Abstract
Alternative splicing is a key process of multi-exonic gene expression during pre-mRNA maturation. In this process, particular exons of a gene will be included within or excluded from the final matured mRNA, and the resulting transcripts generate diverse protein isoforms. Recent evidence demonstrates that approximately 95% of human genes with multiple exons undergo alternative splicing during pre-mRNA maturation. Thus, alternative splicing plays a critical role in physiological processes and cell development programs, and.dysregulation of alternative splicing is highly associated with human diseases, such as cancer, diabetes and neurodegenerative diseases. In this review, we discuss the regulation of alternative splicing, examine the relationship between alternative splicing and human diseases, and describe several approaches that modify alternative splicing, which could aid in human disease diagnosis and therapy.
Keywords: mRNA, alternative splicing, human diseases, cancer, diabetes, neurodegenerative diseases, biomarker, drug discovery
Introduction
Alternative splicing (AS) is a mechanism for the generation of multiple mRNAs during pre-mRNA maturation, resulting in the generation of multiple proteins with distinct functions from a single gene. During this process, particular exons of a gene will be included within or excluded from the final matured mRNA, and the resulting transcripts generate diverse protein isoforms. Recent studies using mRNA-Sequence and Expression Sequence Tag (EST-cDNA) technology indicate that approximately 95% of human genes with multiple exons undergo alternative splicing during pre-mRNA maturation1,2, highlighting the important role of alternative splicing in determining gene function. Specifically, dysfunction of alternative splicing has been implicated in different disease pathophysiology, specificity and severity1,3,4,5.
AS plays a critical role in physiological processes and cell development programs. For example, alternative splicing plays a role in stem cell renewal and differentiation. Recently, Han et al showed that the muscle blind-like RNA-binding proteins, MBNL1 and MBNL2, differentially regulate cassette exon alternative splicing events in embryonic stem cells and other cell types6. The inhibition of these proteins in differentiated cells induces embryonic stem cell-like patterns of alternative splicing; in contrast, the overexpression of these proteins in embryonic stem cells induces differentiated cell-like patterns of alternative splicing. These findings suggest that regulatory proteins play critical roles in regulating alternative splicing, associated with physiological processes and cell development programs. The disruption of the alternative splicing likely affects cell differentiation, resulting in the development of cancer and other diseases7.
In this review, we highlight the pervasive pathophysiological consequences of alternative splicing. We also discuss the regulation of alternative splicing and the diverse roles of this mechanism in tumorigenesis. Moreover, we examine the potential use of alternative splicing as a biomarker and target for drug discovery.
Alternative splicing and its regulation
An estimated 90 000 proteins can be produced in human cells, however, only approximately 25 000 genes encoding proteins have been identified in the human genome8. Thus, it has been suggested that alternative splicing is one of the major mechanisms contributing to protein diversity9.
Alternative splicing is a key process that regulates pre-mRNA maturation. During this process, particular exons of a gene could be included or excluded from the maturated messenger RNA (mRNA) transcribed from that gene10. High-throughput sequencing of the human genome, particularly the mRNA sequences and expressed sequence tags (EST-cDNA), revealed that approximately 95% of the genes with multiple exons undergo alternative splicing during pre-mRNA maturation in a majority of human tissues2.
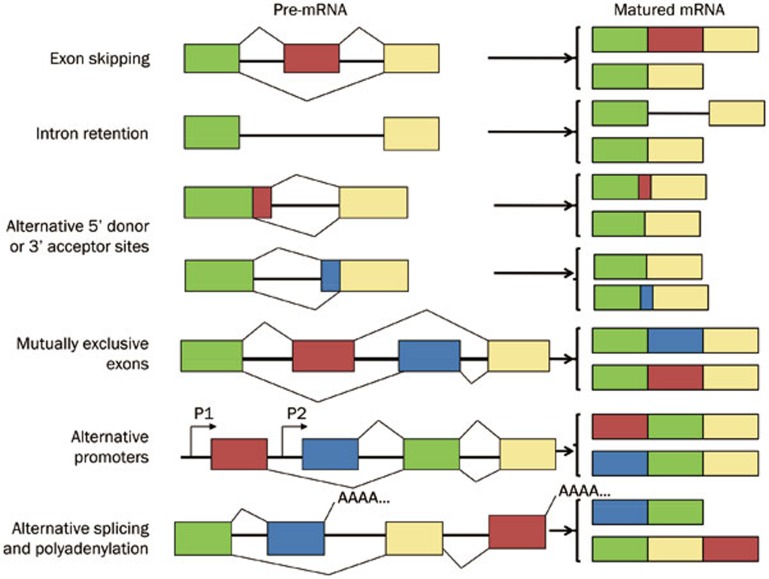
The completion of genome sequencing has provided important insights into the various modalities of pre-mRNA splicing. The different modes of alternative splicing can be grouped into the following categories (Figure 1). Exon skipping is the most common event in alternative splicing in mammals accounting for 38.4% of alternative splicing events in humans11; in this mode, an exon is excised from the matured mRNA along with the flanking introns. Alternative 5'/3' donor/acceptor sites, which occur when two or more splice sites at one end of an exon are present, are the next most frequently observed events, accounting for 7.9% and 18.4%, respectively11. Intron retention, whereby an intron is retained in the resulting mature mRNA, accounts for approximately 2.8% of the alternative splicing events11 Other classes of alternative splicing7,12,13 include mutually exclusive exons, alternative promoters and alternative splicing and polyadenylation; these modes account for approximately 32.4% of the alternative splicing events11.
Figure 1.
Commonly observed alternative splicing patterns, summarized and modified from Blencowe13.
The basic elements regulate alternative splicing, including cis- and trans-acting elements. The cis-acting elements, including consensus splice site sequences and auxiliary elements, determine the outcome of alternative splicing, while the trans-acting elements, comprising a group of serine/arginine-rich (SR) proteins, heterogeneous nuclear ribonucleoproteins (hnRNPs), and small nuclear ribonucleoprotein (snRNP)s, precisely regulate the process of alternative splicing14. This process is achieved through the precise and dynamic assembly of the spliceosome, which comprises both cis- and trans-acting elements. Understanding the specific functions and properties of the participating proteins will provide a better understanding of the splicing regulatory network (SRN). Moreover, this information will help to determine how the dysregulation of this process contributes to the pathophysiology of the resulting disease.
Alternative splicing and human diseases
Increasing evidence has shown that the disruption of alternative splicing negatively impacts health and contributes to human diseases, including cancer, diabetes and neurodegenerative diseases. Here, we will discuss some of the evidence relating alternative splicing with diseases in the context of the expression patterns resulting from alternative splicing events, the association of alternative splicing with disease severity, and the extent to which alternative splicing affects disease progression.
Alternative splicing occurs in a tissue-specific manner, associated with specific genetic mutations in diseases
Approximately 0%–30% of alternative splicing events occur in a tissue-specific manner15. Studies have shown that the majority of alternative splicing events are differently regulated in different tissues, suggesting that alternative splicing is a major contributor to the evolution of phenotypic complexity in mammals. Evidence for the tissue-specific regulation of alternative splicing comes from a comparison of the ratio of the specific exons of a gene included in the mRNA expressed in each tissue relative to the other tissues. A minimum of 10% change in the inclusion ratio was indicative of tissue specificity. These analyses have resulted in the identification of over 22 000 tissue-specific alternative transcripts1.
An excellent example of tissue-specific alternative splicing is transcription factor 7-like 2 (TCF7L2)3, which has been strongly associated with type 2 diabetes. This study examined the mRNA expression of multiple TCF7L2 splicing isoforms in eight different human tissues (ie, pancreas, pancreatic islets, colon, liver, monocytes, skeletal muscle, subcutaneous adipose tissue and lymphoblastoid cell lines), and the results indicated a tissue-specific pattern of alternative splicing. Furthermore, these studies showed significant differences in the expression of TCF7L2 'exons 7–8' in other human tissues relative to that in the pancreas.
Another example of tissue-specific alternative splicing is IG20/MADD. Through alternative splicing, this gene produces six splicing variants, namely IG20pa, MADD, IG20-SV2, DENN-SV, IG20-SV4, and KIAA0358. Previous studies16,17 have examined the expression of IG20/MADD splicing variants using RT-PCR and showed that MADD and DENN-SV were constitutively expressed in all tissues tested, including brain, breast, kidney, lung, ovary, pancreatic, testis, uterus, stomach, and thyroid, while the expression of the other IG20/MADD isoforms varied in different tissues17, particularly KIAA0358 and IG20-SV4, which showed enriched expression primarily in neuronal tissues16.
It has been reported that many diseases are associated with the abnormal function of pre-mRNA, reflecting either alternative splicing defects or genetic mutations. As changes in alternative splicing patterns account for many human disorders, we observed that the MADD isoform of the IG20 gene is overexpressed in thyroid, breast, and ovarian cancer tissues and cell lines compared with non-tumor tissues18,19,20. Disease-associated or disease-specific mutation/s in a given gene is the leading cause of changes in the alternative splicing patterns of pre-mRNA. Nearly 15% of diseases result from abnormal alternative splicing due to mutations within cis- and/or trans-elements 21.
Cis-acting mutations that disrupt constitutive splice sites
Mutations can disrupt the classical splicing sites in an exon, resulting in the expression of unnatural mRNAs, with the loss of function of the mutated allele due to nonsense-mediated decay (NMD), expression of proteins with internal deletions, a shift in the reading frame or C-terminal truncations. A point mutation in the muscular dystrophy gene disrupted constitutive splicing sites, causing an in-frame skipping of exon 3122.
Cis-effects: mutations that disrupt the use of alternative splice sites
Pre-mRNA mutations that affect the use of an alternative splice sites can result in aberrant protein expression. Such an event can shift the ratio of natural protein isoforms to the mutated forms and cause functional disruption. This outcome is different from an aberrant splice variant with the usual associated loss of function. Familial-isolated growth hormone deficiency type II (IGHD II), Frasier syndrome, fronto-temporal dementia, Parkinsonism linked to chromosome 17 (FTDP-17), and atypical cystic fibrosis are diseases in which splicing mutations have been common identified21.
Trans-effects: mutations that affect the basal splicing machinery
Trans-effect mutations affect the components of the basic splicing machinery through either the assembly of the constitutive components of the spliceosome or other factors that regulate alternative splicing. Null mutations in spliceosome components are typically lethal at the cellular level in metazoans. Retinitis pigmentosa and spinal muscular atrophy are caused by these types of splicing mutations21.
Trans-effects: mutations that affect regulators of alternative splicing
In trans-effect mutations, instead of the components of the spliceosomes, the regulators of splicing are silenced. The inactivation of a splicing regulator in mice specifically affects the function of natural pre-mRNA targets23. A similar phenomenon is expected to occur in human diseases, reflecting the functional disruption of alternative splicing regulators24,25.
Alternative splicing is associated with the severity of the disease stage
Previous studies have shown that alternative splicing mutations are highly correlated with various types of cancer. Through alternative splicing, a gene might produce multiple isoforms, and each isoform might be differentially expressed during different stages of cancer progression. CXCL12 and IG20/MADD are good examples of genes whose isoform expression patterns change with disease progression.
CXCL12-CXCR4-CXCR7 is a signaling pathway that promotes tumor growth and cancer metastasis4. Previous studies have shown that CXCL12-α, -β, and -γ are highly co-expressed in breast cancers, with lower levels of expression correlating with more aggressive subtypes, increased disease stage, and worse clinical outcomes. Additional studies have also shown that the expression of CXCL12-α, -β, and –γ isoforms significantly varies at different stages of tumor progression.
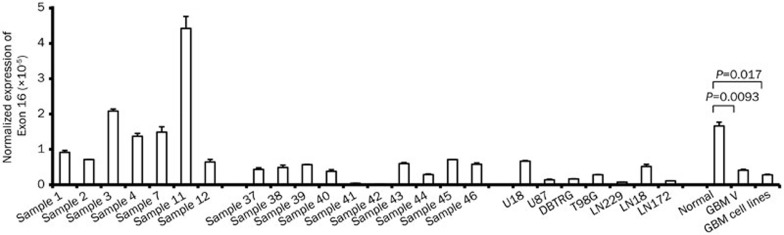
We examined the expression of IG20pa and MADD in non-tumor and malignant glioblastoma tumor tissues using q-RT-PCR and observed that the pro-apoptotic isoform of the IG20 gene, IG20pa, is downregulated in malignant glioblastoma and glioblastoma cancer cell lines compared with non-tumor tissues (Figure 2), likely associated with the severity of glioblastoma. Similar result was also observed by Lefave, et al26.
Figure 2.
Expression of IG20pa in malignant glioblastoma and non-tumor tissues, determined using real-time RT-PCR. Samples 1–12, non-tumor tissues; samples 37–46 malignant glioblastoma tissues and glioblastoma cell lines, U18, U87, DBTRG, T98G, LN229, LN18, and LN172.
These findings indicate that alternative splicing is differently regulated during cancer progression. Additional studies5 have also shown evidence supporting the argument that alternative splicing patterns are altered during cancer progression, associated with the acquisition of cancerous features, such as high proliferation rates, angiogenesis, extra-cellular matrix invasion and survival under extreme stress condition.
Splicing variants from same gene primarily have antagonistic functions
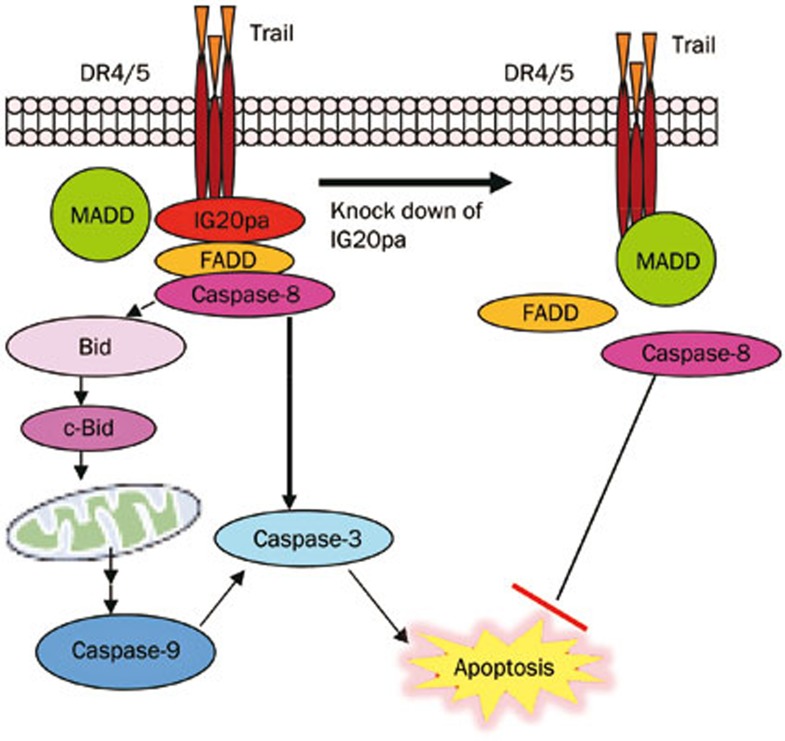
Alternative splicing primarily regulates disease progression through the antagonizing functions of splice variants. One variant from alternative splicing might affect the regular function of another variant. In previous studies, we have shown that the isoforms IG20pa and MADD display antagonistic functions in the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptotic pathway, which is directly associated with tumor apoptosis. Among the six IG20 splicing variants, MADD is expressed at high levels in all cancer cells tested. This protein confers resistance to TRAIL-induced apoptosis in cancer cell lines, particularly in the absence of IG20pa expression27. MADD directly interacts with the death receptor DR4/DR5 and subsequently inhibits apoptosis by preventing the recruitment of the Fas-Associated Death Domain (FADD)-containing protein procaspase-8, required to form the death-inducing signaling complex (DISC), which mediates apoptosis. The deletion of MADD, but not the other IG20 splice variants, induces susceptibility to spontaneous and TRAIL-induced apoptosis in cancer cells. Thus, it is reasonable to propose that the MADD isoform of the IG20 gene is oncogenic, playing an important role in controlling cancer cell apoptosis and conferring resistance to TRAIL-induced apoptosis. However, other studies have shown that another isoform of the IG20 gene, IG20pa (ie, pro-apoptotic isoform), acts as a dominant-negative MADD, which enhances DISC formation and apoptosis. DISC formation induces caspase-8 activation, which is inhibited in the presence of MADD alone. The activation of caspase-8 can either lead to the activation of caspase-3 and extrinsic apoptosis or the cytoplasmic cleavage of Bid, which is subsequently translocated to mitochondria, leading to the activation of the mitochondrial pathway, resulting in the activation of caspase-9 and caspse-3 and the induction of apoptosis28 (Figure 3).
Figure 3.
IG20pa and MADD as antagonists in the TRAIL-induced apoptotic pathway.
An interesting phenomenon has been observed for another pair of IG20/MADD isoforms, KIAA0358 and IG20-SV4. We demonstrated that KIAA0358 is a pro-survival factor, while IG20-SV4 is a pro-apoptotic factor in certain neuronal cells. The function of the KIAA0358 isoform dominates the function of IG20-SV4. The expression of IG20-SV4 alone, in the absence of KIAA0358, overcomes the transcriptional inhibition of caspase-8 gene expression, resulting in the upregulation of the expression of this gene. However, the co-expression of both KIAA0358 and IG20-SV4 suppresses caspase-8 gene transcription16.
The antagonistic effects of different isoforms of the same gene have been widely observed. The Bcl-x gene encodes two protein isoforms with antagonistic functions, the anti-apoptotic protein — Bcl-xL and the pro-apoptotic protein — Bcl-xS. Altering the ratio between these two proteins using an antisense oligonucleotide can be used to sensitize the cells to chemotherapeutic drug-induced apoptosis29. Similarly, TNFR2 also encodes two splicing variants with antagonistic functions through the alternative splicing of exons 7 and 8, namely wild type (TNFR2) and the deleted form (DS-TNFR2) of TNFR2. While TNFR2 mediates TNF-alpha induced apoptosis, the DS-TNFR2 isoform blocks TNF-alpha induced apoptosis30.
Alternative splicing as a potential biomarker and therapeutic target
Alternative splicing is tissue specific, and changes in alternative splicing occur in a disease-specific manner during disease progression, suggesting that this mechanism could serve as a disease-specific or disease stage-specific biomarker. Additionally, regulating alternative splicing might affect the disease outcome, implying the potential targeting for new drug development.
Traditional genetic biomarkers present a great challenge for the diagnosis of complex diseases such as cancer, as many of these diseases arise from multiple distinct molecular mechanisms. The exon-specific detection of alternative splicing might serve as a reliable biomarker and provide a novel approach to diagnose and monitor disease progression. This approach is likely to be more successful than traditional methods31. Recent studies have identified some splicing variants of common regulatory proteins in diseased cells that are not present in normal cells, suggesting the use of alternative splicing as a potential biomarker. For example, alternative splicing isoforms of many genes have been detected in cancer diagnosis. Using a high-throughput reverse transcription PCR-based system for splicing annotation, the alternative splicing profiles of 600 cancer-associated genes were determined from a panel of 21 normal and 26 cancerous breast tissues32. A total of 41 alternative splicing events that significantly differed in breast tumors relative to normal breast tissues were validated. Most cancer-specific changes in splicing disrupt protein domains and increase cell proliferation or cell survival, consistent with a functional role for alternative splicing in cancer. This observation could serve as a biomarker for the diagnosis of complicated diseases, such as cancer.
Approaches used to regulate alternative splicing as potential therapies
Because specific alternative splicing is highly associated with the specificity and severity of diseases, modulating this process might prevent the development and/or alter the course of diseases. Thus, modulating alternative splicing could offer a useful strategy for new drug discovery. Several approaches, from conventional small-molecule compounds33,34 to oligonucleotide and RNAi-based gene therapies, have been proposed for drug development (Table 1).
Table 1. Approaches for the regulation of alternative splicing as a potential therapy.
| Approaches | Examples/targeted genes | References | |
|---|---|---|---|
| Small molecule | Targeting protein isoforms | Etoricoxib/COX2 | 35 |
| Targeting expression | Aclarubicin/SMA | 36 | |
| Targeting trans-acting element | Toyocamycin/XBP1 | 37 | |
| Antisense oligonucleotide-mediated regulation | tDNA-AONs/DMD | 38,39 | |
| RNAi-based isoform specific modulation | SMaRT/MAPT | 40 | |
Common conventional therapeutics
Targeting protein isoforms
Alternative splicing can produce a diverse range of protein isoforms with a unique function and the ability to react differently to various pharmaceutical products. Targeting each individual protein isoform might represent a novel splicing isoform-specific therapy. For example, cyclooxygenases (COXs) play critical roles in platelet activation and inflammation. COX-1 is constitutively expressed in most cells, whereas COX-2 is an inducible isoform highly expressed under inflammatory conditions. Etoricoxib, a COX-2 specific inhibitor, has been developed for the treatment of inflammatory conditions35.
Targeting expression
Instead of targeting the protein isoform, small molecules that directly target gene expression can be developed. For example, spinal muscular atrophy (SMA) reflects the loss of the transcription of exon 7 due to allelic mutation. Several potential treatments, including the topoisomerase II inhibitor aclarubicin, increase the level of SMN protein by increasing the inclusion of exon 7 in SMN2 mRNA36.
Targeting alternative splicing through trans-acting elements
Pharmaceutical agents designed to modulate alternative splicing either target splicing factors (trans-acting elements) or splicing factor-related proteins. XBP1 is a basic region/leucine zipper (bZIP) transcription factor of the CREB-ATF family that plays an important pro-survival role in multiple myeloma (MM) cells. Toyocamycin inhibits IRE1a-induced ATP-dependent XBP1 mRNA cleavage in vitro, with no apparent effect on IRE1a auto-phosphorylation. Therefore, this agent can be used to modulate MM cell death37. However, the therapeutic targeting of splicing factors might affect multiple transcripts, thereby disrupting normal intracellular function and generating undesirable side effects. To overcome this challenge, oligonucleotide and RNA-based gene therapies have been proposed.
Antisense oligonucleotide-mediated regulation of alternative splicing
This approach targets oligonucleotides to specific cis-acting elements within the transcript and has been used to inhibit or activate specific splicing events. The oligonucleotides bind to an element on the targeted transcript and directly block its activity or indirectly affect transcription by recruiting effectors to this site. Chemically modified oligonucleotides, such as a 2'-O-methyl groups or morpholino backbones, are used to modify alternative splicing and restore the desired splicing outcomes. The most successful example is the antisense oligonucleotide therapy used in Duchenne muscular dystrophy (DMD). A series of Phase II/III clinical trials have been completed using antisense oligonucleotides, including tricyclo-DNA antisense oligonucleotides (tDNA-AONs), to induce exon skipping for the restoration of the open reading frame (ORF) of the DMD gene38,39.
RNAi-based isoform specific modulation for corrective therapy
This represents a novel therapeutic approach based on mRNA reprograming techniques. Mutations in the MAPT gene, which encodes the tau protein, result in excess exon 10 (E10) inclusion in tau mRNA, leading to frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17). Spliceosome-mediated RNA trans-splicing (SMaRT) could reprogram and correct aberrant E10 splicing resulting from FTDP-17 mutations40, highlighting a novel approach for the development of new therapies that facilitate the restoration of the desired endogenous mRNA and protein function in diseased cells.
Conclusion and future perspectives
In the last decade, significant progress has been made in understanding the complexity and regulation of alternative splicing and the relationship between this mechanism and human disease. Alternative splicing is a tissue-specific mechanism, and particular splice variants are strongly associated with specific diseases. Moreover, the pattern of expression of splice variants changes with disease progression, and specific splice variants have been associated with the severity and/or stage of the disease. Targeting alternative splicing through the modulation of either cis- or trans-acting elements could serve as a basis for developing disease-specific biomarkers and novel therapeutics for treating genetic diseases.
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities (Grant No 0250/ZK1019), the 985 Project from Xiamen University (Grant No 0250-X170100101), and the Science Foundation of Fujian Province, China (Grant No 2015J01347).
References
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature 2008; 456: 470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40: 1413–5. [DOI] [PubMed] [Google Scholar]
- Prokunina-Olsson L, Welch C, Hansson O, Adhikari N, Scott LJ, Usher N, et al. Tissue-specific alternative splicing of TCF7L2. Hum Mol Genet 2009; 18: 3795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Chang SL, Linderman JJ, Feng FY, Luker GD. A comprehensive analysis of CXCL12 isoforms in breast cancer. Transl Oncol 2014; May 13. doi: 10.1016/j.tranon.2014.04.001. [DOI] [PMC free article] [PubMed]
- Biamonti G, Catillo M, Pignataro D, Montecucco A, Ghigna C. The alternative splicing side of cancer. Semin Cell Dev Biol 2014; 32: 30–6. [DOI] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 2013; 498: 241–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Stranger BE. Genomics of alternative splicing: evolution, development and pathophysiology. Hum Genet 2014; 133: 679–87. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science 2001; 291: 1304–51. [DOI] [PubMed] [Google Scholar]
- Roy B, Haupt LM, Griffiths LR. Alternative splicing (AS) of genes as an approach for generating protein complexity. Curr Genomics 2013; 14: 182–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet 2002; 30: 13–9. [DOI] [PubMed] [Google Scholar]
- Sugnet CW, Kent WJ, Ares M Jr, Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac Symp Biocomput 2004: 66–77. [DOI] [PubMed]
- Blaustein M, Pelisch F, Srebrow A. Signals, pathways and splicing regulation. Int J Biochem Cell Biol 2007; 39: 2031–48. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell 2006; 126: 37–47. [DOI] [PubMed] [Google Scholar]
- Coelho MB, Smith CW. Regulation of alternative pre-mRNA splicing. Methods Mol Biol 2014; 1126: 55–82. [DOI] [PubMed] [Google Scholar]
- Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res 2002; 30: 3754–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Sheng JR, Mulherkar N, Prabhakar BS, Meriggioli MN. Regulation of apoptosis and caspase-8 expression in neuroblastoma cells by isoforms of the IG20 gene. Cancer Res 2008; 68: 7352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimova EV, Al-Zoubi AM, Martinez O, Kaithamana S, Lu S, Arima T, et al. IG20, in contrast to DENN-SV, (MADD splice variants) suppresses tumor cell survival, and enhances their susceptibility to apoptosis and cancer drugs. Oncogene 2004; 23: 1076–87. [DOI] [PubMed] [Google Scholar]
- Turner A, Li LC, Pilli T, Qian L, Wiley EL, Setty S, et al. MADD knock-down enhances doxorubicin and TRAIL induced apoptosis in breast cancer cells. PLoS One 2013; 8: e 56817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta A, Malena A, Tibaldi E, Rocchi L, Loro E, Pena E, et al. MBNL142 and MBNL143 gene isoforms, overexpressed in DM1-patient muscle, encode for nuclear proteins interacting with Src family kinases. Cell Death Dis 2013; 4: e770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Jayaram S, Ganesh L, Qian L, Rotmensch J, Maker AV, et al. Knockdown of MADD and c-FLIP overcomes resistance to TRAIL-induced apoptosis in ovarian cancer cells. Am J Obstet Gynecol 2011; 205: 362. e12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev 2003; 17: 419–37. [DOI] [PubMed] [Google Scholar]
- Disset A, Bourgeois CF, Benmalek N, Claustres M, Stevenin J, Tuffery-Giraud S. An exon skipping-associated nonsense mutation in the dystrophin gene uncovers a complex interplay between multiple antagonistic splicing elements. Hum Mol Genet 2006; 15: 999–1013. [DOI] [PubMed] [Google Scholar]
- O'Bryan MK, Clark BJ, McLaughlin EA, D'Sylva RJ, O'Donnell L, Wilce JA, et al. RBM5 is a male germ cell splicing factor and is required for spermatid differentiation and male fertility. PLoS Genet 2013; 9: e 1003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton M, Voss TC, Misteli T. Identification by high-throughput imaging of the histone methyltransferase EHMT2 as an epigenetic regulator of VEGFA alternative splicing. Nucleic Acids Res 2014; 42: 13662–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cheng C. Alternative RNA splicing and cancer. Wiley Interdiscip Rev RNA 2013; 4: 547–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefave CV, Squatrito M, Vorlova S, Rocco GL, Brennan CW, Holland EC, et al. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J 2011; 30: 4084–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar BS, Mulherkar N, Prasad KV. Role of IG20 splice variants in TRAIL resistance. Clin Cancer Res 2008; 14: 347–51. [DOI] [PubMed] [Google Scholar]
- Ramaswamy M, Efimova EV, Martinez O, Mulherkar NU, Singh SP, Prabhakar BS. IG20 (MADD splice variant-5), a proapoptotic protein, interacts with DR4/DR5 and enhances TRAIL-induced apoptosis by increasing recruitment of FADD and caspase-8 to the DISC. Oncogene 2004; 23: 6083–94. [DOI] [PubMed] [Google Scholar]
- Taylor JK, Zhang QQ, Wyatt JR, Dean NM. Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nat Biotechnol 1999; 17: 1097–100. [DOI] [PubMed] [Google Scholar]
- Lainez B, Fernandez-Real JM, Romero X, Esplugues E, Canete JD, Ricart W, et al. Identification and characterization of a novel spliced variant that encodes human soluble tumor necrosis factor receptor 2. Int Immunol 2004; 16: 169–77. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang M, Michael T, Drabier R. Novel alternative splicing isoform biomarkers identification from high-throughput plasma proteomics profiling of breast cancer. BMC Syst Biol 2013; 7 Suppl 5: S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Koh C, et al. Identification of alternative splicing markers for breast cancer. Cancer Res 2008; 68: 9525–31. [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol 2004; 22: 535–46. [DOI] [PubMed] [Google Scholar]
- Effenberger KA, Anderson DD, Bray WM, Prichard BE, Ma N, Adams MS, et al. Coherence between cellular responses and in vitro splicing inhibition for the anti-tumor drug pladienolide B and its analogs. J Biol Chem 2014; 289: 1938–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrignani P, Capone ML, Tacconelli S. Clinical pharmacology of etoricoxib: a novel selective COX2 inhibitor. Expert Opin Pharmacother 2003; 4: 265–84. [DOI] [PubMed] [Google Scholar]
- Andreassi C, Jarecki J, Zhou J, Coovert DD, Monani UR, Chen X, et al. Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum Mol Genet 2001; 10: 2841–9. [DOI] [PubMed] [Google Scholar]
- Ri M, Tashiro E, Oikawa D, Shinjo S, Tokuda M, Yokouchi Y, et al. Identification of Toyocamycin, an agent cytotoxic for multiple myeloma cells, as a potent inhibitor of ER stress-induced XBP1 mRNA splicing. Blood Cancer J 2012; 2: e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmin S, Kymalainen H, Popplewell L, Dickson G. New developments in the use of gene therapy to treat Duchenne muscular dystrophy. Expert Opin Biol Ther 2014; 14: 209–30. [DOI] [PubMed] [Google Scholar]
- Goyenvalle A, Griffith G, Babbs A, Andaloussi SE, Ezzat K, Avril A, et al. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat Med 2015; 21: 270–5. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Martin T, Anthony K, Garcia-Blanco MA, Mansfield SG, Anderton BH, Gallo JM. Correction of tau mis-splicing caused by FTDP-17 MAPT mutations by spliceosome-mediated RNA trans-splicing. Hum Mol Genet 2009; 18: 3266–73. [DOI] [PMC free article] [PubMed] [Google Scholar]