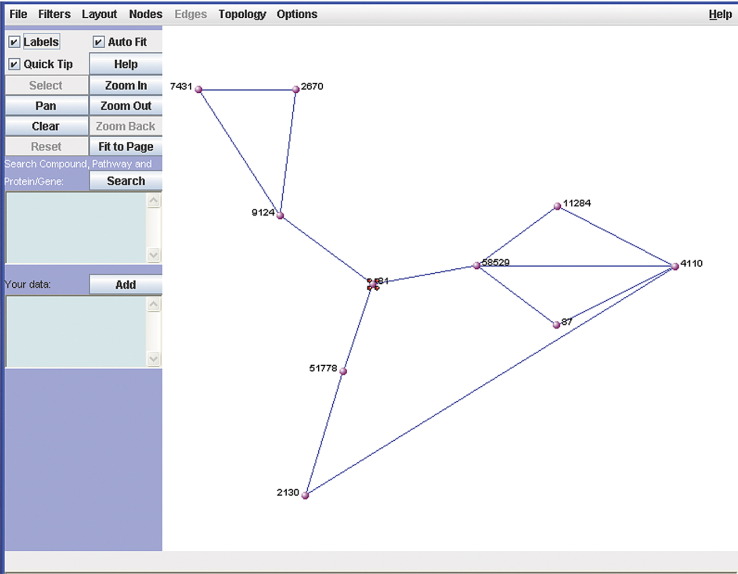
Any yeast two-hybrid interactions based on the human interaction dataset produced by Rual et al. [24] are also listed. These interactions can be visualized using the network visualization tool VisANT [36]. If the queried protein has been detected as bait or prey in the above-mentioned interaction dataset, hORFDB links directly to a first-level interaction network (proteins that interact directly with the queried protein) and a second-level interaction network (proteins that interact with the interaction partners of the queried protein). The user can expand the visible network by clicking on each node of interest, thereby revealing the next level of interactors. Each protein in the network contains links back to its corresponding hORFeome v3.1 Web page, as well as to its corresponding pages on the NCBI EntrezGene, NCBI Nucleotide, and KEGG Web sites.
All ORFs labeled as cloned in hORFDB are part of the physical resource of ORF Entry minipools and are available from Open Biosystems, Inc. (http://www.openbiosystems.com). The complete list of cloned human ORFs is also available as a downloadable Fasta file on our home page.
Discussion
hORFeome v3.1 greatly expands the human ORFeome collection. Unique MGC cDNAs, initially generated largely by random cDNA library screening, were used individually as templates to clone successfully 4111 additional ORFs, generating a consolidated collection of 12,212 ORFs representing 10,214 genes. Although random library screening followed by PCR amplification and Gateway cloning is an excellent method to clone ORFs corresponding to more than half of the well-defined RefSeq predictions, this approach would be less efficient for the identification of “rare” ORFs. Strategies to overcome this hurdle are to generate normalized cDNA libraries or to presubtract cDNAs retrieved in previous screens. An alternative approach is to perform directed PCR from cDNA using primers that have been designed based on ORF predictions, as has been successful for C. elegans [21].
Recently, the MGC, Integrated Molecular Analysis of Genomes and Their Expression Consortium, Wellcome Trust Sanger Institute, DFCI–CCSB (Dana Farber Cancer Institute–Center for Cancer Systems Biology), Harvard Institute of Proteomics, Deutsches Krebsforschungszentrum, Kazusa DNA Research Institute, and RIKEN Yokohama Institute initiated the human “ORFeome Collaboration” with the aim of sharing existing resources and dividing the task of completing the human ORFeome [37]. This effort is using directed PCR to clone missing ORFs whose exon–intron structure is annotated based on literature or full-length cDNAs. About 4700 ORFs that meet these criteria are currently being processed. In addition to library screening and directed PCR, direct ORF synthesis is a third approach to expand the human ORFeome and will be particularly valuable for ORFs that prove difficult to clone. In a small pilot project to demonstrate the feasibility of the synthetic approach, the MGC recently contracted for the successful synthesis and cloning of 72 ORF sequences, ranging in size from several hundred nucleotides to over 11 kb (Gary Temple, personal communication).
In addition to gene coverage, future versions of the human ORFeome will increase coverage of alternatively spliced genes. While recent estimates predict that up to 80% of all human genes code for multiple isoforms, only 1160 ORFs correspond to splice variants in hORFeome v3.1. Finally, while the current ORFeome is a collection of minipools, each initially derived from a single, fully sequenced cDNA template, we ultimately want to generate a resource of wild-type clones, which will require the isolation and full-length sequencing of single colonies for each ORF in the minipools.
Materials and methods
Gateway cloning of the human ORFeome v3.1
For PCR amplification, we designed primers using the automatic primer design program OSP [38]. Although this program is no longer publicly available, we suggest using Primer3 [39] as an alternative primer design program. Forward primers start from the A of the ATG, whereas the reverse primers start from the second nucleotide in the stop codon. Consequently, the reverse attb2.1 primers do not contain the last nucleotide of the termination codon, so as to allow subsequent generation of C-terminal fusion proteins. For ORFs that failed in the first ORFeome project and that we reattempted to clone in ORFeome version 3, we did not synthesize new primers but instead used the primers generated for the previous project. To generate hORFeome v3.1 we closely followed the protocol of Reboul et al. [21], except that we applied the improved PCR conditions and used the improved donor vector pDONR223 [23].
All nonredundant MGC clones were consolidated into a unique set (some MGC clones exist in duplicates) and arranged by size of the ORF and by antibiotic resistance marker. Plasmid preps were obtained using a Qiagen Biorobot 8000. PCR was performed in 25-μl reactions containing 1 unit of KOD Hot Start DNA polymerase according to the manufacturer (Novagen). Gateway BP reactions were performed as described [23] using 2 μl of unpurified PCR product in 10 μl final volume. A 2-μl aliquot of the BP reaction was used to transform Escherichia coli DH5α to spectinomycin resistance (50 μg/ml). Plasmid preps were obtained from 1.0-ml overnight cultures and then used for PCR with M13-based Fwd and Rev primers to generate templates for cycle-sequencing reactions [23]. PCR products were sequenced at the 5′ end using the M13-Fwd primer, generating an OST.
Sequence analysis of the initial MGC cDNAs
For this ORFeome project, we attempted to clone ORFs from 9236 MGC cDNA clones that either were not yet available or remained uncloned in hORFeome v1.1. The coding sequences of all these cDNAs were retrieved from the NCBI Web site and compared to one another to eliminate any cDNAs containing redundant open reading frames (this includes duplicate clones as well as those cDNAs with different 5′ and/or 3′ UTRs but otherwise identical ORF sequences). Next, we aligned the set of unique coding sequences to the human genome (Golden Path hg35) and identified ORFs that were splice variants or polymorphic clones of the same gene.
Sequence analysis of OSTs from minipools
First, OSTs were used as queries for BLASTN searches against our internal database containing all coding sequences that we attempted to clone. In a second step, aligned OSTs were truncated after the first 400 nucleotides (or fewer if the sequence read was short or of low quality before the 400th nucleotide) and a BLAST (blast2seq) was performed between each OST sequence and its best hit. Based on these results, OSTs were grouped into the following classes: (1) good, (2) good but potential polymorphism detected, (3) good but not full length, (4) wrong identity, and (5) empty clones. Only OSTs of categories (1) and (2) were retained for further analysis.
Sequence analysis of OSTs from isolated colonies
Five hundred sixty-four ORFs (six 96-well plates) were selected from the ORFeome 3.1 collection to represent a variety of insert sizes, including the smallest and largest ORFs. Minipools were streaked to single colonies on LB agar containing 100 μg/ml spectinomycin and incubated at 37 °C for 16 h. Six colonies were selected for further analysis. Individual colonies were picked into 0.8-ml 96-well plates (ABgene AB-0859) containing 0.5 ml of selective growth medium (Circlegrow supplemented with 100 μg/ml spectinomycin) and grown in a shaking incubator at 37 °C for 16 h. The sequencing template was prepared for successfully cultivated colonies by standard alkaline-lysis plasmid purification. Initial end sequencing was performed with BigDye terminator v3 Cycle Sequencing Kits (Applied Biosystems) using M13 forward (TGTAAAACGACGGCCAGT) and reverse (CAGGAAACAGCTATGACC) primers and primers designed to pDONR223 (CCCAGTCACGACGTTGTAAAACG; GTAACATCAGAGATTTTGAGACAC) on ABI 3730 sequencing machines. Reads were analyzed for the presence of a complete att site, the correct insert sequence, and the presence of the gene-specific oligonucleotide using crossmatch (Green P, http://www.phrap.org/phredphrap/general.html) and Blastn.
Analysis of successful ORF clones on chromosomes
Sequences of the RefSeq set (June 2005), NCBI's consensus set of nonredundant transcripts, were used as queries to perform a BLAT alignment to the human genome build hg35.1. We chose only those RefSeq models that fulfill the following requirements: (1) RefSeqs are of the “NM” category, which corresponds to sequences that have been validated by one or more cDNAs and (2) they are known as “protein-coding” by NCBI. Using their genomic coordinates, cloned ORFs and RefSeqs were grouped into 1-Mb bins on all chromosomes. The distribution of RefSeq models and the ORF cloning success rate on the chromosomes were plotted using Matlab 6.
In the scatter graph of Fig. 2B, we find that the ORF density is linearly proportional to the RefSeq density, as described by the function NORF = 0.49 NRefSeq + 0.42 (the standard errors are 0.006 and 0.32 for the slope and the intercept of the regression function, respectively) for the given binning and considering every chromosome.
Analysis of ORF distribution by functional classes
Gene ontology functional classification was obtained from the EntrezGene database at ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/gene2go (April 6, 2006). Each gene-to-GO term association was mapped to a GO Slim association as defined in ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/goslim/goaslim.map (February 27, 2006). The frequency distribution of ORFs in each GO Slim class was then calculated for ORFs in hORFeome v3.1 as well as for the entire proteome.
Analysis of ORF distribution by disease category
The list of human diseases and their associated genes was obtained from the OMIM database at ftp://ftp.ncbi.nih.gov/repository/OMIM/morbidmap. Similar diseases were collapsed into just one disease. We then manually curated these diseases and divided them into 22 classes mostly based on the type of disease (such as cancer) and the physiological system affected.
Acknowledgments
We thank Ed Benz, Stan Korsmeyer, David Livingston, Priya McCue, Jane Song, and the DFCI Strategic Planning Initiative for support; the NIH Mammalian Gene Collection Program and Open Biosystems for making the MGC collection available; Gary Temple and Lukas Wagner for all the valuable information they provided on the MGC datasets; Charles Delisi for making the VisANT software available and Joe Mellor for advice on integrating VisANT with hORFDB; members of the Vidal Lab and the participants of the ORFeome meeting for discussions; and Carlene Fraughton for technical support. This work was supported by the High-Tech Fund of the Dana-Farber Cancer Institute (S. Korsmeyer) and by an Ellison Foundation grant awarded to M.V.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ygeno.2006.11.012.
Contributor Information
David E. Hill, Email: david_hill@dfci.harvard.edu.
Marc Vidal, Email: marc_vidal@dfci.harvard.edu.
Appendix A. Supplementary data
Summary of the information available on the hORFDB 3.1 website
Supplementary Fig. 1.
Correlation between ORF size and cloning success rate. As expected, the cloning success rate decreased with increasing ORF size.
Supplementary Fig. 2.
Distribution of cloned ORFs within each chromosome. See Fig. 2 in main text for details.
Supplementary Fig. 3.
Comparison between the distributions of local cloning success rates and the aggregated distribution for each of the chromosomes. The red curves show the cumulative probability distribution function for the cloning success rates as measured in 1 Mb bins for each of the chromosomes, i.e., what fraction of the bins (Y axis) have smaller success rates than a specific value (X axis). Success rate was measured as the ratio of the number of cloned ORFs to that of RefSeq sequences in a given bin, as described in the text. While the success rate may occasionally be greater than 1 (there were more ORFs cloned than there were RefSeq models in a bin), these events are very rare and thus we only show success rates between 0 and 1. The identical blue curves serve as reference in each plot, and correspond to the cumulative probability distribution function of the local cloning success rates if {all} chromosomes are taken into account in calculating the statistics. Additional explanation:Table 2 shows that on most of the chromosomes the number of cloned ORFs is 42% to 53% of the total number of RefSeq sequences identified on the respective chromosome (except for chromosome 21). While this suggests that cloning of ORFs is carried out with a nearly uniform success rate for every chromosome, there may be loci on chromosomes where ORFs are under-or overrepresented. To check this, we performed a Kolmogorov-Smirnov goodness-of-fit test for each of the chromosomes: the test decides if the local cloning success rates of a chromosome may come from the reference distribution at a specified level of significance. For the reference distribution, we chose the distribution of local cloning success rates in 1 Mb bins, taking {every} chromosome into account. Calculating the largest absolute difference between the reference cumulative distribution and the cumulative distributions determined for each chromosome as required by the test (Supplementary figure 3), we found that the distributions for chromosomes 19, 20, 21, X, and Y were different from the overall distribution at the 0.05 significance level.
Supplementary Fig. 4.
VisANT vizualisation tool showing a protein-protein interaction sub-network. This figure shows a screen-shot of VisANT displaying a 2nd-level interaction network for human actinin alpha 4 (highlighted with red dots) containing three first-level interactors (proteins that interact directly with actinin) and six second-level interactors (proteins that interact with the interaction partners of actinin). The user can expand the visible network by clicking on each node of interest, thereby revealing the next level of interactors. Each protein in the network contains links back to its corresponding hORFeome v3.1 webpage, as well as to its corresponding pages on the NCBI EntrezGene, NCBI Nucleotide and KEGG websites.
References
- 1.Adams M.D. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 2.Gibbs R.A. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 3.A. Goffeau, et al., Life with 6000 genes, Science 274 (1996) 546, 563–567. [DOI] [PubMed]
- 4.Lander E.S. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Venter J.C. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 6.C. elegans Sequencing Consortium, Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 7.International Human Genome Sequencing Consortium, Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 8.Rounsley S.D. The construction of Arabidopsis expressed sequence tag assemblies: a new resource to facilitate gene identification. Plant Physiol. 1996;112:1177–1183. doi: 10.1104/pp.112.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stapleton M. The Drosophila gene collection: identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 2002;12:1294–1300. doi: 10.1101/gr.269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerhard D.S. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ota T. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 12.Walhout A.J. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 13.Strausberg R.L., Feingold E.A., Klausner R.D., Collins F.S. The mammalian gene collection. Science. 1999;286:455–457. doi: 10.1126/science.286.5439.455. [DOI] [PubMed] [Google Scholar]
- 14.Hartley J.L., Temple G.F., Brasch M.A. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brasch M.A., Hartley J.L., Vidal M. ORFeome cloning and systems biology: standardized mass production of the parts from the parts-list. Genome Res. 2004;14:2001–2009. doi: 10.1101/gr.2769804. [DOI] [PubMed] [Google Scholar]
- 16.Liu Q., Li M.Z., Leibham D., Cortez D., Elledge S.J. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 1998;8:1300–1309. doi: 10.1016/s0960-9822(07)00560-x. [DOI] [PubMed] [Google Scholar]
- 17.Marsischky G., LaBaer J. Many paths to many clones: a comparative look at high-throughput cloning methods. Genome Res. 2004;14:2020–2028. doi: 10.1101/gr.2528804. [DOI] [PubMed] [Google Scholar]
- 18.Dricot A. Generation of the Brucella melitensis ORFeome version 1.1. Genome Res. 2004;14:2201–2206. doi: 10.1101/gr.2456204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gelperin D.M. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamesch P. C. elegans ORFeome version 3.1: increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. 2004;14:2049–2064. doi: 10.1101/gr.2496804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reboul J. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- 22.Reboul J. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 2001;27:332–336. doi: 10.1038/85913. [DOI] [PubMed] [Google Scholar]
- 23.Rual J.F. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rual J.F. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 25.Zhu H. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 26.Harada J.N. Identification of novel mammalian growth regulatory factors by genome-scale quantitative image analysis. Genome Res. 2005;15:1136–1144. doi: 10.1101/gr.3889305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunsalus K.C. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436:861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- 28.Takagi M. Characterization of DNA polymerase from Pyrococcus sp. strain KOD1 and its application to PCR. Appl. Environ. Microbiol. 1997;63:4504–4510. doi: 10.1128/aem.63.11.4504-4510.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shevchenko Y. Systematic sequencing of cDNA clones using the transposon Tn5. Nucleic Acids Res. 2002;30:2469–2477. doi: 10.1093/nar/30.11.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strausberg R.L. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinrichs A.S. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karolchik D. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashburner M. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler D.L. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Z., Mellor J., Wu J., DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Temple G. From genome to proteome: developing expression clone resources for the human genome. Hum. Mol. Genet. 2006;1:R31–R43. doi: 10.1093/hmg/ddl048. [DOI] [PubMed] [Google Scholar]
- 38.Hillier L., Green P. OSP: a computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl. 1991;1:124–128. doi: 10.1101/gr.1.2.124. [DOI] [PubMed] [Google Scholar]
- 39.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of the information available on the hORFDB 3.1 website