Abstract
During the last decade, high-throughput technologies including genomic, epigenomic, transcriptomic and proteomic have been applied to further our understanding of the molecular pathogenesis of this heterogeneous disease, and to develop strategies that aim to improve the management of patients with lung cancer. Ultimately, these approaches should lead to sensitive, specific and noninvasive methods for early diagnosis, and facilitate the prediction of response to therapy and outcome, as well as the identification of potential novel therapeutic targets.
Genomic studies were the first to move this field forward by providing novel insights into the molecular biology of lung cancer and by generating candidate biomarkers of disease progression. Lung carcinogenesis is driven by genetic and epigenetic alterations that cause aberrant gene function; however, the challenge remains to pinpoint the key regulatory control mechanisms and to distinguish driver from passenger alterations that may have a small but additive effect on cancer development.
Epigenetic regulation by DNA methylation and histone modifications modulate chromatin structure and, in turn, either activate or silence gene expression. Proteomic approaches critically complement these molecular studies, as the phenotype of a cancer cell is determined by proteins and cannot be predicted by genomics or transcriptomics alone.
The present article focuses on the technological platforms available and some proposed clinical applications. We illustrate herein how the “-omics” have revolutionised our approach to lung cancer biology and hold promise for personalised management of lung cancer.
Keywords: Biomarkers, high throughput, lung cancer, -omics, translational
Lung cancer is the leading cause of cancer-related mortality worldwide among both males and females, with >1 million deaths annually [1]. Despite recent advances made in diagnosis and treatment strategies, nonsmall cell lung cancer (NSCLC) prognosis remains poor, with a 5-yr overall survival of 15% [1]. More than 60% of patients are diagnosed with advanced or metastatic disease, mainly due to a lack of early diagnosis tools, and are subsequently not eligible for surgical resection. Although surgery offers the best chance for a cure, 5-yr overall survival remains disappointing at 50% [2]. Recent studies have shown that survival might be improved by platinum-based adjuvant chemotherapy [3, 4], but which patients might benefit from it is not clear. In recent years, some new methods have been developed for high-throughput molecular analysis of tumours and have provided markers as powerful tools for the development of innovative diagnostic and therapeutic strategies in cancer. Genomic studies have been pioneering in providing information on lung cancer molecular biology [5], followed by clear evidence that both genetic and epigenetic alterations are driving carcinogenesis. More recently, proteomic approaches were revealed to be crucial, as the phenotype of a cell is determined by proteins and cannot be predicted by genomics or transcriptomics alone. Indeed, expression levels of proteins are poorly correlated with those of messenger RNA [6], and proteins are commonly subject to post-translational modifications (e.g. phosphorylation, glycosylation) that may modify their functions. This article evaluates the four main categories of high-throughput molecular analysis (genomics, epigenomics, transcriptomics and proteomics) and summarises lessons learned in lung cancer biology and their upcoming new clinical applications.
METHODOLOGICAL ASPECTS
A summary of selected high-throughput methodologies is presented in table 1.
TABLE 1.
Selected high-throughput technologies
| Nature of analytes | Number of analytes | Detected abnormalities | |
|---|---|---|---|
| Genomics | |||
| Global genome sequencing | Genomic DNA | Unlimited | Mutations |
| CGH arrays | Sequences of genomic DNA (bacterial artificial chromosomes, PCR products or cloned cDNA) | Up to 2 million probes | Copy number alterations |
| SNP arrays | Oligonucleotides (sequences of 25 bases characteristic of a gene) | Up to 2 million probes | Polymorphisms, loss of heterozygosity, copy number alterations |
| Epigenomics | |||
| DNA methylation arrays | Oligonucleotides covering whole genome or CpG islands +/− promoters | Up to 385000 probes | DNA hyper- or hypo-methylation |
| Transcriptomics | |||
| Gene expression arrays | Oligonucleotides or cDNA | Up to 385000 probes | Modification of gene expression levels |
| miRNA expression arrays | Oligonucleotides (sequences of 8 to 9 bases) | Up to 1300 probes | Modification of miRNA levels |
| Proteomics | |||
| Two-dimensional gel electrophoresis | Proteins, peptides | 100–1000 proteins | Modification of proteins relative |
| MALDI-TOF MS | Proteins, peptides | 100–300 features | abundance and activity |
| Tandem MS | Peptide sequences | 500–5000 proteins | |
| Protein arrays | Lysates, proteins, peptides | Hundreds of probes | |
| Tissue microarrays | Tissue biopsies | Hundreds of tissue specimens | |
CGH: comparative genomic hybridisation; SNP: single-nucleotide polymorphism; miRNA: microRNA; MALDI-TOF MS: matrix-assisted laser desorption ionisation time-of-flight mass spectrometry.
Genomics
Genomics is the study of an organism’s entire genome. The most important of the related technologies are high-throughput capillary sequencing, comparative genomic hybridisation (CGH) arrays and single-nucleotide polymorphism (SNP) arrays.
Global genome sequencing
Global genome sequencing has made major improvements from gel-based sequencing to automated reading of the TGCA oligonucleotides [7–9]. A better separation system, called capillary electrophoresis, allows DNA to be sorted inside capillary tubes instead of in a gel, which allows the automation of the DNA loading system, leading to an increase in throughput and higher speeds. The new sequencers do not use the old chain-termination paradigm; instead sequencing-by-synthesis technology binds short fragments of DNA to small beads that are dropped into wells in a fibreoptic chip. The DNA adds another molecule to its chain. When that happens, the sequencer identifies which wells used a T or G molecule, indicating which base is next in the sequence. The assembly of these pieces of DNA is a major challenge and may require multiple runs through a sequencer before the complete sequence of the genome can be assembled.
CGH arrays
CGH arrays are used to study genomic copy number variations at high resolution [10–12]. Pieces of genomic DNA (bacterial artificial chromosome, PCR products representative of genomic sequences or cloned cDNA) with known chromosomal location and serving as target DNA are spotted on a glass slide. Test (cancer) and reference (normal) DNA are labelled with different fluorophores and simultaneously hybridised to clone the target chromosomal fragments. The ratio of fluorescence intensity of cancer to normal DNA is calculated to measure the copy number changes for a particular genomic location.
SNP arrays
SNP arrays allow accurate measurement of cancer-specific loss of heterozygosity in a high-throughput manner. Detecting loss of heterozygosity across the genome allows identification of patterns of allelic imbalance with potential prognostic and diagnostic utilities. It can also detect small regions of copy number alterations by producing genomic maps of high resolution [13, 14]. SNP arrays are commercially available in 10,000, 100,000, 500,000 and up to 2 million SNP loci formats. They are synthesised by photolithography and contain up to 40 separate oligonucleotide probes for each SNP locus, representing both mismatch and perfect match probes. After DNA labelling and hybridisation, fluorescence intensities are measured for each allele of each SNP.
Epigenomics
Epigenomics is the large-scale study of epigenetic modifications, i.e. heritable changes in gene expression without DNA sequence alterations, mainly DNA methylation and histone post-translational modifications.
Different techniques are available for the detection of DNA methylation, based on the ability to distinguish cytosine from 5-methylcytosine in the DNA sequence, as follows: DNA digestion by a methylation-sensitive or -insensitive restriction endonuclease, DNA chemical modification by sodium bisulfite or metabisulfite, and 5-methylcytosine immunoprecipitation to separate the methylated and unmethylated fractions of the genome. All these techniques are now coupled to high-throughput technologies. Early epigenomic approaches used custom-made, slide-based arrays of CpG-rich regions corresponding to methylated or unmethylated DNA [15]. Later, because of their better precision and quantitative potential, the commercially available high-density oligonucleotide arrays were used [16].
So far, there is no high-throughput method to study histone modifications. However, chromatin immunoprecipitation-on-chip technology is a microarray platform upon which immunoprecipitated DNA is hybridised against known probes; it allows the assessment of chromatin states [17].
Transcriptomics
Transcriptomics or global analysis of gene expression is the study of the transcriptome, the complete set of mRNA transcripts produced by the genome. The most common related high-throughput technologies are gene expression arrays (oligonucleotide and cDNA arrays) and microRNA (miRNA) expression arrays.
Gene expression arrays
Over the past 15 yrs, methods have been developed for the purpose of measuring semiquantitatively the relative abundance of transcribed genes in a given tumour sample [18]. Gene expression arrays mostly make use of matrix-bound probes to which processed mRNA templates of the analysed specimens will hybridise. Two major types of arrays have been developed for that purpose: oligonucleotide and cDNA arrays. While the former makes use of short oligonucleotides synthesised on the array matrix, the latter type of array employs probes of copy-DNA. Following hybridisation of a pre-processed and fluorescently labelled mRNA sample extracted from the tumour specimen, the arrays are scanned and transcript abundance is measured as a direct correlate of signal intensity. Following data normalisation, data can be analysed using a virtually unlimited array of computational and statistical methods. The two general approaches most commonly used for the study of cancer specimens involve methods for unsupervised clustering and supervised learning methods. While unsupervised methods allow self-organisation of data matrices according to similarity of features, supervised learning methods can yield gene sets or signatures of genes that can distinguish between a priori defined subsets of tumours. For example, one may define two groups of tumours according to clinical outcome (“poor survival” and “good survival” groups). Supervised learning methods, such as those involving K-nearest neighbour-based prediction methods can now help to identify those sets of genes whose transcriptional levels can best distinguish between the two groups [19]. In order to minimise the risk of overfitting the predictive signature, such approaches typically involve validation of the predictor in a separate dataset or by splitting the original dataset into a learning and test set. In the latter case, the predictor is built using the learning set and then validated in the test set. Also, the stability of the predictor is typically tested by random permutation of the data. By contrast, hierarchical clustering methods can help identify subgroups of tumours not known a priori that are characterised by a typical transcriptional signature.
miRNA expression arrays
miRNAs are single-stranded, small (18 to 24 nucleotides in length), noncoding RNAs that negatively regulate gene expression by binding to and modulating the translation of specific mRNAs. Each miRNA appears to regulate the translation of multiple genes, and many genes appear to be regulated by multiple miRNAs. miRNA microarrays enable the comparison of miRNA expression profiles of different tissues [20]. Total RNA from tumour and normal tissues are isolated and then gel fractionated using an electrophoresis system designed to speed up small RNA fractionation and isolation. miRNA fractions for each sample are labelled with different fluorophores. Labelled miRNAs are hybridised with slides arrayed with up to 1,300 distinct probes of eight to nine nucleotides. The ratio of fluorescence intensity of cancer to normal miRNA is calculated to measure the changes in expression levels for a particular miRNA sequence.
Proteomics
Proteomics is the large-scale study of proteins, particularly their structure and functions. Several high-throughput technologies have been developed.
Two-dimensional gel electrophoresis
This technique relies on polyacrylamide gels that separate proteins based first on their charge and then on their molecular weight. Gels are scanned with laser densitometers and analysed with software allowing the semiquantitative visualisation of >500–1,000 proteins per gel [21]. Individual protein spots of interest can be digested into peptides for sequence analysis by mass spectrometry (MS). A recent modification of this technique is the differential in gel-electrophoresis (DIGE), used to compare two protein mixtures from two different cell types directly on the same gel. The two pools are labelled with different fluorescent dyes, mixed together and run on the same gel [22]. Identical proteins from the two pools co-migrate and are independently detected by quantitative fluorometry. Differentially expressed proteins of interest are identified by alterations in the ratios of the two fluorescent signals. When samples are run on different gels, a third fluorescent dye is used as an internal standard allowing the normalisation of all spots across all gels. The standard is composed of a pooled sample containing equal amounts of both test and control samples.
Matrix-assisted laser desorption ionisation time-of-flight MS
Matrix-assisted laser desorption ionisation time-of-flight MS (MALDI-TOF MS) is a high-throughput technique that analyses with high sensitivity and specificity proteins expressed in complex biological mixtures, such as serum, urine and tissues [23, 24]. It requires sample co-crystallisation with a matrix that absorbs laser energy and subsequently ejects and ionises molecules into the gas phase. Ions are then accelerated from the ion source by a fixed potential difference and travel a fixed-length, field-free distance before reaching the detector at a speed inversely proportional to their m/z ratios (lighter ions are faster to reach the detector than heavier ions with the same charge). The time taken by each ion to hit the detector creates a signal and indicates an m/z ratio of a given ion with a particular intensity. This methodology has been extensively applied to proteomic profiling of biological specimens as described below.
Liquid chromatography tandem MS
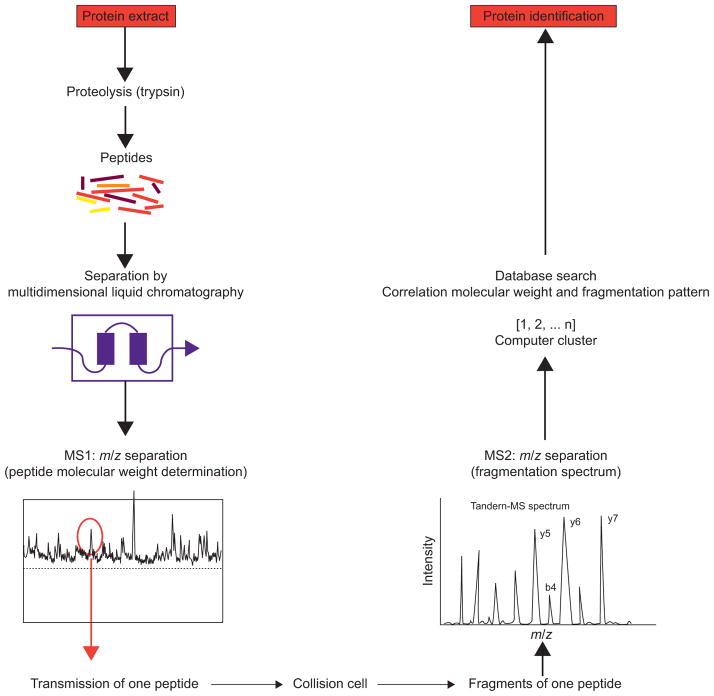
This technique combines high-performance liquid chromatography (LC) with electrospray ionisation MS, ionising and vaporising proteins from liquid solutions [25]. The multidimensional fractionation of biological samples coupled with this technique allows deeper interrogation of the proteome. The shotgun proteomic analysis platform uses digestion of the sample with site-specific proteases, multidimensional separation of peptides by strong cation exchange chromatography, such as in the multidimensional protein identification (MudPIT) technology [26–28] or isoelectric focusing [29, 30], followed by reverse phase (RP) LC separation coupled directly to a tandem MS instrument (MS/MS) (fig. 1). The most abundant peptides are sequentially selected for MS/MS analyses. Resulting fragment ions are then analysed in a second MS scan according to their m/z ratios. Based on the fragments produced in the collision cell and their precise molecular weight, the peptides sequence can be derived [31]. Through comparisons with predicted sequences of same nominal mass in databases, peptides are identified and the proteins from which they came are deduced.
FIGURE 1.
Schematic representation of “bottom up” or shotgun analysis. A protein mixture is first digested (by trypsin) and the resulting peptides are separated by multidimensional liquid chromatography (typically strong cation exchange followed by reverse-phase separation) coupled online to a mass spectrometer. As they elute, the m/z ratios of the peptides are first determined, followed by one or several mass spectrometry (MS)/MS scans from the most abundant peptide signals (y5, y6 and y7 are m/z values for “y-ions” and b4 is the m/z value for a “b-ion”. The fragment peaks that appear to extend from the amino terminus are termed “b” ions, and those that appear to extend from the C-terminus are termed “y ions”). This cycle is repeated until all of the peptides have eluted from the chromatography column. For each precursor peptide selected for MS/MS, peptides of similar nominal mass are extracted from sequence databases and predicted fragmentation patterns are derived in silico. These patterns are then compared with the experimental fragmentation spectrum to generate correlation scores. Positive identification of a protein is based on the observation of two or more peptides issued from its sequence.
Protein arrays
Protein arrays offer an efficient way of simultaneously analysing multiple samples or proteins in a high-throughput manner. The two main forms are forward- and reverse-phase arrays. In forward-phase arrays, hundreds of specific antibodies, or bait proteins, are arrayed on a glass slide and one protein extract sample, representing one specific condition, is placed on the array. Expression levels or post-translational modifications of hundreds of proteins are analysed in a single experiment [32]. In reverse phase arrays, hundreds of tissue protein extracts or recombinant proteins are placed on glass slides and probed with a single antibody [33] or a drug candidate for drug development [34]. Expression levels of a given protein are evaluated in multiple samples.
Tissue microarrays
In tissue microarrays (TMAs), up to 3,000 cylindrical tissue biopsies (0.6 mm in diameter) from individual tumours are distributed in a single paraffin block. Sections of the block provide targets for parallel in situ detection of DNA, RNA and proteins in each specimen on the array, and consecutive sections allow the analysis of hundreds of molecular markers in the same set of specimens [35, 36].
Considerations for tissue collection
The majority of the discovery efforts are based on the collection, storage and processing of tissue specimens obtained at the time of surgery, bronchoscopy or other diagnostic procedures. After informed consent, all biological specimens need to be collected under a standard operating procedure and quality control strategies must be in place to guarantee adequacy of the samples. This requires a concerted effort between clinicians, pathologists and research coordinators. Profiling using high-throughput technologies in biological fluids or tissue samples is best served by the use of fresh frozen materials. Tumour-derived markers are likely to be present at lower levels in blood and other biological specimens. Thus, the dynamic range of analytes concentration in biological specimens adds a critical dimension of technical considerations to the successful molecular analysis. Difficulties related to the lack of standardisation of the methodology, result in large variations between the results of different studies and making the translation of most discovery efforts to clinical practice difficult. To address these issues, huge efforts have been undertaken [37, 38], including the one at the Office of Biorepositories and Biospecimen Research (National Cancer Institute, Bethesda, MD, USA).
The use of formalin-fixed, paraffin-embedded tissue has many advantages. It allows the use of large collections of tissue available, which are fully annotated and easier to handle. Methods of extraction allow the recovery of quality DNA, RNA and proteins for high-throughput discovery and validation strategies. However, issues around the methods of preservation and the small amount of tissue available still render these efforts challenging [39]. Promising single-molecule sequencing and high-throughput oncogene mutation profiling represent strategies that may be applicable to small clinical samples in the future to address personalised medicine. However, before the next generation of sequencing enters clinical use, issues of cost, data analysis and interpretation will have to be resolved.
BIOLOGICAL INFERENCES FROM HIGH-THROUGHPUT MOLECULAR ANALYSES OF LUNG CANCER
Discovery of critical mutations by systematic re-sequencing of genes in tumours
An area that has recently provided a rich source of clinically relevant information is the systematic re-sequencing of genes in tumour specimens. These studies were inspired by the observation that genetically activated kinases represent excellent therapeutic targets. Kinases were among the most heavily sequenced genes in systematic cancer gene re-sequencing efforts. Early discoveries from these projects include the discovery of BRAF mutations in thyroid, colorectal and lung cancers, as well as in a majority of malignant melanomas [40]. The fraction of BRAF-mutant lung cancers appears to be <5% and to be exclusively restricted to lung adenocarcinomas [41]. This observation has led numerous pharmaceutical companies to launch drug discovery programmes dedicated to specific targeting of BRAF-dependent tumours. Trials investigating the efficacy of these agents are ongoing in several tumour types [42]. Whether BRAF-mutant lung cancer can be successfully treated with inhibitors targeting downstream pathway members of BRAF remains to be established. In another study, mutations in the gene encoding the catalytic subunit of the class-1a phosphoinositide-(3,4,5)-kinase (PIK3CA), were found in a large fraction of epithelial cancers [43]. While frequency of these mutations was found to be <5% in lung cancer, PIK3CA is the most frequently mutated oncogene in breast cancer and is also frequently mutated in colorectal cancer. In several functional analyses, these mutations were found to be oncogenic by activating the Akt survival pathway [44]. Most recently, pre-clinical studies involving new generation phosphoinositide (PI)3-kinase inhibitors suggest that tumours carrying these mutations might be exclusively sensitive to such treatment [45]. Since some of these agents are currently being developed in early clinical trials, there is considerable hope that they might be clinically efficient in multiple solid tumours. This hope is supported by the observation that tumours driven by oncogenically activated receptor tyrosine kinases frequently exhibit activation of the PI3-kinase/Akt pathway.
The most groundbreaking discovery made in these projects was the finding that the receptor tyrosine kinase epidermal growth factor receptor (EGFR) is mutated in ~10% of Caucasian and up to 40% of East-Asian patients with NSCLC [46–49]. These mutations were enriched in patients who were of East-Asian ethnicity, who had never smoked and whose tumours were of lung adenocarcinoma histology. These features were closely related to those of patients who had experienced responses when treated with the EGFR tyrosine kinase inhibitors erlotinib or gefitinib. In fact, exon re-sequencing revealed mutations in the EGFR kinase domain in responders to gefitinib or erlotinib but not in nonresponders. Thus, mutant EGFR appeared to be an additional example of successful therapeutic targeting of oncogenically activated tyrosine kinases. Unfortunately, all patients initially responding to EGFR inhibitors will eventually relapse. Systematic genetic analyses of tumour biopsies obtained at the time of relapse revealed a second site mutation of EGFR as the causative mechanism. This mutation, T790M, is analogous to the T315I mutation of ABL, causing acquired resistance in chronic myeloid leukaemia patients who initially responded to imatinib [50–52]. Functional cell and structural biology studies have now proven, on a formal level, the causative nature of the T790M mutation in acquisition of resistance. An additional mechanism of acquired resistance is the de novo amplification of the MET receptor tyrosine kinase gene [53, 54]. Importantly, this mechanism was identified using in-depth cell biology and cancer genome analyses in cell line experiments and subsequently validated in patient specimens. This discovery, therefore, further supports the use of cell line experiment as pre-clinical proxies with substantive clinical relevance.
Numerous trials have been performed since the discovery of EGFR mutations in lung cancer with the goal of defining the role of EGFR mutations’ detection as a tool to select patients for treatment with EGFR inhibitors. Although a detailed discussion of the results from these trials is beyond the scope of this review, we would like to highlight some central results. Importantly, trials selecting patients based on the presence of EGFR mutations or on clinical characteristics of EGFR mutations (Asians, never-smokers, adenocarcinomas) consistently yielded response rates exceeding 70% and led to median overall survival exceeding 20 months [55–59]. Although these results led to widespread excitement in the community, as they suggested an effective treatment for a significant subgroup of lung cancer patients, it was suggested that EGFR mutations were prognostic rather than predictive. These results were initially supported by a study by Eberhard et al. [60] suggesting that EGFR-mutant patients had a better overall survival independent of treatment. The study by Eberhard et al. [60] was recently confirmed by findings of the Iressa Non-small cell lung cancer Trial Evaluating REsponse and Survival against Taxotere (INTEREST) trial [61], which failed to show a benefit in overall survival for EGFR-mutant patients with gefitinib. In fact, progression-free survival in this trial was significantly better for EGFR-mutant patients receiving gefitinib. Most recently, the results from the IRESSA Pan Asia Study (IPASS) trial [62] comparing gefitinib and carboplatinum plus docetaxel in East-Asian never- or ex light-smokers with adenocarcinomas were reported. This trial not only confirmed a clear survival benefit for EGFR-mutant patients treated with gefitinib but also showed that EGFR mutation-negative patients receiving gefitinib had the worst prognosis. Altogether, these findings show that EGFR mutations are in fact predictive for treatment outcome in patients receiving EGFR inhibitors and that patients without EGFR mutations should not be treated with these agents; thus, EGFR mutation testing should be mandatory before treating lung cancer patients with EGFR inhibitors.
In independent studies, EGFR copy number gain and amplification were found to be better markers of treatment outcome in patients receiving gefitinib or erlotinib than the presence of EGFR mutations [55, 63–65]. However, concerns were raised with respect to the methods used for mutation detection and for criteria of copy number assessment. Furthermore, mutation detection using conventional sequencing methods is hampered by low sensitivity, in particular in samples with high admixture of nontumoural cells (see following section). In contrast, detection of chromosomal copy number gains is far less sensitive to sample impurity. Given the fact that EGFR mutations are highly correlated with EGFR copy number gains and amplifications ([66] and R.K. Thomas, Max Planck Institute for Neurological Research, Köln, Germany; personal communication), it is tempting to speculate that findings suggesting a predictive role for EGFR copy number gains/amplifications might be reflective of the low sensitivity in EGFR-mutation detection.
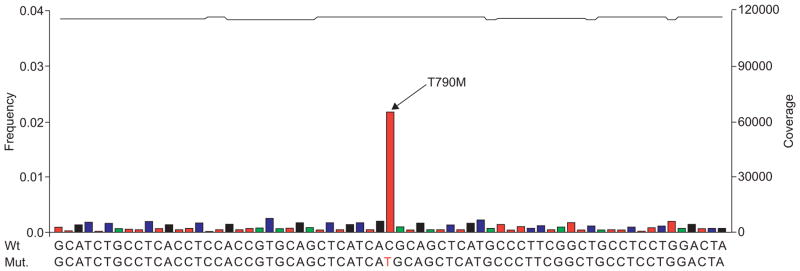
One important issue to consider when interpreting results from studies reporting the frequency and clinical relevance of somatic oncogene mutations is the fact that the most widely used method for mutation detection in cancer specimens, di-deoxy nucleotide sequencing [67], is hampered by low sensitivity. In particular, this applies to samples with an admixture of nontumoural cells, a typical feature of most biopsy specimens analysed in the clinic. For example, in a patient series of 22 specimens enriched for the presence of EGFR mutations that had been previously heavily sequenced using the Sanger method [47], definite determination of EGFR mutation prevalence showed that conventional sequencing had missed three mutations [68]. By contrast, recently developed genetic analyses methods that were originally designed to enable rapid genome sequencing afford accurate and independent sampling of all allele species present within a given tumour specimen. Using the array-based picotitre pyrosequencing-by-synthesis technology developed by 454 LifeSciences (Roche Diagnostics, Base, Switzerland), oncogene mutations were discovered in samples where conventional sequencing had failed to do so. This approach accurately detected mutations in formalin-fixed, paraffin-embedded tissue specimens [68]. Furthermore, it yielded accurate mutation diagnosis at extreme sensitivity (e.g. down to allelic representations of 1%) and allowed the identification of the gatekeeper mutation T790M, associated with resistance to EGFR inhibitors, in malignant pleural effusion specimen (fig. 2). Thus, the dramatic limitations of conventional sequencing can easily be overcome employing novel genome analytics when used in the context of clinical genetic diagnostics.
FIGURE 2.
Sequencing of DNA from cells recovered in a pleural effusion specimen obtained at the time of acquired erlotinib resistance. After PCR amplification of exon 20, 454 sequencing revealed the presence of the T790M resistance mutation of epidermal growth factor receptor (EGFR) [49] at ~2%. Robust calling of T790M carrying EGFR alleles was ensured by >60,000 × over-sampling. Wt: weight; Mut.: mutation. ——: coverage; green: A; blue: C; black: G; red: T. Reprinted from [68], with permission from the publisher.
In another project termed OncoMap, genotyping assays were designed for >200 known oncogene mutations in 17 oncogenes covering multiple classes (e.g. receptor tyrosine kinases, intracellular tyrosine kinases, serine-threonine kinases and small GTPases). A mass spectrometric genotyping method (Sequenom) was applied to genotype this panel of oncogene mutations across 1,000 human cancer specimens, covering multiple tumour types. This approach was highly accurate, sensitive and specific. Most importantly, due to the scalability and rapidity of this method, we were able to profile all 1,000 tumours within a few weeks of the time of the experiment and at a reasonable cost [69].
The efforts to fully characterise the genomes of major human cancer types [70–74] revealed that beyond known cancer-related genes, many additional genes are mutated in individual cancers. However, most of these mutations occurred only in a single tumour, suggesting that each tumour is composed of an individual set of mutations contributing to tumorigenesis. The average number of mutations per tumour of a given entity ranged from 48 in pancreatic cancer to 101 in breast cancer [71]. Ultimately, functional cell biology experiments will be required to establish the individual contributions of these mutations to oncogenic transformation. Such efforts will need to afford assaying cell biology at high-throughput in order to cope with the multitude of genetic lesions discovered by high-throughput cancer genome profiling.
Finally, technologies are being developed that will help to potentiate the throughput and sensitivity in genome sequencing. When considering that in contrast to discovery-oriented genome profiling projects that employ specimens enriched for high tumour content, the prototypical diagnostic specimen in the clinic is a small piece of formalin-fixed transbronchial biopsy with large amounts of necrosis and numbers of inflammatory cells. Technologies such as single-molecule sequencing or high-throughput oncogene mutation profiling represent good examples of how these problems can be tackled in order to ensure accurate diagnosis and optimal patient stratification for targeted therapy.
Stable genomic alterations in lung tumours measured by array CGH tell us about tumour behaviour
Over the last 10 yrs, we have learned that somatic molecular alterations in cancers yield signatures that can be used for sub-classification [75–77], and that they provide information relevant to predicting patient survival [75, 78], risk of recurrence [79] and response to therapy [76, 80]. Nevertheless, NSCLC is still largely managed as a single major entity using similar preventive, diagnostic and therapeutic approaches. Chromosome abnormalities often correlate with molecular abnormalities and provide a starting point for gene discovery and characterisation in the context of a specific disorder [81]. In cancer biology, chromosomal abnormalities carry diagnostic, prognostic and predictive value of response to treatment. The Cancer Gene Consensus of the Cancer Genome Project at the Sanger Institute [82] contains 363 cancer genes, among which 292 are oncogenes otherwise commonly activated by translocation. Recent studies report 138 hot spots of genomic amplification (across 104 human cancer cell lines) [83]. The authors identified ~50% of putative oncogenes in the previous study showing genomic amplification. In lung cancer cell lines, an average of 32 regions of amplification was detected per genome. Taken together, these data suggest that genomic amplification may be a common mechanism of oncogene activation.
The use of array CGH based on high density of bacterial artificial chromosome clones, cDNA microarrays, oligonucleotide arrays or SNP arrays combined with mRNA expression arrays has greatly improved the resolution of traditional CGH and has facilitated identification of new candidate genes across the genome [84–87]. Among the prevalent chromosomal changes in lung cancer, chromosome 3q amplification is one of the most frequent and is an early event in lung as well as in aero-digestive tract tumours [88, 89]. The amplification of the distal portion of chromosome 3q in lung cancer is a major signature of neoplastic transformation [90]. It is found in early stages of lung cancer development, including severe bronchial dysplasia, and is maintained throughout the progression of cancer as well as in metastatic stages [91]. A causal relationship between smoking history and 3q amplification has been suggested but has not yet been proven. The size of the amplicon varies greatly between tumours and spans from chromosome 3q22 to 3qter, with a most frequent region of amplification in squamous cell carcinoma between 3q26 and 3q29 (~35 Mb). High-throughput technologies will eventually allow the integration of complex molecular analyses and shed light on the role of this amplicon in lung cancer.
Airway epithelium in the field of carcinogenesis: the oncogenic battleground
Carcinogen exposure induces diffuse epithelial injury, with genetic changes and premalignant/malignant lesions in one region of the field representing an increased risk of cancer development in the entire field. The impact of smoking on airway transcriptome has been the subject of recent investigations.
One of these investigations, using Affymetrix HG-U133A and HG-U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA, USA) to study small airway bronchial epithelium, found 300 genes significantly up- or down-regulated in phenotypically normal smokers (n=16) compared to matched nonsmokers (n=17), including genes coding for response to oxidants and xenobiotics, immunity and apoptosis [92]. Moreover, variability in small airway epithelium gene expression among phenotypically normal smokers was shown, suggesting a genetic factor in response to smoking [93, 94].
Patterns of gene expression that occur in bronchial airway epithelial cells obtained via bronchoscopy from healthy current, former and never-smokers have also been explored by high-density oligonucleotide microarray analysis [95]. Airway epithelial cells were obtained by brushing of the main stem bronchus. RNA was isolated, processed and hybridised onto the Affymetrix HG-U133A array. In that study, Spira et al. [95] identified gene expression changes that occur in response to smoke exposure and demonstrated that the expression of a number of genes correlated with cumulative tobacco exposure. Using Affymetrix HG-U133A microarrays, the same group performed gene expression profiling of histologically normal large airway (main stem bronchus) epithelial cell brushings obtained from current and former smokers undergoing flexible bronchoscopy as a diagnostic study for the suspicion of lung cancer at four medical centres [96]. Each subject was followed until a final diagnosis of lung cancer or an alternate diagnosis was made. In a training set of 77 samples, they identified an 80-gene biomarker that distinguishes smokers with and without lung cancer. The biomarker was tested on an independent test set (n=52), with an accuracy of 83% (80% sensitive, 84% specific), and subsequently validated on prospective series independently obtained from five medical centres (n=35). These results suggest that tobacco smoke induces a cancer-specific field of injury throughout the airway epithelium, with potential value as a marker of diagnosis and/or risk. More recently, the same group used a clinical and combined genomic model that proved the biomarker independence from other clinical factors and showed its higher prediction accuracy (fig. 3) [97]. In patients with suspicion of lung cancer, this marker may improve the diagnostic sensitivity of histological assessment and reduce the use of additional diagnostic tests that are invasive and expensive. Moreover, it could be tested as a candidate biomarker of risk of lung cancer development and as intermediate end-point biomarker of response to chemopreventive strategies.
FIGURE 3.
Added value of genomic to a clinical model predictive of lung cancer. Receiver operating characteristic curves of the combined training and test sets (n=118), consisting of smokers undergoing bronchoscopy for suspicion of lung cancer. The clinical model (······) includes three variables: age, mass size and lymphadenopathy; the clinicogenomic model (——) includes the previous variables and the biomarker score. The area under the curve for the clinical and clinicogenomic models is 0.89 and 0.94, respectively, which represents a significant difference between the two curves (p<0.05). Reproduced from [97], with permission from the publisher.
Potential of gene expression profiles to analyse oncogenic pathways
Carcinogenesis is a complex process characterised by the accumulation of multiple independent genetic alterations, often involving overexpression of oncogenes and loss of tumour suppressor genes. These genetic alterations disrupt the normal regulation of cell signalling pathways, essential for the control of cell growth, differentiation and apoptosis. Several studies have demonstrated the potential of gene expression profiles to analyse oncogenic pathways and to describe the complexity of cancer phenotype [98–102].
In a recent study, gene expression profiles characterising activated pathways were found after infection of quiescent primary mammary epithelial cells with adenovirus to express relevant genes and artificially activate pathways of interest [103]. The ability of each signature to accurately assess the corresponding pathway status was verified by internal validation measures, by the use of three mouse tumour models (mouse mammary tumour virus-Myc, Ras, E2F3) and by use of adult mouse tumours in which Ras activity was activated by homologous recombination. Pathway activities were then evaluated in NSCLC samples. Ras pathway deregulation was found in most adenocarcinomas as opposed to squamous cell carcinoma, suggesting its role in adenocarcinoma development. Tumours with high Ras activity were also predicted to have low levels of Myc, E2F3, β-catenin and Src activity, and vice versa. Patients with deregulation of Ras associated with β-catenin, Myc and Src constituted a poor survival population. Finally, investigators successfully used the pathway deregulation signatures to predict sensitivity to therapeutic agents targeting the corresponding pathways in cancer cell lines. This study is a good illustration of how integrated pathway analysis develops our understanding of carcinogenesis. Moreover, this approach may be helpful for a better categorisation of patients with lung cancer, in terms of histology and also outcome, in an attempt to address the need for personalised therapy. Finally, this strategy represents a promising tool to guide the use of combined targeted therapies.
Role of epigenetic events in tumour progression
Recently, it is becoming clear that epigenetic events, such as DNA methylation and histone modifications, are also central to tumour progression.
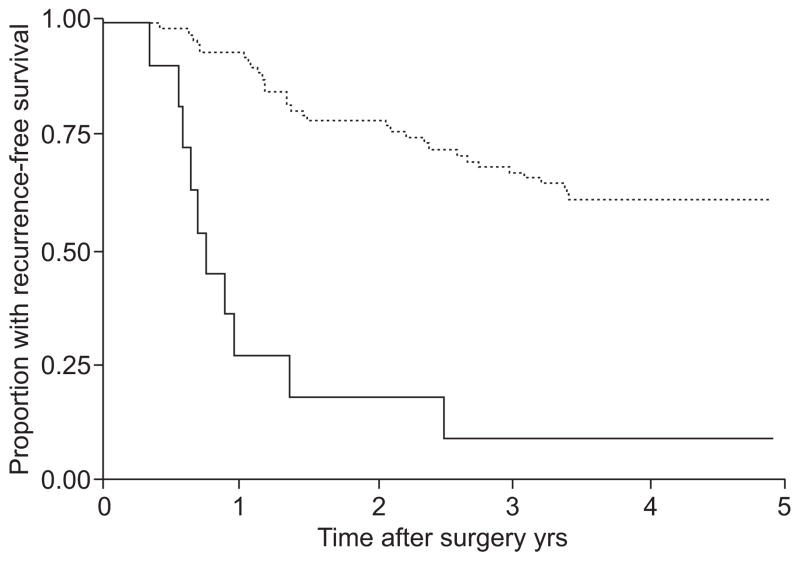
Genomic DNA hypomethylation, leading to genomic instability, as well as aberrant promoter hypermethylation, leading to inactivation of tumour suppressor genes [104], have been shown to be common events in human cancers. In patients with lung cancer, promoter hypermethylation has been detected in blood [105], bronchial lavages [106], induced sputum [107] and pleural fluid [108]. TP16 promoter methylation was found in sputum of smokers up to 3 yrs before their clinical diagnosis of squamous cell carcinoma [109], and has thus been proposed as a biomarker for early detection of lung cancer and monitoring of prevention trials [110, 111]. Furthermore, methylation of the promoter region of four genes (TP16, CDH13, RASSFIA and APC) in patients with stage I NSCLC was associated with early recurrence (fig. 4) [112]. With the development of high-throughput technologies, novel target genes for aberrant methylation have been identified [113, 114]. Protein expression of one of them, OLIG1, correlated significantly with survival in lung cancer patients [115].
FIGURE 4.
Methylation of the promoter region of TP16 and CDH13 in patients with stage I nonsmall cell lung cancer is associated with early recurrence. When both TP16 and CDH13 were methylated in the tumour and the mediastinal nodes, there was a significantly lower rate of recurrence-free survival (9.1% (95% CI 0.5–33.3)) than if TP16 and CDH13 were unmethylated (61.2% (95% CI, 49.7–70.9); p<0.001). ——: methylated (n=11); ······: unmethylated (n=80). Reproduced from [112], with permission from the publisher.
Chromatin is an important player in gene expression regulation, and alterations in its structure have been linked to cancer development, through DNA methylation and also histone hyper- or de-acetylation (depending on the target gene). The acetylated state of histones is associated with transcriptional activity, and it has been shown that active histone acetylation plays a role in re-expression of silenced tumour suppressor genes [116]. Recent studies indicate an antitumour activity of histone deacetylase inhibitors against NSCLC [117–119].
Alterations of miRNA expression may deregulate cancer-related genes
Through gene expression regulation, miRNAs seem to be involved in diverse cellular functions, such as proliferation, differentiation, death and stress resistance [120]. Expression levels of miRNAs have been reported to be dysregulated in human cancer, suggesting a role in oncogenesis.
In a study analysing 104 pairs of NSCLC and corresponding normal lung tissues, an expression profile of 43 miRNAs discriminated lung cancers from noncancerous lung tissues [121]. Six miRNAs (hsa-mir-205, hsa-mir-99b, hsa-mir-203, hsa-mir-202, hsa-mir-102 and hsa-mir-204-prec) were differentially expressed in adenocarcinomas and squamous cell carcinomas, with higher expression levels of hsa-mir-99b and hsa-mir-102 in adenocarcinomas. Furthermore, high hsa-mir-155 and low hsa-let-7a-2 expression correlated with poor survival in lung adenocarcinomas (p=0.033). This observation was confirmed by RT-PCR analysis and cross-validated with an independent set of adenocarcinomas. In another study of 143 surgically resected NSCLC, low let-7 expression was also significantly associated to shorter survival (p=0.0003); and overexpression of let-7 in A549 lung adenocarcinoma cell line inhibited lung cancer cell growth in vitro [122].
Differentially expressed miRNA genes in NSCLC are frequently located in fragile sites and/or chromosomal regions with frequent copy number alterations, suggesting that differences in miRNA expression may be induced by genomic alterations. However, since>50% of miRNAs are located at cancer-related chromosomal regions, miRNAs are also suspected to play a role as oncogenes or tumour suppressor genes. miRNA expression profiles represent potential markers for lung cancer diagnosis, classification and prognosis. However, inconsistencies between studies are observed, probably due to technical and analytical differences, as well as lack of standardisation.
Importance of proteomics to detect post-translational modifications
Post-translational modifications of proteins, such as phosphorylation, glycosylation and proteolytic processing, are common events and have the potential to significantly modify protein functions as well as confer cellular or tissue specificity. Unlike genomic analysis, proteomic analysis has the ability to detect these modifications.
An illustration of the importance of post-translational modifications is provided by tyrosine kinase signalling, which is often deregulated in cancer. In a study using a phosphoproteomic approach based on phosphopeptide immunoprecipitation and analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS), the tyrosine kinase signalling was characterised across 41 NSCLC cell lines and 150 NSCLC tumours [123]. Kinases already known as oncogenes (e.g. EGFR and c-MET), as well as kinases never previously implicated in NSCLC (PDGFRα and DDR1) were identified. In the actual era of tyrosine kinase-based targeted therapies, the insights provided by this study are particularly interesting.
As discussed before, histone modifications represent another example of post-translational modifications contributing to carcinogenesis. Since post-translational modifications are reversible, drugs inhibiting these modifications are developed and hold great promise for lung cancer therapy. Proteomic strategies have an important role by allowing not only the identification of post-translational modifications, but also the quantification and monitoring of the changes induced by their inhibitors.
Involvement of glycans in cancer biology
The technique of glycomics addresses sugars, whether free or present in complex molecules such as proteins or lipids. Sugars drive a series of molecular processes that are related to cancer development. The following few examples illustrate this point.
Mucins are highly O-glycosylated proteins synthesised by epithelial cells, and their glycosylation patterns can be altered during inflammation or neoplastic transformation. Peritumoural epithelium and squamous metaplasia show an abnormal pattern of mucin expression. However, squamous cell carcinomas and adenocarcinomas display a similar pattern of mucin gene expression, supporting the concept of a common cellular origin [124]. Recent results have uncovered a new link between death-receptor O-glycosylation and apoptotic signalling. O-glycosylation of DR4 and DR5 promotes ligand-stimulated clustering of DR4 and DR5, which mediates recruitment and activation of caspase-8 [125].
N-glycans of receptors or adhesion molecules are considered to be involved in cellular functions by altering their functions such as cell signalling and cell adhesion, which are implicated in cancer invasion and metastasis [126]. For example, lack of core fucosylation of the EGFR leads to the suppression of epidermal growth factor (EGF) signalling and cell growth [127]. N-glycosylation profiling of blood proteins by MS has been shown to be valuable in proof of concept studies in, for example, pancreatic cancer [128]. Defining serum glycan profiles for normal, invasive and preinvasive lesions might have important diagnostic, prognostic and therapeutic implications. Lectins represent highly versatile carbohydrate-binding proteins that are employed for the profiling of glycoproteins [129]. This is an emerging field that currently suffers in its applications (for glycoprotein enrichment and glycan profiling) from great variability across platforms and laboratories.
Glycoproteins in human serum play critical roles in many biological processes [126, 130, 131] and also have clinical value as biomarkers for disease progression and treatment [132]. For example, glycoproteins have been isolated from the sera using multilectin affinity chromatography, digested first by peptide-N-glycosidase F and then by trypsin. Peptides have been analysed by nano-LC-MS/MS and the majority of identified proteins were found to contain more than one potential glycosylation site. Comparison of the serum glycoproteome of healthy and adenocarcinoma individuals revealed 38 cancer-selective proteins [133]. Their value as candidate biomarkers needs to be tested in prospective studies. In another dataset from 180 patients with unresectable and metastatic NSCLC enrolled in six docetaxel phase II studies at 100 mg·m−2, alpha1-acid glycoprotein was found to be an independent predictor of response with an odds ratio of 0.44 (p=0.0039) [134]. Sialylated glycoprotein IL-6 has been shown to predict poor prognosis in patients with EGFR-mutated NSCLC treated with gefitinib [135]. Tumours expressing a high level of certain types of tumour-associated carbohydrate antigens (TACA) exhibit greater metastasis and progression than those expressing low level of TACA, as reflected in decreased patient survival rate [136].
Glycomics is a new field of interest in lung cancer research, holding great promise for the development of biomarkers and therapeutic targets. However, this area of research is challenged by an inherent level of complexity and requires improvements in molecular sequencing and bioinformatics.
Lipidomics as a promising area of cancer research
The progress made in genomics and proteomics has been led by novel analytical approaches, which have been slower to appear for the analysis of lipids and their biology. LC and MS are now making this field a promising area of biomedical research [137]. Lipids have a prominent biology in lungs as surfactants [138] and prostanoids [139], and in phospholipid signalling-related cancer biology (e.g. PI3-kinase [140]). Immunohistochemical studies have shown that human bronchial dysplasia and atypical adenomatous hyperplasia express high levels of fatty acid synthase (FAS) when compared with normal lung tissues [141]. This suggests that FAS might be a target for intervention in lung carcinogenesis. Thus, lipidomics represents a promising area of biomedical research, with a variety of applications in drug and biomarker development.
SELECTED APPLICATIONS OF HIGH-THROUGHPUT TECHNOLOGIES TO ADDRESS CLINICAL QUESTIONS
Lung cancer risk assessment
While 80–90% of lung cancers are attributable to cigarette smoking, only a minority of smokers will develop lung cancer [142, 143]. In addition, 10–20% of cases occur among never-smokers and a familial risk has been described [144]. This suggests gene–environment interactions in disease development, as well as different molecular mechanisms in smokers and never-smokers. Selective expression of EGFR mutations in never-smokers with NSCLC supports this hypothesis.
The familial occurrence of lung cancer has been investigated in a genome-wide linkage analysis of 52 extended pedigrees of lung cancer patients with several affected relatives, which localised a lung cancer susceptibility locus at 6q23-q25, containing many genes of interest (SASH1, LATS1, IGF2R, PARK2 and TCF21) [145], some of which seem to be frequently inactivated by methylation [146, 147]; however, no tumour suppressor genes inactivated by mutation have been identified in this locus.
Several case–control studies that focused on genes with susceptibility to modify the risk were conducted to address why only a minority of smokers develop lung cancer, and found polymorphisms in carcinogen-metabolising enzymes [148] and DNA repair enzymes [149, 150], as well as in genes with impact on smoking behaviour [151]. Recently, a genome-wide association study was conducted to identify common low-penetrance alleles influencing NSCLC risk [152]. Investigators analysed 315,450 tagging SNP in 1,154 ever-smoker lung cancer patients and 1,137 matched ever-smoker controls. The 10 SNP most significantly linked with lung cancer were tested in two additional large datasets. They found two SNP significantly associated with lung cancer risk, both of them located in the chromosomal region 15q25.1, and homing three genes: CHRNA3 and CHRNA5 (nicotinic acetylcholine receptor alpha subunits 3 and 5) and PMSA4 (proteasome alpha 4 subunit isoform 1). While there is no current evidence for a role of PMSA4 in lung cancer, the nicotinic acetylcholine receptor pathway has been implicated in lung cancer pathogenesis and progression [153–155]. Two other large genetic epidemiological studies reported very similar results, illustrating the gene–environment interaction in lung cancer and further incriminating this genomic region in the pathogenesis of lung cancer [156, 157]. Taken together, these data suggest nicotinic acetylcholine receptors to be potential chemopreventive targets.
Further studies are needed to elucidate the mechanisms responsible for lung cancer development, and subsequently permit the identification of patients at risk of developing lung cancer. This selected population may benefit from chemoprevention and careful surveillance, which may ultimately improve outcome.
Blood-based early detection and noninvasive diagnosis of lung cancer
The discovery of biomarker signatures or panels for lung cancer diagnosis is crucial, as a single biomarker is unlikely to be specific and sensitive enough on its own to demonstrate clinical utility. These new emerging technologies, combined with the assumption that circulating proteins/peptides are deriving from the perfusion of tumours, have revived a long-term interest in analysing the blood proteome of cancer patients. Since blood access is easy, fast and amenable to repetitive measurements, this biospecimen appears very attractive for the application of biomarkers to early diagnosis of lung cancer, monitoring of disease status, development of targeted therapies, evaluation of response to therapy and survival. It may improve our diagnostic accuracy and decrease the number of thoracotomies currently required for pathological evidence of malignant cells.
Several serum biomarkers have already been investigated in lung cancer but have not been proven useful in clinical practice because of their limited sensitivity and/or specificity, especially in early-stage disease [158–160].
A team of investigators assayed six serum proteins, four discovered by proteomics (2DIGE and MALDI-MS) and two previously known to be cancer-associated, on a training set of sera from 100 patients (50 lung cancers, 50 controls) [161]. They found that four of these proteins (carcinoembryonic antigen, retinol binding protein, α1-antitrypsin and squamous cell carcinoma antigen) were able to distinguish lung cancer cases from controls with 89.3% sensitivity and 84.7% specificity. When applied to an independent validation set (50 lung cancers, 50 controls), the four-protein signature achieved 77.8% sensitivity and 75.4% specificity, while none of the four markers had sufficient diagnostic power when used alone. MALDI-MS was also used to analyse undepleted and unfractionated sera from a total of 288 NSCLC patients and controls divided into training (92 cases, 92 controls) and test (50 cases, 56 controls) sets [162]. A seven-signal proteomic signature was found, distinguishing lung cancer cases and controls with 78% accuracy, 67.4% sensitivity and 88.9% specificity in the training set; and 72.6% accuracy, 58% sensitivity and 85.7% specificity in the test set. Moreover, the serum signature was associated with lung cancer diagnosis independently of the smoking history and levels of C-reactive protein, a marker of inflammation. Although MS-based proteomic analysis of lung cancer allowed the discovery of novel diagnostic biomarkers, their application is still limited to laboratory use. Therefore, population studies are required for their translation into the clinic.
Lung tumour class discovery and class prediction
Microarray analysis of NSCLC samples combined with a class discovery approach showed that gene expression profiles were able to accurately classify tumours into their classical histological groups, and also to identify histological subgroups [163–166]. This information is important as therapeutic options can be different among the histological groups and even subgroups. Using Affymetrix U133A gene chips (Affymetrix, Santa Clara, CA, USA) to analyse 129 squamous cell carcinomas, a study defined two squamous cell carcinoma subgroups, with different overall survival [167]. Three other studies found adenocarcinoma subgroups [76, 164, 168]. Each of these three studies defined a different number of adenocarcinoma subgroups, questioning the reproducibility and consistency of tumour classification by gene expression microarrays. Sample heterogeneity, sample size differences and analytical platform differences can explain some of these results [169].
Proteomics tumour profiling also allowed tumour class prediction and discovery. When profiling 79 NSCLC and 14 normal frozen lung tissue sections by use of MALDI-TOF MS, differentially expressed MS signals were selected and a class prediction model using established methods was defined [170]. Investigators found a 75% accurate signature allowing lung tumours classification by histology [171]. They extended this approach to the analysis of preinvasive lesions to distinguish low-grade from high-grade preinvasive lesions [172]. These efforts have not yet lead to applications in clinical practice. Validation of these biomarker candidates will require prospective validation in larger studies, across institutions and laboratories, and will have to prove clinical utility in the current management of lung cancer.
Prediction of prognosis
Prediction of prognosis may improve the management of patients with lung cancer by identifying those who are more likely to benefit from treatment.
Gene expression profiles have shown the potential to predict lung cancer patient survival. Indeed, in a study analysing 86 adenocarcinomas, 50 genes were differentially expressed between low- and high-risk patient groups [75]. The results were validated in an independent dataset made of 62 adenocarcinomas. Several stage I lung cancer patients were clustered with stage III patients into the poor prognosis group, showing that the gene expression profile was independent of the pathological stage at time of diagnosis, therefore providing an added prognostic value. A 37-gene signature predicting prognosis was also identified in a cohort of 86 adenocarcinomas [173]. When applied to an independent cohort of 84 adenocarcinomas, the signature separated patients into three prognostic groups (good, moderate and poor) with 96% accuracy. In another report using oligonucleotide microarrays to define ratios of gene expression to evaluate risk of recurrence in resected stage I adenocarcinomas (36 in the test set and 60 in the validation test), a three-ratio test predicting recurrence with >90% accuracy was found [174]. Moreover, gene expression microarrays performed on 89 early-stage NSCLC identified profiles predicting recurrence after surgical resection better than the clinical predictor factors [80]. When applied to two independent cohorts of 25 and 84 early-stage NSCLC, the prediction accuracy was 72% and 79%, respectively. A subgroup of stage IA patients at high risk for recurrence was also identified, which is useful as these patients are probably those who might best benefit from adjuvant chemotherapy. Meta-analyses also identified predictive markers [169, 175]. In a cross-study comparison, gene expression profiles correlated to prognosis were poorly reproducible, altough 14 genes accurately predicted survival in the three studies being compared [169].
Recently, a pooled analysis of 442 lung adenocarcinomas from multiple institutions established the performance of gene expression signatures across different patient populations and different laboratories, which is necessary before considering any clinical application [176]. Investigators analysed whether gene expression data either alone or combined with clinical data could be used to predict overall survival in patients with lung cancer; several models were tested. When using gene expression data alone, only two models (out of eight) performed with consistent statistical significance and gave hazard ratios (HR) >1 for all pathological stages in the two validation datasets. When combining clinical and gene expression data the outcome prediction was improved, with HR >2 in two models. This study identified predictors of survival based on clinical and gene expression microarray data, with a better accuracy to predict survival when combining clinical and molecular data.
Proteomic studies have also been used to identify prognostic markers [171, 177]. In the most recent study [177], MALDI-MS was used to analyse protein profiles of surgically resected NSCLC. In the training set (116 NSCLC and 20 controls), a 25-signal signature differentially expressed between patients with high and low risk of recurrence was found, associated with both relapse-free and overall survival. In the independent validation set (58 NSCLC and seven controls), the signature was also significantly associated with overall survival and, among patients with stage I disease only, to relapse-free survival. By predicting more accurately than the prognostic factors currently used (e.g. histology, and tumour, node, metatasis (TNM) classification), this signature indicates which patients are likely to relapse after surgery, and may help to decide when systemic adjuvant therapy will be beneficial. Recently, protein members of a candidate signature of prognosis previously identified by MALDI-MS were validated by immunohistochemistry (IHC) on lung cancer TMAs [178]. Combined IHC scores of calmodulin, thymosin β4 and thymosin β10 were correlated to survival. This combined strategy consisting of identification by MALDI-MS followed by validation by IHC represents key steps prior to bringing such candidates to the test in the clinical setting.
Prediction of response to therapy
The overall response to chemotherapy in NSCLC is <30%. Therefore, tremendous efforts are made to find biological markers that could identify patients who will actually benefit from a specific treatment and not suffer from its side-effects. Such markers could help to identify the best therapeutic strategies.
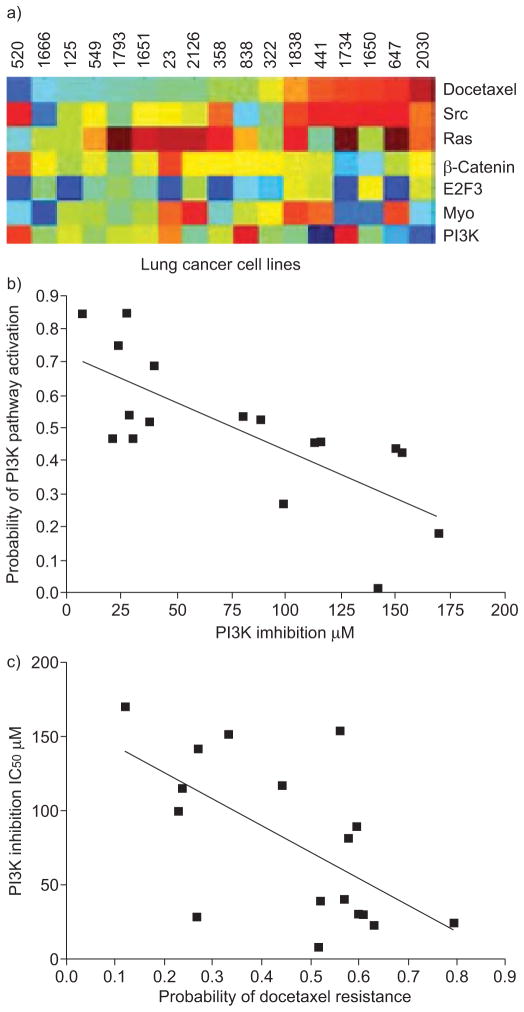
In a study using oligonucleotide microarrays to analyse 16 NSCLC specimens (eight in a training set and eight in a validation set), lysosomal protease inhibitors Serpin B3 and Cystatin C predicted clinical response to platinum-based chemotherapy with 72% accuracy [179]. Another group compared microarray data to in vitro drug sensitivity data of various cancer cell lines and found gene expression profiles predicting sensitivity to individual chemotherapeutic drugs (topotecan, adriamycin, etoposide, 5-fluorouracil, paclitaxel, cyclophosphamide), validated in an independent set with >80% accuracy [180]. Using published datasets, investigators showed that the in vitro-generated profiles were able to predict clinical response to individual drugs with >81% accuracy, and also to multidrug regimens. In lung cancer cases, docetaxel-sensitive individuals were likely to be resistant to etoposide. Investigators also linked gene expression signatures of chemotherapy sensitivity to those of known oncogenic pathways and found a significant association between PI3-kinase pathway deregulation and docetaxel resistance, suggesting a potential benefit to use PI3-kinase inhibitors in this subgroup (fig. 5).
FIGURE 5.
Relationship between predicted chemotherapeutic sensitivity and oncogenic pathway deregulation. a) Probability of oncogenic pathway deregulation as a function of predicted docetaxel sensitivity in a series of lung cancer cell lines (red: sensitive; blue: resistant). b) Lung cancer cell lines showing an increased probability of phosphoinositide 3-kinase (PI3K) activation were more likely to respond to a PI3K inhibitor (p=0.001, log-rank test), as measured by sensitivity to the drug in cell proliferation assays. c) Furthermore, cell lines predicted to be resistant to docetaxel were more likely to be sensitive to PI3K inhibition (p<0.001, log-rank test). Src: avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homologue; Ras: rat sarcoma viral oncogene homologue; E2F3: E2F transcription factor 3; Myc: myelocytomatosis viral oncogene homologue; IC50: half maximal inhibitory concentration. Reproduced from [180], with permission from the publisher.
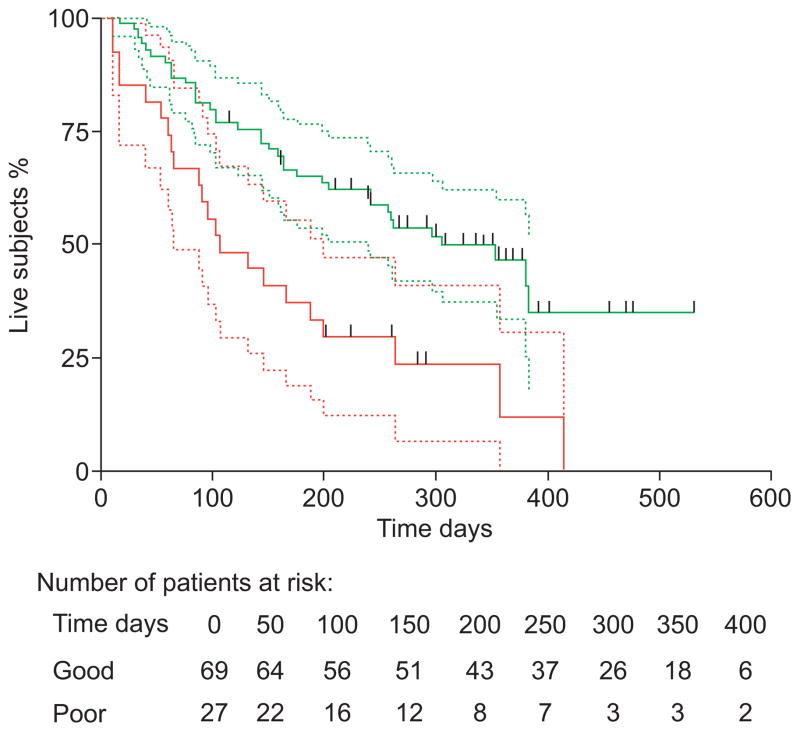
To identify NSCLC patients likely to benefit from EGFR tyrosine kinase inhibitors (TKI) treatment, MALDI-MS was performed on pre-treatment sera of 302 patients treated with gefitinib or erlotinib (139 patients from three cohorts assigned into a training and 163 from two cohorts assigned into a validation set), as well as sera from 158 patients not treated with EGFR TKI [181]. An algorithm based on eight MS signals successfully identified patients with improved survival and time to progression after EGFR TKI treatment, independently of clinical factors associated with sensitivity to EGFR TKI (fig. 6). The algorithm did not accurately classify outcome of patients not treated with EGFR TKI.
FIGURE 6.
Kaplan–Meier analysis of overall survival in the Eastern Cooperative Oncology Group validation cohort (n=96). These patients had advanced nonsmall cell lung cancer and had been treated first line with erlotinib alone. red; poor event-free fraction; green: good event-free fraction; ------: 95% confidence intervals; |: censored patients. Reproduced from [181], with permission from the publisher.
Several studies found markers predicting response to therapy. However, for all of them, prospective validation studies are required to confirm their utility in the clinical setting.
CONCLUSIONS AND FUTURE PERSPECTIVES
This review highlights the impact of new global molecular approaches on our understanding of lung cancer biology and on the personalised management of patients with lung cancer. The studies presented here illustrate how the “-omics” era has revolutionised our approach to cancer biology. Genomics has been pioneering and has provided a solid base to our knowledge. Most importantly, genomics has led to discoveries with clinical implications. Even though genomics is still leading the field, there are growing interests in epigenetics and proteomics. Recently, the dynamics of proteins in response to anti-cancer drugs have been shown to differ between cancer cells according to their outcome (either cell death or survival), highlighting the importance of proteomics to help us understand the individual molecular responses to drugs [182]. Early fields of glycomics and lipidomics also hold promise to improve our understanding of lung cancer biology.
However, multiple challenges lie ahead. While these high-throughput technologies hold the promise of bringing personalised lung cancer care to the clinic, the field is in need of careful validation of candidate biomarkers emerging from large discovery strategies. The integration of these highly dimensional and complex data will require major efforts in bioinformatics and biostatistics. It will take time before models showing how complementary these techniques are, are put to the test. Another major challenge is to study disease process rather than disease state and to do so in a high-throughput manner. We need to refine our methods of approaching the dynamics of the carcinogenic process. From the biomarker point of view, repetitive measurements of biomarkers in cohorts at risk will be critical in the future. For example, the identification of plasma DNA [183], circulating tumour antigens or their related auto-antibodies [184, 185] provides attractive means for early cancer diagnosis as well as a lead for therapy. A test based on the demonstration of auto-antibodies to tumour antigens in the sera of patients could be of great importance for early detection of cancer, since antibodies against a carcinogen stimulus could be detected well before the tumour phenotype arises. The future of a new area of investigation such as metabolomics rests with its ability to monitor subtle changes in the metabolome that occur prior to the detection of a gross phenotypic change reflecting disease. The integrated analysis of the “-omics” may provide more sensitive ways to detect changes related to the disease process and discover useful novel biomarkers.
Acknowledgments
SUPPORT STATEMENT
This work was supported by National Institutes of Health grant CA102353, Lung SPORE CA90949, the Damon Runyon Cancer Research Foundation (New York, NY, USA; Ci no. 19-03), and a Merit Review grant from the US Department of Veterans Affairs to P.P. Massion. S. Ocak was supported by a grant from the Université Catholique de Louvain (Belgium) and an IASLC Lung Cancer Fellowship Award.
Footnotes
STATEMENT OF INTEREST
A statement of interest for R.K. Thomas can be found at www.erj.ersjournals.com/misc/statements.dtl
References
- 1.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Giaccone G. Clinical impact of novel treatment strategies. Oncogene. 2002;21:6970–6981. doi: 10.1038/sj.onc.1205565. [DOI] [PubMed] [Google Scholar]
- 3.Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 4.Pisters KM, Le Chevalier T. Adjuvant chemotherapy in completely resected non-small-cell lung cancer. J Clin Oncol. 2005;23:3270–3278. doi: 10.1200/JCO.2005.11.478. [DOI] [PubMed] [Google Scholar]
- 5.Chanin TD, Merrick DT, Franklin WA, et al. Recent developments in biomarkers for the early detection of lung cancer: perspectives based on publications 2003 to present. Curr Opin Pulm Med. 2004;10:242–247. doi: 10.1097/01.mcp.0000130321.11513.13. [DOI] [PubMed] [Google Scholar]
- 6.Chen G, Gharib TG, Huang CC, et al. Proteomic analysis of lung adenocarcinoma: identification of a highly expressed set of proteins in tumors. Clin Cancer Res. 2002;8:2298–2305. [PubMed] [Google Scholar]
- 7.Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Wang W, Li R, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy S, Sutton G, Ng PC, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 11.Pollack JR, Perou CM, Alizadeh AA, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 12.Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 13.Janne PA, Li C, Zhao X, et al. High-resolution single-nucleotide polymorphism array and clustering analysis of loss of heterozygosity in human lung cancer cell lines. Oncogene. 2004;23:2716–2726. doi: 10.1038/sj.onc.1207329. [DOI] [PubMed] [Google Scholar]
- 14.Lindblad-Toh K, Tanenbaum DM, Daly MJ, et al. Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat Biotechnol. 2000;18:1001–1005. doi: 10.1038/79269. [DOI] [PubMed] [Google Scholar]
- 15.Adorjan P, Distler J, Lipscher E, et al. Tumour class prediction and discovery by microarray-based DNA methylation analysis. Nucleic Acids Res. 2002;30:e21. doi: 10.1093/nar/30.5.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan PS, Wei SH, Huang TH. Methylation-specific oligonucleotide microarray. Methods Mol Biol. 2004;287:251–260. doi: 10.1385/1-59259-828-5:251. [DOI] [PubMed] [Google Scholar]
- 17.Ren B, Robert F, Wyrick JJ, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 18.Schena M, Shalon D, Davis RW, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 19.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 20.Liu CG, Calin GA, Meloon B, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergman AC, Benjamin T, Alaiya A, et al. Identification of gel-separated tumor marker proteins by mass spectrometry. Electrophoresis. 2000;21:679–686. doi: 10.1002/(SICI)1522-2683(20000201)21:3<679::AID-ELPS679>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 22.Patton WF. Detection technologies in proteome analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;771:3–31. doi: 10.1016/s1570-0232(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 23.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 24.Farmer TB, Caprioli RM. Assessing the multimeric states of proteins: studies using laser desorption mass spectrometry. Biol Mass Spectrom. 1991;20:796–800. doi: 10.1002/bms.1200201209. [DOI] [PubMed] [Google Scholar]
- 25.McCormack AL, Schieltz DM, Goode B, et al. Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal Chem. 1997;69:767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- 26.Link AJ, Eng J, Schieltz DM, et al. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 27.Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 28.Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 29.Cargile BJ, Bundy JL, Stephenson JL., Jr Potential for false positive identifications from large databases through tandem mass spectrometry. J Proteome Res. 2004;3:1082–1085. doi: 10.1021/pr049946o. [DOI] [PubMed] [Google Scholar]
- 30.Essader AS, Cargile BJ, Bundy JL, et al. A comparison of immobilized pH gradient isoelectric focusing and strong-cation-exchange chromatography as a first dimension in shotgun proteomics. Proteomics. 2005;5:24–34. doi: 10.1002/pmic.200400888. [DOI] [PubMed] [Google Scholar]
- 31.Liebler DC. Shotgun mass spec goes independent. Nat Methods. 2004;1:16–17. doi: 10.1038/nmeth1004-16. [DOI] [PubMed] [Google Scholar]
- 32.Liotta LA, Espina V, Mehta AI, et al. Protein microarrays: meeting analytical challenges for clinical applications. Cancer Cell. 2003;3:317–325. doi: 10.1016/s1535-6108(03)00086-2. [DOI] [PubMed] [Google Scholar]
- 33.Wulfkuhle JD, Aquino JA, Calvert VS, et al. Signal pathway profiling of ovarian cancer from human tissue specimens using reverse-phase protein microarrays. Proteomics. 2003;3:2085–2090. doi: 10.1002/pmic.200300591. [DOI] [PubMed] [Google Scholar]
- 34.Huang J, Zhu H, Haggarty SJ, et al. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc Natl Acad Sci USA. 2004;101:16594–16599. doi: 10.1073/pnas.0407117101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 36.Shen J, Behrens C, Wistuba II, et al. Identification and validation of differences in protein levels in normal, premalignant, and malignant lung cells and tissues using high-throughput Western Array and immunohistochemistry. Cancer Res. 2006;66:11194–11206. doi: 10.1158/0008-5472.CAN-04-1444. [DOI] [PubMed] [Google Scholar]
- 37.Morente MM, Mager R, Alonso S, et al. TuBaFrost 2: standardising tissue collection and quality control procedures for a European virtual frozen tissue bank network. Eur J Cancer. 2006;42:2684–2691. doi: 10.1016/j.ejca.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 38.Orr S, Alexandre E, Clark B, et al. The establishment of a network of European human research tissue banks. Cell Tissue Bank. 2002;3:133–137. doi: 10.1023/A:1022811232250. [DOI] [PubMed] [Google Scholar]
- 39.Blow N. Tissue preparation: tissue issues. Nature. 2007;448:959–963. doi: 10.1038/448959a. [DOI] [PubMed] [Google Scholar]
- 40.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 41.Naoki K, Chen TH, Richards WG, et al. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res. 2002;62:7001–7003. [PubMed] [Google Scholar]
- 42.Adjei AA, Cohen RB, Franklin W, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 44.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serra V, Markman B, Scaltriti M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 46.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 47.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 48.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 50.Carter TA, Wodicka LM, Shah NP, et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad Sci USA. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 52.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 55.Cappuzzo F, Ligorio C, Janne PA, et al. Prospective study of gefitinib in epidermal growth factor receptor fluorescence in situ hybridization-positive/phospho-Akt-positive or never smoker patients with advanced non-small-cell lung cancer: the ONCOBELL trial. J Clin Oncol. 2007;25:2248–2255. doi: 10.1200/JCO.2006.09.4300. [DOI] [PubMed] [Google Scholar]
- 56.Paz-Ares LJMS, Garcia-Velasco A, Massuti B, et al. A prospective phase II trial of erlotinib in advanced non-small cell lung cancer (NSCLC) patients (p) with mutations in the tyrosine kinase (TK) domain of the epidermal growth factor receptor (EGFR) J Clin Oncol 2006 ASCO Annual Meeting Proceedings Part I. 2006;24:18S. [Google Scholar]
- 57.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 58.Sugio UH, Oyama T, Onizuka T, et al. A prospective phase II study of gefitinib in non-small cell lung cancer patients with epidermal growth factor receptor gene (EGFR) mutations. J Clin Oncol 2006 ASCO Annual Meeting Proceedings Part I. 2007;25:18S. [Google Scholar]
- 59.Yamamoto H, Toyooka S, Mitsudomi T. Impact of EGFR mutation analysis in non-small cell lung cancer. Lung Cancer. 2009;63:315–321. doi: 10.1016/j.lungcan.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 60.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 61.Douillard HV, Mok TS, Socinski A, et al. Molecular and clinical subgroup analyses from a phase III trial comparing gefitinib with docetaxel in previously treated non-small cell lung cancer (INTEREST). J Clin Oncol; 2008 ASCO Annual Meeting Proceedings (post-meeting edition); 2008. p. abstract 8001. [Google Scholar]
- 62.Mok T, Wu Y-L, Thongprasert S, et al. Phase III, randomized, open-label, first line study of gefitinib vs carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer (NSCLC) (IPASS) [abstract LBA2] Ann Oncol. 2008;19(Suppl 8):VIII1. [Google Scholar]
- 63.Eberhard DA, Giaccone G, Johnson BE. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: standardization for use in the clinical trial setting. J Clin Oncol. 2008;26:983–994. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- 64.Hirsch FR, Varella-Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–6845. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 65.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer: molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 66.Yatabe Y, Takahashi T, Mitsudomi T. Epidermal growth factor receptor gene amplification is acquired in association with tumor progression of EGFR-mutated lung cancer. Cancer Res. 2008;68:2106–2111. doi: 10.1158/0008-5472.CAN-07-5211. [DOI] [PubMed] [Google Scholar]
- 67.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas RK, Nickerson E, Simons JF, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 69.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 70.Campbell PJ, Stephens PJ, Pleasance ED, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 75.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 76.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hayes DN, Monti S, Parmigiani G, et al. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol. 2006;24:5079–5090. doi: 10.1200/JCO.2005.05.1748. [DOI] [PubMed] [Google Scholar]
- 78.Lu Y, Lemon W, Liu PY, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xi L, Lyons-Weiler J, Coello MC, et al. Prediction of lymph node metastasis by analysis of gene expression profiles in primary lung adenocarcinomas. Clin Cancer Res. 2005;11:4128–4135. doi: 10.1158/1078-0432.CCR-04-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Potti A, Mukherjee S, Petersen R, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 81.Gray JW, Collins C. Genome changes and gene expression in human solid tumors. Carcinogenesis. 2000;21:443–452. doi: 10.1093/carcin/21.3.443. [DOI] [PubMed] [Google Scholar]
- 82.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garnis C, Lockwood WW, Vucic E, et al. High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer. 2006;118:1556–1564. doi: 10.1002/ijc.21491. [DOI] [PubMed] [Google Scholar]
- 84.Massion PP, Kuo WL, Stokoe D, et al. Genomic copy number analysis of non-small cell lung cancer using array comparative genomic hybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res. 2002;62:3636–3640. [PubMed] [Google Scholar]
- 85.Tonon G, Wong KK, Maulik G, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dehan E, Ben-Dor A, Liao W, et al. Chromosomal aberrations and gene expression profiles in non-small cell lung cancer. Lung Cancer. 2007;56:175–184. doi: 10.1016/j.lungcan.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 87.Zhao X, Weir BA, LaFramboise T, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 88.Imoto I, Yuki Y, Sonoda I, et al. Identification of ZASC1 encoding a Kruppel-like zinc finger protein as a novel target for 3q26 amplification in esophageal squamous cell carcinomas. Cancer Res. 2003;63:5691–5696. [PubMed] [Google Scholar]
- 89.Singh B, Gogineni SK, Sacks PG, et al. Molecular cytogenetic characterization of head and neck squamous cell carcinoma and refinement of 3q amplification. Cancer Res. 2001;61:4506–4513. [PubMed] [Google Scholar]
- 90.Qian J, Massion PP. Role of chromosome 3q amplification in lung cancer. J Thorac Oncol. 2008;3:212–215. doi: 10.1097/JTO.0b013e3181663544. [DOI] [PubMed] [Google Scholar]
- 91.Levin NA, Brzoska PM, Warnock ML, et al. Identification of novel regions of altered DNA copy number in small cell lung tumors. Genes Chromosomes Cancer. 1995;13:175–185. doi: 10.1002/gcc.2870130307. [DOI] [PubMed] [Google Scholar]
- 92.Harvey BG, Heguy A, Leopold PL, et al. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med. 2007;85:39–53. doi: 10.1007/s00109-006-0103-z. [DOI] [PubMed] [Google Scholar]
- 93.Ammous Z, Hackett NR, Butler MW, et al. Variability in small airway epithelial gene expression among normal smokers. Chest. 2008;133:1344–1353. doi: 10.1378/chest.07-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hackett NR, Heguy A, Harvey BG, et al. Variability of antioxidant-related gene expression in the airway epithelium of cigarette smokers. Am J Respir Cell Mol Biol. 2003;29:331–343. doi: 10.1165/rcmb.2002-0321OC. [DOI] [PubMed] [Google Scholar]
- 95.Spira A, Beane J, Shah V, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA. 2004;101:10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Spira A, Beane JE, Shah V, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 97.Beane J, Sebastiani P, Whitfield TH, et al. A prediction model for lung cancer diagnosis that integrates genomic and clinical features. Cancer Prev Res. 2008;1:56–64. doi: 10.1158/1940-6207.CAPR-08-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang E, Ishida S, Pittman J, et al. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat Genet. 2003;34:226–230. doi: 10.1038/ng1167. [DOI] [PubMed] [Google Scholar]
- 99.Black EP, Huang E, Dressman H, et al. Distinct gene expression phenotypes of cells lacking Rb and Rb family members. Cancer Res. 2003;63:3716–3723. [PubMed] [Google Scholar]
- 100.Segal E, Friedman N, Koller D, et al. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 101.Rhodes DR, Yu J, Shanker K, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ramaswamy S, Ross KN, Lander ES, et al. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 103.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 104.Esteller M, Fraga MF, Paz MF, et al. Cancer epigenetics and methylation. Science. 2002;297:1807–1808. doi: 10.1126/science.297.5588.1807d. [DOI] [PubMed] [Google Scholar]
- 105.Esteller M, Sanchez-Cespedes M, Rosell R, et al. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59:67–70. [PubMed] [Google Scholar]
- 106.Ahrendt SA, Chow JT, Xu LH, et al. Molecular detection of tumor cells in bronchoalveolar lavage fluid from patients with early stage lung cancer. J Natl Cancer Inst. 1999;91:332–339. doi: 10.1093/jnci/91.4.332. [DOI] [PubMed] [Google Scholar]
- 107.Belinsky SA, Palmisano WA, Gilliland FD, et al. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62:2370–2377. [PubMed] [Google Scholar]
- 108.Ng CS, Zhang J, Wan S, et al. Tumor p16M is a possible marker of advanced stage in non-small cell lung cancer. J Surg Oncol. 2002;79:101–106. doi: 10.1002/jso.10046. [DOI] [PubMed] [Google Scholar]
- 109.Palmisano WA, Divine KK, Saccomanno G, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 110.Belinsky SA, Nikula KJ, Palmisano WA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nuovo GJ, Plaia TW, Belinsky SA, et al. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc Natl Acad Sci USA. 1999;96:12754–12759. doi: 10.1073/pnas.96.22.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 113.Zhong S, Fields CR, Su N, et al. Pharmacologic inhibition of epigenetic modifications, coupled with gene expression profiling, reveals novel targets of aberrant DNA methylation and histone deacetylation in lung cancer. Oncogene. 2007;26:2621–2634. doi: 10.1038/sj.onc.1210041. [DOI] [PubMed] [Google Scholar]
- 114.Rauch TA, Zhong X, Wu X, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci USA. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brena RM, Morrison C, Liyanarachchi S, et al. Aberrant DNA methylation of OLIG1, a novel prognostic factor in non-small cell lung cancer. PLoS Med. 2007;4:e108. doi: 10.1371/journal.pmed.0040108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brehm A, Miska EA, McCance DJ, et al. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 117.Mukhopadhyay NK, Weisberg E, Gilchrist D, et al. Effectiveness of trichostatin A as a potential candidate for anticancer therapy in non-small-cell lung cancer. Ann Thorac Surg. 2006;81:1034–1042. doi: 10.1016/j.athoracsur.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 118.Kim SM, Kee HJ, Eom GH, et al. Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem Biophys Res Commun. 2006;345:318–323. doi: 10.1016/j.bbrc.2006.04.095. [DOI] [PubMed] [Google Scholar]
- 119.Loprevite M, Tiseo M, Grossi F, et al. In vitro study of CI-994, a histone deacetylase inhibitor, in non-small cell lung cancer cell lines. Oncol Res. 2005;15:39–48. doi: 10.3727/096504005775082066. [DOI] [PubMed] [Google Scholar]
- 120.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 121.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 122.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 123.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 124.Lopez-Ferrer A, Curull V, Barranco C, et al. Mucins as differentiation markers in bronchial epithelium. Squamous cell carcinoma and adenocarcinoma display similar expression patterns. Am J Respir Cell Mol Biol. 2001;24:22–29. doi: 10.1165/ajrcmb.24.1.4294. [DOI] [PubMed] [Google Scholar]
- 125.Wagner KW, Punnoose EA, Januario T, et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. 2007;13:1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- 126.Zhao YY, Takahashi M, Gu JG, et al. Functional roles of N-glycans in cell signaling and cell adhesion in cancer. Cancer Sci. 2008;99:1304–1310. doi: 10.1111/j.1349-7006.2008.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang X, Gu J, Ihara H, et al. Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J Biol Chem. 2006;281:2572–2577. doi: 10.1074/jbc.M510893200. [DOI] [PubMed] [Google Scholar]
- 128.Zhao J, Qiu W, Simeone DM, et al. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J Proteome Res. 2007;6:1126–1138. doi: 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- 129.Hirabayashi J. Concept, strategy and realization of lectin-based glycan profiling. J Biochem. 2008;144:139–147. doi: 10.1093/jb/mvn043. [DOI] [PubMed] [Google Scholar]
- 130.Allahverdian S, Patchell BJ, Dorscheid DR. Carbohydrates and epithelial repair–more than just post-translational modification. Curr Drug Targets. 2006;7:597–606. doi: 10.2174/138945006776818719. [DOI] [PubMed] [Google Scholar]
- 131.Handa K, Kojima N, Hakomori S. Analysis of glycolipid-dependent cell adhesion based on carbohydrate-carbohydrate interaction. Methods Enzymol. 2000;312:447–458. doi: 10.1016/s0076-6879(00)12930-1. [DOI] [PubMed] [Google Scholar]
- 132.Pettijohn DE, Pfenninger O, Brown J, et al. Tumorigenic human squamous lung cancer cells have defined cell surface carbohydrates that are absent from nontumorigenic cells. Proc Natl Acad Sci USA. 1988;85:802–806. doi: 10.1073/pnas.85.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Heo SH, Lee SJ, Ryoo HM, et al. Identification of putative serum glycoprotein biomarkers for human lung adenocarcinoma by multilectin affinity chromatography and LC-MS/MS. Proteomics. 2007;7:4292–4302. doi: 10.1002/pmic.200700433. [DOI] [PubMed] [Google Scholar]
- 134.Bruno R, Olivares R, Berille J, et al. Alpha-1-acid glycoprotein as an independent predictor for treatment effects and a prognostic factor of survival in patients with non-small cell lung cancer treated with docetaxel. Clin Cancer Res. 2003;9:1077–1082. [PubMed] [Google Scholar]
- 135.Fujiwara Y, Kiura K, Toyooka S, et al. Elevated serum level of sialylated glycoprotein KL-6 predicts a poor prognosis in patients with non-small cell lung cancer treated with gefitinib. Lung Cancer. 2008;59:81–87. doi: 10.1016/j.lungcan.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 136.Hakomori S. Tumor-associated carbohydrate antigens defining tumor malignancy: basis for development of anti-cancer vaccines. Adv Exp Med Biol. 2001;491:369–402. doi: 10.1007/978-1-4615-1267-7_24. [DOI] [PubMed] [Google Scholar]
- 137.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 138.Serrano AG, Perez-Gil J. Protein-lipid interactions and surface activity in the pulmonary surfactant system. Chem Phys Lipids. 2006;141:105–118. doi: 10.1016/j.chemphyslip.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 139.Keith RL, Geraci MW. Prostacyclin in lung cancer. J Thorac Oncol. 2006;1:503–505. [PubMed] [Google Scholar]
- 140.Kong D, Yamori T. Phosphatidylinositol 3-kinase inhibitors: promising drug candidates for cancer therapy. Cancer Sci. 2008;99:1734–1740. doi: 10.1111/j.1349-7006.2008.00891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Orita H, Coulter J, Lemmon C, et al. Selective inhibition of fatty acid synthase for lung cancer treatment. Clin Cancer Res. 2007;13:7139–7145. doi: 10.1158/1078-0432.CCR-07-1186. [DOI] [PubMed] [Google Scholar]
- 142.Peto R, Darby S, Deo H, et al. Smoking, smoking cessation, and lung cancer in the UK since 1950: combination of national statistics with two case-control studies. BMJ. 2000;321:323–329. doi: 10.1136/bmj.321.7257.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mattson ME, Pollack ES, Cullen JW. What are the odds that smoking will kill you? Am J Public Health. 1987;77:425–431. doi: 10.2105/ajph.77.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tokuhata GK, Lilienfeld AM. Familial aggregation of lung cancer in humans. J Natl Cancer Inst. 1963;30:289–312. [PubMed] [Google Scholar]
- 145.Bailey-Wilson JE, Amos CI, Pinney SM, et al. A major lung cancer susceptibility locus maps to chromosome 6q23-25. Am J Hum Genet. 2004;75:460–474. doi: 10.1086/423857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Smith LT, Lin M, Brena RM, et al. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proc Natl Acad Sci USA. 2006;103:982–987. doi: 10.1073/pnas.0510171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tessema M, Willink R, Do K, et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res. 2008;68:1707–1714. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- 148.Ye Z, Song H, Higgins JP, et al. Five glutathione s-transferase gene variants in 23,452 cases of lung cancer and 30,397 controls: meta-analysis of 130 studies. PLoS Med. 2006;3:e91. doi: 10.1371/journal.pmed.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Manuguerra M, Saletta F, Karagas MR, et al. XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the risk of cancer: a HuGE review. Am J Epidemiol. 2006;164:297–302. doi: 10.1093/aje/kwj189. [DOI] [PubMed] [Google Scholar]
- 150.Hung RJ, Hall J, Brennan P, et al. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162:925–942. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 151.Campa D, Zienolddiny S, Lind H, et al. Polymorphisms of dopamine receptor/transporter genes and risk of non-small cell lung cancer. Lung Cancer. 2007;56:17–23. doi: 10.1016/j.lungcan.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 152.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25. 1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Minna JD. Nicotine exposure and bronchial epithelial cell nicotinic acetylcholine receptor expression in the pathogenesis of lung cancer. J Clin Invest. 2003;111:31–33. doi: 10.1172/JCI17492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lam DC, Girard L, Ramirez R, et al. Expression of nicotinic acetylcholine receptor subunit genes in non-small-cell lung cancer reveals differences between smokers and nonsmokers. Cancer Res. 2007;67:4638–4647. doi: 10.1158/0008-5472.CAN-06-4628. [DOI] [PubMed] [Google Scholar]
- 155.Zhang Q, Tang X, Zhang ZF, et al. Nicotine induces hypoxia-inducible factor-1alpha expression in human lung cancer cells via nicotinic acetylcholine receptor-mediated signaling pathways. Clin Cancer Res. 2007;13:4686–4694. doi: 10.1158/1078-0432.CCR-06-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 157.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Seemann MD, Beinert T, Furst H, et al. An evaluation of the tumour markers, carcinoembryonic antigen (CEA), cytokeratin marker (CYFRA 21-1) and neuron-specific enolase (NSE) in the differentiation of malignant from benign solitary pulmonary lesions. Lung Cancer. 1999;26:149–155. doi: 10.1016/s0169-5002(99)00084-7. [DOI] [PubMed] [Google Scholar]
- 159.Pastor A, Menendez R, Cremades MJ, et al. Diagnostic value of SCC, CEA and CYFRA 21. 1 in lung cancer: a Bayesian analysis. Eur Respir J. 1997;10:603–609. [PubMed] [Google Scholar]
- 160.Kulpa J, Wojcik E, Reinfuss M, et al. Carcinoembryonic antigen, squamous cell carcinoma antigen, CYFRA 21-1, and neuron-specific enolase in squamous cell lung cancer patients. Clin Chem. 2002;48:1931–1937. [PubMed] [Google Scholar]
- 161.Patz EF, Jr, Campa MJ, Gottlin EB, et al. Panel of serum biomarkers for the diagnosis of lung cancer. J Clin Oncol. 2007;25:5578–5583. doi: 10.1200/JCO.2007.13.5392. [DOI] [PubMed] [Google Scholar]
- 162.Yildiz PB, Shyr Y, Rahman JS, et al. Diagnostic accuracy of MALDI mass spectrometric analysis of unfractionated serum in lung cancer. J Thorac Oncol. 2007;2:893–901. doi: 10.1097/JTO.0b013e31814b8be7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fujii T, Dracheva T, Player A, et al. A preliminary transcriptome map of non-small cell lung cancer. Cancer Res. 2002;62:3340–3346. [PubMed] [Google Scholar]
- 164.Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nacht M, Dracheva T, Gao Y, et al. Molecular characteristics of non-small cell lung cancer. Proc Natl Acad Sci USA. 2001;98:15203–15208. doi: 10.1073/pnas.261414598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sugita M, Geraci M, Gao B, et al. Combined use of oligonucleotide and tissue microarrays identifies cancer/testis antigens as biomarkers in lung carcinoma. Cancer Res. 2002;62:3971–3979. [PubMed] [Google Scholar]
- 167.Raponi M, Zhang Y, Yu J, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 168.Borczuk AC, Gorenstein L, Walter KL, et al. Non-small-cell lung cancer molecular signatures recapitulate lung developmental pathways. Am J Pathol. 2003;163:1949–1960. doi: 10.1016/S0002-9440(10)63553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Parmigiani G, Garrett-Mayer ES, Anbazhagan R, et al. A cross-study comparison of gene expression studies for the molecular classification of lung cancer. Clin Cancer Res. 2004;10:2922–2927. doi: 10.1158/1078-0432.ccr-03-0490. [DOI] [PubMed] [Google Scholar]
- 170.Shyr Y, Kim K. Weighted flexible compound covariate method for classifying microarray data. In: Berrar D, editor. A Practical Approach to Microarray Data Analysis. New York: Kluwer Academic; 2003. pp. 186–200. [Google Scholar]
- 171.Yanagisawa K, Shyr Y, Xu BJ, et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362:433–439. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- 172.Rahman SM, Shyr Y, Yildiz PB, et al. Proteomic patterns of preinvasive bronchial lesions. Am J Respir Crit Care Med. 2005;172:1556–1562. doi: 10.1164/rccm.200502-274OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Guo L, Ma Y, Ward R, et al. Constructing molecular classifiers for the accurate prognosis of lung adenocarcinoma. Clin Cancer Res. 2006;12:3344–3354. doi: 10.1158/1078-0432.CCR-05-2336. [DOI] [PubMed] [Google Scholar]
- 174.Gordon GJ, Jensen RV, Hsiao LL, et al. Using gene expression ratios to predict outcome among patients with mesothelioma. J Natl Cancer Inst. 2003;95:598–605. doi: 10.1093/jnci/95.8.598. [DOI] [PubMed] [Google Scholar]
- 175.Jiang H, Deng Y, Chen HS, et al. Joint analysis of two microarray gene-expression data sets to select lung adenocarcinoma marker genes. BMC Bioinformatics. 2004;5:81. doi: 10.1186/1471-2105-5-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yanagisawa K, Tomida S, Shimada Y, et al. A 25-signal proteomic signature and outcome for patients with resected non-small-cell lung cancer. J Natl Cancer Inst. 2007;99:858–867. doi: 10.1093/jnci/djk197. [DOI] [PubMed] [Google Scholar]
- 178.Xu BJ, Gonzalez AL, Kikuchi T, et al. MALDI-MS derived prognostic protein markers for resected non-small cell lung cancer. Proteomics ClinAppl. 2008;2:1508–1517. doi: 10.1002/prca.200800094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Petty RD, Kerr KM, Murray GI, et al. Tumor transcriptome reveals the predictive and prognostic impact of lysosomal protease inhibitors in non-small-cell lung cancer. J Clin Oncol. 2006;24:1729–1744. doi: 10.1200/JCO.2005.03.3399. [DOI] [PubMed] [Google Scholar]
- 180.Potti A, Dressman HK, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 181.Taguchi F, Solomon B, Gregorc V, et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst. 2007;99:838–846. doi: 10.1093/jnci/djk195. [DOI] [PubMed] [Google Scholar]
- 182.Cohen AA, Geva-Zatorsky N, Eden E, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 183.Sozzi G, Roz L, Conte D, et al. Plasma DNA quantification in lung cancer computed tomography screening: five-year results of a prospective study. Am J Respir Crit Care Med. 2009;179:69–74. doi: 10.1164/rccm.200807-1068OC. [DOI] [PubMed] [Google Scholar]
- 184.Zhong L, Hidalgo GE, Stromberg AJ, et al. Using protein microarray as a diagnostic assay for non-small cell lung cancer. Am J Respir Crit Care Med. 2005;172:1308–1314. doi: 10.1164/rccm.200505-830OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhong L, Peng X, Hidalgo GE, et al. Identification of circulating antibodies to tumor-associated proteins for combined use as markers of non-small cell lung cancer. Proteomics. 2004;4:1216–1225. doi: 10.1002/pmic.200200679. [DOI] [PubMed] [Google Scholar]