SUMMARY
Understanding cells as integrated systems requires that we systematically decipher how single genes affect multiple biological processes and how processes are functionally linked. Here, we used multi-process phenotypic profiling, combining high-resolution 3D confocal microscopy and multi-parametric image analysis, to simultaneously survey the fission yeast genome with respect to three key cellular processes: cell shape, microtubule organisation and cell cycle progression. We identify, validate and functionally annotate 262 genes controlling specific aspects of those processes. Of these 62% had not been linked to these processes before and 35% are implicated in multiple processes. Importantly, we identify a conserved role for DNA-damage responses in controlling microtubule stability. In addition, we investigate how the processes are functionally linked. We show unexpectedly that disruption of cell cycle progression does not necessarily impact on cell size control and that distinct aspects of cell shape regulate microtubules and vice-versa, identifying important systems-level links across these processes.
INTRODUCTION
In many ways the genomes of most organisms remain black boxes, with the function of the majority of genes and gene products still unknown. This is the case foremost in humans, where a decade after publication of the human genome sequence we still have no direct experimental evidence of the function of over half of all the proteins it encodes (www.ebi.ac.uk/QuickGO/GAnnotation). Yet this is just the tip of the iceberg, as many genes and proteins play roles in multiple biological processes, themselves functionally linked, with most of those multiple roles and links awaiting discovery.
Fission yeast (Schizosaccharomyces pombe) is excellently placed for that discovery, with a genome of ~4900 protein coding genes (26.1% essential) of which 40% currently have a function only inferred from homology and a further 20% are completely uncharacterised (Aslett and Wood, 2006; Wood et al., 2002). Over the last four decades, classical genetic screening using S. pombe has allowed the discovery of numerous molecules and pathways controlling many essential eukaryotic processes thanks to the genetic tractability, simple morphology and uniform growth and division pattern of S. pombe cells (Forsburg, 2003). Recently a genome-wide library of knockout (KO) haploid strains - where each of 3004 non-essential genes across the S. pombe genome was systematically deleted - became commercially available (Kim et al., 2010), opening the possibility to potentiate that discovery power using ultrasensitive image-based phenotypic screening strategies (Chia et al., 2012; Collinet et al., 2010; Cotta-Ramusino et al., 2011; Laufer et al., 2013; Mercer et al., 2012; Neumann et al., 2010; Rohn et al., 2011; Simpson et al., 2012; Yin et al., 2013).
Here, we used fission yeast to carry out a 3D image-based genomic screen monitoring cell shape, microtubule organisation and cell cycle progression in order to find genes involved in these processes, identify genes controlling multiple processes and determine how processes are functionally linked. We describe the identification, large-scale validation and quantitative annotation of 262 putative regulators, with 62% newly implicated in the processes studied and 35% implicated in more than one. As a result of in-depth validation of one hit class, we identify a conserved role of the DNA damage response in controlling microtubule stability, revealing a previously unappreciated link between those two therapeutically-relevant cell biological machineries. Moreover, by exploiting the richness of the multidimensional feature sets obtained from the screen, we investigate statistically and in detail the functional links across processes. We show that disruption of cell cycle progression does not necessarily impact on cell size control, and demonstrate that the causal links between cell shape and microtubule regulation in S. pombe are directional and complex, with distinct cell shape and microtubule features having defined epistatic relationships in this species.
The multi-process screen images and gene annotations are available online as a resource for the community at www.sysgro.org as well as linked to the centralized fission yeast repository PomBase www.pombase.org.
RESULTS AND DISCUSSION
Establishment of a 3D image-based, yeast phenotypic profiling pipeline
In order to carry out a multi-process phenotypic screen in fission yeast we developed a live cell, 3D fluorescence image-based phenotypic profiling pipeline combining automated high-resolution spinning disk confocal microscopy and large-scale, quantitative multiparametric image analysis. We used confocal microscopy and 3D (xyz) imaging in order to extract high-resolution subcellular information from individual yeast cells, allowing us both to screen with high sensitivity and obtain refined phenotypic cell biological annotations. Details of the experimental and computational implementation of the pipeline are described in the Experimental Procedures.
We chose to screen for genes controlling cell shape, microtubules and cell cycle progression because they are fundamental, well-studied processes for which an extensive yet likely not exhaustive list of regulators is known. In addition, all three processes can be monitored simultaneously in live cells expressing only fluorescently-labelled tubulin, minimizing manipulation of their genetic background. Indeed, microtubules can be used as bona fide reporters of cell cycle state, as they take defined stereotypical patterns across the cell cycle (Hagan, 1998); in turn, cell shape can be simply monitored using extracellular fluorescent dyes (see below). Thus, we generated a version of the genome-wide KO library expressing GFP-tagged endogenous alpha tubulin 2 (GFP-Atb2; Figure 1 and Figure S1A), allowing us to visualize microtubules and cell cycle stage ‘live’ in all mutants. As the different KO mutants arrayed in 96-well plates had different growth proficiencies compared to the wild-type (Kim et al., 2010), prior to imaging we used a serial dilution and manual re-pooling strategy to ensure all mutants grew exponentially and were hence physiologically comparable (Figure S1B). Then, in preparation for high-throughput imaging cells were immersed in Cascade blue dextran-containing fluorescent growth medium. This allowed visualisation of live cell morphology without the need to express a cytoplasmic fluorophore (Figure 1). Thereafter, mutants in the 96-well plates were filmed by two-colour (405nm, 488nm) automated high-throughput confocal microscopy at high magnification (60× 1.2NA) and in 3D (xy and 16 z-planes), and their images computationally analysed and phenotyped using custom-made image analysis software.
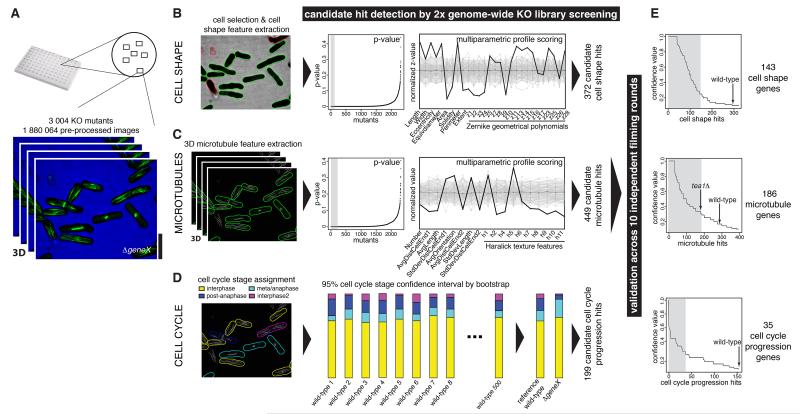
Figure 1. 3D image-based multi-process screening pipeline and multiparametric hit detection.
(A) 2× genomic screen input. 3004 gene KO strains were imaged in 96-well microplates. 3D z-stack images were collected from 6 locations per well in two fluorescence channels to detect cell outlines and GFP-microtubules. (B) and (C) Cell shape (B) and microtubule (C) hit detection strategy, using single feature p-value measurements to detect extreme hits in one feature (left) or multi-feature profile analysis to detect subtle changes across many features (right). (D) Cell cycle progression hit identification, by comparison for each mutant of the proportion of its cells assigned to each cell cycle stage compared to a bootstrapped reference wild-type. (E) 10-fold high-throughput hit validation using the strategies in (B), (C) and (D). Hits were ranked based on the fraction of independent screening rounds where they were coincidently-identified as hits (‘confidence value’). Hits with >35% confidence value were subsequently analysed. Scalebar: 10μm. See also Figures S1-S6 and Tables S1-S3.
Quantitative phenotyping of cell shape, microtubules and cell cycle stage
First, we segmented images in the Cascade blue channel, and extracted from each 2D cell object 57 shape and grey-level features (length, width, area, convexity, concavity, topological skeleton, fluorescence intensity along the object’s contour, etc; Figures 1B and 2A, Figure S2 and Table S1). Then, using Machine Learning (Jones et al., 2009; Sommer and Gerlich, 2013), specifically a Random Forest classifier trained with both wild-type shaped and strongly misshapen mutant cells, we identified and rejected poorly segmented objects and kept only well-segmented cells for further analysis (9.28% out-of-bag error rate).
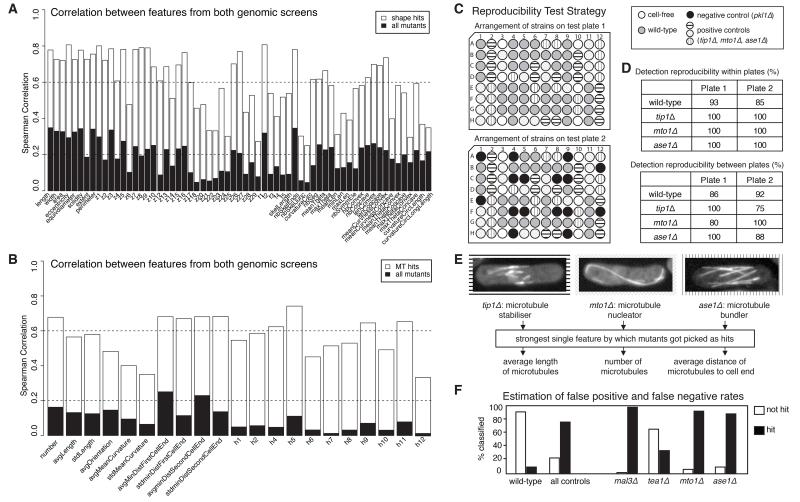
Figure 2. Quality Control.
(A) Spearman correlation of cell shape features for all mutants and for cell shape hits between the two genomic screening rounds. Only features consistent between both screens were used for analysis. (B) Similar to (A) for microtubule features. (C) Arrangement of known microtubule mutants and wild-type cells in plates used in proof-of-method experiments, designed to test for plate location effects and reproducibility of positive and negative control phenotypes. (D) Percentage of correct identification of mutant vs. wild-type populations within and between test plates. (E) Known mutants used and features for which they were picked. (F) Rates of incorrect identification of a mutant as ‘wild-type’ (false negative rate) for four mutants versus the rate of incorrect identification of wild-type controls as ‘mutant’ (false positive rate).
Subsequently, we detected microtubules in the GFP channel xyz image stacks, reconstructed microtubule orientation within every cell in 3D and extracted 24 geometrical and greylevel microtubule features (number, length, fluorescence intensity, orientation, etc; Figures 1C and 2B, Figure S3A-D and Table S2).
Finally, we identified cell cycle stage for each cell based on 3D microtubule pattern, using a four-class Support Vector Machine (SVM) classifier (Jones et al., 2009; Sommer and Gerlich, 2013),. The classifier, trained with wild-type cells and cells from 4 known microtubule mutants (lacking Tip1/CLIP170 (Brunner and Nurse, 2000), Mto1/Centrosomin (Sawin et al., 2004), Ase1/PRC1 (Loiodice et al., 2005) and Pkl1/Kinesin-14A (Troxell et al., 2001)) distinguished 4 cell cycle-related microtubule patterns: interphase microtubule array (IP; characteristic of G2 phase in our exponential growth conditions), metaphase spindle (SP; characteristic of M phase), post-anaphase array (PAA; reflective of G1 phase) and post-mitotic interphase microtubule array (called IP2 for ‘interphase2’; likely reflective of G1-S phases). The classifier accuracy was 93.78% across the wild-type and 4 mutants combined, indicating that we could achieve robust cell cycle stage assignment even when microtubules had an abnormal phenotype. This was partly thanks to the use of 3D microtubule features, which allowed a more accurate assignment of cell cycle stage than 2D features (see Extended Experimental Procedures). The output was a signature of 4 scores reflecting the proportion of cells assigned to each cell cycle stage, for each wild-type (typically 70% IP, 10% SP, 10% PAA and 10% IP2) and mutant cell population, indicative of their cell cycle progression characteristics (Figure 1D and Figure S3E-F).
Hit detection, 10-fold validation and selection
We used two complementary strategies for detecting KO mutants with aberrant cell shape or microtubules (‘hits’, Figure 1B-C, Figure S4). The first strategy identified mutants with a prominent alteration in a single feature (‘p-value’, Figure 1). The second strategy identified mutants with multiple subtle feature alterations (‘multiparametric profile scoring’, Figure 1). In proof-of-principle experiments prior to screening, the use of both strategies combined led to highly consistent detection of the wild-type and of 4 known microtubule mutants within and across 96-well microplates (Figure 2C-F), validating the quality and reproducibility of our hit detection strategy. The combined hit detection procedure was optimized independently for cell shape and microtubules based on the results of visual screening by a human observer of one genomic image dataset (Figures S5-S6).
To detect KO mutants (‘hits’) with altered cell cycle progression, we used bootstrap statistics to estimate the typical proportions of wild-type cells in each cell cycle stage, scoring as hits KOs where at least 2 cell cycle stages were statistically disproportionate with respect to the wild-type (i.e. under- or over-represented, Figure 1D). That criterion ensured only detection of hits where general cell cycle progression was affected. In particular, this allowed us to screen for genes distinct from classical cell cycle regulators which, when mutated, often lead to checkpoint-mediated delay in just one cell cycle stage or transition.
We grew, imaged and computationally screened independently the entire library twice (Figure S1C-F), analyzing 1 880 064 images and making and analyzing 1 707 870 cell, 5 597 165 microtubule and 1 607 406 cell cycle stage assignments. This identified 372 cell shape hits, 449 microtubule hits and 199 cell cycle progression hits (note: hit identification for each process was independent of the others). To generate a high-confidence hit list, we then rescreened all hits at large-scale to obtain 10 biologically-independent screening rounds and ranked hits according to the fraction of repeats in which they were detected (‘confidence value’; Figure 1E). Only hits with ≥35% confidence - the percentage corresponding to a well-established but weak phenotype hit (tea1Δ; Figure 2F) added as positive control in all repeats - were selected for further analysis. Altogether, this led to identification of 143 cell shape, 186 microtubule and 35 cell cycle progression high-confidence hit genes (Table S3) described next.
Genes and pathways controlling cell shape and cell size control
Genes whose KO affected cell shape (‘cell shape genes’, Figure 1E) included expected regulators of cell morphology, cell polarity and growth (Tea3, Pom1, Arf6, Rga2, Tea2, Sla2/End4, Myo1, Efc25, Scd2; for genes’ annotations see www.pombase.org/) but also many genes related to a wide range of other processes like trafficking or cell cycle control, and 17 altogether unannotated genes. 41% (58/143) of genes implicated in cell shape regulation had not been previously reported as such, to our knowledge. Importantly, they were not identified in a recent visual screen of the S. pombe KO library (Hayles et al., 2013). This is likely due to the very different physiological conditions used in that study (non-exponential growth on solid medium) and our use of sensitive computational hit detection.
Similarly to previously published studies (Bakal et al., 2007; Fuchs et al., 2010), cell shape hit classification was done using 8 SVM classifiers trained to recognize 8 basic phenotypic classes on an individual cell basis (Figure 3A-B and Figure S4A): ‘stubby’ (wide), ‘banana’ (curved), ‘orb’ (round), ‘kinky’ (S-shaped), ‘long’ (elongated), ‘skittle’ (with one side wider than the other) and ‘T-shaped’ (branched). Classically, cell shape mutants are thought to display only one aberrant shape phenotype, like being round or curved (Hayles et al., 2013). Surprisingly we found that, instead, all strains including the wild-type did not display only a single shape phenotype but rather could be defined as a mixture of those eight phenotypes (Figure 3A-B and Figure S4A). Thus, even within a genotypically uniform cell population, the genome allows S. pombe cells to explore multiple morphogenetic states. These might be brought about by cell-to-cell differences in the content of key shape-controlling proteins due e.g. to non-exact equipartition of cellular material - polarity landmarks, secretory machinery, cell wall composition/properties, etc - between daughter cells at cell division, or from stochastic gene expression.
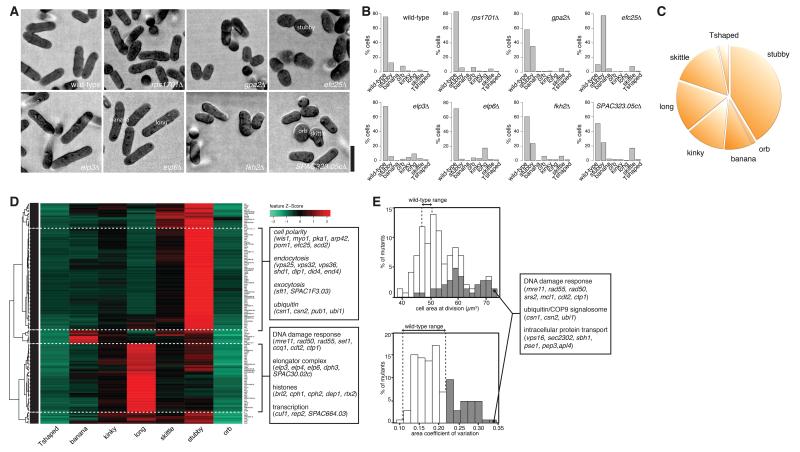
Figure 3. Cell shape genes and deregulation of cell size control.
(A) Example cell shape hits. (B) Corresponding cell shape hit shape profiles illustrating the co-manifestation of multiple cell shape phenotypes within the genotypically uniform mutant populations. (C) Phenotypic trait abundance (pie chart) illustrating the proportion of cells from all shape hit populations combined into stereotyped categories. (D) Phenoprint clustering of cell shape hits based on the shape profiles in (B) and major functional gene ontology (GO) groups. (E) Distributions of cell size at division (top) and its coefficient of variation (lower) for cell shape mutants. Mutants with greater coefficient of variation than wild-type are shown in grey with selected gene names. Scalebar: 10μm. See also Figures S2 and S4 and Tables S1 and S3.
Quantitatively the most common aberrant cell phenotype was stubby (Figure 3C), indicating it may be the most general manifestation of compromised cell shape; conversely the least common was orb (i.e. completely non-polarised), consistent with the finding that known genes whose disruption leads to complete rounding are essential (Hayles et al., 2013) and with the notion that complete loss of polarity may be incompatible with viability.
We clustered mutants based on their shape phenoprint and found that KOs of specific pathways shared characteristic morphological signatures (Figure 3D). One major cluster of predominantly stubby mutants comprised regulators of endocytosis and exocytosis (Vps25, Vps32, Vps36, Shd1, Dip1, Did4, Sla2/End4, Sft1; likely involved in apical restriction of cellular growth zones), genes involved in ubiquitin/COP9 signalosome-mediated protein degradation (Csn1, Csn2, Pub1, Ubi1) and several uncharacterized factors. Another major cluster comprised significantly longer mutants (note: cell elongation, usually associated with cell cycle deregulation, was scored in our screen as a cell shape defect), corresponding to factors involved in the DNA damage response (DDR) (Mre11, Rad50, Rad55, Set1, Ccq1, Cdt2, Ctp1; the DDR leads to cell cycle delay and cell elongation), transcriptional regulators (Cuf1, Rep2), elongator complex subunits (Elp3, Elp4, Elp6, Dph3; this complex has been involved in negatively regulating exocytosis), histone modifiers (Brl2, Cph1, Cph2, Dep1, Rtx2) and other putative regulators.
We next asked whether, given their geometrical disruption, cell shape mutants properly control cell size. S. pombe cells are thought to need to reach a critical cell size at the G1/S and most importantly the G2/M cell cycle transition, when cells engage in cell division only after reaching twice their original size at birth (Mitchison, 2003). We calculated the average cell area (as a proxy for size) at mitosis by looking at cells containing a mitotic spindle, and plotted distributions of the average area at mitosis for all hits (Figure 3E, top) and its coefficient of variation (Figure 3E, bottom). 90% of cell shape hits had an area at division lower or higher than wild-type cells, which divide at an area of ~48 μm2.
This included KOs of factors known to be involved in cell size control such as Pom1 (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). Strikingly, 30% had a higher coefficient of variation of the cell area than the 0.12-0.22 coefficient of the wild-type (grey, Figure 3E), indicative of lack of precision in cell size control at division. Interestingly, the latter was enriched for mutants in the ubiquitin/COP9 signalosome complex (implicated in cell cycle and cell size control in D. melanogaster (Bjorklund et al., 2006)), DDR regulators and various factors involved in intracellular protein transport. As the COP9 complex regulates cullin activity in mammals and cullin (Cul-4) has been implicated in both cell cycle control and the DDR (Hu et al., 2004), one possibility is that ubiquitin/COP9 and the DDR act on cell size control via the same pathway, possibly via their role in cell cycle regulation. Alternatively, each may play a distinct role that needs to be further explored. Similarly, the role in size control of other factors identified needs to be clarified.
Genes and pathways associated with characteristic microtubule signatures
Genes whose KO affected microtubules (‘microtubule genes’; Figure 1E) included known microtubule regulators (Tea2, Tip1, Mal3), mitochondrial factors, trafficking-related genes and 19 altogether unannotated factors. Notably, 93.5% (174/186) of the genes implicated in microtubule regulation had not, to our knowledge, been previously reported as such. Mutants in those genes primarily led to deregulation of microtubule number, length or orientation, with most KOs affecting several features simultaneously albeit in different proportions (Figure 4A). Microtubule length (encompassing the features: ‘length’, ‘length variance’, ‘occupancy’, ‘occupancy variance’; Figures 4B-C) was by far the most common quantitatively affected microtubule property, demonstrating that microtubule length per se is not essential for cell viability. By contrast, low microtubule number was a very infrequent feature, consistent with the fact that microtubule nucleation is essential for cell viability.
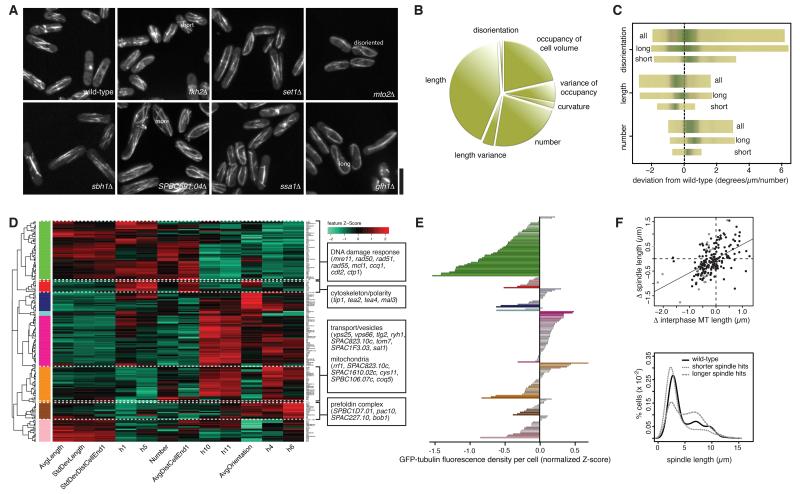
Figure 4. Genes associated with characteristic microtubule signatures.
(A) Example microtubule hits. (B) Phenotypic trait abundance (pie chart) illustrating the proportion of cells from all microtubule mutant cell populations combined displaying a significant phenotype in the features shown. (C) Three significant features for all (top), only short (bottom) or only long (middle) mutants. (D) Phenoprint clustering of microtubule hits based on extracted microtubule features and major functional GO groups. (E) GFP-tubulin content. Average GFP-Atb2 fluorescence level per cell for each mutant shown in (D), as a proxy for tubulin concentration with respect to the average wild-type value. Colours of the gene groups indicate which clusters from (D) they correspond to. (F) Correspondence between interphase and mitotic phenotype. Top: Comparison of interphase and spindle microtubule length changes for all microtubule hits. Grey: KOs with significantly altered spindle length (‘spindle hits’). Bottom: Percentage of cells with a given spindle length, for spindle hits with a spindle shorter/longer than wild-type (‘shorter’/’longer’ spindle hits). In all three classes of populations shown, cells accumulate with a short spindle, likely corresponding to pro-metaphase state during chromosome capture. Long spindle mutants (grey dashed) also accumulate cells with a longer spindle, which may indicate a delay during or on exiting anaphase B. Scalebar: 10μm. See also Figures S3 and S4 and Tables S2-S3.
Clustering of microtubule hits was done using a subset of 12 features selected by visual quality control to optimize for high inter-class variability and low intra-class variability (i.e. to optimally group together KOs judged visually to have the same phenotype and assign to separate groups KOs with visually different phenotypes) and identified various pathways, each associated with a specific microtubule phenoprint (Figure 4D). Among the most prominent pathways we found: cytoskeleton/cell polarity (Tea2, Tip1, Tea4, Mal3; whose KO leads to short, disoriented microtubules), DDR (Mre11, Rad50, Rad51, Rad55, Mcl1, Ccq1, Cdt2, Ctp1; slightly elongated, hyper-oriented microtubules), transport/vesicles and mitochondria (Vps25, Vps66, Tlg2, Ryh1, SPAC823.10c, Tom7, SPAC1F3.03, Sat1, and Rrf1, SPAC823.10c, SPAC1610.02c, Cys11, SPBC106.07c, Coq5; slightly more microtubules) and tubulin folding (the Prefoldin complex subunits SPBC1D7.01, Pac10, SPAC227.10, Bob1; fewer microtubules). We next assessed whether differences in tubulin content could account for the mutants‘ microtubule feature signatures by quantitating their intracellular GFP-Atb2 fluorescence. We found that, although many mutants displayed substantial differences in tubulin content compared to the wild-type, there was no obvious correlation between their microtubule feature signatures and GFP-tubulin fluorescence, suggesting that their microtubule phenotype arises from deregulation of microtubule assembly rather than tubulin content (Figure 4E). Interestingly, analysis of the hits’ microtubule length in interphase versus mitosis revealed a correlation between the two in ~80% of cases (Figure 4F), indicating that many genes identified may also play a role in mitotic spindle control.
Cell cycle progression genes reveal temporal linkage between non-consecutive cell cycle phases
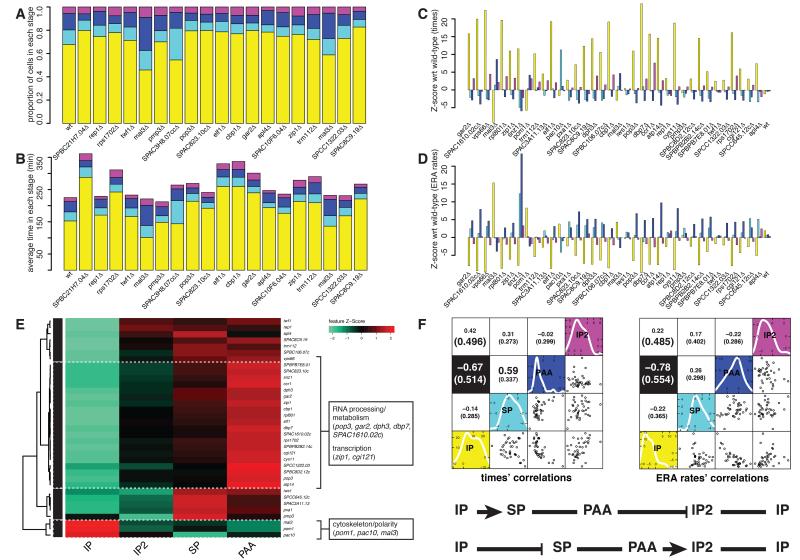
Genes whose KO affected cell cycle progression (‘cell cycle progression genes’; Figure 1E) comprised a diverse range of factors and, as expected, did not include classical cell cycle regulators. To look for interesting functional groups, we measured experimentally the cell cycle duration of all cell cycle progression hits. This allowed us to convert for each KO the proportions of cells in each cell cycle stage into average times spent in each stage (Figures 5A-B). We then calculated the Z-score of all four stage times (durations) with respect to the wild-type, for all hits (Figure 5C). Subsequently, we used Ergodic Rate Analysis (ERA (Kafri et al., 2013)) to estimate the average rate of progression from each cell cycle stage to the next (i.e. the rate of exit from each stage) and calculated the ERA rates’ Z-score with respect to the wild-type rates for all cell cycle progression hits (Figure 5D).
Figure 5. Cell cycle progression genes and cell cycle stage correlation analysis.
(A) Proportional representation of the cell cycle phases - interphase (yellow), spindle (light blue), PAA (blue), interphase 2 (purple) - for a selection of cell cycle progression hits and a reference wild-type. (B) As in (A) but with distributions scaled by the doubling time of each mutant. (C) z-score plot showing the significance of deviation from wild-type of cell cycle stage durations for all hits. (D) As (C) for ergodic rate analysis (ERA) derived progression rates. (E) Phenoprint clustering based on hits’ ERA rates and major functional GO groups. (F) Correlation analysis among the four cell cycle stages’ durations and ERA progression rates. The graphs show correlation plots of the duration (left graph, bottom half) and rate (right graph, bottom half) of each cell cycle stage against every other, for all hits. Spearman (and MIC, in parenthesis) correlation coefficients are shown for each combination in the top half of the graphs. Black boxes indicate significant anti-correlation between the duration and ERA rate of IP and PAA. The diagonal shows the distribution of z-score values for the durations (left) and ERA rates (right) across all hits, for each cell cycle stage quantitated. A diagram representation of the implications of that anti-correlation for the overall cell cycle is illustrated beneath (pointy arrows: accelerated progression; blunt arrows: delayed progression). See also Figure S3 and Table S3.
Clustering of the KOs based on their ERA rate Z-scores (Figure 5E) revealed groups of factors whose KO mainly results in accelerated exit from: IP into SP (Mal3, Pom1, Pac10: cytoskeleton/polarity related); SP into PAA (for example Pxa1: intracellular trafficking; Tos4, Iws1: transcription); PAA into IP2 (the largest group, including Pop3, Gar2, Dph3, Dbp7; RNA processing and metabolism); and IP2 into IP (including Twf1, Apl4: actin and endocytosis). Genes from the first cluster could be involved in regulating the IP-SP transition via the Pom1 cell size control pathway (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). Some genes from the fourth cluster could control the IP2-IP transition via regulation of the cytokinetic actomyosin ring (Rincon and Paoletti, 2012). It will be interesting to determine how all other genes, known and uncharacterized, are linked to cell cycle progression control.
Analysis of correlations in durations/rates among the four cell cycle stages (Spearman or MIC (Reshef et al., 2011); Figure 5F) for our hits revealed a strong anti-correlation between IP and PAA, i.e. KOs with an extended IP tend to have a shorter PAA and vice versa. Since IP and PAA reflect G2- and G1-phase respectively, this suggests that an accelerated G1/S transition (PAA-IP2 exit) causes delayed G2/M transition (IP-SP exit) and vice versa. As S. pombe has been shown to possess two apparently independent cell size control mechanisms acting at G2/M (the dominant one) and G1/S (revealed in small wee1-50 mutants) (Fantes, 1977; Fantes and Nurse, 1978; Mitchison, 2003), which secure that nuclear division does not occur before cells reach a critical size, the observed anti-correlation might reflect compensation in cell cycle progression caused by one of the mechanisms when the other fails. Interestingly, as the cell cycle progression mutants identified here did not display altered size at cell division (Figure 6B and next section; contrary for example to wee1-50 mutants), this suggests that both mechanisms exist and are active even in normally-sized cells.
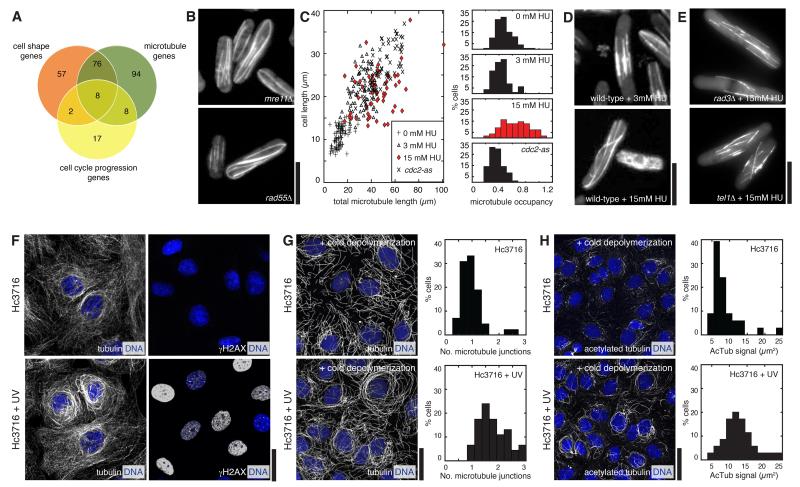
Figure 6. Genes regulating multiple processes and role of the DDR in inducing interphase microtubule stabilisation.
(A) Venn diagram illustrating cell shape, microtubule and cell cycle progression hits. (B) The DNA repair defective mutants mre11Δ and rad55Δ display highly elongated microtubules. (C) Hydroxyurea (HU) induces disproportionate microtubule elongation in wild-type cells. Microtubule occupancy is the ratio of average microtubule length to cell length. Contrary to 15 mM HU-treated cells, 3mM HU-treated cells and cell cycle-arrested cdc2-as cells do not disproportionately elongate microtubules. (D) Images of 3 mM HU- (top) and 15 mM HU-treated cells (bottom). (E) 15 mM HU treatment does not induce enhanced microtubule stabilisation in the DNA damage checkpoint mutants rad3Δ and tel1Δ. (F) Left: Effect on microtubules of UV-induced DNA damage in human cultured cells (Hc3716-hTERT). Right: γH2AX staining showing extent of DNA damage. (G) Contrary to UV-untreated cells, UV-treated cells contain denser (right: quantitations) microtubule bundles that resist cold-induced depolymerisation indicative of microtubule stabilisation. (H) UV-treated cells contain significantly higher acetylated tubulin levels (right: quantitations) than UV-untreated cells. Scalebars: 10μm.
Genes involved in multiple processes
Of the 262 genes identified for the three processes investigated 168 (64%) were associated to a single process and 94 (36%) had multiple functional assignments (Figure 6A). Only 10 (4%) genes were shared between cell shape and cell cycle progression and 16 (6%) between microtubules and cell cycle progression, implying that those processes are largely independent.
By contrast, 84 (32%) genes were shared between cell shape and microtubules implying a potentially significant co-regulation of both processes, as expected. These included factors involved in cell morphogenesis and polarity (Aah3, Efc25, Tea2, Tip1), endocytosis (Did4, Vps25, Vps36, Snf7), but also transcriptional regulation (Pof3, Pmc6, Rep2, Rxt2, Tup12), chromatin remodelling (Rsc4, Set1, Arp42, Spp1, Swd1) and DDR (Mre11, Rad50, Rad55, Ctp1, Set1, Mcl1, Ubi1). Notably, 60/143 (42%) cell shape genes and 106/186 (57%) microtubule genes had a role in a single process alone. This implies that a link between cell shape and microtubule deregulation is not obligatory in cells and may be specific to particular machineries.
Future work will be needed to clarify which genes associated here with multiple processes actually correspond to factors that actively couple those processes.
The DDR induces stabilisation of interphase microtubules
The largest functional group of KOs co-deregulating cell shape and microtubules was that of genes related to the DNA damage response (DDR). While the role of DDR genes in cell shape control (specifically in cell length and, hence, size control; Figures 3D-E) could be understood as resulting from DDR-induced cell cycle arrest and elongation (Melo and Toczyski, 2002; Zhou and Elledge, 2000), a link between DDR and microtubules (Figure 4D) was unexpected and raised the question of whether DDR gene KOs lead to microtubule deregulation indirectly (via cell shape deregulation) or directly.
Therefore, we decided to investigate this link further and use this as a means to validate the predictions of our phenoprint clustering analysis. Inspection of cells lacking the DNA repair factors Mre11 and Rad55 revealed that in those mutants interphase microtubules are elongated compared to wild-type cells, often curling around cell ends indicating increased microtubule stability (Figure 6B, quantitation not shown). By contrast, in cells lacking the ATR and ATM checkpoint kinases Rad3 and Tel1 microtubules were of wild-type length (not shown). This suggested that impaired DNA repair functions, and ensuing DDR arising from unrepaired DNA damage, may induce microtubule stabilisation via the ATM/ATR pathway. To test this directly, we treated wild-type cells with different doses of hydroxyurea (HU) to induce the DDR in a primarily ATR dependent manner, and we quantitated the effect of HU dosage on the average length of cellular microtubules and how it scales with cell length (‘microtubule occupancy’). Cells untreated with HU displayed a range of microtubule lengths that varied proportionately with the wild-type range of cell lengths (Figure 6C, 0 mM HU). Similarly, following treatment with low dose of HU cells became elongated due to cell cycle arrest and their microtubules became longer proportionally with cell length increase (Figures 6C and 6D top, 3 mM HU). By contrast, at higher dose of HU microtubules became disproportionately longer than cells, indicating an increase in microtubule stability induced specifically by the DDR (Figures 6C and 6D bottom, 15 mM HU). In agreement with this, in elongated G1- and G2-arrested cdc2-as mutant cells - where in principle no DDR activation occurs - microtubule elongation was also proportional to cell elongation (Figure 6C quantitations). Furthermore, the enhanced HU-induced microtubule stabilization was specifically DDR dependent, as microtubules did not become stabilized in cells lacking Rad3 or Tel1 treated with 15 mM HU (Figure 6E). Altogether these data suggest that DNA damage induces microtubule lengthening in an ATR-dependent manner. To test whether this functional link is conserved, we then asked whether induction of DNA damage in human (Hc3716-hTERT) cells by UV treatment (30J/m2) also elicited a similar effect. We found that in those cells induction of DDR often causes microtubules to organise in dense bundles around the cell nucleus (Figure 6F and not shown), suggestive of microtubule stabilisation. To test this further, we then induced partial microtubule depolymerisation by cold (4°C) treatment in both UV-untreated and UV-treated Hc3716 cells. We found that while hardly any UV-untreated cells contain thick bundles after cold depolymerisation (1.22%, n=575 cells; 15% of cell fields scored) many UV-treated cells still contain thick bundles (5.89%, n=577 cells; 45% of cell fields scored) of much more connected microtubules (Figure 6G). In addition, microtubules in DDR-activated cells contain a much higher amount of acetylated tubulin (Figure 6H), indicative of increased microtubule stability (Hammond et al., 2008). We conclude that the DDR specifically induces stabilisation of interphase microtubules, revealing a conserved link between the DDR and cytoskeletal control in cells. Though some links between those two machineries have been reported (Baschal et al., 2006; Lee et al., 2010; Lee et al., 2011; Xie et al., 2011), the conserved link described here points to a more general connection, whose exact physiological role and mechanistic details - in particular the cytoskeletal DDR target(s) involved - will need to be clarified. This might be of particular therapeutic relevance as a combination of cytostatic doses of DNA-damaging drugs with microtubule drugs has been shown to result in selective cytotoxicity and radio-/chemo-sensitisation in some cancer cells (Baumgart et al., 2012; Blagosklonny et al., 2000; Lee et al., 2011).
Systems-level functional relationships between processes
One important aspect of our quantitative, multi-process screening strategy is that it allowed us, beyond hit identification and phenotypic clustering, to obtain rich multi-dimensional feature sets characterizing all three processes simultaneously in cells, for hundreds of different KO conditions. Hence, we exploited the richness of those multidimensional feature sets to investigate statistically the systems-level functional relationships across processes.
We first turned to cell cycle progression hits - selected as having at least two disproportionate cell cycle stages with respect to the wild-type (Figure 1D) – and asked whether their cell cycle progression defect impacted quantitatively on their cell size. Remarkably, we found that their cell size distribution – as assessed by measuring cells undergoing division - was indistinguishable from that of wild-type cells (Figure 7A). This is in contrast with mutants in conventional cell cycle genes, which have traditionally been identified by virtue of their abnormal cell size phenotype, and indicates that the cell cycle progression genes uncovered here likely constitute a mechanistically distinct class of cell cycle regulators. Likewise and importantly, it demonstrates that disruption of cell cycle progression does not necessarily impact on cell size control.
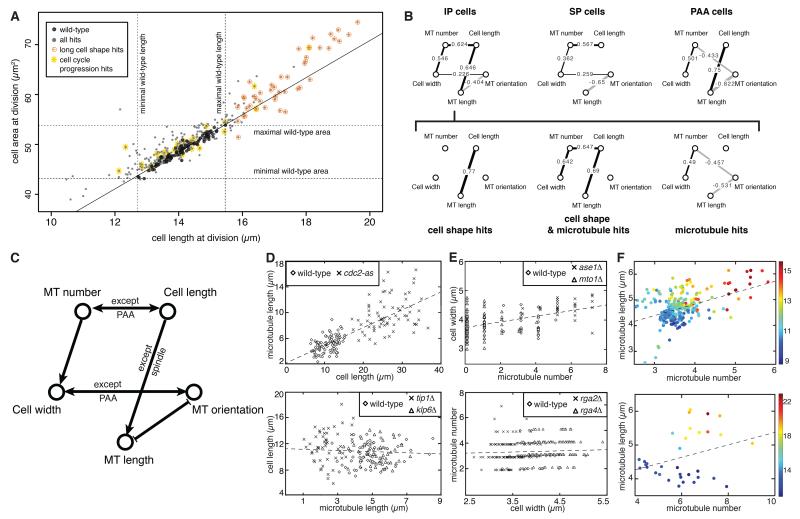
Figure 7. Systems-level functional interactions between processes.
(A) Distribution of cell length and area at division for long cell shape hits (orange) versus cell cycle progression hits (yellow). Note their lack of intersection. Wild-type controls (black) and other hits (grey) are shown, for comparison. (B) Bayesian network inference graphs of the conditional correlation between cell shape and microtubule features across hits, filtered by cell cycle stage (top row) or, for interphase cells only, subdivided by hitlist (cell shape and/or microtubule; bottom row). Correlation coefficients between variables are displayed on top of network edges (note that correlations were not directly used to obtain the networks). (C) Diagram summarising significant Bayesian causal links between cell shape and microtubule regulation in S. pombe. (D) Causation between cell length and microtubule length. Top: Plot of interphase cell length and microtubule length for wild-type and cdc2-as cells. Bottom: The inverse plot for cells with shorter (tip1Δ) or longer (klp6Δ) microtubules. Note that cell length influences specifically microtubule length, not the inverse. (E) Causation between microtubule number and cell width. Top: Plot of microtubule number against cell width for cells with more (ase1Δ) and fewer (mto1Δ) microtubules. Bottom: Inverse plot for wider (rga4Δ) and narrower (rga2Δ) cells. To aid visualisation, points have been artificially displaced along the microtubule number axis for different genotypes. Note that microtubule number influences specifically cell width, not the inverse. (F) Plots of microtubule number against microtubule length coloured by cell length (different colours signify different cell lengths), illustrating that correlation between these features is dependent upon cell length. Top: Data from genomic screens, each point represents the average of all cells analysed for a given mutant. Bottom: Low-throughput validation using cells artificially elongated by cell cycle arrest. See also Figure S7.
We next turned to cell shape and microtubule hits and investigated dependencies between their features using Bayesian network analysis (Collinet et al., 2010; Yu et al., 2004). Like correlation analysis, Bayesian network analysis allows graphical representation of the probabilistic relationships between variables in a dataset. However, in addition Bayesian networks allow inferring conditional dependencies between variables. Thus, two variables are disconnected in a Bayesian network graph if they are either independent or conditionally independent knowing one or several other variables. This allows inference of direct links between two variables, as opposed to indirect effects mediated or caused by a third variable.
We focused on five key features quantitated in our screen: microtubule number, microtubule length, microtubule (dis)orientation, cell length and cell width. Using data from all cell shape and microtubule hits together, we constructed three Bayesian networks corresponding to three different cell cycle stages (IP, SP, PAA), by using for each network only cells in the corresponding cell cycle stage, and compared edges between the networks. We found that, while specific pairs of features are interdependent in a consistently correlated or anti-correlated manner, many of those dependencies are cell cycle stage specific (Figure 7B, top three graphs; + sign: correlation, − sign: anti-correlation; Figure S7 illustrates the robustness of the analysis). For example, we found that cell length and microtubule number are always linked in cells except during post-anaphase (PAA), when a profuse, radial microtubule network assembles in the cell middle. Similarly, cell width and microtubule (dis)orientation are always linked except in post-anaphase, indicating that in this species not only interphase microtubule alignment but also mitotic spindle orientation relies mostly on cellular geometry (Thery et al., 2007). Instead, cell length and microtubule length are interdependent except during mitosis, indicating that mitotic spindle size is independent of cell size in this species (Wilbur and Heald, 2013).
We then constructed three different Bayesian networks using features from KOs deregulated in cell shape, microtubules or both (Figure 7B, bottom three graphs), and visually compared the networks to seek to infer directionality in the dependencies among features. We found that cell length and microtubule length are linked in hits deregulated in cell shape alone and in hits deregulated in both cell shape and microtubules. Instead, the two features are not linked in hits deregulated in microtubules only. We interpret this as implying that the causality is from cell length to microtubule length (e.g. increased cell length promotes increased microtubule length). Conversely and unexpectedly, we found that microtubule number and cell width are linked in microtubule only hits and in cell shape and microtubule hits, but not in hits only deregulated in cell shape, leading us to interpret that the causality in this case is from microtubule number to cell width (e.g. increased microtubule number in cells promotes increased cell width). These results are summarized in Figure 7C.
To corroborate the inferred statistical relationships, we sought to externally validate some of its predictions. First, we looked at cdc2-as cells - which have altered cell length due to cell cycle arrest - and found (Figure 6C) that in this mutant cell length changes induce proportional microtubule length changes (Figure 7D, top). By contrast, in mutants lacking the microtubule stabilizer Tip1/CLIP170 or the catastrophe-promoting kinesin-like protein Klp6/Kinesin-8 - with, respectively, shorter and longer microtubules - microtubule length changes do not induce cell length changes (Figure 7D, bottom). Thus, cell length controls microtubule length and not the reverse, as predicted.
Secondly, we looked at mutants lacking the microtubule bundling protein Ase1/PRC1 or the microtubule nucleation factor Mto1/Centrosomin - with, respectively, a higher and lower number of microtubule bundles - and found that in those mutants microtubule number changes induce cell width changes (Figure 7E, top). Instead and by contrast, in mutants lacking the Rho2 GTPase-activating protein (GAP) Rga2 or the Cdc42/Rho2 GAP Rga4 - narrower and wider than wild-type cells, respectively - cell width changes do not induce microtubule number changes (Figure 7E, bottom). Hence, microtubule number controls cell width and not the reverse, as inferred.
Thirdly we asked whether, as implied by the analysis, microtubule number and microtubule length are not directly linked. The absence of that direct link is interesting as (despite the fact that microtubule number and length are known to be regulated by different machineries in cells) one could expect both features to be somewhat dependent on one another, given that both rely on the common pool of tubulin available in each cell.
In fact, plotting microtubule length versus number - both using the screen dataset (Figure 7F, top) and external data from wild-type, 3mM HU-treated and cdc2-as cells for validation (Figure 7F, bottom) - showed a positive correlation between both features, implying a link between them. However, the correlation was absent amongst cells of similar length (Figure 7F). This demonstrates that, as predicted, microtubule number is not directly linked to microtubule length but rather both features are conditionally independent knowing cell length, i.e. they are coupled indirectly via the length of the cell.
Thus, the causal relationship between cell shape and microtubule regulation in S. pombe is directional and complex, with specific cell shape and microtubule features having defined epistatic relationships. Importantly, although our preliminary validation from Figures 7D-F suggests that Bayesian analysis is indeed a useful predictor, given the partial and therefore potentially misleading nature of the parametrical description we have used, it will be crucial in the future to carefully validate the other predictions obtained using this approach.
CONCLUSIONS
In an effort to begin deciphering systematically how genes affect multiple cell biological processes and how processes are functionally linked, we carried out a quantitative multi-process screen simultaneously monitoring three fundamental cell biological processes: cell shape, microtubule organisation and cell cycle progression.
Our screen identified and annotated 262 genes of which 131 were uncharacterised in S. pombe (of those, 29 had no predicted function in any species) and 131 had been described in the context of a different process, including: 6 genes of the ubiquitin/elongator pathways not implicated in cell shape control, 39 membrane trafficking/mitochondrial genes, which we show are involved in microtubule regulation, and 9 RNA processing/metabolism genes implicated in cell cycle progression. We also probed in depth the systemic relationships between those three processes - identifying 94 genes regulating multiple processes and functional relationships linking defined aspects of such processes, in particular of cell shape and microtubule control. This provides a rich, quantitative genomic dataset available for further mining, validation and mechanistic analysis by the scientific community (available at www.sysgro.org and www.pombase.org).
We extrapolate that an important gain in unexpected mechanistic insights could be obtained from multi-process screening across an increasing range of cell biological processes, using simple unicellular systems with standardised/optimised genomic KO or siRNA/RNAi libraries (Collinet et al., 2010). A future challenge therefore lies in developing graphical and statistical modelling approaches (Nicolau et al., 2011; Singh et al., 2007) suited to cope with the very high dimensionality information generated by such screens, in particular to combine information from independent studies (Kemmer et al., 2009; Nicolau et al., 2011; Spalding et al., 2000; Troyanskaya et al., 2003) in a representable and interpretable manner. Such developments will be key to obtain a systemic picture of cell function and to better understand and predict synergistic or antagonistic interactions, including those among clinically-relevant compounds (Horn et al., 2011).
EXPERIMENTAL PROCEDURES
Full methods are available in Extended Experimental Procedures.
Knockout screen for cell shape, microtubule & cell cycle progression genes
The Bioneer haploid deletion (knockout, KO) library v.2 (Bioneer, Korea) was modified to generate a GFP-tubulin expressing library (Dixon et al., 2008). KO mutants were grown exponentially for >48 hours and imaged in 96-well microplates (lectin-coated glass bottom, 10μg/well) containing Cascade Blue Dextran-labelled YES medium (0.1 mg/ml). Two-colour images were acquired using an automated OperaLX spinning-disk confocal microscope (Perkin Elmer) with 60× water-immersion objective (NA 1.2). Six stacks of 16 z-planes 0.4 μm separation were collected for each well. The entire genomic KO library was filmed twice. Customized software was used for image analysis and feature extraction.
Hit detection, large-scale validation and Bayesian analysis
Cell shape or microtubule hits were KO strains with a: a) low Kolmogorov-Smirnov p-value relative to a mean wild-type for any one feature, or b) significant Euclidean distance deviation from a mean wild-type across many features. Cell cycle progression hits were KOs with the proportion of cells in >2 two cell cycle stages outside the 95% bootstrap confidence interval of the corresponding wild-type stages. Genotypic and visual quality controls were done for corroboration. Hits were large-scale validated by 10-times independent rescreening and if picked in >35% of cases kept as high-confidence hits for analysis. SVM classifiers assigned 8 phenotypic classes to cell shape hits. ERA rates (Kafri et al., 2013) were calculated for cell cycle progression hits. Clustering used R, functional GO assignments DAVID, Bayesian analysis the R package bnlearn (Scutari, 2010).
Investigation of DNA damage response & microtubules
Yeast were treated with Hydroxyurea 9 hours before filming and every 3 hours after, and were imaged on lectin-coated MatTek dishes using a DeltaVision system (GE Healthcare). Hc3716-hTERT cells were grown to 70% confluence in Hepatocyte Medium Bullet Kit, exposed to 30J/m2 UVC and grown for 8 hours before cold treatment or fixing. Immunostaining for β-tubulin or α-acetylated tubulin was done with ALEXAfluor-conjugated secondary antibodies, DNA was DAPI-labelled. Cells were imaged on a Leica SP5 confocal.
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. Barral, M. Peter, C. Wilson-Zbinden, D. Gerlich, G. Székely, P. Horvath, M. Held, D. Brunner, T. Makushok, J. Ellenberg, P. Nurse, J. Hayles, A. Sossick, C. Bradshaw, N. Samusik, S. Tooze, A. Csikász-Nagy, F. Vaggi, M. Sato, C. Schmidt, S. Jackson, J. Ahringer, D. St Johnston, V. Wood, P. Zegerman, E. Khabirova, C. Stocker, J. Pines and the Carazo-Salas group for help and comments, A. Sossick and N. Lawrence for assistance with imaging, S. Jackson for antibodies (gamma-H2AX) and use of the UVC box, M. Sato for S. pombe strains, and J. Pines, S. Jackson, P. Horvath, F. Navarro and E. Miska for critical reading of the manuscript. This work was supported by an European Research Council (ERC) Starting Researcher Investigator Grant (R.E.C.-S., M.G., A.C.; SYSGRO), a SystemsX.ch Interdisciplinary PhD (IPhD) Grant (R.E.C.-S., V.G.; 2009-026), a Swiss National Foundation (SNF) Project Grant (R.E.C.-S., X.S.; 3100A0_120656/1), a Human Frontier Science Program (HFSP) Young Investigator Grant (R.E.C.-S., A.C.; HFSP RGY0066/2009-C), a Biological Sciences Research Council (BBSRC) Responsive Mode grant (R.E.C.-S., M.G., A.C.; BB/K006320/1), an Isaac Newton Trust research grant (R.E.C.-S.; 10.44(n)), a Wellcome Trust PhD studentship (J.L.D.L.), a University Research Fellowship (URF) from the Royal Society (E.P.; UF0905080) and a Cancer Research UK Programme Grant (E.P. and L.W.; A12460).
Footnotes
Supplemental Information includes Extended Experimental Procedures, 7 figures, and 3 tables and can be found online at developmental-cell.edmgr.com/.
Financial conflict of interest statement
The authors declare no financial conflict of interest.
REFERENCES
- Aslett M, Wood V. Gene Ontology annotation status of the fission yeast genome: preliminary coverage approaches 100% Yeast. 2006;23:913–919. doi: 10.1002/yea.1420. [DOI] [PubMed] [Google Scholar]
- Bakal C, Aach J, Church G, Perrimon N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 2007;316:1753–1756. doi: 10.1126/science.1140324. [DOI] [PubMed] [Google Scholar]
- Baschal EE, Chen KJ, Elliott LG, Herring MJ, Verde SC, Wolkow TD. The fission yeast DNA structure checkpoint protein Rad26ATRIP/LCD1/UVSD accumulates in the cytoplasm following microtubule destabilization. BMC cell biology. 2006;7:32. doi: 10.1186/1471-2121-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart T, Klautke G, Kriesen S, Kuznetsov SA, Weiss DG, Fietkau R, Hildebrandt G, Manda K. Radiosensitizing effect of epothilone B on human epithelial cancer cells. Strahlentherapie und Onkologie: Organ der Deutschen Rontgengesellschaft [et al] 2012;188:177–184. doi: 10.1007/s00066-011-0029-4. [DOI] [PubMed] [Google Scholar]
- Bjorklund M, Taipale M, Varjosalo M, Saharinen J, Lahdenpera J, Taipale J. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature. 2006;439:1009–1013. doi: 10.1038/nature04469. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV, Robey R, Bates S, Fojo T. Pretreatment with DNA-damaging agents permits selective killing of checkpoint-deficient cells by microtubule-active drugs. The Journal of clinical investigation. 2000;105:533–539. doi: 10.1172/JCI8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D, Nurse P. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell. 2000;102:695–704. doi: 10.1016/s0092-8674(00)00091-x. [DOI] [PubMed] [Google Scholar]
- Chia J, Goh G, Racine V, Ng S, Kumar P, Bard F. RNAi screening reveals a large signaling network controlling the Golgi apparatus in human cells. Molecular systems biology. 2012;8:629. doi: 10.1038/msb.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinet C, Stoter M, Bradshaw CR, Samusik N, Rink JC, Kenski D, Habermann B, Buchholz F, Henschel R, Mueller MS, et al. Systems survey of endocytosis by multiparametric image analysis. Nature. 2010;464:243–249. doi: 10.1038/nature08779. [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C, McDonald ER, 3rd, Hurov K, Sowa ME, Harper JW, Elledge SJ. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011;332:1313–1317. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Fedyshyn Y, Koh JL, Prasad TS, Chahwan C, Chua G, Toufighi K, Baryshnikova A, Hayles J, Hoe KL, et al. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16653–16658. doi: 10.1073/pnas.0806261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes PA. Control of cell size and cycle time in Schizosaccharomyces pombe. Journal of cell science. 1977;24:51–67. doi: 10.1242/jcs.24.1.51. [DOI] [PubMed] [Google Scholar]
- Fantes PA, Nurse P. Control of the timing of cell division in fission yeast. Cell size mutants reveal a second control pathway. Experimental cell research. 1978;115:317–329. doi: 10.1016/0014-4827(78)90286-0. [DOI] [PubMed] [Google Scholar]
- Forsburg SL. Overview of Schizosaccharomyces pombe. In: Ausubel Frederick M, , et al. Current protocols in molecular biology. Chapter 13 2003. Unit 13 14. [DOI] [PubMed] [Google Scholar]
- Fuchs F, Pau G, Kranz D, Sklyar O, Budjan C, Steinbrink S, Horn T, Pedal A, Huber W, Boutros M. Clustering phenotype populations by genome-wide RNAi and multiparametric imaging. Molecular systems biology. 2010;6:370. doi: 10.1038/msb.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan IM. The fission yeast microtubule cytoskeleton. Journal of cell science. 1998;111(Pt 12):1603–1612. doi: 10.1242/jcs.111.12.1603. [DOI] [PubMed] [Google Scholar]
- Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Current opinion in cell biology. 2008;20:71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J, Wood V, Jeffery L, Hoe KL, Kim DU, Park HO, Salas-Pino S, Heichinger C, Nurse P. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open biology. 2013;3:130053. doi: 10.1098/rsob.130053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn T, Sandmann T, Fischer B, Axelsson E, Huber W, Boutros M. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nature Methods. 2011;8:341–U391. doi: 10.1038/nmeth.1581. [DOI] [PubMed] [Google Scholar]
- Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nature cell biology. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, et al. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1826–1831. doi: 10.1073/pnas.0808843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Levy J, Ginzberg MB, Oh S, Lahav G, Kirschner MW. Dynamics extracted from fixed cells reveal feedback linking cell growth to cell cycle. Nature. 2013;494:480–483. doi: 10.1038/nature11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmer D, McHardy LM, Hoon S, Reberioux D, Giaever G, Nislow C, Roskelley CD, Roberge M. Combining chemical genomics screens in yeast to reveal spectrum of effects of chemical inhibition of sphingolipid biosynthesis. BMC microbiology. 2009;9:9. doi: 10.1186/1471-2180-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nature biotechnology. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer C, Fischer B, Billmann M, Huber W, Boutros M. Mapping genetic interactions in human cancer cells with RNAi and multiparametric phenotyping. Nature methods. 2013;10:427–431. doi: 10.1038/nmeth.2436. [DOI] [PubMed] [Google Scholar]
- Lee H, Jeon J, Ryu YS, Jeong JE, Shin S, Zhang T, Kang SW, Hong JH, Hur GM. Disruption of microtubules sensitizes the DNA damage-induced apoptosis through inhibiting nuclear factor kappaB (NF-kappaB) DNA-binding activity. Journal of Korean medical science. 2010;25:1574–1581. doi: 10.3346/jkms.2010.25.11.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJ, Lin YF, Chou HY, Yajima H, Fattah KR, Lee SC, Chen BP. Involvement of DNA-dependent protein kinase in normal cell cycle progression through mitosis. The Journal of biological chemistry. 2011;286:12796–12802. doi: 10.1074/jbc.M110.212969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I, Staub J, Setty TG, Nguyen NP, Paoletti A, Tran PT. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Molecular biology of the cell. 2005;16:1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Current opinion in cell biology. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- Mercer J, Snijder B, Sacher R, Burkard C, Bleck CK, Stahlberg H, Pelkmans L, Helenius A. RNAi screening reveals proteasome- and Cullin3-dependent stages in vaccinia virus infection. Cell reports. 2012;2:1036–1047. doi: 10.1016/j.celrep.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Mitchison JM. Growth during the cell cycle. International review of cytology. 2003;226:165–258. doi: 10.1016/s0074-7696(03)01004-0. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolau M, Levine AJ, Carlsson G. Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7265–7270. doi: 10.1073/pnas.1102826108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, Lander ES, Mitzenmacher M, Sabeti PC. Detecting novel associations in large data sets. Science. 2011;334:1518–1524. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon SA, Paoletti A. Mid1/anillin and the spatial regulation of cytokinesis in fission yeast. Cytoskeleton. 2012;69:764–777. doi: 10.1002/cm.21056. [DOI] [PubMed] [Google Scholar]
- Rohn JL, Sims D, Liu T, Fedorova M, Schock F, Dopie J, Vartiainen MK, Kiger AA, Perrimon N, Baum B. Comparative RNAi screening identifies a conserved core metazoan actinome by phenotype. The Journal of cell biology. 2011;194:789–805. doi: 10.1083/jcb.201103168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Lourenco PC, Snaith HA. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Current biology: CB. 2004;14:763–775. doi: 10.1016/j.cub.2004.03.042. [DOI] [PubMed] [Google Scholar]
- Scutari M. Learning Bayesian Networks with the bnlearn R Package. Journal of Statistical Software. 2010;35:1–22. [Google Scholar]
- Simpson JC, Joggerst B, Laketa V, Verissimo F, Cetin C, Erfle H, Bexiga MG, Singan VR, Heriche JK, Neumann B, et al. Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway. Nature cell biology. 2012;14:764–774. doi: 10.1038/ncb2510. [DOI] [PubMed] [Google Scholar]
- Singh G, Mémoli F, Carlsson GE. Topological Methods for the Analysis of High Dimensional Data Sets and 3D Object Recognition; Paper presented at: SPBG; 2007. [Google Scholar]
- Sommer C, Gerlich DW. Machine learning in cell biology - teaching computers to recognize phenotypes. Journal of cell science. 2013;126:5529–5539. doi: 10.1242/jcs.123604. [DOI] [PubMed] [Google Scholar]
- Spalding DJM, Harker AJ, Bayliss MK. Combining high-throughput pharmacokinetic screens at the hits-to-leads stage of drug discovery. Drug Discovery Today. 2000;5:70–76. doi: 10.1016/s1359-6446(00)00061-1. [DOI] [PubMed] [Google Scholar]
- Thery M, Jimenez-Dalmaroni A, Racine V, Bornens M, Julicher F. Experimental and theoretical study of mitotic spindle orientation. Nature. 2007;447:493–U496. doi: 10.1038/nature05786. [DOI] [PubMed] [Google Scholar]
- Troxell CL, Sweezy MA, West RR, Reed KD, Carson BD, Pidoux AL, Cande WZ, McIntosh JR. pkl1(+)and klp2(+): Two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Molecular biology of the cell. 2001;12:3476–3488. doi: 10.1091/mbc.12.11.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae) Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur JD, Heald R. Mitotic spindle scaling during Xenopus development by kif2a and importin alpha. eLife. 2013;2:e00290. doi: 10.7554/eLife.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Xie P, Li L, Xing G, Tian C, Yin Y, He F, Zhang L. ATM-mediated NuSAP phosphorylation induces mitotic arrest. Biochemical and biophysical research communications. 2011;404:413–418. doi: 10.1016/j.bbrc.2010.11.135. [DOI] [PubMed] [Google Scholar]
- Yin Z, Sadok A, Sailem H, McCarthy A, Xia X, Li F, Garcia MA, Evans L, Barr AR, Perrimon N, et al. A screen for morphological complexity identifies regulators of switch-like transitions between discrete cell shapes. Nature cell biology. 2013;15:860–871. doi: 10.1038/ncb2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Smith VA, Wang PP, Hartemink AJ, Jarvis ED. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics. 2004;20:3594–3603. doi: 10.1093/bioinformatics/bth448. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.