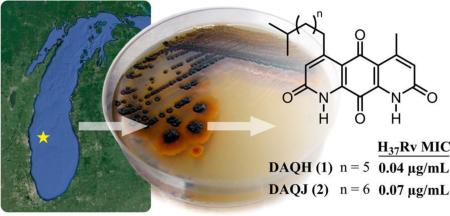
Abstract
Multidrug- and extensively drug-resistant strains of Mycobacterium tuberculosis are resistant to first- and second-line drug regimens and resulted in 210,000 fatalities in 2013. In the current study, we screened a library of aquatic bacterial natural product fractions for their ability to inhibit this pathogen. A fraction from a Lake Michigan bacterium exhibited significant inhibitory activity, from which we characterized novel diazaquinomycins H and J. This antibiotic class displayed an in vitro activity profile similar or superior to clinically used anti-tuberculosis agents and maintained this potency against a panel of drug-resistant M. tuberculosis strains. Importantly, these are among the only freshwater-derived actinomycete bacterial metabolites described to date. Further in vitro profiling against a broad panel of bacteria indicated that this antibiotic class selectively targets M. tuberculosis. Additionally, in the case of this pathogen we present evidence counter to previous reports that claim the diazaquinomycins target thymidylate synthase in Gram-positive bacteria. Thus, we establish freshwater environments as potential sources for novel antibiotic leads and present the diazaquinomycins as potent and selective inhibitors of M. tuberculosis.
Keywords: natural products, Great Lakes, actinobacteria, antibiotic, diazaquinomycin, drug discovery
Graphical Abstract
In 2013 the World Health Organization estimated that there were 1.5 million deaths as a result of tuberculosis (TB) infection, with 9.0 million new cases reported.1 The most significant threat to our population is multidrug- and extensively drug-resistant strains of Mycobacterium tuberculosis (MDR- and XDR-TB), which are resistant to first- and second-line drug regimens and resulted in 210,000 fatalities in 2013 (of 480,000 reported infections).1–3 A major deficiency of current TB treatment is its long duration, which is necessary to eliminate a persistent subpopulation of slow-growing or nonreplicating cells. Importantly, TB is a disease that affects predominantly underprivileged populations in the developing world, and the emergence of drug-resistant forms coupled with the ease with which TB spreads between humans has solidified it as a pathogen of global concern.4
Natural products have been essential components of antibacterial drug discovery; they serve both as a direct source of small molecule therapies and as an inspiration for biologically active synthetic derivatives.5 In particular, actinomycete bacteria have been an abundant source of these bioactive secondary metabolites, providing us with greater than half of the antibiotics on the market today.5,6 However, in previous decades extensive antibiotic screening efforts have exhausted the repertoire of unique terrestrial actinobacteria, resulting in the continuous re-isolation of known antibiotic scaffolds. As a result, researchers prospected new sources for drug lead discovery, such as the ocean. Libraries of organisms (including bacteria) and their resulting secondary metabolites were created that were not incorporated in the biological screening efforts of previous decades.6,7 This investment had a considerable effect toward progress in natural product drug discovery, affording the development of several drugs from marine sources (Prialt, Yondelis, Halaven).8–10
The next logical step is to expand this paradigm to freshwater environments, which harbor distinct environmental selection pressures and growth conditions and to date are virtually unexplored for their capacity to afford unique actinomycete bacteria. Furthermore, despite several cultivation-independent studies aimed at characterizing lake actinomycete populations,11 to the best of our knowledge few efforts (including one study from our laboratory) have identified secondary metabolites from freshwater-derived actinomycetes, and included in this gap is a notable absence of studies aimed specifically at generating anti-TB drug leads.12–14 Thus, a major focus of our antibiotic discovery program is to study actinobacteria derived from the Great Lakes and other freshwater bodies. We have created an extensive library of these bacteria and their resulting secondary metabolite fractions. A preliminary in vitro growth inhibition screening of this fraction library against M. tuberculosis H37Rv led to the identification of a Micromonospora sp. isolated from Lake Michigan sediment, whose fraction exhibited submicromolar inhibitory activity. From this strain we isolated and characterized two novel secondary metabolites, diazaquinomycins H and J (DAQH and DAQJ), which to our knowledge are among the only freshwater-derived actinomycete metabolites described to date.12,13 Further in vitro profiling suggested that this group of diaza-anthracene antibiotics selectively targets M. tuberculosis over other bacteria and is active against several forms of drug-resistant TB. Herein we present the identification and in vitro biological characterization of this unique antibiotic class.
RESULTS AND DISCUSSION
Isolation and Identification of DAQH (1) and DAQJ (2)
Screening of our actinomycete secondary metabolite library against M. tuberculosis in the microplate Alamar Blue assay (MABA) and low-oxygen-recovery assay (LORA) led to the selection of Lake Michigan-derived strain B026 for further chemical investigation. A 28 L fermentation of B026 was performed, and following the extraction of secondary metabolites from the fermentation broth and several chromatographic steps using bioassay-guided fractionation, 0.3 mg each of 1 and 2 was purified using RP-C18 semipreparative HPLC (2.4 mL min−1, gradient of 50% aqueous ACN to 100% ACN for 25 min, followed by an isocratic flow of 100% ACN for 15 min; tR 18.6 and 22.0 min, respectively).
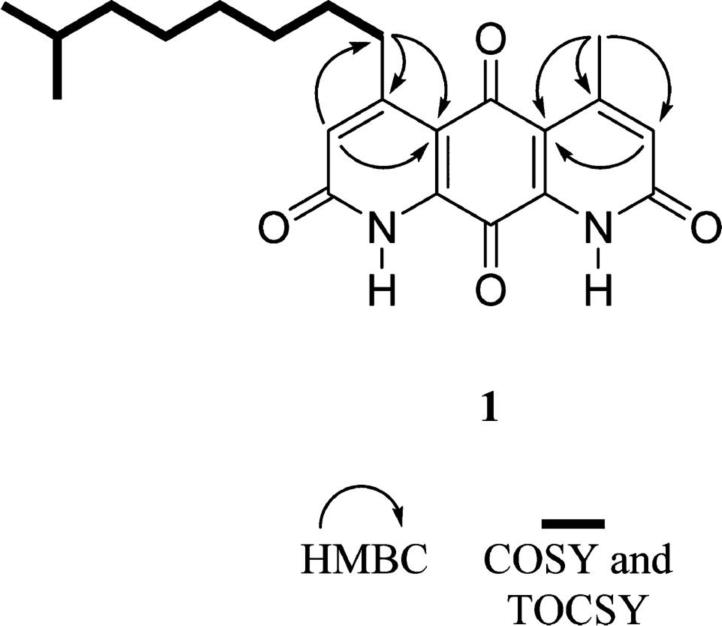
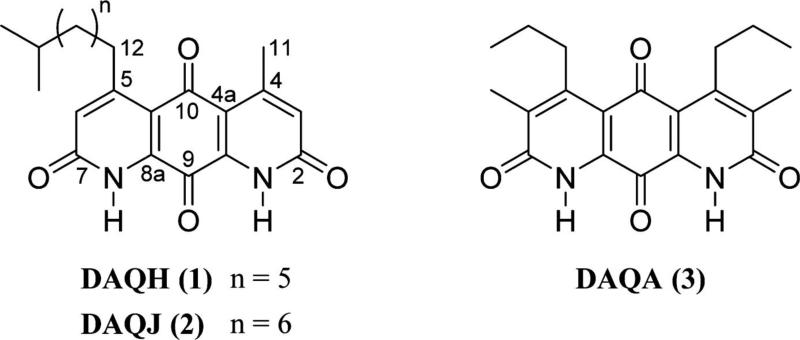
Detailed structure elucidation analysis for compounds 1 and 2, including full 1H and 13C NMR assignments in addition to 2D NMR data and MS experiments, is located in the Supporting Information. Combined NMR and high-resolution IT-TOF mass spectrometry (MS) experiments of 1 established the molecular formula as C22H26N2O4, whereas the UV spectrum displayed a characteristic absorption profile of molecules from the diazaquinomycin class, thus helping to confirm the fused diaza-anthracene core skeleton. 1H NMR resonances for aromatic (H3, H6) and aliphatic (H11, H12) hydrogens and their associated correlations from a heteronuclear multiple-bond correlation (HMBC) NMR experiment suggested that each lactam ring contained α-unsaturated, β-alkylated carbonyl moieties (Table S1). 1H NMR data suggested there was a clear asymmetry to the ring system, and further exploration of the HMBC and MS data confirmed the presence of methyl and isononyl β-substituents on either lactam ring. The connectivity of each set of hydrogens in the isononyl group was determined using correlation NMR spectroscopy (COSY) and a series of one-dimensional selective total correlation spectroscopy (TOCSY) experiments (Figure 2). Therefore, the structure of 1 is as shown (Figure 1).
Figure 2.
Key 2D NMR correlations of 1.
Figure 1.
Structures of diazaquinomycins H (1), J (2), and A (3).
Structure elucidation of DAQJ was executed in the same fashion as DAQH, using a similar series of MS and one- and two-dimensional NMR experiments to identify the molecule. A 14 Da increase in molecular weight and an extra resonance in the 1H and 13C spectra suggested an additional methylene was present in the aliphatic side chain.
In Vitro Evaluation of 1–3 in M. tuberculosis Whole Cell Assays
Once compounds 1 and 2 were identified, we tested their ability to inhibit replicating M. tuberculosis in vitro using the MABA. The MABA is a phenotypic whole cell-based microplate dilution assay that employs a fluorometric readout, relying on the correlation of resazurin dye reduction to bacterial proliferation.15,16 Compounds 1 and 2 exhibited minimum inhibitory concentrations (MICs; defined as the lowest concentration resulting in ≥90% growth inhibition of H37Rv and averaged from triplicates) of 0.04 and 0.07 μg/mL, respectively. A luminescence reporter gene assay (LuxABCDE driven by hsp60) was used to confirm that the activities of 1 and 2 were not readout dependent. Further biological characterization of 1 and 2 was difficult, because a 28 L fermentation afforded only 0.3 mg of each. Fortunately, in a previous study, we isolated and identified four analogues of the diazaquinomycin antibiotic class from a marine-derived Streptomyces sp.17 DAQs F and G were a coeluting isomeric mixture, but DAQA (3) and DAQE were purified and screened in the MABA, exhibiting MICs of 0.1 and 0.04 μg/mL, respectively. From this strain, compound 3 was produced in relatively large amounts; thus, all further biological experiments were carried out using this molecule. Moving forward, the solubility of DAQA was an important consideration in all biological testing. DAQA exhibited a maximum room temperature concentration of 0.15 μg/mL in water in a previous study and 600 μg/mL in DMSO in the current study.18
We also tested 3 for its ability to inhibit nonreplicating M. tuberculosis in the LORA.19 Low-oxygen adapted M. tuberculosis carrying the luxABCDE plasmid was exposed for 10 days to serially diluted 3 in 96-well microplates in a low-oxygen environment created with an Anoxomat commercial system. After 28 h of normoxic “recovery,” activity was assessed using the ability to block recovery of the production of a luminescent signal. The MIC value of 3 was 0.72 μg/mL.
As a confirmatory approach for its anti-TB activity, the minimum bactericidal concentration (MBC99; defined as the lowest concentration that reduces cfu by 99% relative to the zero time inoculum) was determined for 3 by subculture onto 7H11 agar just prior to addition of the Alamar Blue and Tween 80 for MABA MBC99 and reading of luminescence on day 10 from a lux reporter strain for LORA MBC99. Compound 3 exhibited an MBC99 of 0.37 μg/mL under normoxic conditions, but did not exhibit a significant MBC99 under hypoxic conditions.
Cytotoxicity Evaluation of 1–3
To assess the cytotoxicity of the DAQ class, compounds 1–3 were tested in vitro against Vero cells (ATCC CRL-1586). They did not exhibit cytotoxicity at 28 μM, the highest testing concentration. Insufficient yields of 1 and 2 prevented further cytotoxicity screening of these compounds; however, compound 3 exhibited a range of LC50 values when screened against a panel of human cancerous [MDA-MB-435 (0.09 μM), MDA-MB-231 (3.6 μM), HT-29 (5.7 μM), OVCAR3 (0.48 μM), OVACR4 (4.3 μM), Kuramochi (9.4 μM)] and noncancerous cell lines [MOSE (22 μM), MOE (>28 μM)]. Further details describing these experiments can be found in the Supporting Information. Compound 3 was also tested in a previous study against ovarian cancer cell line OVCAR5 and exhibited an LC50 value of 8.8 μM.17 After further investigation, we determined that the moderate cytotoxicity in this cell line was due to DNA damage via the induction of apoptosis.17 A previous paper indicated that compound 3 and DAQC exhibited no significant inhibition when screened for antifungal activity against Mucor miehei and Candida albicans.20 We also assessed the ability of 3 to inhibit the growth of C. albicans (ATCC90028), but no significant activity was exhibited when tested at the highest concentration of 10 μg/mL.
We performed a tolerance assessment of 3 in vivo. It was previously published that 3 deposited solid residues and exhibited acute toxicity when dosed at 100 mg/kg intraperitoneally,18,21 whereas in our study the compound exhibited no qualitatively detectable ill effect and was well tolerated when dosed daily by oral gavage at 100 mg/kg for 5 days in uninfected mice. Although it is possible this difference is due to limited oral bioavailability, our Caco-2 data predict otherwise.
Antibiotic Specificity of 3 toward M. tuberculosis
To assess the ability of DAQA to overcome antibiotic resistance in TB and to gain potential insight into the mechanism of action for this compound class, we screened 3 against a panel of mono-drug-resistant strains using the MABA (Table 1). Compound 3 maintained potency across the panel, which suggested the absence of cross-resistance with current anti-TB agents.
Table 1.
In Vitro Activity of 3 against a Drug-Resistant M. tuberculosis Panel
| MIC (μg/mL) |
|||||||
|---|---|---|---|---|---|---|---|
| 3 | RMPa | INHb | SMc | KMd | PA824e | ||
| M. tuberculosis H37Rv | MABA | 0.10 | <0.08 | 0.05 | 0.12 | 0.65 | 0.01 |
| LORA | 0.72 | 0.85 | >13.7 | 1.10 | |||
| Drug-resistant M. tuberculosis strains | rRMP | 0.27 | >2.00 | 0.02 | 0.27 | 0.92 | 0.03 |
| rINH | 0.13 | 0.01 | >2.00 | 0.46 | 0.95 | 0.03 | |
| rSM | 0.17 | 0.03 | 0.02 | >2.00 | 0.95 | 0.11 | |
| rKM | 0.06 | <0.01 | 0.03 | 1.20 | >2.00 | 0.19 | |
| rCS | 0.14 | <0.01 | 0.01 | 0.41 | 0.98 | 0.12 | |
RMP, rifampicin.
INH, isoniazid.
SM, streptomycin.
KM, kanamycin.
PA824, experimental nitroimidazole antibiotic. rRMP (ATCC 35838); rINH (ATCC 35822); rSM (ATCC 35820); rCS (cycloserine; ATCC35826); rKM (ATCC 35827).
To assess the specificity of 3 toward M. tuberculosis, we screened 3 against a panel of non-tuberculosis mycobacteria using broth microdilution with a spectrophotometric readout at A570 (Table 2). Interestingly, 3 exhibited selectivity toward M. tuberculosis and the closely related M. bovis BCG (MIC = 0.12 μg/mL).
Table 2.
Antimicrobial Spectrum of 3a
| MIC (μg/mL) | ||
|---|---|---|
| mycobacteria | M. abscessus | >7.5 |
| M. chelonae | >7.5 | |
| M. marinum | >7.5 | |
| M. kansasii | >7.5 | |
| M. avium | 3.85 | |
| M. smegmatis | 4.56 | |
| M. bovis | 0.12 | |
| G+e | MSSAb | 4 |
| MRSAc | 16 | |
| E. faecalis | 8 | |
| E. faecium | 8 | |
| VREd | 32 | |
| S. pyogenes | >25 | |
| S. pneumoniae | >10 | |
| B. thuringiensis | 25 | |
| B. cereus 14579 | 25 | |
| B. cereus 10987 | 12.5 | |
| B. anthracis | 6.25 | |
| B. subtilis | 100 | |
| G–f | A. baumannii | 128 |
| E. coli | 128 | |
| E. cloacae | 64 | |
| K. pneumoniae | 128 | |
| P. aeruginosa | 128 | |
| P. mirabilis | 64 |
ATCC designations are listed in the Supporting Information.
Methicillin-sensitive S. aureus.
Methicillin-resistant S. aureus.
Vancomycin-resistant E. faecium.
Gram-positive bacteria.
Gram-negative bacteria.
In the first reports of the diazaquinomycin class, screening of 3 and its 9,10-dihydro derivative, DAQB, against a panel of bacteria revealed a relatively weak MIC of 6.25 μg/mL against three Staphylococcus aureus strains (FDA209P, ATCC6538P, KB199) and Streptococcus faecium IFO3181 while exhibiting a MIC of 1.13 μg/mL against Micrococcus luteus.21,22 In the current study, we determined the MICs of 3 against a diverse panel of Gram-negative and Gram-positive pathogens.23 Compound 3 did not exhibit significant inhibitory activity toward Gram-negative bacteria and showed weak inhibitory activity against the Gram-positive bacteria tested. The results of this and previous studies indicate that 3 possesses a narrow spectrum of activity and shows greatest inhibition toward the pathogen M. tuberculosis.
Caco-2 Permeability and Liver Microsome Stability of 3
Compound 3 was incubated at 1 μM for 30 min with 0.5 mg/mL of human and mouse liver microsomes and resulted in 96 and 98% of the parent compound remaining, respectively. In addition, to predict absorption via oral administration, Caco-2 bidirectional permeability was performed by incubating 5 μM 3 (with vinblastine and propranolol controls) and measuring apical and basolateral concentrations using tandem LC-MS. Compared to control compounds, 3 exhibited moderate absorption showing permeabilities of 44 nm/s with 82% recovery (from apical to basolateral) and 9 nm/s with 98% recovery (from basolateral to apical) and no efflux substrate potential (0.2 efflux ratio).
Studies toward Mechanism of Action of 3
Given its mild reported antibiotic activity, observation of potency against M. tuberculosis provided sufficient motivation for us to further explore its biological mechanism of action. It was reported that the antibacterial activity of 3 was reversed when folate, dihydrofolate, leucovorin, and thymidine were added to the growth medium, suggesting that the target was within the folate pathway.24 Shortly after its discovery, previous studies claimed that 3 inhibited thymidylate synthase competitively with its coenzyme 5,10-methylenetetrahydrofolate in both E. faecium and Ehrlich ascites carcinoma, with Ki values of 36 and 14 μM, respectively.25 Thymidylate synthase is an attractive target as it plays a crucial role in DNA replication and repair by synthesizing de novo deoxythymidine monophosphate (dTMP) from deoxyuridine monophosphate (dUMP). M. tuberculosis encodes for two structurally and mechanistically unrelated thymidylate synthase enzymes (ThyA and ThyX); therefore, we attempted to determine whether either was the target of 3.26,27
Using a cell-free enzyme assay, we found that 3 did not significantly inhibit the activity of purified human ThyA (HsThyA), M. tuberculosis ThyA (MtThyA), or MtThyX when compared to the positive control, folate analogue and known ThyA inhibitor 1843U89 (Figures S1 and S2).28 To rule out the possibility of 3 being metabolized to a biologically active species within the cell, we performed in vitro testing against M. tuberculosis thyA and thyX overexpression mutants. We did not observe a significant differential in the MIC when 3 was screened against M. tuberculosis thyA and thyX over-expression mutants under induced (+ anhydrotetracycline) and uninduced (− anhydrotetracycline) conditions, compared to the H37Rv wild-type strain (Figure S3). Additionally, we observed no reversal of the antibiotic properties of 3 when the culture medium was supplemented with thymidine, indicating that there was likely no disruption in the thymidylate synthase related enzymatic processes of the folate pathway.
Despite decades of research into actinomycete small molecules from terrestrial and marine environments, to the best of our knowledge identification of secondary metabolites from freshwater actinobacteria has been virtually absent from the peer-reviewed literature. The unique structures and activity of the diazaquinomycins provide first evidence that the Great Lakes and, more broadly, freshwater environments are a relatively unexplored resource for novel biologically active molecules.
From a Micromonospora sp. in Lake Michigan sediment, we isolated novel antibiotic molecules of the diazaquinomycin class. An analogue, compound 3, displayed an in vitro activity profile similar or superior to those of clinically used TB agents and maintained potent inhibitory activity against a panel of drug-resistant TB strains. This compound displayed a selectivity profile targeted toward M. tuberculosis, even within the genus Mycobacterium. Since the 1980s, other research groups have reported that members of the diazaquinomycin class exhibited weak antibacterial activity by targeting thymidylate synthase, although no reports of their anti-TB activity existed and our studies have suggested an alternate mechanism of action in M. tuberculosis. Preliminary in vitro analysis of 3 predicts that DAQA will exhibit high metabolic stability and, therefore, a long predicted serum half-life in vivo. Further studies are underway in our laboratories including mutant generation experiments, screening of 3 against a large overexpression library of essential M. tuberculosis genes, and determination of the pharmacokinetic properties of 3 in mice. These studies aim to further assess the potential of 3 as a TB drug lead and identify the molecular target of the diazaquinomycins in M. tuberculosis.
METHODS
Collection and Identification of Actinomycete Strain B026
Strain B026 was isolated from a sediment sample collected by PONAR at a depth of 56 m, from ca. 16.5 miles off the coast north of Milwaukee, WI, USA, in Lake Michigan (43°13′27″ N, 87°34′12″ W) on August 23, 2010. Strain B026 (GenBank accession no. KP009553) shared 100% 16S rRNA gene sequence identity with the type strain Micromonospora maritima strain D10-9-5 (GenBank accession no. NR109311).29
Isolation and Characterization of DAQH (1) and DAQJ (2) from Strain B026 Fermentation Broth
Strain B026 was grown in 28 L for 5 days at 21 °C while shaking at 220 rpm. The extracellular secondary metabolites were absorbed from the fermentation broth using Amberlite XAD-16 resin, followed by extraction with acetone and partitioning between water and ethyl acetate. The organic layer was dried under vacuum to afford 1.1 g of extract.
Using a series of bioassay-guided steps involving preparative and semipreparative normal phase (NP) and reversed phase (RP) high-performance liquid chromatography (HPLC), the organic layer from the liquid–liquid partition was fractionated and purified to afford diazaquinomycin H (1, 0.3 mg, 0.00026% yield) and diazaquinomycin J (2, 0.3 mg, 0.00026% yield). See the Supporting Information for more detailed descriptions of fermentation and extraction methods for B026 and subsequent isolation and purification methods for 1 and 2.
Diazaquinomycin H (1): red solid (0.3 mg); UV (MeOH) λmax (log ε) = 278 (3.83), 301 (3.72), 357 (3.26), and a broad absorption with maximum at 472 (2.59) nm; 1H NMR (900 MHz, CDCl3–1% CF3CO2D) and 13C NMR (226.2 MHz, CDCl3–1% CF3CO2D), see Table S1; HRESI-ITTOF MS m/z 383.1993 [M + H]+ (calcd for C22H27N2O4, 383.1971), m/z 381.1771 [M − H]− (calcd for C22H25N2O4, 381.1820), m/z 765.3778 [2 M + H]+ (calcd for C44H53N4O8, 765.3863), and m/z 763.3639 [2M − H]− (calcd for C44H51N4O8, 763.3712).
Diazaquinomycin J (2): red solid (0.3 mg); UV (MeOH) λmax (log ε) = 278 (3.37), 300 (3.26), 356 (2.90), and a broad absorption with maximum at 472 (2.44) nm; 1H NMR (900 MHz, CDCl3–1% CF3CO2D) and 13C NMR (226.2 MHz, CDCl3–1% CF3CO2D), see Table S1; HRESI-ITTOF MS m/z 397.2162 [M + H]+ (calcd for C23H29N2O4, 397.2127), m/z 395.1924 [M − H]− (calcd for C23H27N2O4, 395.1976), m/z 793.4129 [2M + H]+ (calcd for C46H57N4O8, 793.4176), and m/z 791.3825 [2M − H]− (calcd for C46H55N4O8, 791.4025).
In Vivo Tolerance Assessment of 3
Compound 3 was prepared in a dosing vehicle of 10% polyethylene glycol 400 (PEG 400), 10% polyoxyl 35 castor oil (Kolliphor EL, BASF), and 80% oleic acid. The formulation was then administered by oral gavage at 100 mg/kg of 3 to a pair of female BALB/c mice once daily for 5 days followed by observation for qualitative indicators of toxicity (e.g., weight loss, ragged fur, huddling). Approval for animal studies was provided by the Office of Animal Care and Institutional Biosafety (OACIB) Institutional Biosafety Committee (IBC), case 12-183.
Cell-free ThyA and ThyX Enzyme Inhibition Assay
Overexpression and purification of M. tuberculosis ThyA and ThyX were performed as detailed in Hunter et al.28 Human thymidylate synthase A gene (commonly referred to as TYMS) in pET17xb was transformed into BL21(DE3)pLysS. The enzyme was overexpressed in LB media supplemented with 40 μg/mL chloramphenicol and 100 μg/mL ampicillin at 37 °C until OD600 0.6, when the culture was inoculated with 1 mM IPTG and incubated further at 18 °C for 18 h. Cultures were spun down, and the cell-pellet from a 1.5 L culture was lysed in 30 mL of 20 mM potassium phosphate, 500 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, and 1 mM 2-mercaptoethanol at pH 7.9 on ice by sonication (5 cycles, 1 min, 90 W). The crude extract was spun down at 4 °C and 30597 RCF. A total of 27.5 mL of clarified lysate was obtained; 5.4 g of ammonium sulfate (around 35% saturation) was added in three parts and allowed to dissolve at 4 °C. The precipitate obtained was spun-down at 4 °C and 11952 RCF. Thirty milliliters of supernatant was obtained, to which 10.75 g of ammonium sulfate (approximately 80% saturation) was added slowly at 4 °C and allowed to dissolve. The precipitate obtained was again spun-down at 4 °C and 11952 RCF. It was redissolved in 15 mL of 20 mM Tris, 100 mM NaCl, 2 mM 2-mercaptoethanol, and 10% glycerol at pH 7.8, concentrated to 7 mL, and buffer exchanged into 20 mM Tris, 100 mM NaCl, 2 mM 2-mercaptoethanol, and 10% glycerol at pH 7.8. The sample was purified on a 5 mL HiTrap Q column (low-salt buffer, 20 mM Tris, 100 mM NaCl, 2 mM 2-mercaptoethanol, and 10% glycerol at pH 7.8; high-salt buffer, 20 mM Tris, 1 M NaCl, 2 mM 2-mercaptoethanol, and 10% glycerol at pH 7.8). The protein eluted out before 40% high-salt buffer. Further purification was carried out using a Hiload 16/200 Superdex 200 pg size exclusion column using 20 mM Tris, 100 mM NaCl, 2 mM 2-mercaptoethanol, and 10% glycerol at pH 7.8. HsThyA-containing fractions were pooled and buffer exchanged into 50 mM Tris, 150 mM NaCl, 2 mM TCEP, and 30% glycerol at pH 7.8 and stored at −80 °C until further use.
MtThyA (562.8 ng), MtThyX (8.4 μg), and HsThyA (638.4 ng) were pre-incubated with inhibitors (13 μM) for 15 min at room temperature in 125 mM TES, 60 mM MgCl2, 2.5 mM EDTA, and 2 mM 2-mercaptoethanol at pH 7.9. The control contained DMSO without inhibitors. FAD (10.5 μM) was included in the pre-incubation in the case of MtThyX. The total reaction volume was 151 μL for MtThyA and HsThyA and 160 μL for MtThyX.
Radiolabeled dUMP solution was made by mixing 20 μL of 5′-3H-dUMP and 5 μL of 10 mM dUMP in 475 μL of water. After 15 min, mTHF (67 μM for MtThyA, 5.1 μM for MtThyX, and 5.5 μM for HsThyA) and radiolabeled dUMP (5 μL) were added to 18 μL of the enzyme/inhibitor samples. NADPH at 145 μM was also added to the MtThyX samples. All samples were incubated at room temperature for 30 min. Total reaction volume in all cases was 26 μL. Reaction was stopped after 30 min by the addition of 20 μL of stop solution (1.5 N TCA, 1.1 mM nonradioactive dUMP). A volume of 200 μL of 10% charcoal (w/v) was added, and samples were incubated on ice for 15 min before being spun-down at 4 °C and 14100 RCF for 15 min. A 100 μL aliquot of the supernatant was assayed by liquid scintillation counting to determine the amount of tritium-containing water produced by the reaction.
In Vitro Screening of 3 against M. tuberculosis thyA and thyX Overexpression Mutants
Recombinant strains were constructed in which each gene (thyA or thyX) was under the control of a tetracycline inducible promoter.30 Plasmids were transformed into M. tuberculosis by electroporation.31 Recombinant and wild-type strains were grown to late log phase in roller bottles with the presence of inducer (150 ng/mL anhydrotetracycline). The MIC of compound 3 was determined by measuring bacterial growth after 5 days in the presence of the test compound and inducer.32 Compounds were prepared as a 10-point 2-fold serial dilution in DMSO and diluted into 7H9-Tw-OADC medium in 96-well plates with a final DMSO concentration of 2%. Each plate included assay controls for background (medium/DMSO only, no bacterial cells), zero growth (2% 100 μM rifampicin), and maximum growth (DMSO only), as well as a rifampicin dose response curve. Cultures were filtered through a 5 μm filter and inoculated to a starting OD590 of 0.02. Plates were incubated for 5 days, and growth was measured by OD590. The percent growth was calculated and fitted to the Gompertz model.33 MIC was defined as the minimum concentration at which growth was completely inhibited and was calculated from the inflection point of the fitted curve to the lower asymptote (zero growth).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Dr. Russel Cuhel and the crew of the RV Neeskay from the Great Lakes WATER Institute at the University of Wisconsin—Milwaukee for assistance in sample collection; Mark Sadek, Anam Shaikh, and Maryam Elfeki for fraction library development and preliminary bioactivity screening of strain B026; Thomas Speltz for assistance in the large-scale fermentation of strain B026; Jhewelle Fitz-Henley for assistance in determination of DAQA solubility; the University of Illinois at Chicago Research Resources Center (UIC RRC) for assistance in acquisition of mass spectra; Dr. David Lankin for assistance in acquiring 13C DEPT-Q NMR and 1D TOCSY NMR data; Dr. Benjamin E. Ramirez of the University of Illinois at Chicago Center for Structural Biology for assistance in acquiring remaining NMR data; and Biotranex LLC for in vitro ADME and PK assessment. This project was funded under Department of Defense Grant W81XWH-13-1-0171 and American Cancer Society (Illinois Division) Grant 254871. In the Burdette laboratory, this research was supported by Grant PO1 CA125066 from NCI, NIH. The Rathod laboratory activities at the University of Washington were supported by NIH Grant AI099280. This project was also supported by the Office of the Director, National Institutes of Health (OD), and National Center for Complementary and Integrative Health (NCCIH) (5T32AT007533).
Footnotes
The authors declare no competing financial interest.
Supporting Information
The following file is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.5b00005.
Complete NMR assignments, 1D and 2D NMR spectra, HRMS data, and UV data for 1 and 2, in addition to bioassay methods and figures from investigations of the bioactivity and mechanism of action of 3 (PDF)
REFERENCES
- 1.Global Tuberculosis Report – 2014. World Health Organization; Geneva, Switzerland: 2014. www.who.int/tb. [Google Scholar]
- 2.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorman SE, Chaisson RE. From magic bullets back to the magic mountain: the rise of extensively drug-resistant tuberculosis. Nat. Med. 2007;13:295–298. doi: 10.1038/nm0307-295. [DOI] [PubMed] [Google Scholar]
- 4.Glaziou P, Falzon D, Floyd K, Raviglione M. Global epidemiology of tuberculosis. Semin. Respir. Crit. Care Med. 2013;34:3–16. doi: 10.1055/s-0032-1333467. [DOI] [PubMed] [Google Scholar]
- 5.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenical W, Jensen PR. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat. Chem. Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 7.Murphy BT, Maloney K, Fenical W. Phytochemistry and Pharmacognosy. Eolss Publishers; Oxford, UK: 2011. Marine microorganisms. [Google Scholar]
- 8.Olivera BM, Cruz LJ, de Santos V, LeCheminant GW, Griffin D, Zeikus R, McIntosh JM, Galyean R, Varga J, Gray WR, Rivier J. Neuronal calcium channel antagonists. Discrimination between calcium channel subtypes using omega-conotoxin from Conus magus venom. Biochemistry. 1987;26:2086–2090. doi: 10.1021/bi00382a004. [DOI] [PubMed] [Google Scholar]
- 9.Rinehart KL, Holt TG, Fregeau NL, Stroh JG, Keifer PA, Sun F, Li LH, Martin DG. Ecteinascidins 729, 743, 745, 759A, 759B, and 770: potent antitumor agents from the Caribbean tunicate Ecteinascidia turbinata. J. Org. Chem. 1990;55:4512–4515. [Google Scholar]
- 10.Pettit GR, Herald CL, Boyd MR, Leet JE, Dufresne C, Doubek DL, Schmidt JM, Cerny RL, Hooper JN, Rützler KC. Isolation and structure of the cell growth inhibitory constituents from the western Pacific marine sponge Axinella sp. J. Med. Chem. 1991;34:3339–3340. doi: 10.1021/jm00115a027. [DOI] [PubMed] [Google Scholar]
- 11.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carballeira NM, Pagan M, Shalabi F, Nechev JT, Lahtchev K, Ivanova A, Stefanov K. Two novel iso-branched octadecenoic acids from a Micrococcus species. J. Nat. Prod. 2000;63:1573–1575. doi: 10.1021/np000305r. [DOI] [PubMed] [Google Scholar]
- 13.Carlson S, Tanouye U, Ómarsdóttir S, Murphy BT. Phylum-specific regulation of resistomycin production in a Streptomyces sp. via microbial coculture. [Jan 7, 2015];J. Nat. Prod. 2015 doi: 10.1021/np500767u. DOI: 10.1021/np500767u. [DOI] [PubMed] [Google Scholar]
- 14.Ashforth EJ, Fu C, Liu X, Dai H, Song F, Guo H, Zhang L. Bioprospecting for antituberculosis leads from microbial metabolites. Nat. Prod. Rep. 2010;27:1709–1719. doi: 10.1039/c0np00008f. [DOI] [PubMed] [Google Scholar]
- 15.Collins L, Franzblau SG. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franzblau SG, Witzig RS, McLaughlin JC, Torres P, Madico G, Hernandez A, Degnan MT, Cook MB, Quenzer VK, Ferguson RM, Gilman RH. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J. Clin. Microbiol. 1998;36:362–366. doi: 10.1128/jcm.36.2.362-366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullowney MW, Ó hAinmhire E, Shaikh A, Wei X, Tanouye U, Santarsiero BD, Burdette JE, Murphy BT. Diazaquinomycins E-G, novel diaza-anthracene analogs from a marine-derived Streptomyces sp. Mar. Drugs. 2014;12:3574–3586. doi: 10.3390/md12063574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuzuki K, Yokozuka T, Murata M, Tanaka H, Omura S. Synthesis and biological activity of analogues of diazaquinomycin A, a new thymidylate synthase inhibitor. J. Antibiot. 1989;42:727–737. doi: 10.7164/antibiotics.42.727. [DOI] [PubMed] [Google Scholar]
- 19.Cho SH, Warit S, Wan B, Hwang CH, Pauli GF, Franzblau SG. Low-oxygen-recovery assay for high-throughput screening of compounds against nonreplicating Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2007;51:1380–1385. doi: 10.1128/AAC.00055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maskey RP, Grun-Wollny I, Laatsch H. Isolation and structure elucidation of diazaquinomycin C from a terrestrial Streptomyces sp. and confirmation of the akashin structure. Nat. Prod. Res. 2005;19:137–142. doi: 10.1080/14786410410001704741. [DOI] [PubMed] [Google Scholar]
- 21.Omura S, Iwai Y, Hinotozawa K, Tanaka H, Takahashi Y, Nakagawa A. OM-704 A, a new antibiotic active against gram-positive bacteria produced by Streptomyces sp. J. Antibiot. 1982;35:1425–1429. doi: 10.7164/antibiotics.35.1425. [DOI] [PubMed] [Google Scholar]
- 22.Omura S, Nakagawa A, Aoyama H, Hinotozawa K, Sano H. The structures of diazaquinomycins A and B, new antibiotic metabolites. Tetrahedron Lett. 1983;24:3643–3646. [Google Scholar]
- 23.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. CLSI; Wayne, PA, USA: 2014. 24th Informational Supplement, CLSI Document M100-S24. [Google Scholar]
- 24.Omura S, Murata M, Kimura K, Matsukura S, Nishihara T, Tanaka H. Screening for new antifolates of microbial origin and a new antifolate AM-8402. J. Antibiot. 1985;38:1016–1024. doi: 10.7164/antibiotics.38.1016. [DOI] [PubMed] [Google Scholar]
- 25.Murata M, Miyasaka T, Tanaka H, Omura S. Diazaquinomycin A, a new antifolate antibiotic, inhibits thymidylate synthase. J. Antibiot. 1985;38:1025–1033. doi: 10.7164/antibiotics.38.1025. [DOI] [PubMed] [Google Scholar]
- 26.Myllykallio H, Lipowski G, Leduc D, Filee J, Forterre P, Liebl U. An alternative flavin-dependent mechanism for thymidylate synthesis. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 27.Chernyshev A, Fleischmann T, Kohen A. Thymidyl biosynthesis enzymes as antibiotic targets. Appl. Microbiol. Biotechnol. 2007;74:282–289. doi: 10.1007/s00253-006-0763-1. [DOI] [PubMed] [Google Scholar]
- 28.Hunter JH, Gujjar R, Pang CK, Rathod PK. Kinetics and ligand-binding preferences of Mycobacterium tuberculosis thymidylate synthases, ThyA and ThyX. PLoS One. 2008;3(e2237) doi: 10.1371/journal.pone.0002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Songsumanus A, Tanasupawat S, Igarashi Y, Kudo T. Micromonospora maritima sp. nov., isolated from mangrove soil. Int. J. Syst. Evol. Microbiol. 2013;63:554–559. doi: 10.1099/ijs.0.039180-0. [DOI] [PubMed] [Google Scholar]
- 30.Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, Gomes A, Rustad T, Dolganov G, Glotova I, Abeel T, Mahwinney C, Kennedy AD, Allard R, Brabant W, Krueger A, Jaini S, Honda B, Yu WH, Hickey MJ, Zucker J, Garay C, Weiner B, Sisk P, Stolte C, Winkler JK, Van de Peer Y, Iazzetti P, Camacho D, Dreyfuss J, Liu Y, Dorhoi A, Mollenkopf HJ, Drogaris P, Lamontagne J, Zhou Y, Piquenot J, Park ST, Raman S, Kaufmann SH, Mohney RP, Chelsky D, Moody DB, Sherman DR, Schoolnik GK. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 2013;499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goude R, Parish T. Electroporation of mycobacteria. J. Vis. Exp. 2008;15(e761) doi: 10.3791/761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ollinger J, Bailey MA, Moraski GC, Casey A, Florio S, Alling T, Miller MJ, Parish T. A dual read-out assay to evaluate the potency of compounds active against Mycobacterium tuberculosis. PLoS One. 2013;8(e60531) doi: 10.1371/journal.pone.0060531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sirgel FA, Wiid IJ, van Helden PD. Measuring minimum inhibitory concentrations in mycobacteria. Methods Mol. Biol. 2009;465:173–186. doi: 10.1007/978-1-59745-207-6_11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.