Abstract
Purpose
To determine whether uveal melanoma, the most common primary intraocular malignancy in adults, requires Notch activity for growth and metastasis.
Experimental Design
Expression of Notch pathway members was characterized in primary tumor samples and in cell lines, along with the effects of Notch inhibition or activation on tumor growth and invasion.
Results
Notch receptors, ligands, and targets were expressed in all five cell lines examined and in 30 primary uveal melanoma samples. Interestingly, the three lines with high levels of baseline pathway activity (OCM1, OCM3, and OCM8) had their growth reduced by pharmacologic Notch blockade using the γ-secretase inhibitor (GSI) MRK003. In contrast, two uveal melanoma lines (Mel285 and Mel290) with very low expression of Notch targets were insensitive to the GSI. Constitutively active forms of Notch1 and Notch2 promoted growth of uveal melanoma cultures and were able to rescue the inhibitory effects of GSI. MRK003 treatment also inhibited anchorage-independent clonogenic growth and cell invasion and reduced phosphorylation levels of STAT3 and extracellular signal-regulated kinase (Erk)1/2. Suppression of canonical Notch activity using short hairpin RNA targeting Notch2 or CBF1 was also able to reduce tumor growth and invasion. Finally, intraocular xenograft growth was significantly decreased by GSI treatment.
Conclusion
Our findings suggest that Notch plays an important role in inducing proliferation and invasion in uveal melanoma and that inhibiting this pathway may be effective in preventing tumor growth and metastasis.
Introduction
Uveal melanoma is the most common malignant intraocular tumor in adults, and up to 50% of patients die from metastatic disease within 10 years of initial diagnosis (1, 2). Prognostic molecular factors are beginning to be identified, and monosomy of chromosome 3 represents the genomic alteration most commonly associated with poor clinical outcome (3–5). Gene expression profiling has also been used to generate prognostic groups in several studies (6–8). Primary uveal melanomas can be separated using gene expression profiling into class 1 tumors associated with gain of chromosome 6p, low metastatic risk and better prognosis, and class 2 tumors with loss of chromosome 3 and 8p, higher likelihood of distant spread, and poor prognosis (1, 6). Although initial studies were based on these class distinctions on large data sets, a more focused panel of 12 discriminating genes has also been developed (9).
Although prognostic information is useful, effective therapies for metastatic disease are needed if patients with high-risk uveal melanomas are to experience improved survival. Treatments affecting specific signal transduction cascades required for tumor growth and invasion represent promising options. Pathways activated by the insulin-like growth factor (IGF), hepatocyte growth factor (HGF), and VEGF, and the signaling cascades downstream of G-protein–coupled receptors, have all been implicated in uveal melanoma initiation and spread (10–12), but it is unclear whether targeting these pathways will have an impact on metastatic disease. Mutations in the tumor suppressor BAP1 were recently reported to occur almost exclusively in metastasizing class 2 tumors (13). Furthermore, a recent study in zebrafish linked BAP1 to the Notch pathway (14). Consistent with these reports, we found that several Notch pathway members were preferentially upregulated in class 2 uveal melanomas. We have also recently shown that Notch activation can induce formation of pigmented, invasive uveal tumors in mice (15). On the basis of this, we explored the Notch pathway as a therapeutic target in metastatic uveal melanoma.
The Notch pathway controls diverse processes such as stem cell self-renewal, differentiation, and cell fate decisions in many organs, including pigmented and nonpigmented cells in the eye (16–18). Signaling is initiated by the interaction of cell surface Jagged and Delta ligands with Notch receptors on adjacent cells, which activates the pathway through several successive proteolytic cleavages (reviewed in ref. 17). The final cleavage occurs within the transmembrane domain and is catalyzed by the enzyme γ-secretase, which induces release of the intracellular domain (ICD) of Notch receptor and permits its translocation to the nucleus, where it forms a complex with CBF1 and MAML. These proteins form a heteromeric complex that induces the transcription of target genes including several in the Hairy and enhancer of split (Hes) and Hes-related repressor protein (Hey) families (17).
Aberrant Notch signaling has been identified in numerous tumor types, but the effects of the pathway depend upon the tissue and cellular context (19–21). The mechanism and role of Notch activation is perhaps best described in T-cell acute lymphoblastic leukemia, in which point mutations activating the receptor are commonly identified (20). Receptor amplification has also been found in brain tumors and a few other types of cancer (22), but in many tumor types, pathway activity is driven by canonical ligands or is triggered by unknown mechanisms (19, 21). In human cutaneous melanoma, activation of Notch1 promotes cell growth and tumor invasion in 3-dimensional spheroids (23); Notch1 activation also induces a transformed cellular phenotype in cutaneous melanocytes in vitro (24). Here, we examine the role of Notch signaling in uveal melanoma and show that it promotes proliferation, clonogenic growth, and invasion in tumor cells.
Materials and Methods
Cell culture and plasmids
Human uveal melanoma cell lines (OCM1, OCM3, OCM8, Mel285, Mel290), kindly provided by Dr. J. Niederkorn (UT Southwestern Medical Center, Dallas, TX), were cultured in RPMI-1640 medium as previously described (25), and their identity authenticated at the Johns Hopkins Molecular Core Laboratory through short tandem repeat (STR) analysis. With local Institutional Research Board (IRB) approval, excess tumor tissues not required for diagnosis were obtained from primary uveal melanoma tumors removed by enucleation at the Wilmer Eye Institute (Baltimore, MD) from 2004 to 2006 and snap frozen. Normal epidermal melanocytes, kindly provided by Dr. Meenhard Herlyn, via Dr. Akrit Sodhi, were maintained in 254 CF medium, supplemented with HMGS-2 (Cascade Biologics). Retroviruses were generated from CLEN1 and CLEN2 plasmids provided by Dr. Nicholas Gaiano and previously described (26). Lentiviruses encoding short hairpin RNAs (shRNA) against Notch2 and CBF1 (Thermo Fisher Scientific) were prepared in the PLKO.1 vector as previously described (27), with target sequences shown in Supplementary Table S1. The γ-secretase inhibitor (GSI) MRK003 was provided by Merck & Co., Inc. (28).
RNA extraction and quantitative real-time PCR
RNA extraction from cell lines or snap-frozen tumor tissues was carried out using the RNeasy Mini Kit (Qiagen) with on-column DNA digestion. Quantitative real-time PCR (qPCR) was carried out as previously described (29), with primer sequences shown in Supplementary Table S2. All reactions were carried out in triplicate on an iQ5 Multicolor real-time PCR detection system (Bio-Rad), using SYBR Green (Applied Biosystems) fluorescent dye, and normalized to β-actin mRNA levels.
Protein analysis
Cells were lysed in TNE buffer including protease inhibitor cocktail diluted 1:100 (Cytoskeleton Inc.). Nitro-cellulose membrane filters (Invitrogen) were incubated overnight at 4°C with the following antibodies: Hes1 1:700 (Aviva Systems Biology, #ARP32372; 1:700), GAPDH (1:5,000; RDI, #TRK5G4-6C5), cleaved caspase-9 (1:1,000; Cell Signaling Technology, #9505), β-actin (1:1,000; Sigma-Aldrich), and phospho-Erk1-2Thr202/Tyr204, phospho-AktSer473, STAT3Tyr705, Erk1/2, Akt (Cell Signaling Technology), all diluted 1:1,000. Immunohistochemical staining using anti-Hes1 rabbit polyclonal antibody diluted 1:100 (Santa Cruz Biotechnology, #sc-13844) was carried out on a tissue microarray as previously described (22) in tumor cores isolated from primary uveal melanomas after local IRB approval. Nuclear Hes1 immunoreactivity was scored in 73 samples by a board certified pathologist (C.G. Eberhart), and the intensity of staining scored as 0, no expression; 1+, mild/moderate expression; 2+, strong expression. Human fetal brain was used as a positive control for Hes1 staining.
Luciferase activity
CBF1 activity was measured in OCM3 cells transfected with the CBF-RE-luciferase plasmid containing a CBF1 responsive element. E1α-β-galactosidase construct was cotransfected to measure the efficiency of the transfection, whereas the CBF-BM (binding mutant)-luciferase construct was used as negative control. All these plasmids were kindly provided by Dr. Nicholas Gaiano. Transfection was conducted with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Two days after transfection and treatment with MRK003 at 2, 5, and 10 μmol/L, luciferase activity was measured using the Luciferase Reporter Assay System (Promega), according to the manufacturer’s instructions.
Cell growth, invasion, and migration assays
Cell growth was determined by MTS colorimetric assays. For soft agar clonogenic assays, cells were mixed with medium containing 0.5% agar (Invitrogen) and placed over 1% basal agar in 6-well plates; MRK003 (2 μmol/L) or dimethyl sulfoxide (DMSO) was added to each layer. Colonies were counted with the Artek Counter Model 880 (Artek Systems Corporation), after being stained overnight with 1 mg/mL p-Nitro Blue Tetrazolium Chloride (NBT; Usb) solution. Cellular invasion was analyzed over 14 hours using 6.5-mm diameter Falcon cell culture inserts (8 μm pore size; Becton Dickinson), precoated with Matrigel (diluted 1:100 in medium) in 24-well plates, with medium containing 10% FBS in the lower chamber and cells in serum-free medium in the upper part of the chamber. Data represent the mean (±SEM) of the number of cells counted in 8 random high-power field (HPF) in each of 3 independent experiments. Exposure to γ-irradiation was carried out using a Gammacell 3000 Elan (MDS Nordion). Viable cells were counted using Guava ViaCount, according to the manufacturer’s protocol (Guava Technologies Inc.). Cell-cycle and Annexin V analysis were carried out using the Guava instrument with Guava Cell Cycle and Nexin Reagent solutions and softwares.
In vivo tumorigenicity
Female athymic 5-week-old nude mice (NU/J, Jackson Laboratories) were deeply anesthetized with ketamine hydrochloride (Sigma-Aldrich) and 5 × 104 OCM1 cells were inoculated intravitreally in the right eye using a 33-gauge needle. Either 25 μg of MRK003 (1% of the 100 mg/kg dose recommended for systemic administration) or DMSO was coinjected with the cellular suspension. Mice were sacrificed after 3 weeks, and the eyes, lungs, and liver were analyzed microscopically for the presence of tumor by an experienced pathologist (C.G. Eberhart). Microphthalmia-associated transcription factor (MITF) immunostaining was carried out in the Johns Hopkins Pathology Department clinical laboratory using standard techniques. Cross-sectional tumor area was measured using a Spot Insight4 camera and Spot Advanced software (Diagnostic Instruments Inc.).
Statistical analysis
Unless otherwise noted, experiments were carried out in triplicate and data are presented as the mean ± SEM. Levels of significance were determined by 2-sided Student t test, with P values lower than 0.05 considered statistically significant. Statistical calculations were carried out using GraphPad Prism4 software.
Results
The Notch pathway is active in uveal melanoma tumors and cell lines
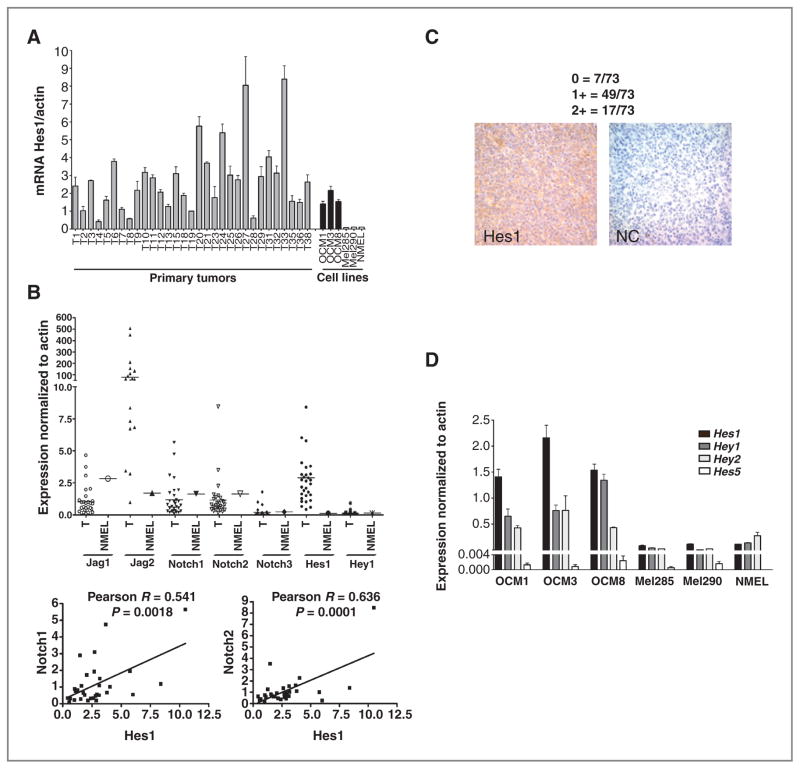
To investigate the role of Notch signaling in uveal melanoma, we first examined mRNA levels of the receptors Notch1, Notch2, and Notch3, the ligands Jag1 and Jag2, and the targets Hes1 and Hey1 in 30 snap-frozen tumors (Fig. 1A and B). These Notch pathway components were detected in almost all primary tumors, particularly the Notch1 and Notch2 receptors and Jag2 ligand. Consistent with activation of the pathway in vivo by receptor-mediated signaling, a statistically significant positive correlation was found between the expression of Hes1 and levels of the Notch1 (Pearson R =0.54, P =0.002) and Notch2 (Pearson R = 0.64, P = 0.0001) receptors (Fig. 1B). Clinical and pathologic information was only available for a minority of these tumors, thus we were not able to directly correlate Notch activity with outcome or other clinical variables in this cohort. However, examination of our previously published data sets (6) revealed a number of Notch pathway genes, as defined by KEGG, which were significantly altered in the metastasis-prone class 2 tumors as compared with class 1. These transcripts were primarily upregulated in the most aggressive tumors and included Notch3 (2.0-fold increase), Jag2 (1.9-fold increase), and DLL4 (1.8-fold increase). Baseline expression of Notch pathway members in untransformed melanocytes was established using primary epidermal melanocytes (Fig. 1A, B, and D). Expression of Notch pathway targets was also confirmed by immunohistochemistry, using surgical uveal melanoma samples collected on a tissue microarray (Fig. 1C). Nuclear Hes1 immunoreactivity was examined in 73 tumors, with mild to moderate expression observed in 67% (49 of 73), strong expression in 23% (17 of 73), and no staining in 10% (7 of 73). These data show Notch pathway activation in most of the primary uveal melanoma tissues that we analyzed.
Figure 1.
Notch pathway components are expressed in uveal melanoma cell lines and primary tumors. A, mRNA levels of Hes1 were analyzed by quantitative PCR in 30 snap-frozen primary tumors, 5 uveal melanoma cell lines, and normal cutaneous melanocytes (NMEL). B, mRNA transcripts of additional Notch receptors, ligands, and target genes were detected in the 30 primary uveal melanomas (T) and in normal epidermal melanocytes (NMEL; top); Pearson correlation coefficient R was determined between Hes1 and Notch1, Notch2 mRNA levels in the primary uveal melanomas (bottom). C, nuclear Hes1 immunoreactivity was examined in 73 tumors collected on a tissue microarray with strong Hes1 immunostaining shown in the left and a no primary antibody negative control on the right. D, examination of mRNA expression of the Hes1, Hes5, Hey1, Hey2 genes in 5 uveal melanoma cell lines and normal cutaneous melanocytes revealed that three uveal melanoma lines had elevated levels of multiple Notch targets.
We also examined expression of Notch pathway members in 5 established uveal melanoma cell lines (Fig. 1D and Supplementary Fig. S1A and S1B). As in the primary tumors, Notch1 and Notch2 were in general more highly expressed than Notch3 (Supplementary Fig. S1A). Notch4 was either not present or showed extremely low levels in the 5 cell lines and the subset of primary tumors tested (data not shown). Interestingly, the Notch ligand Jag1 and the target genes Hes1, Hey1, and Hey2 were all significantly elevated in OCM1, OCM3, and OCM8 cell lines as compared with Mel285 and Mel290 (Fig. 1D and Supplementary Fig. S1B), suggesting that the first 3 lines have higher levels of pathway activity. Primary epidermal melanocytes contained low levels of Hes1, Hey1, and Hey2 similar to those found in the Mel285 and Mel290 cell lines, whereas Hes5 was absent in these normal cells (Fig. 1D).
Notch blockade inhibits proliferation and invasion in uveal melanoma cell lines with high pathway activity
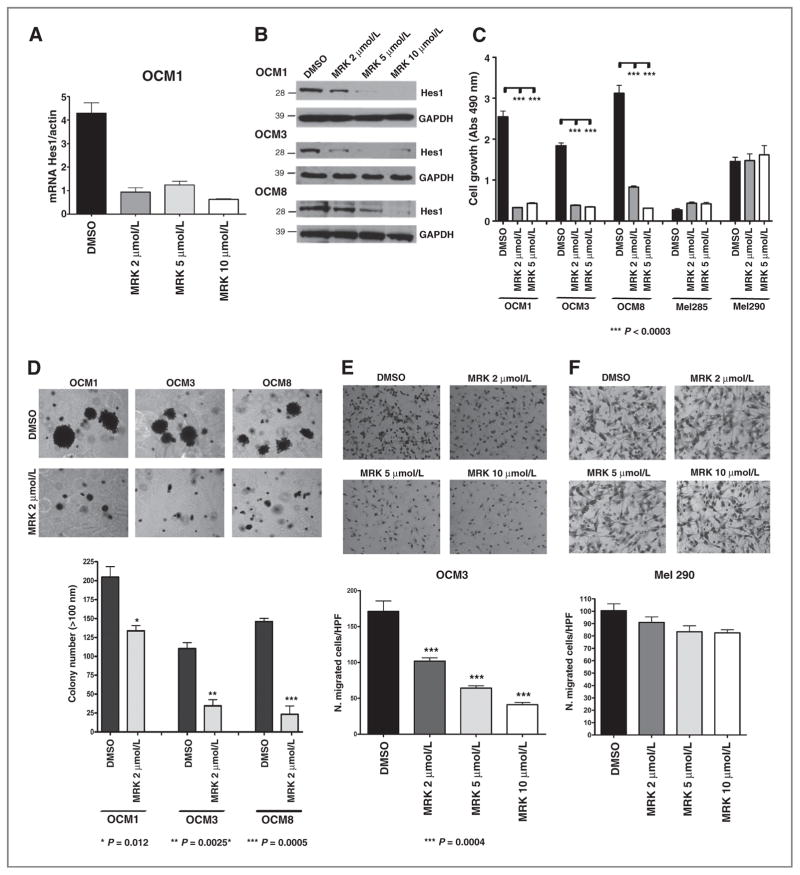
To address the functional role of Notch in our tumor lines, we conducted pharmacologic loss-of-function studies using MRK003 to repress the pathway by preventing the intramembranous proteolytic cleavage of Notch receptors (17, 28). A profound reduction in expression of pathway target transcripts was identified after 48 hours of treatment, including suppression of Hes1 in OCM1, OCM3, and OCM8 cells (Fig. 2A and Supplementary Fig. S2A and S2B) and Hey1 in OCM1 and OCM3 cells (Supplementary Fig. S2C and S2D). These data were confirmed by Western blot analysis, with partial loss of Hes1 protein in OCM1, OCM3, and OCM8 cells using 2 μmol/L MRK003 and complete suppression at higher levels of the compound (Fig. 2B). Interestingly, the low baseline levels of Hes1 mRNA in Mel285 and Mel290 cells were not reduced after treatment with GSI (data not shown). To further confirm suppression of canonical Notch activity by the GSI, we analyzed CBF1 activity in OCM3 cells treated for 48 hours with MRK003 using a luciferase reporter system. We observed that GSI reduced in a dose-dependent manner the baseline level of luciferase expression driven by the CBF-RE-luciferase construct (Supplementary Fig. S2E), whereas no expression was detected in cells transfected with CBF-BM, which contains a point mutation that renders the CBF1 responsive element inactive.
Figure 2.
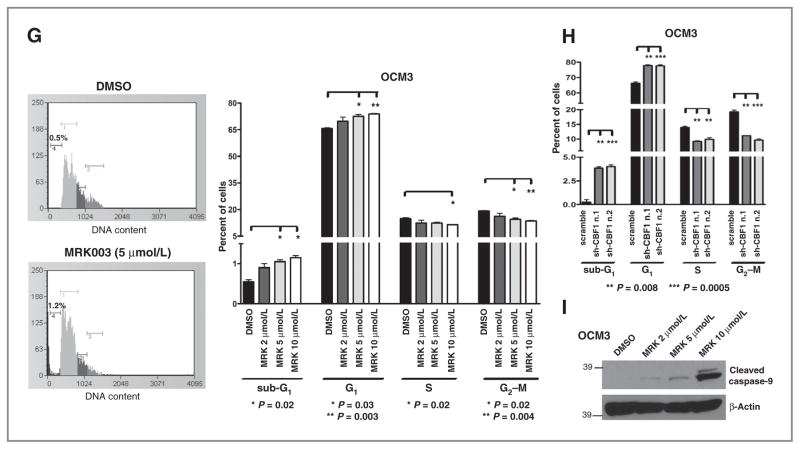
Treatment with MRK003 reduces Hes1 mRNA and protein expression and suppresses uveal melanoma growth and invasion. A and B, Hes1 mRNA and protein levels were reduced in OCM1 cells treated with MRK003 at 2, 5, and 10 μmol/L for 48 hours. C, MTS assay of 5 uveal melanoma cell lines treated with MRK003 at 2 and 5 μmol/L for 7 days revealed that growth was only inhibited in cells with high baseline Notch activity. D, clonogenic growth in soft agar assay was significantly reduced by 2 μmol/L MRK003 (*, P = 0.01; **, P = 0.003; ***, P = 0.0005). E and F, transwell assays revealed significantly decreased invasion of OCM3 cells treated for 24 hours with MRK003 as compared with DMSO at the indicated doses (***, P < 0.0004), but no significant changes in Mel290 cells with lower Notch activity. G, cell-cycle analysis of OCM3 cells after 48-hour treatment with MRK003 at 2, 5, and 10 μmol/L, with data from triplicate experiments in right. H, cell-cycle analysis of OCM3 cells 72 hours after infection with shCBF1. I, Western blot analysis reveals increased levels of cleaved caspase-9 in OCM3 cells treated with MRK003 for 96 hours. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To determine whether GSI treatment might also affect cell growth, uveal melanoma lines were treated for 7 days using 2 and 5 μmol/L MRK003. The OCM1, OCM3, and OCM8 lines with high baseline levels of Notch target expression (Fig. 1C) had their growth inhibited by pharmacologic pathway blockade (Fig. 2C). In contrast, growth of Mel285 and Mel290 cells, which had low levels of Hes1 and Hey1 mRNA, was not affected by the treatment, although the slower baseline growth of these lines could make them less sensitive.
Notch blockade also suppressed the clonogenicity and invasive capacity of the cell lines with high levels of pathway activity. OCM1, OCM3, and OCM8 cells were treated with 2 μmol/L MRK003 or DMSO for 24 hours and equal numbers of viable cells were seeded in soft agar and grown for 3 weeks in the presence of GSI or DMSO. We observed significant reductions in the number of colonies formed after treatment with 2 μmol/L MRK003, with 40% inhibition in OCM1 (P = 0.01), 70% in OCM3 (P = 0.002), and 75% in OCM8 (P = 0.0005; Fig. 2D). In a separate experiment, additional further reductions were seen at higher doses in OCM8 cells (Supplementary Fig. S2F). Mel285 and Mel290 cells did not form colonies in soft agar.
Notch pathway inhibition using MRK003 also significantly suppressed invasion (P < 0.0004) in a dose-dependent manner in OCM3 cells as measured using Transwell assays (Fig. 2E). Invasion of OCM1 and OCM8 cultures was also inhibited by MRK003 (data not shown), whereas no inhibition in cell invasion was observed in Mel290 cells after treatment with MRK003 (Fig. 2F).
To investigate a potential mechanism of growth inhibition, we conducted cell-cycle analysis in OCM3 cells treated for 48 hours with MRK003 (Fig. 2G). We found a significant increase in the sub-G1 fraction at 5 and 10 μmol/L (P = 0.02), suggesting an increase in cell death. Similar data were obtained in OCM1 cells (data not shown). To confirm that Notch pathway blockade was inducing cell death, we infected OCM3 and OCM1 cells with shRNAs targeting CBF1, the DNA-binding mediator of canonical Notch signaling. Two separate constructs which significantly reduced both CBF1 and Hey1 mRNA levels (Supplementary Fig. S3A), substantially induced the sub-G1 fraction (Fig. 2H) in OCM3 cells and also significantly increased the fraction of cells positive for Annexin V, a marker of early apoptosis, both in OCM1 and in OCM3 cells (Supplementary Fig. S3B). A dose-dependent increase in the active cleaved form of caspase-9 was noted after treating OCM3 cells for 96 hours with MRK003, suggesting that Notch signaling blockade induces activation of the intrinsic pathway of apoptosis (Fig. 2I). However, some nonspecific toxic effects may be present at the highest dose used (10 μmol/L). Finally, both GSI (Fig. 2G) and CBF1 knockdown (Fig. 2H) resulted in a modest induction of the G1 fraction with a corresponding reduction in S-phase and G2–M fractions.
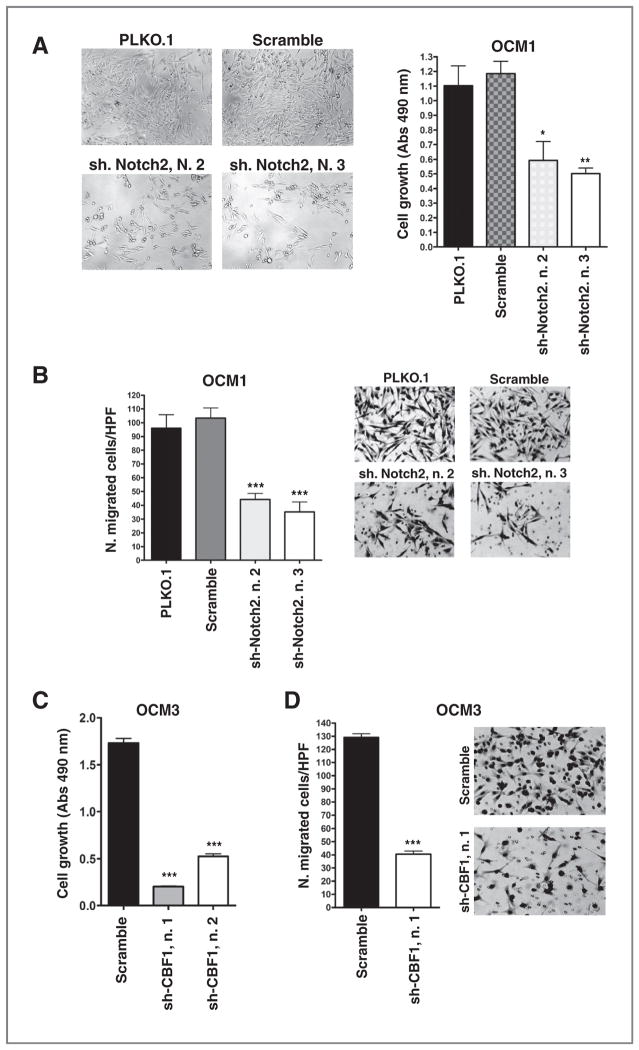
To further validate the role of Notch signaling in promoting uveal melanoma growth and invasion, we used shRNA to knockdown the expression of Notch2, the most abundant Notch receptor in our lines (Supplementary Fig. S1A). Notch2 and Hey1 mRNA levels were reduced by 50% or more in OCM1 cells by 2 separate constructs as compared with scrambled shRNA or PLKO.1 empty vector controls (Supplementary Fig. S3C). No effect was seen on Notch1 mRNA levels (Supplementary Fig. S3C), further supporting the specificity of the effect. A statistically significant reduction in cell growth (Fig. 3A) and invasion (Fig. 3B) was noted in the cells with Notch2 suppression caused by either of these constructs. Moreover, the shCBF1 constructs significantly suppressed growth of OCM3 cells (Fig. 3C) measured by MTS assay, as well their invasive capacity in Transwell assays (Fig. 3D).
Figure 3.
Notch signaling inhibition reduces cell growth and invasion. A and C, MTS assay revealed cell growth inhibition in shNotch2 (A) or shCBF1 (C) infected cells compared with scrambled shRNA or PLKO.1 empty vector infected cells after 5 days of culture (*, P = 0.04; **, P = 0.003; ***, P < 0.0001). B and D, transwell assay revealed that cell invasion was reduced in cells infected with shNotch2 (B) or shCBF1 (D) infected cells compared with scrambled shRNA. P values were calculated versus scrambled shRNA (***, P < 0.0001).
Constitutive Notch activation promotes uveal melanoma cell growth and rescues the inhibitory effects of GSI
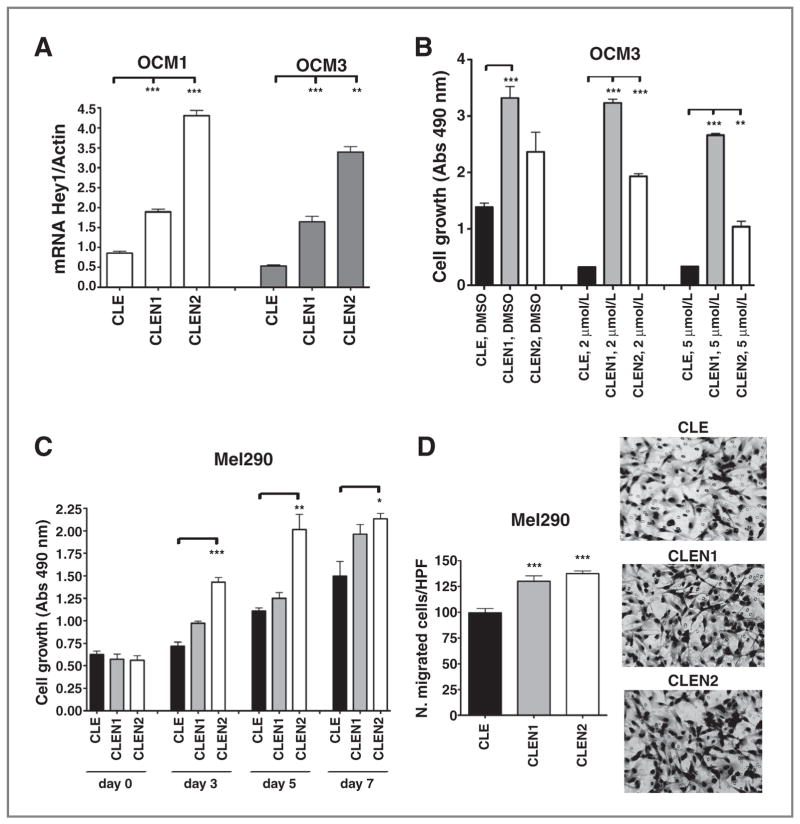
To examine the effects of increased Notch signaling in uveal melanoma cells, we infected OCM1 and OCM3 cultures with retroviral CLEN1 or CLEN2 constructs encoding the constitutively active ICD of Notch1 (NICD1) or Notch2 (NICD2), respectively. Empty CLE vector was used as a control. Upregulation of the Notch pathway target Hey1 was confirmed in CLEN1- and CLEN2-expressing cells (Fig. 4A), and cell growth was significantly induced by both constructs in the OCM3 line (Fig. 4B). A less pronounced growth-promoting effect was seen in OCM1 cells (Supplementary Fig. S4A). We also used these cells expressing NICD to verify that the biologic effects of γ-secretase inhibition were due to regulation of Notch signaling, as the γ-secretase complex can process other targets as well. Both CLEN1 and CLEN2 could largely rescue the inhibitory effect of MRK003 on cell growth in OCM3 (Fig. 4B) and partially rescued growth in OCM1 cells (Supplementary Fig. S4A). We also observed that the overexpression of NICD2 totally rescued the inhibitory effect of MRK003 on cellular invasion in OCM3 cells whereas NICD1 produced a partial rescue (Supplementary Fig. S4B). The fact that these truncated Notch receptors were able to abrogate the inhibition of growth and invasion because of GSI treatment suggests that MRK003 is acting at least in part through Notch inhibition.
Figure 4.
Constitutive Notch activation promotes growth and rescues MRK003 inhibitory effects. A, Hey1 mRNA levels were increased in OCM1 and OCM3 cells after infection with CLEN1 or CLEN2 constructs (**, P = 0.003; ***, P = 0.0005). B, MTS assay conducted in OCM3 cells infected with CLE, CLEN1, or CLEN2 vectors, after 7 days of treatment with MRK003, shows that growth inhibition induced by GSI is rescued by Notch pathway activation (**, P = 0.002; ***, P < 0.0008). C and D, CLEN1 and CLEN2 constructs also promote cell growth and invasion as determined respectively by MTS (C) and Transwell (D) assays in Mel290 cells (*, P = 0.02; **, P < 0.006; ***, P = 0.0009).
We confirmed the potency of Notch pathway in enhancing cell growth and invasion in uveal melanoma cells, through gain-of-function experiments carried out in Mel290 cells, which show minimal Notch activity (Fig. 1C). Infection with retroviral CLEN1 or CLEN2 constructs induced Notch pathway activation, as found by measuring Hey1 mRNA levels, which were increased by 3 and 6 times in Mel290 infected respectively with CLEN1 or CLEN2 constructs (Supplementary Fig. S4C). Cellular growth and invasion were analyzed by MTS and Transwell assays, respectively. The constitutively active form of Notch2 receptor strongly increased cellular growth in adherent conditions, as shown by MTS assay conducted after 3, 5, and 7 days of culture (Fig. 4C). Both the constructs were able to significantly enhance cell invasion by 30% and 38%, respectively (P = 0.0009), as shown in Fig. 4D.
Notch blockade affects Akt, Erk, and STAT3 activity in uveal melanoma cells
Because Notch pathway suppression by GSI treatment or shRNA inhibited uveal melanoma growth, we analyzed several potential downstream targets associated with proliferation and survival of tumor cells previously shown by our group and others to be affected by Notch blockade in various types of cancer (19, 21, 30, 31). We observed a substantial reduction in phospho-AktSer473, phospho-Erk1-2Thr202/Tyr204, and phospho-STAT3Tyr705 following MRK003 treatment at 2 and 5 μmol/L for 48 hours (Supplementary Fig. S5A), suggesting that the phosphoinositide 3-kinase (PI3K), mitogen-activated protein kinase (MAPK), and JAK/STAT pathways may be regulated by Notch in uveal melanoma. Because inhibition of γ-secretase can affect pathways other than Notch, we also examined these proteins following knockdown of CBF1 in OCM3 cells using shRNA and found that phosphorylation of extracellular signal-regulated kinase (Erk)1/2 and STAT3 was greatly reduced (Supplementary Fig. S5B). In contrast, Akt phosphorylation was reduced in some experiments by shCBF1, but not in others (data not shown), and it may not be regulated by the canonical Notch cascade in these cells.
Modulation of orthotopic xenograft growth and metastasis by pharmacologic Notch blockade
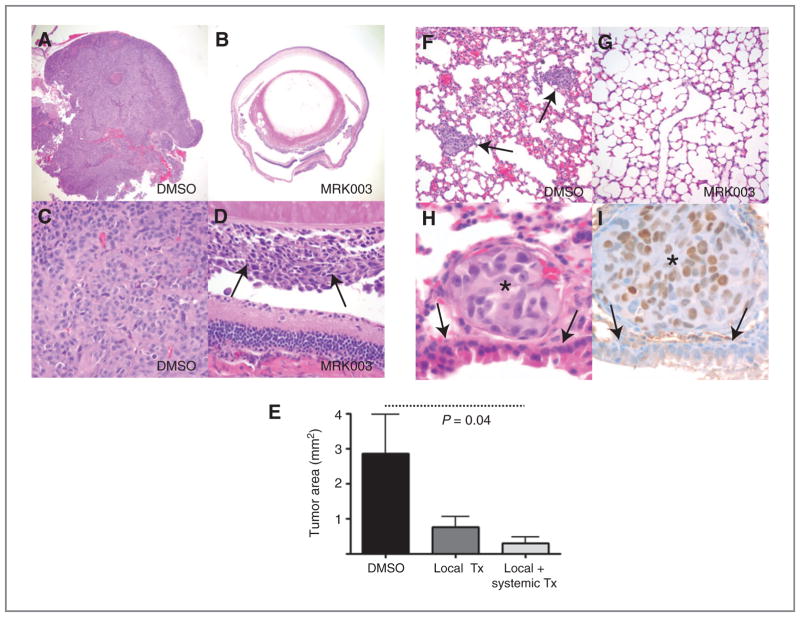
Given the effects of Notch blockade on the growth and invasion of uveal melanoma cells in vitro, we examined the ability of MRK003 to inhibit in vivo tumor formation and metastasis. Xenografts were established by intravitreal injection of OCM1 cells in the right eye of immunocompromised (nude) mice. Two treatment groups of 10 mice each were tested. In the first, animals received intraocular therapy with MRK003 at the time of xenograft establishment, whereas the second cohort received both this local therapy and subsequent systemic weekly GSI treatments via oral gavage. A control group of 10 animals received only local vehicle (DMSO) treatment. All animals were sacrificed after 3 weeks because of the development of very large masses in the eyes of several mice in the control group. Microscopic examination confirmed the presence of large pigmented melanoma xenografts filling the eyes of most vehicle-treated animals, whereas tumors in both treatment groups were smaller—significantly so in those receiving both local and systemic therapy (P = 0.04; Fig. 5A–E). Representative sections from the liver and lungs were also examined, and multiple lung metastasis were present in 2 of 10 vehicle-treated mice but none from the 20 animals receiving GSI (Fig. 5F and G). Melanoma metastasis in the mouse lung tissue were immunostained with an antibody recognizing the human form of the MITF, which is known to regulate the differentiation and development of the melanocytes (32). We observed selective nuclear staining in the tumor metastases, confirming their human origin (Fig. 5H and I).
Figure 5.
Notch pathway inhibition affects lung metastasis. A and B, a large tumor filled the entire eye 3 weeks after intravitreal injection of OCM1 cells treated with vehicle (DMSO), whereas only microscopic disease was present in most eyes receiving MRK003 at the time of xenograft initiation (original magnification 40× for both panels). C and D, higher magnification views showing solid sheets of melanoma cells in control eyes, but only focal tumor growth (D, arrows) following local MRK003 treatment (original magnification 400×). E, intraocular xenograft area was significantly reduced (P = 0.04) by combined local and systemic MRK003 treatment, whereas the changes in tumors treated locally alone was not significant. F and G, multiple melanoma metastases were identified in the lungs of 2 of 10 control animals (F, arrows), but none in the 20 mice treated with MRK003 (G, original magnification 200×). H and I, the metastatic tumor nodules were composed of cells with enlarged nuclei and prominent nucleoli (asterisks) as compared with normal adjacent bronchial cells (arrows, original magnification 1,000×). Immunostains for the MITF were positive in tumor cells (I).
Effect of the combination of GSI treatment with γ-radiation
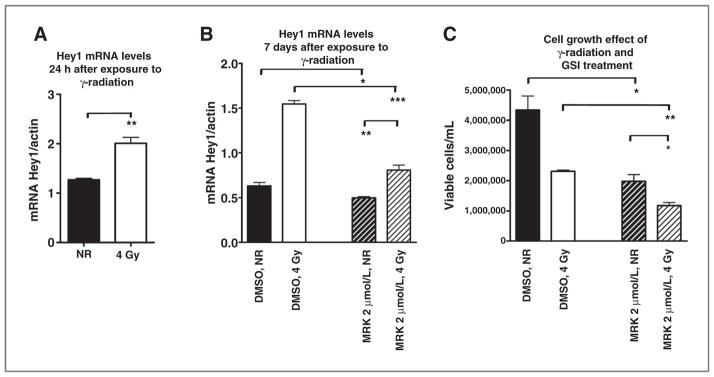
Because radiotherapy represents a mainstay of uveal melanoma treatment and Notch has been suggested to modulate resistance to radiation in other tumor types (33), we investigated the effects of combined γ-radiation and GSI treatment in OCM1 cells. Consistent with the prior report that Notch signaling is induced by radiation in gliomas, we found that exposure to γ-radiation at 4 Gy increased the expression of the Notch pathway target Hey1 after 24 hours (Fig. 6A). Interestingly, the increase in Hey1 levels persisted 7 days after radiation exposure (Fig. 6B) but could be suppressed by 2 μmol/L MRK003 (P = 0.0004). We also compared the combination of γ-radiation and GSI treatment with the effects of γ-radiation alone on cell growth. OCM1 cells were treated with 2 μmol/L MRK003 or DMSO and exposed to 0 or 4 Gy of γ-radiation in a single dose, and viable cells were counted 7 days after exposure to radiation. Combined treatment was more effective in suppressing growth than either agent alone, suggesting the possibility of a combined clinical regimen (Fig. 6C).
Figure 6.
Combination of MRK003 treatment with γ-radiation. A, Hey1 mRNA levels were significantly increased in OCM1 cells 24 hours after exposure to 4 Gy of γ-radiation as compared with nonirradiated cells (NR) cells (**, P < 0.005). B, Hey1 mRNA levels remained elevated 7 days after radiation, but could be suppressed by MRK003 (*, P = 003; **, P = 0.006; ***, P = 0.0004). C, OCM1 cells were treated with 2 μmol/L MRK003 or DMSO for 5 hours and then exposed to 0 or 4 Gy of γ-radiation in a single dose. Viable cells were counted 7 days after radiation exposure in triplicate experiments (*, P = 0.045; **, P = 0.01).
Discussion
Up to half of patients with uveal melanoma die from hematogenous metastases of the liver and other organs, thus improved treatments for metastatic disease are clearly needed (1, 2). Prompted by the upregulation of Notch pathway members in metastasis-prone class 2 tumors and the association between Notch and BAP1 in normal development (13, 14), we examined whether Notch activity was required in uveal melanoma. We found that inhibition of Notch signaling using small-molecule inhibitors of the γ-secretase complex, or shRNA targeting the receptor Notch2 or the transcriptional cofactor CBF1, slowed tumor growth and reduced clonogenic capacity and invasion in vitro. In contrast, introduction of constitutively active forms of Notch1 and Notch2 promoted growth of uveal melanoma cell lines and rescued the effects of the GSI MRK003.
Interestingly, only cell lines with higher levels of pathway target mRNAs showed this ongoing requirement for Notch signaling. These GSI-sensitive lines had levels of Notch pathway targets similar to those seen in snap-frozen primary tumors, suggesting that they represent biologically relevant models in terms of their degree of Notch activity, although it will be important to confirm this at the protein level. Our analysis of previously published oligonucleotide expression array data from primary uveal melanomas revealed significant elevations of Notch ligands (Jag2) and receptors (Notch3) in the metastatic tumor cohort, providing additional support for the notion that this pathway represents a novel potential therapeutic target. Finally, our preliminary in vivo analysis suggests that Notch inhibition using MRK003 significantly slows tumor growth in orthotopic xenograft models and may block hematogeneous spread, although models which metastasize more efficiently will need to be tested to establish the statistical significance of the latter finding. It will also be important to initiate treatment at later time points to more accurately model human clinical disease.
Notch activation has previously been implicated in the growth and survival of cutaneous melanoma. Early reports described increased expression of Notch pathway members in melanoma cells as compared with normal melanocytes (34) and revealed that a tripeptide GSI could induce apoptosis in melanoma cells (35). Subsequent studies showed that the Notch target Hes1 promotes survival of melanocyte stem cells (36), that active Notch1 can transform primary human melanocytes (24), and that transcription factors such as BRN2 and MITF, which play an important role in melanoma lineage specification, can regulate the Notch pathway (32). Interactions between the embryonic morphogens Nodal and Notch have also been suggested to induce the aggressive melanoma phenotype (37–38). Uveal melanoma, however, are distinct from cutaneous melanoma in many ways, and little is known about the potential role of Notch in tumors arising in the eye. Thus, although our data are broadly consistent with prior studies of cutaneous melanomas, they are important in suggesting that melanocytic tumors arising from other sites may also require ongoing Notch activity for their growth and metastatic spread.
In summary, we have shown that Notch signaling promotes the malignant phenotype of uveal melanoma cells and that pharmacologic or genetic blockade of the pathway can inhibit tumor growth and invasion. The invasive behavior associated with Notch activity may be due in part to the induction of Akt, Erk, and JAK/STAT pathways. These in vitro and in vivo studies also suggest that therapies targeting Notch may be useful in new treatments for the most common intraocular tumors in adults.
Supplementary Material
Translational Relevance.
Approximately half of patients diagnosed with uveal melanoma will die from hematogeneous metastases of the liver and other organs, and no curative therapies exist for tumors which have spread outside the eye. We show that the Notch cascade is active in uveal melanomas and that inhibition of the pathway using genetic manipulation or small molecules suitable for clinical translation can slow tumor growth and invasion. Our studies suggest that γ-secretase inhibitors or other agents targeting Notch may represent effective new therapies for patients with uveal melanoma, the most common primary intra-ocular malignancy in adults.
Acknowledgments
The authors thank Dr. Jerry Niederkorn for providing OCM1, OCM3, OCM8 (Kan-Mitchell and colleagues, 1989) and Mel285, Mel290 cell lines (Verbik and colleagues, 1997). The authors also thank Naheed Gul and Antoinette Price for technical assistance and Drs. Meenhard Herlyn and Akrit Sodhi for providing normal epidermal melanocytes.
Grant Support
This research was supported in part by Research to Prevent Blindness and the ABB Foundation.
Footnotes
Disclosure of Potential Conflicts of Interest
C.G. Eberhart discloses a patent licensed to Stemline Therapeutics. No potential conflicts of interest were disclosed by other authors.
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
References
- 1.Landreville S, Agapova OA, Harbour JW. Emerging insights into the molecular pathogenesis of uveal melanoma. Future Oncol. 2008;4:629–36. doi: 10.2217/14796694.4.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shields CL, Furuta M, Thangappan A, Nagori S, Mashayekhi A, Lally DR, et al. Metastasis of uveal melanoma millimeter-by-millimeter in 8033 consecutive eyes. Arch Ophthalmol. 2009;127:989–98. doi: 10.1001/archophthalmol.2009.208. [DOI] [PubMed] [Google Scholar]
- 3.Prescher G, Bornfeld N, Hirche H, Horsthemke B, Jockel KH, Becher R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347:1222–5. doi: 10.1016/s0140-6736(96)90736-9. [DOI] [PubMed] [Google Scholar]
- 4.Damato B, Dopierala JA, Coupland SE. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin Cancer Res. 2010;16:6083–92. doi: 10.1158/1078-0432.CCR-10-2076. [DOI] [PubMed] [Google Scholar]
- 5.Shields CL, Ganguly A, Bianciotto CG, Turaka K, Tavallali A, Shields JA. Prognosis of uveal melanoma in 500 cases using genetic testing of fine-needle aspiration biopsy specimens. Ophthalmology. 2011;118:396–401. doi: 10.1016/j.ophtha.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 6.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–9. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrausch U, Martus P, Tonnies H, Bechrakis NE, Lenze D, Wansel S, et al. Significance of gene expression analysis in uveal melanoma in comparison to standard risk factors for risk assessment of subsequent metastases. Eye (Lond) 2008;22:997–1007. doi: 10.1038/sj.eye.6702779. [DOI] [PubMed] [Google Scholar]
- 8.van Gils W, Lodder EM, Mensink HW, Kiliç E, Naus NC, Brüggenwirth HT, et al. Gene expression profiling in uveal melanoma: two regions on 3p related to prognosis. Invest Ophthalmol Vis Sci. 2008;49:4254–62. doi: 10.1167/iovs.08-2033. [DOI] [PubMed] [Google Scholar]
- 9.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12:461–8. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakalian S, Marshall JC, Logan P, Faingold D, Maloney S, Di Cesare S, et al. Molecular pathways mediating liver metastasis in patients with uveal melanoma. Clin Cancer Res. 2008;14:951–6. doi: 10.1158/1078-0432.CCR-06-2630. [DOI] [PubMed] [Google Scholar]
- 11.Crosby MB, Yang H, Gao W, Zhang L, Grossniklaus HE. Serum vascular endothelial growth factor (VEGF) levels correlate with number and location of micrometastases in a murine model of uveal melanoma. Br J Ophthalmol. 2011;95:112–7. doi: 10.1136/bjo.2010.182402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tse WK, Eisenhaber B, Ho SH, Ng Q, Eisenhaber F, Jiang YJ. Genome-wide loss-of-function analysis of deubiquitylating enzymes for zebra-fish development. BMC Genom. 2009;10:637. doi: 10.1186/1471-2164-10-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierfelice TJ, Schreck KC, Dang L, Asnaghi L, Gaiano N, Eberhart CG. Notch3 activation promotes invasive glioma formation in a tissue site-specific manner. Cancer Res. 2011;71:1115–25. doi: 10.1158/0008-5472.CAN-10-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schouwey K, Aydin IT, Radtke F, Beermann F. RBP-Jkappa-dependent Notch signaling enhances retinal pigment epithelial cell proliferation in transgenic mice. Oncogene. 2011;30:313–22. doi: 10.1038/onc.2010.428. [DOI] [PubMed] [Google Scholar]
- 17.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Austin CP, Feldman DE, Ida JA, Jr, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–50. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- 19.Pannuti A, Foreman K, Rizzo P, Osipo C, Golde T, Osborne B, et al. Targeting Notch to target cancer stem cells. Clin Cancer Res. 2010;16:3141–52. doi: 10.1158/1078-0432.CCR-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aster JC, Blacklow SC, Pear WS. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol. 2011;223:262–73. doi: 10.1002/path.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch U, Radtke F. Notch signaling in solid tumors. Curr Top Dev Biol. 2010;92:411–55. doi: 10.1016/S0070-2153(10)92013-9. [DOI] [PubMed] [Google Scholar]
- 22.Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, et al. Notch1 and Notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–93. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 23.Liu ZJ, Xiao M, Balint K, Smalley KS, Brafford P, Qiu R, et al. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer Res. 2006;66:4182–90. doi: 10.1158/0008-5472.CAN-05-3589. [DOI] [PubMed] [Google Scholar]
- 24.Pinnix CC, Lee JT, Liu ZJ, McDaid R, Balint K, Beverly LJ, et al. Active Notch1 confers a transformed phenotype to primary human melanocytes. Cancer Res. 2009;69:5312–20. doi: 10.1158/0008-5472.CAN-08-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Folberg R, Kadkol SS, Frenkel S, Valyi-Nagy K, Jager MJ, Pe’er J, et al. Authenticating cell lines in ophthalmic research laboratories. Invest Ophthalmol Vis Sci. 2008;49:4697–701. doi: 10.1167/iovs.08-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaiano N, Kohtz JD, Turnbull DH, Fishell G. A method for rapid gain-of-function studies in the mouse embryonic nervous system. Nat Neurosci. 1999;2:812–9. doi: 10.1038/12186. [DOI] [PubMed] [Google Scholar]
- 27.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–71. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis HD, Leveridge M, Strack PR, Haldon CD, O’neil J, Kim H, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem Biol. 2007;14:209–19. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 29.Schreck KC, Taylor P, Marchionni L, Gopalakrishnan V, Bar EE, Gaiano N, et al. The Notch target Hes1 directly modulates Gli1 expression and Hedgehog signaling: a potential mechanism of therapeutic resistance. Clin Cancer Res. 2011;16:6060–70. doi: 10.1158/1078-0432.CCR-10-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li YM, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–52. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 31.Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thurber AE, Douglas G, Sturm EC, Zabierowski SE, Smit DJ, Ramakrishnan SN, et al. Inverse expression states of the BRN2 and MITF transcription factors in melanoma spheres and tumour xenografts regulate the NOTCH pathway. Oncogene. 2011;30:3036–48. doi: 10.1038/onc.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Wakeman TP, Lathia JD, Hjelmeland AB, Wang XF, White RR, et al. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28:17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–82. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 35.Qin JZ, Stennett L, Bacon P, Bodner B, Hendrix MJ, Seftor RE, et al. p53-independent NOXA induction overcomes apoptotic resistance of malignant melanomas. Mol Cancer Ther. 2004;3:895–902. [PubMed] [Google Scholar]
- 36.Moriyama M, Osawa M, Mak SS, Ohtsuka T, Yamamoto N, Han H, et al. Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. J Cell Biol. 2006;173:333–9. doi: 10.1083/jcb.200509084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardy KM, Kirschmann DA, Seftor EA, Margaryan NV, Postovit LM, Strizzi L, et al. Regulation of the embryonic morphogen Nodal by Notch4 facilitates manifestation of the aggressive melanoma phenotype. Cancer Res. 2010;70:10340–50. doi: 10.1158/0008-5472.CAN-10-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strizzi L, Hardy KM, Kirsammer GT, Gerami P, Hendrix MJ. Embryonic signaling in melanoma: potential for diagnosis and therapy. Lab Invest. 2011;91:819–24. doi: 10.1038/labinvest.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.