Abstract
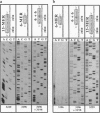
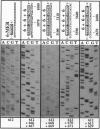
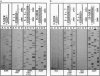
Here we report a striking effect displayed by "modular primers," which consist of hexamer or pentamer oligonucleotide modules base-stacked to each other upon annealing to a DNA template. Such a combination of modules is found to prime DNA sequencing reactions uniquely, unlike either of the modules alone. We attribute this effect in part to the increase in the affinity of an oligonucleotide for the template in the presence of an adjacent module. All possible pentamer (or hexamer) sequences total 1024 (or 4096) samples, a manageable size for a presynthesized library. This approach can replace the synthesis of primers, which is the current bottleneck in time and cost of the primer walking sequencing, and can allow full automation of the closed cycle of walking.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brumley R. L., Jr, Smith L. M. Rapid DNA sequencing by horizontal ultrathin gel electrophoresis. Nucleic Acids Res. 1991 Aug 11;19(15):4121–4126. doi: 10.1093/nar/19.15.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A. S., Najarian D. R., Karger B. L. Separation and analysis of DNA sequence reaction products by capillary gel electrophoresis. J Chromatogr. 1990 Sep 7;516(1):49–60. doi: 10.1016/s0021-9673(01)90203-1. [DOI] [PubMed] [Google Scholar]
- Kieleczawa J., Dunn J. J., Studier F. W. DNA sequencing by primer walking with strings of contiguous hexamers. Science. 1992 Dec 11;258(5089):1787–1791. doi: 10.1126/science.1465615. [DOI] [PubMed] [Google Scholar]
- Kostichka A. J., Marchbanks M. L., Brumley R. L., Jr, Drossman H., Smith L. M. High speed automated DNA sequencing in ultrathin slab gels. Biotechnology (N Y) 1992 Jan;10(1):78–81. doi: 10.1038/nbt0192-78. [DOI] [PubMed] [Google Scholar]
- Lin S. B., Blake K. R., Miller P. S., Ts'o P. O. Use of EDTA derivatization to characterize interactions between oligodeoxyribonucleoside methylphosphonates and nucleic acids. Biochemistry. 1989 Feb 7;28(3):1054–1061. doi: 10.1021/bi00429a020. [DOI] [PubMed] [Google Scholar]
- Nevinsky G. A., Veniaminova A. G., Levina A. S., Podust V. N., Lavrik O. I., Holler E. Structure-function analysis of mononucleotides and short oligonucleotides in the priming of enzymatic DNA synthesis. Biochemistry. 1990 Feb 6;29(5):1200–1207. doi: 10.1021/bi00457a016. [DOI] [PubMed] [Google Scholar]
- Smith L. M. High-speed DNA sequencing by capillary gel electrophoresis. Nature. 1991 Feb 28;349(6312):812–813. doi: 10.1038/349812a0. [DOI] [PubMed] [Google Scholar]
- Stegemann J., Schwager C., Erfle H., Hewitt N., Voss H., Zimmermann J., Ansorge W. High speed on-line DNA sequencing on ultrathin slab gels. Nucleic Acids Res. 1991 Feb 11;19(3):675–676. doi: 10.1093/nar/19.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. A strategy for high-volume sequencing of cosmid DNAs: random and directed priming with a library of oligonucleotides. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6917–6921. doi: 10.1073/pnas.86.18.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szybalski W. Proposal for sequencing DNA using ligation of hexamers to generate sequential elongation primers (SPEL-6). Gene. 1990 May 31;90(1):177–178. doi: 10.1016/0378-1119(90)90458-4. [DOI] [PubMed] [Google Scholar]