Abstract
Background
The aim of this study was to investigate associations of 3 common polymorphisms in the VEGF gene, −2578C>A, −634C>G, and 936C>T, with risk of tetralogy of Fallot (TOF) in Chinese Han children.
Material/Methods
From January 2010 to June 2013, a total of 400 pediatric subjects were recruited, including 160 cases with TOF (TOF group) and 240 healthy controls (control group). The genotypes of 3 common VEGF polymorphisms, −2578C>A, −634C>G, and 936C>T, were analyzed by polymerase chain reaction restriction fragment length polymorphism. All data were analyzed with SPSS 18.0 software.
Results
No significant differences were observed in body mass index or sex between TOF patients and controls (both P>0.05), but significant differences in age and family history of TOF were observed between the 2 groups (both P<0.05). The AA genotype in -2578C>A of VEGF was correlated with a significantly increased risk of TOF, and TOF risk in A allele carrier was 1.54-fold higher than that of C allele carrier (OR=1.54, 95%CI=1.14–2.09, P=0.005); the statistical significance was still present after Bonferroni correction (Pc=0.045). GG genotype in −634C>G of VEGF gene was also associated with an increased risk of TOF, and TOF risk in patients with G allele was 1.62-fold higher compared to patients with C allele (OR=1.62, 95%CI=1.19–2.21, P=0.002); the statistical significance was still present after Bonferroni correction (Pc=0.018). Interestingly, T allele in VEGF 936C>T polymorphism is associated with a decreased TOF risk (OR=0.65, 95%CI=0.49–0.87, P=0.003, the statistical significance was still present after Bonferroni correction (Pc=0.027). The result of logistic regression analysis revealed that −2578C>A, −634C>G, and 936C>T genotypes are independently related to the prevalence of TOF (all P<0.05).
Conclusions
Our results confirmed that VEGF genetic polymorphisms, −2578C>A and −634C>G, may be associated with an increased TOF risk, while 936C>T polymorphism may be associated with decreased TOF risk.
MeSH Keywords: Amplified Fragment Length Polymorphism Analysis; Anticipation, Genetic; Tetralogy of Fallot; Vascular Endothelial Growth Factor A
Background
Tetralogy of Fallot (TOF) is the most frequently form of cyanotic heart defect or cyanotic congenital heart disease, and is the major cause of blue baby syndrome [1]. TOF is characterized by a misalignment of the conal septum, resulting in a rightward deviation of the aorta, which may lead to a large ventricular septal defect and varying degrees of right ventricular outflow tract narrowing [2]. The incidence of TOF is estimated at 5~7 per 10 000 live births and TOF accounts for 5~7% of all congenital heart lesions [3]. Importantly, patients with TOF generally receive surgical repair in the first year of life, with additional requirement of surgical intervention as the child grows [4]. Since the first description of procedures in the 1950s, the diagnosis, surgical treatment, and postoperative care of TOF have significantly improved; hence, patients born with TOF can now expect to survive to adulthood [5–7]. However, despite excellent short- and medium-term survival rates, the 30-year actuarial survival for patients repaired before their 5th birthday is 90% of the expected survival rate and the annualized risk of death triples in the third postoperative decade [8]. The exact pathophysiology of TOF is not well understood, but both genetic and environmental factors play major roles in TOF [9,10]. Viral infections, urticarial vasculitis, radiation exposure, lifestyle, and other environmental factors are associated with a high risk of TOF [11]. Recent evidence suggests that VEGF gene is a key player in the etiology of TOF [12,13].
Vascular endothelial growth factor (VEGF) belongs to a sub-family of the platelet-derived growth factors and is a prominent endothelial cell-specific mitogen and inducer of vascular permeability, and is an important signaling molecule involved in vasculogenesis and angiogenesis [14]. In the angiogenic network, the VEGF are by far the best characterized and has been implicated in the pathogenesis of coronary artery disease and in its complication, acute myocardial infarction [15]. As a multifunctional cytokine, VEGF exerts a variety of essential biological effects on vascular endothelium [16]. VEGF induces angiogenesis in various physiological and pathological conditions, such as normal embryogenesis, compensatory angiogenesis, wound-healing, and in tumor growth and metastasis [14,17]. Hypoxia up-regulates VEGF gene expression, which has tremendous physiological implications for a wide variety of pathological conditions in humans [18]. Adequate VEGF serum levels are essential for heart development, and alterations in VEGF levels may contribute to cardiovascular developmental defects [19,20]. In this context, up-regulated VEGF levels, during the development of right ventricular outflow tract, result in abnormal development of both cushion and myocardial structures [21,22]. Therefore, genetic polymorphisms in VEGF are an important way to understand the role of the VEGF gene in heart development. The human VEGF gene is located on chromosome 6p21.3 and consists of 8 exons and 7 introns [23]. Earlier studies have demonstrated that there are 5 common genetic polymorphisms involved in various human diseases: −2578C>A (rs699947), −460T/C (rs833061), −634G>C (rs2010963), +405C/G (rs2010963), and +936C>T (rs3025039) [24–26]. Takuya Awata et al. demonstrated the correlation of the VEGF 634CC genotype with higher VEGF production is consistent with the genetic association of the 634C allele [27]. VEGF gene polymorphism of −2549 (rs3034659) have been reported to be associated with variations in VEGF plasma concentrations and with a susceptibility to disorders, such as diabetic retinopathy, diabetic nephropathy, and cardiovascular diseases [28]. In general, genetic variations in VEGF may influence VEGF expression and activity, and variously confer susceptibility to congenital heart diseases [20,29]. We hypothesized that VEGF polymorphisms may be associated with the risk of TOF. We carried out this study to gather evidence in relation to our hypothesis and thus investigated the relationship between VEGF polymorphisms and the risk of TOF in Chinese Han children.
Material and Methods
Study subjects and ethics statement
A total of 160 Chinese Han children with TOF were recruited for this study as the case group (TOF group) at the Department of Cardiovascular, Affiliated Hospital of Jining Medical University between January 2010 and June 2013. The TOF patients exhibited typical clinical symptoms and were all diagnosed with color Doppler echocardiography and surgery. Patients with abnormalities other than cardiovascular abnormalities were excluded. There were no abnormalities found by karyotype analysis of the included TOF patients. Among the 160 TOF children, 103 patients were males and 57 patients were females, with an average age of 4.27±1.93 years old (age range, 1 to 8 years). Another group of 240 healthy Han children, who received health examination at the medical examination center of our hospital during the same period, were selected as the control group. The Han children who suffered from congenital heart disease or other cardiac structural abnormalities, as confirmed by clinical manifestations and echocardiography, were excluded from this study. The control group included 148 males and 92 females, with an average age of 5.68±2.17 years old (age range, 2 to 10 years). There were no significant differences in age or sex between the TOF group and control group (P>0.05). This study was approved by the Ethics Committee of the Affiliated Hospital of Jining Medical University. All parents of all included Han children signed the informed consent prior to the study and the study conformed to the Declaration of Helsinki [30].
DNA extraction
A total of 2 ml peripheral venous blood from all included subjects was drawn in the morning, following an overnight fast, and was placed in ethylenediamine tetraacetic acid (EDTA)-containing tubes for anticoagulation, and stored at −20°C until further use. The 2-ml blood samples were centrifuged at 2500r/m for 5 min to serum and cells, and the collected serum was evenly split into 3-ml cryotubes and stored at −20°C for later use. Genomic DNA was extracted from leukocytes using the conventional saturated phenol-chloroform extraction method. The concentration and purity of DNA was measured and the isolated genomic DNA was stored at −20°C for further use.
SNP selection and primer design
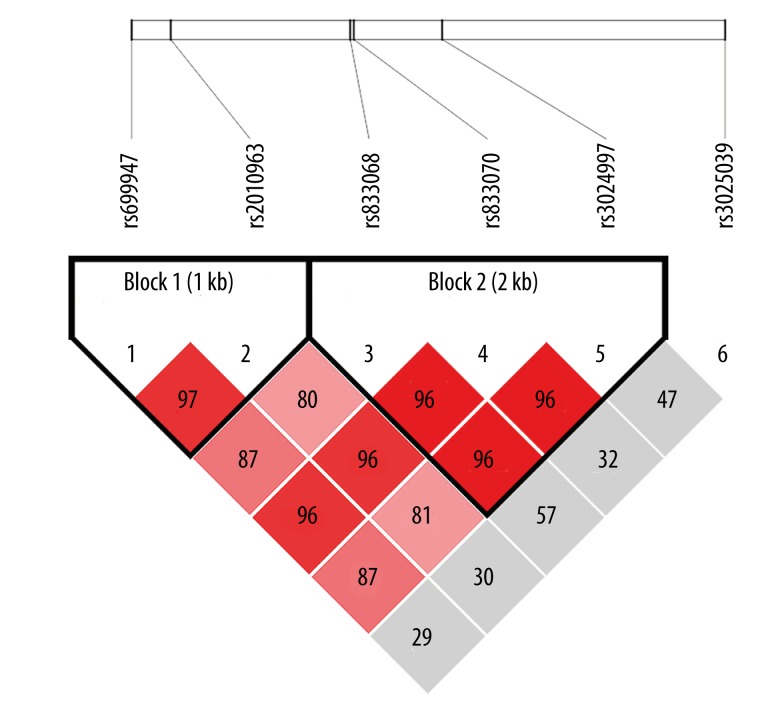
We investigated 3 relevant SNP candidates in the VEGF gene. All SNP sequences selected for genotyping were initially identified using the HapMap database (http://www.hapmap.org/). The linkage disequilibrium of VEGF polymorphisms is illustrated in Figure 1. The SNP genotypes of the VEGF functional polymorphic loci, −2578C>A, −634C>G, and 936C>T, were detected by polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). The full-length VEGF gene sequence was retrieved from GenBank (Accession Number AF 437895) and, based on the gene sequence, the PCR amplification primers for each polymorphic loci were designed using Primer 5.0 software (Takara, Dalian), as shown in Table 1.
Figure 1.
Linkage disequilibrium (LD) analysis of SNPs [−2578C>A (rs699947), −634C>G (rs2010963), and +936C>T (rs3025039)] in the VEGF gene. Values in squares are LD between single markers.
Table 1.
Primers used for amplification of the VEGF genetic polymorphisms.
| SNP | Alias name | Annealing Temperature (°C) | Primers for PCR amplification | Restriction enzyme |
|---|---|---|---|---|
| rs699947 | −2578C>A | 53 | F: 5′-CATACCGATGGAACTGG-3′ | Bg1II |
| R: 5′-GTTTCTGACCTGGCTATTT-3′ | ||||
| rs2010963 | −634C>G | 67 | F: 5′-CGACGGCTTGGGGAGATTGC-3′ | BsmFI |
| R: 5′-GGGCGGTGTCTGTCTGTCTG-3′ | ||||
| rs3025039 | 936C>T | 53 | F: 5′-AGGGTTTCGGGAACCACATC-3′ | NlaIII |
| F: 5′-AGGGTTTCGGGAACCACATC-3′ |
SNP – single nucleotide polymorphism; PCR – polymerase chain reaction; F – forward; R – reward.
Polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP)
The gradient PCR was performed using a Thermocycler MJPTC-200 PCR System. The PCR amplification of the genetic polymorphisms of VEGF was performed after determining the optimal annealing temperature. The PCR reaction system in a 15-μl volume contained the following components: 50 ng genomic DNA, 12.5 pmol of each primer, 0.1 mml of each nucleotide, l0×PCR buffer (50 mml KCl, 10 mml TrisHCl, 0.1% Triton X-100, 1.5 mml MgCl2, and 1.0 U Taq polymerase). Reaction conditions of PCR were as follows: initial denaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at annealing temperature, which were 53°C −2578>A), 67°C (−634C>G), and 53°C (936C>T), respectively, for 40 s, and extension at 72°C for 45 s. After 34 cycles of amplification, final extension was at 72°C for 10 min. The PCR amplification product were first purified and 5 μl of the purified PCR amplification products were digested with 5 U of restriction enzymes Bg1II, BsmFI, and NlaIII (Shanghai Sangon Biotech, Shanghai, China) to distinguish between the genetic polymorphisms of VEGF. Restriction digestions were performed under standard digestion conditions, with an overnight incubation at 37°C. The results were analyzed by 3.0% agarose gel electrophoresis to detect PCR fragment pattern, and individual genotypes were determined from the size of the bands. The restriction enzymes for each locus are shown in Table 1.
Genotyping
Restriction fragment length polymorphism (RFLP) was employed to detect the genetic polymorphisms of −2578C>A, −634C>G, and 936C>T in VEGF gene. The restriction enzyme Bg1II distinguished the A and C alleles and CC, CA, and AA genotypes were observed in the −2578C>A polymorphism of VEGF gene. The mutant AA genotype showed 2 fragments of 194 bp and 102 bp, and the CA genotype showed 3 fragments of 296 bp, 194 bp, and 102 bp, while wild genotype (CC) showed only 1 band of 296 bp. With restriction enzyme BsmFI, 2 alleles (C and G) and 3 genotypes) CC, CG, and GG) were observed in the −634C>G polymorphism of VEGF gene. The mutant GG genotype showed 166 bp and 108 bp bands, and the CG genotype showed 274 bp, 166 bp and 108 bp bands, while the wild CC genotype showed only 1 band of 274 bp. With restriction enzyme NlaIII, 2 alleles (C and T) and 3 genotypes (CC, CT and TT) were observed in the 936C>T polymorphism of VEGF gene. The mutant TT genotype showed 2 fragments of 211 bp and 55 bp, and CT genotype showed 3 fragments of 266 bp, 211 bp, and 55 bp, while the wild CC genotype showed only 1 band of 266 bp. The results were further confirmed by sequencing and by alignment of the sequencing results with GenBank sequences using CLUSTAL method.
Statistical analysis
All data were analyzed with SPSS17.0 software (SPSS, Chicago, IL, USA). Measurement data are expressed as mean ± standard deviation (Mean ±SD) and t test were applied to compare the general data of TOF group and control group. We adopted χ2 test for the differences of genotypes and allele frequencies. The Bonferroni correction was conducted and the statistical differences were presented if Pc<0.05. We also used χ2 test to confirm if the allele frequency of VEGF polymorphisms were in Hardy-Weinberg equilibrium (HWE). Linkage disequilibrium was evaluated by D and r2 values (r2 >0.33 showed disequilibrium). A non-conditional logistic regression was used to calculate the odds ratios (OR) and 95% confidence interval (CI) represented the relative risk. All statistical tests were 2-sided and a value of P<0.05 indicated statistical significance.
Results
Baseline characteristics
The baseline characteristics of TOF patients and normal controls are illustrated in Table 2. No statistically significant differences in body mass index (BMI) or sex distribution were observed between the TOF group and the healthy control group (both P>0.05). Importantly, there was a significant difference in age and in the family history of TOF between the 2 groups (both P<0.05).
Table 2.
The comparison of baseline characteristics between Chinese Han children with tetralogy of Fallot (TOF patients) and normal Chinese Han children (healthy controls).
| Clinical characteristics | TOF patients (n=160) | Healthy controls (n=240) | P value |
|---|---|---|---|
| Age | 4.27±1.93 | 5.68±2.17 | <0.0001a |
| BMI | 16.03±3.36 | 16.28±2.63 | 0.406a |
| Gender (male/female) | 103/57 | 148/92 | 0.583b |
| Family history of TOF | 38/122(23.8%) | 23/217(9.6%) | 0.0001b |
TOF – tetralogy of Fallot; BMI – body mass index;
p value of student’s t test;
chi-square test.
Distribution of genotypes and allele frequencies of VEGF genetic polymorphisms
The electrophoresis patterns for the genotypes of VEGF genetic polymorphisms are as illustrated in Figures 2–4. The PCR-RFLP results of VEGF rs699947 (−2578C>A) polymorphism showed that AA genotype showed 2 fragments of 202 bp and 122 bp, and the CA genotype showed 3 fragments of 324 bp, 202 bp, and 122 bp, while CC genotype remained undigested. The VEGF rs2010963 (−634C>G) polymorphism showed 3 patterns: CC genotype with a single band of 180 bp, CG genotype with 180 bp, 120 bp, and 60 bp fragments, and GG genotype with 60 bp and 120 bp fragments. Further, the CC genotype of VEGF rs3025039 (936C>T) remained undigested, while the TT genotype showed 2 fragments of 86 bp and 112 bp fragments and CT genotype showed 3 fragments of 198 bp, 86 bp, and 112 bp.
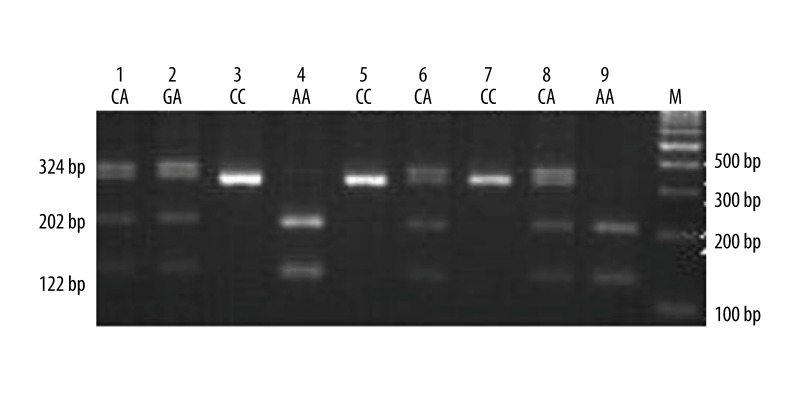
Figure 2.
Electrophoresis patterns for the genotypes of VEGF rs699947 (−2578C>A) polymorphism analyzed by PCR-RFLP-based assay. Lane M shows DNA marker. Lanes 3, 5, and 7 (324bp) show individuals with CC genotype. Lanes 4 and 9 (202bp and 122bp) show those with AA genotype. Lanes 1, 6, and 8 (324bp, 202bp and 122bp) show those with CA genotype.
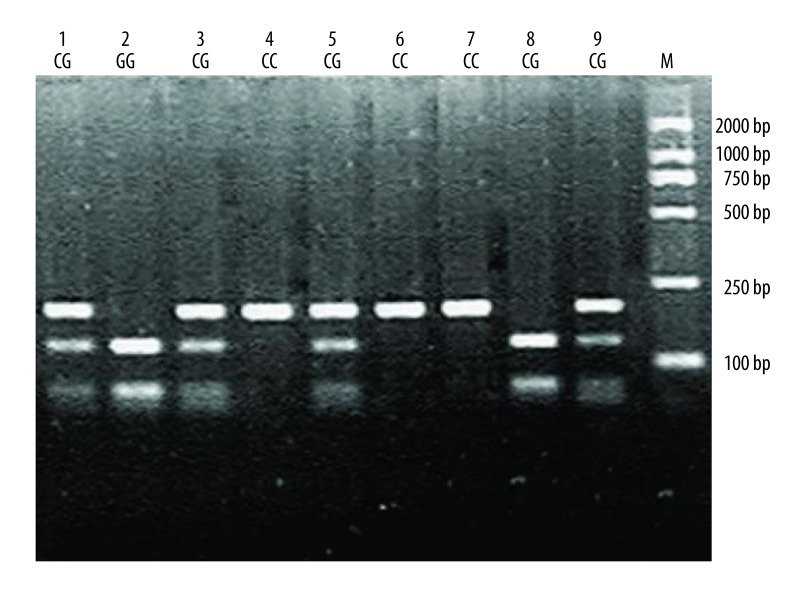
Figure 3.
Electrophoresis patterns for the genotypes of VEGF rs2010963 (−634C>G) polymorphism analyzed by PCR-RFLP-based assay. Lane M shows DNA marker. Lanes 4, 6 and 7 (180bp) show individuals with CC genotype. Lanes 1, 3, 5 and 9 (180bp, 120bp, and 60bp) show those with CG genotype. Lanes 2 and 8 (60bp and 120bp) show those with GG genotype.
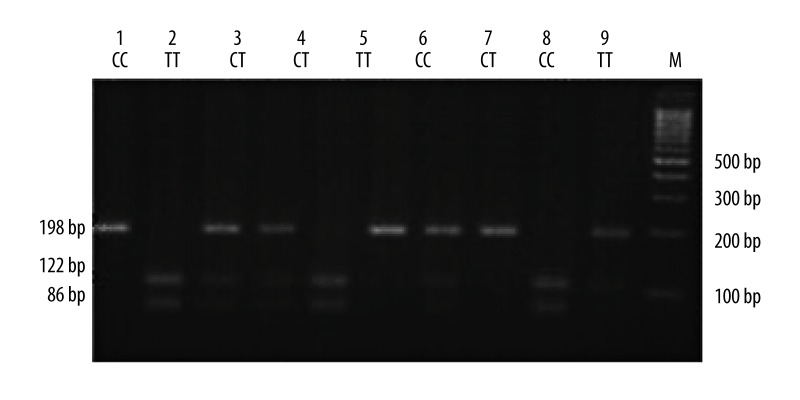
Figure 4.
Electrophoresis patterns for the genotypes of VEGF rs3025039 (+936C>T) polymorphism analyzed by PCR-RFLP-based assay. Lane M shows DNA marker. Lanes 1, 6, and 8 (198bp) show individuals with CC genotype. Lanes 2, 5, and 9 (86bp and 112bp) show those with TT genotype. Lanes 3, 4, 7, and 10 (198bp, 86bp, and 112bp) show those with CT genotype.
The distribution of genotypes and allele frequencies of the 3 VEGF gene polymorphisms, −2578C>A, −634C>G, and 936C>T, are shown in Table 3. The genotypes and allele frequencies of VEGF −2578C>A, −634C>G, and 936C>T polymorphisms were in accordance with HWE, suggesting that each gene frequency had reached equilibrium and the selected sample represented the population (P>0.05). The frequency of mutant AA genotype in VEGF −2578C>A polymorphism in TOF group was significantly higher than the control group (48.8% vs. 32.9%, P=0.042), and the statistical significance was still present after Bonferroni correction (Pc=0.372). The A allele frequency in −2578C>A in the TOF group was markedly higher than that observed in the control group (71.2% vs. 61.7%, P=0.005), and the statistical significance was still present after Bonferroni correction (Pc=0.045). The AA+CA genotype frequency of the TOF group was higher than the control group (93.8% vs. 90.4%), with no statistical difference (P=0.235). Additionally, the GG genotype frequency of VEGF −634C>G in TOF group was significantly higher than the control group (53.8% vs. 36.7%, P=0.031), and the statistical significance was still present after Bonferroni correction (Pc=0.279). The G allele frequency in −634C>G in the TOF group was higher than in the control group (72.8% vs. 62.3%, P=0.002); the statistical significance was still present after Bonferroni correction (Pc=0.018). The GG+CG frequency on −634C>G in the TOF group was higher than in the control group (91.9% vs. 81.9%), but with no statistical difference (P=0.206). Conversely, the frequencies of TT genotype and T allele in VEGF 936C>T in TOF group were both significantly lower than in the control group (TT genotype: 41.9% vs. 54.6%, P=0.030, Pc=0.270; T allele: 48.1% vs. 58.7%, P=0.003, Pc=0.027). However, there was no statistically significant difference in TT+CT genotypes frequencies between the 2 groups (54.4% vs. 62.9%, P=0.088).
Table 3.
The comparisons of genotypes and allele frequency distribution of VEGF genetic polymorphisms (−2578C>A, −634C>G and 936C>T) between case group (TOF group) and control group (healthy controls).
| Genotype | TOF patients (n=160) | Healthy controls (n=240) | OR (95%CI) | χ2 | P value |
|---|---|---|---|---|---|
| −2578C>A | |||||
| CC | 10 (6.2%) | 23 (9.6%) | Reference | ||
| CA | 72 (45%) | 138 (57.5%) | 1.20 (0.54–2.66) | 0.202 | 0.653 |
| AA | 78 (48.8%) | 79 (32.9%) | 2.27 (1.01–5.08) | 4.118 | 0.042 |
| AA+CA | 150 (93.8%) | 217 (90.4%) | 1.59 (0.74–3.44) | 1.409 | 0.235 |
| C allele | 92 (28.8%) | 184 (38.3%) | Reference | ||
| A allele | 228 (71.2%) | 296 (61.7%) | 1.54 (1.14–2.09) | 7.803 | 0.005 |
| −634C>G | |||||
| CC | 13 (8.1%) | 29 (12.0%) | Reference | ||
| CG | 61 (38.1%) | 123 (51.3%) | 1.11 (0.54–2.28) | 0.075 | 0.784 |
| GG | 86 (53.8%) | 88 (36.7%) | 2.18 (1.06–4.47) | 4.651 | 0.031 |
| GG+CG | 147 (91.9%) | 211 (81.9%) | 1.55 (0.78–3.09) | 1.601 | 0.206 |
| C allele | 87 (27.2%) | 181 (37.7%) | Reference | ||
| G allele | 233 (72.8%) | 299 (62.3%) | 1.62 (1.19–2.21) | 9.540 | 0.002 |
| 936C>T | |||||
| CC | 73 (45.6%) | 89 (37.1%) | Reference | ||
| CT | 20 (12.5%) | 20 (8.3%) | 1.22 (0.61–2.44) | 0.315 | 0.575 |
| TT | 67 (41.9%) | 131 (54.6%) | 0.62 (0.41–0.96) | 4.723 | 0.030 |
| TT+CT | 87 (54.4%) | 151 (62.9%) | 1.42 (0.96–2.14) | 2.907 | 0.088 |
| C allele | 166 (51.9%) | 198 (41.3%) | Reference | ||
| T allele | 154 (48.1%) | 282 (58.7%) | 0.65 (0.49–0.87) | 8.741 | 0.003 |
TOF – tetralogy of Fallot; OR – odds ratio; 95%CI – 95% confidence interval.
Linkage analysis of SNP in VEGF gene
Linkage disequilibrium was evaluated by D and r2 values. It is showed linkage disequilibrium when r2 >0.33 (Table 4). The rs699947 and rs2010963 locus presented linkage disequilibrium (r2=0.694), while the rs2010963and rs3025039 locus did not (r2=0.198).
Table 4.
The results of linkage analysis of SNP in VEGF gene.
| Linked genes | D′ | r2 |
|---|---|---|
| rs699947-rs2010963 | 0.138 | 0.627 |
| rs699947-rs3025039 | 0.256 | 0.694 |
| rs2010963-rs3025039 | 0.009 | 0.198 |
Correlation analysis of VEGF polymorphisms with the risk of TOF
Multiple logistic regression analysis was carried out to calculate the adjusted odds ratio. The TOF was the dependent variable, and the AA, CC, and TT genotype of VEGF −2578C>A, −634G>C, and 936C>T were the independent variables. The multivariate logistic regression analysis results showed that the mutable gene of −2578C>A and −634C>G polymorphisms are independently related to the TOF risk (OR=1.951, 95%CI=1.280~2.974, P=0.002; OR=2.144, 95%CI=1.411~3.258, P<0.001), and the TT genotypes of 936C>T are also associated with TOF risk (OR=0.590, 95%CI=0.389–0.894, P=0.013) (Table 5).
Table 5.
The multivariate logistic regression analysis of VEGF gene polymorphisms in the risk of tetralogy of Fallot.
| Variables | S.E | Wald | Sig | Exp(B) | 95%CI |
|---|---|---|---|---|---|
| −2578C>A | 0.215 | 9.662 | 0.002 | 1.951 | 1.280–2.974 |
| −634C>G | 0.213 | 6.181 | <0.001 | 2.144 | 1.411–3.258 |
| 936C>T | 0.213 | 0.339 | 0.013 | 0.590 | 0.389–0.894 |
S.E – standard error of regression; Wald – Wald χ2; Sig – significance, P value; Exp(B) – adjusted odds ratio, OR value; 95%CI – 95% confidence interval.
Discussion
In this study, we examined the correlations of VEGF −2578C>A (rs699947), −634G>C (rs2010963), and +936C>T (rs3025039) polymorphisms with TOF risk. The main results of our study confirmed that VEGF genetic polymorphisms −2578C>A and −634C>G may be associated with an increased TOF risk, while 936C>T polymorphism may be associated with decreased TOF risk. The pathogenesis of TOF is multifactorial and involves various genetic and environmental factors. However, the exact cellular mechanisms leading to TOF are incompletely understood. The heart is the first organ to form and various signaling pathways, including VEGF, Notch, Wnt/β-catenin, and BMP/TGF-β, may be involved in heart development [31–34]. VEGF is an important player in endocardial and epicardial epithelial-mesenchymal transformation (EMT), which results in the establishment of most non-cardiomyocyte lineages of the mature heart during heart valve development [35,36]. VEGF can also play a key role in endocardial cushion development, possibly by regulating cell proliferation, vasculogenesis, vascular permeability, and angiogenesis in embryonic development [36,37]. Yang et al. found that VEGF expression was visualized as nuclear and cytoplasmic localized granular brown-yellow precipitate and their findings indicated that BMSC transplantation in dilated cardiomyopathy could promote the myocardial expression of VEGF [38]. On the other hand, VEGF is also linked to the formation of collateral vessels in ischemic heart disease and cyanotic congenital heart disease [39]. Several studies have also demonstrated that the expression of VEGF is regulated by hypoxia and alterations in VEGF expression level may be involved in the development of congenital heart disease, including TOF [40,41]. Several variant genotypes and haplotypes of VEGF gene alter the basal level of VEGF expression. These alterations in the VEGF gene are also functionally reflected in the altered expression levels of VEGF protein in the serum, and may tightly regulate cardiac morphogenesis, as evidenced by various genetic mouse models [42–44].
The gene coding for VEGF is affected by common, functionally important genetic polymorphisms associated with cardiovascular disease and are early predictors of congenital heart disease. Among the most important polymorphisms are: −2578C>A (rs699947), located in the promoter region; −634G>C (rs2010963), located in exon 1; and +936C>T (rs3025039), located in exon 8, in the 5′ untranslated region [45]. In the current study, we found that the frequency of AA genotype in −2578C>A in the TOF group was significantly higher than in the control group, and the TOF risk of patients with A allele carrier was 1.54-fold higher than that of C allele carrier, suggesting that the AA genotype and A allele in −2578C>A polymorphism may be a potential risk factor for TOF development. Further, the frequency of GG genotype in −634C>G in the TOF group was also significantly higher than the control group, and the risk of TOF of the patients with G allele was 1.62-fold higher than in patients with C allele, implying that the GG genotype and G allele in −634C>G in polymorphism may be correlated with the increased risk of TOF. However, T allele in 936C>T polymorphism decreased TOF risk, indicating that the T allele may be a protective factor for TOF. All of the above results had statistical significance after Bonferroni correction. Douvaras et al. showed that the −634C>G polymorphism of VEGF gene and its co-inheritance with genotypes of other VEGF genetic polymorphisms might be potential risk factors for the development of heart failure after acute myocardial infarction [46]. Further, Lambrechts et al. also showed that the common polymorphisms of −2578C>A, −1154A, and −634C>G in VEGF gene may be linked with the elevated risk of TOF [12]. The polymorphism at position −634 of VEGF gene has been proved to be correlated with the pathological processes of ventricular septal defect in a Chinese population [47]. Additionally, consistent with our study results, the polymorphism at position 936 C/T of VEGF gene has previously been shown to be related to lower VEGF plasma levels in healthy young individuals, which may be associated with reduced risk of human diseases [48,49].
There are some limitations in the present study. First of all, the sample sizes of our study were relatively small, which may reduce the strength of our conclusions. Secondly, the statistical power was not conducted in this study, which also decreased the power of our study.
Conclusions
Our results provide strong evidence that the polymorphisms of VEGF −2578C>A (rs699947) and −634G>C (rs2010963) are correlated to an increased risk of TOF, while the +936C>T (rs3025039) polymorphism may be correlated with a decreased risk of TOF, suggesting these polymorphisms can serve as biomarkers to evaluate the susceptibility to TOF. Nevertheless, due to the small sample size of this study, further studies with larger sample sizes and more integral data are necessary for the development of statistical analysis with general applicability.
Footnotes
Competing interests
The authors have declared that no competing interests exist.
Source of support: Departmental sources
References
- 1.Apitz C, Webb GD, Redington AN. Tetralogy of Fallot. Lancet. 2009;374:1462–71. doi: 10.1016/S0140-6736(09)60657-7. [DOI] [PubMed] [Google Scholar]
- 2.Witters I, De Groot R, Van Loo K, et al. Tetralogy of Fallot with coronary artery to pulmonary artery fistula. Prenat Diagn. 2014;34:1345–46. doi: 10.1002/pd.4481. [DOI] [PubMed] [Google Scholar]
- 3.Bittel DC, Kibiryeva N, Marshall JA, O’Brien JE. MicroRNA-421 Dysregulation is associated with tetralogy of fallot. Cells. 2014;3:713–23. doi: 10.3390/cells3030713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsze DS, Vitberg YM, Berezow J, et al. Treatment of tetralogy of Fallot hypoxic spell with intranasal fentanyl. Pediatrics. 2014;134:e266–69. doi: 10.1542/peds.2013-3183. [DOI] [PubMed] [Google Scholar]
- 5.Miyazaki A, Sakaguchi H, Ohuchi H, et al. Efficacy of hemodynamic-based management of tachyarrhythmia after repair of tetralogy of Fallot. Circ J. 2012;76:2855–62. doi: 10.1253/circj.cj-12-0208. [DOI] [PubMed] [Google Scholar]
- 6.Luijten LW, van den Bosch E, Duppen N, et al. Long-term outcomes of transatrial-transpulmonary repair of tetralogy of Fallot. Eur J Cardiothorac Surg. 2015;47:527–34. doi: 10.1093/ejcts/ezu182. [DOI] [PubMed] [Google Scholar]
- 7.Ylitalo P, Nieminen H, Pitkanen OM, et al. Need of transannular patch in tetralogy of Fallot surgery carries a higher risk of reoperation but has no impact on late survival: results of Fallot repair in Finland. Eur J Cardiothorac Surg. 2015;48(1):91–97. doi: 10.1093/ejcts/ezu401. [DOI] [PubMed] [Google Scholar]
- 8.Pandya B, Quail MA, Cullen S. Clinical issues and outcomes in adults following repair of tetralogy of fallot. Curr Treat Options Cardiovasc Med. 2013;15:602–14. doi: 10.1007/s11936-013-0264-3. [DOI] [PubMed] [Google Scholar]
- 9.Rauch R, Hofbeck M, Zweier C, et al. Comprehensive genotype-phenotype analysis in 230 patients with tetralogy of Fallot. J Med Genet. 2010;47:321–31. doi: 10.1136/jmg.2009.070391. [DOI] [PubMed] [Google Scholar]
- 10.Kuciene R, Dulskiene V. Selected environmental risk factors and congenital heart defects. Medicina (Kaunas) 2008;44:827–32. [PubMed] [Google Scholar]
- 11.Gupta R. Tetralogy of Fallot. Rapid Review Anesthesiology Oral Boards. 2013:117–22. [Google Scholar]
- 12.Lambrechts D, Devriendt K, Driscoll DA, et al. Low expression VEGF haplotype increases the risk for tetralogy of Fallot: a family based association study. J Med Genet. 2005;42:519–22. doi: 10.1136/jmg.2004.026443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silversides CK, Lionel AC, Costain G, et al. Rare copy number variations in adults with tetralogy of Fallot implicate novel risk gene pathways. PLoS Genet. 2012;8:e1002843. doi: 10.1371/journal.pgen.1002843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrara N. Vascular endothelial growth factor. Arterioscler Thromb Vasc Biol. 2009;29:789–91. doi: 10.1161/ATVBAHA.108.179663. [DOI] [PubMed] [Google Scholar]
- 15.Petrovic D. The role of vascular endothelial growth factor gene as the genetic marker of atherothrombotic disorders and in the gene therapy of coronary artery disease. Cardiovasc Hematol Agents Med Chem. 2010;8:47–54. doi: 10.2174/187152510790796183. [DOI] [PubMed] [Google Scholar]
- 16.Poh CK, Shi Z, Lim TY, et al. The effect of VEGF functionalization of titanium on endothelial cells in vitro. Biomaterials. 2010;31:1578–85. doi: 10.1016/j.biomaterials.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 17.Bao P, Kodra A, Tomic-Canic M, et al. The role of vascular endothelial growth factor in wound healing. J Surg Res. 2009;153:347–58. doi: 10.1016/j.jss.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osada-Oka M, Ikeda T, Imaoka S, et al. VEGF-enhanced proliferation under hypoxia by an autocrine mechanism in human vascular smooth muscle cells. J Atheroscler Thromb. 2008;15:26–33. doi: 10.5551/jat.e533. [DOI] [PubMed] [Google Scholar]
- 19.Stankunas K, Ma GK, Kuhnert FJ, et al. VEGF signaling has distinct spatiotemporal roles during heart valve development. Dev Biol. 2010;347:325–36. doi: 10.1016/j.ydbio.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vannay A, Vasarhelyi B, Kornyei M, et al. Single-nucleotide polymorphisms of VEGF gene are associated with risk of congenital valvuloseptal heart defects. Am Heart J. 2006;151:878–81. doi: 10.1016/j.ahj.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh PC, Davis ME, Lisowski LK, Lee RT. Endothelial-cardiomyocyte interactions in cardiac development and repair. Annu Rev Physiol. 2006;68:51–66. doi: 10.1146/annurev.physiol.68.040104.124629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villefranc JA, Nicoli S, Bentley K, et al. A truncation allele in vascular endothelial growth factor c reveals distinct modes of signaling during lymphatic and vascular development. Development. 2013;140:1497–506. doi: 10.1242/dev.084152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vincenti V, Cassano C, Rocchi M, Persico G. Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3. Circulation. 1996;93:1493–95. doi: 10.1161/01.cir.93.8.1493. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs EJ, Feigelson HS, Bain EB, et al. Polymorphisms in the vascular endothelial growth factor gene and breast cancer in the Cancer Prevention Study II cohort. Breast Cancer Res. 2006;8:R22. doi: 10.1186/bcr1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulgu Y, Cetin GO, Caner V, et al. Vascular endothelial growth factor gene polymorphisms in age-related macular degeneration in a Turkish population. Int J Ophthalmol. 2014;7:773–77. doi: 10.3980/j.issn.2222-3959.2014.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smedts HP, Isaacs A, de Costa D, et al. VEGF polymorphisms are associated with endocardial cushion defects: a family-based case-control study. Pediatr Res. 2010;67:23–28. doi: 10.1203/PDR.0b013e3181c1b144. [DOI] [PubMed] [Google Scholar]
- 27.Awata T, Inoue K, Kurihara S, et al. A common polymorphism in the 5′-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes. 2002;51:1635–39. doi: 10.2337/diabetes.51.5.1635. [DOI] [PubMed] [Google Scholar]
- 28.Petrovic D, Verhovec R, Globocnik Petrovic M, et al. Association of vascular endothelial growth factor gene polymorphism with myocardial infarction in patients with type 2 diabetes. Cardiology. 2007;107:291–95. doi: 10.1159/000099064. [DOI] [PubMed] [Google Scholar]
- 29.Richards AA, Garg V. Genetics of congenital heart disease. Curr Cardiol Rev. 2010;6:91–97. doi: 10.2174/157340310791162703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.World Medical A. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–94. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 31.Gaengel K, Genove G, Armulik A, Betsholtz C. Endothelial-mural cell signaling in vascular development and angiogenesis. Arterioscler Thromb Vasc Biol. 2009;29:630–38. doi: 10.1161/ATVBAHA.107.161521. [DOI] [PubMed] [Google Scholar]
- 32.Holderfield MT, Hughes CC. Crosstalk between vascular endothelial growth factor, notch, and transforming growth factor-beta in vascular morphogenesis. Circ Res. 2008;102:637–52. doi: 10.1161/CIRCRESAHA.107.167171. [DOI] [PubMed] [Google Scholar]
- 33.Cohen ED, Tian Y, Morrisey EE. Wnt signaling: an essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development. 2008;135:789–98. doi: 10.1242/dev.016865. [DOI] [PubMed] [Google Scholar]
- 34.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet. 2008;9:49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- 35.Wagner M, Siddiqui MA. Signal transduction in early heart development (II): ventricular chamber specification, trabeculation, and heart valve formation. Exp Biol Med (Maywood) 2007;232:866–80. [PubMed] [Google Scholar]
- 36.Armstrong EJ, Bischoff J. Heart valve development: endothelial cell signaling and differentiation. Circ Res. 2004;95:459–70. doi: 10.1161/01.RES.0000141146.95728.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–76. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 38.Yang S, Piao J, Jin L, Zhou Y. Does pretreatment of bone marrow mesenchymal stem cells with 5-azacytidine or double intravenous infusion improve their therapeutic potential for dilated cardiomyopathy? Med Sci Monit Basic Res. 2013;19:20–31. doi: 10.12659/MSMBR.883737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aydin HI, Yozgat Y, Demirkaya E, et al. Correlation between vascular endothelial growth factor and leptin in children with cyanotic congenital heart disease. Turk J Pediatr. 2007;49:360–64. [PubMed] [Google Scholar]
- 40.Baghdady Y, Hussein Y, Shehata M. Vascular endothelial growth factor in children with cyanotic and acyanotic and congenital heart disease. Arch Med Sci. 2010;6:221–25. doi: 10.5114/aoms.2010.13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamada H, Ebata R, Higashi K, et al. Serum vascular endothelial growth factor in cyanotic congenital heart disease functionally contributes to endothelial cell kinetics in vitro. Int J Cardiol. 2007;120:66–71. doi: 10.1016/j.ijcard.2006.08.106. [DOI] [PubMed] [Google Scholar]
- 42.Hutson MR, Kirby ML. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin Cell Dev Biol. 2007;18:101–10. doi: 10.1016/j.semcdb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Combs MD, Yutzey KE. Heart valve development: regulatory networks in development and disease. Circ Res. 2009;105:408–21. doi: 10.1161/CIRCRESAHA.109.201566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Akker NM, Molin DG, Peters PP, et al. Tetralogy of fallot and alterations in vascular endothelial growth factor-A signaling and notch signaling in mouse embryos solely expressing the VEGF120 isoform. Circ Res. 2007;100:842–49. doi: 10.1161/01.RES.0000261656.04773.39. [DOI] [PubMed] [Google Scholar]
- 45.Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine. 2000;12:1232–35. doi: 10.1006/cyto.2000.0692. [DOI] [PubMed] [Google Scholar]
- 46.Douvaras P, Antonatos DG, Kekou K, et al. Association of VEGF gene polymorphisms with the development of heart failure in patients after myocardial infarction. Cardiology. 2009;114:11–18. doi: 10.1159/000210189. [DOI] [PubMed] [Google Scholar]
- 47.Xie J, Yi L, Xu ZF, et al. VEGF C-634G polymorphism is associated with protection from isolated ventricular septal defect: case-control and TDT studies. Eur J Hum Genet. 2007;15:1246–51. doi: 10.1038/sj.ejhg.5201890. [DOI] [PubMed] [Google Scholar]
- 48.Krippl P, Langsenlehner U, Renner W, et al. A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int J Cancer. 2003;106:468–71. doi: 10.1002/ijc.11238. [DOI] [PubMed] [Google Scholar]
- 49.Renner W, Kotschan S, Hoffmann C, et al. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res. 2000;37:443–48. doi: 10.1159/000054076. [DOI] [PubMed] [Google Scholar]