Abstract
Background
Glioblastoma (GBM) is the most common and aggressive form of brain cancer. Our previous studies demonstrated that combined inhibition of HDAC and KDM1A increases apoptotic cell death in vitro. However, whether this combination also increases death of the glioma stem cell (GSC) population or has an effect in vivo is yet to be determined. Therefore, we evaluated the translational potential of combined HDAC and KDM1A inhibition on patient-derived GSCs and xenograft GBM mouse models. We also investigated the changes in transcriptional programing induced by the combination in an effort to understand the induced molecular mechanisms of GBM cell death.
Methods
Patient-derived GSCs were treated with the combination of vorinostat, a pan-HDAC inhibitor, and tranylcypromine, a KDM1A inhibitor, and viability was measured. To characterize transcriptional profiles associated with cell death, we used RNA-Seq and validated gene changes by RT-qPCR and protein expression via Western blot. Apoptosis was measured using DNA fragmentation assays. Orthotopic xenograft studies were conducted to evaluate the effects of the combination on tumorigenesis and to validate gene changes in vivo.
Results
The combination of vorinostat and tranylcypromine reduced GSC viability and displayed efficacy in the U87 xenograft model. Additionally, the combination led to changes in apoptosis-related genes, particularly TP53 and TP73 in vitro and in vivo.
Conclusions
These data support targeting HDACs and KDM1A in combination as a strategy for GBM and identifies TP53 and TP73 as being altered in response to treatment.
Keywords: epigenetics, glioblastoma, HDAC inhibitors, lysine demethylases, KDM1A, LSD1, orthotopic GBM models, tranylcypromine
In recent years, there has been considerable effort placed on understanding how changes in the epigenome contribute to cancer formation and how enzymes that control epigenetic marks can be targeted for cancer therapy. Histone deacetylases are one family of epigenetic enzymes to be targeted, and three histone deacetylase inhibitors (HDACis), vorinostat, panobinostat, and romidepsin, have been approved by the US Food and Drug Administration for treatment of cutaneous T-cell lymphoma. However, the efficacy of HDACis as single agents is limited, especially in solid tumors, and the full potential of HDACis is most readily observed in combination with other chemotherapeutic agents (reviewed in1).
Another epigenetic enzyme that has been the target of drug discovery is LSD1/KDM1A (lysine-specific demethylase 1). KDM1A is overexpressed in a variety of cancers, including GBM, and tends to correlate with more aggressive cancers with poor prognosis.2–5 Our previous work identified a novel way to enhance the efficacy of HDACis for glioblastoma (GBM) therapy by combining them with inhibitors of KDM1A.2 We demonstrated that simultaneous inhibition of HDACs and KDM1A, through genetic and pharmacological means, leads to enhanced apoptotic cell death in GBM cells but not in normal counterparts.2 The efficacy of this therapeutic strategy is conserved in breast cancer6 and acute myeloid leukemia.7 However, the molecular mechanism by which combined HDAC and KDM1A inhibition causes increased apoptotic cell death is largely unknown.
The p53 tumor suppressor holds important roles in cell death and brain tumor biology. Recent analysis of GBM tumor samples by The Cancer Genome Atlas (TCGA) project revealed that p53 signaling is altered in the majority of GBM patient samples.8 Relevant to GBM therapy, p53 mutation has been shown to be a factor in predicting sensitivity to radiotherapy and temozolomide treatment,9–12 whereas expression of mutant p53 reduces sensitivity.9
Unlike p53, the p73 gene, a member of the p53 family, is rarely mutated in human cancers.13 However, it has been demonstrated that aberrant expression of p73 contributes to tumor formation14,15 and that certain isoforms of p73 are associated with tumor progression.16,17 Isoforms of p73 result from differential promoter usage and alternative splicing.18 The isoforms that retain the transactivation domain, termed TAp73, are able to induce transcription and target many of the same genes as p53; thus, they are generally known to play a role in promoting apoptosis.19–21 Despite the established role of TAp73 as a tumor suppressor, some studies suggest that the overexpression of TAp73 correlates with tumor aggressiveness and poor prognosis for patients with hepatocellular carcinoma.22 However, the overexpression of TAp73 is often accompanied by upregulation of ΔNp73 isoforms that inhibit apoptosis by acting in a dominant negative manner to block p53 and TAp73 function.23 Therefore, developing agents that inhibit the ΔNp73 isoforms while activating the TAp73 isoforms is a rational strategy for cancer therapy.
In this study, we evaluated the effects of combined HDAC and KDM1A inhibition on gene expression and found changes in TP53 and TP73 after combined HDAC and KDM1A inhibition in cell lines and in an orthotopic GBM mouse model. We also examined the therapeutic efficacy of 2 FDA-approved drugs that target HDACs and KDM1A in patient-derived glioma stem cells (GSCs) and a xenograft mouse model.
Materials and Methods
Cell Lines and Chemicals
LN-18 and U87-MG brain tumor cell lines were obtained from American Type Culture Collection. The p53 null GBM cell line, LNZ308,24 are from previously described sources. Brain tumor cell lines were maintained in DMEM)/F12 supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/mL penicillin, and 100 µg/mL streptomycin. All cells were authenticated by the Characterized Cell Line Core Facility at MD Anderson Cancer Center, which conducts short tandem repeat DNA fingerprinting within 6 months of use of the cell lines. Patient-derived GSCs were isolated in accordance with the Declaration of Helsinki, and the protocol was approved by the Institutional Review Board at the University of Texas MD Anderson Cancer Center (LAB04-0001) as previously described.25 GSCs were grown in DMEM/F12 containing 20 ng/mL EGF (Sigma) and bFGF (Invitrogen). All cells were grown at 37°C in a humidified atmosphere containing 5% CO2. The Flag-p53 overexpression construct was generated by cloning wild-type p53 into the pCMV-FLAG vector (Clontech Laboratories). The KDM1A shRNA construct was previously described.2 Constructs were transfected into LNZ308 or LN-18 cells according to the manufacturer's instructions (Lonza). Cells (1 × 106) were resuspended in Buffer V and nucleofected using program T-20. Vorinostat (suberoylanilide hydroxamic acid ]SAHA])) was purchased from Cayman Chemical, and all other chemicals were purchased from Sigma-Aldrich. Vorinostat was resuspended in DMSO and tranylcypromine in PBS. For all experiments, vehicle-labeled lanes indicate treatment with DMSO/PBS at equivalent volumes to those used for the inhibitors. Antibodies used for this study were as follows: p53 (Invitrogen), p73 (Imgenex), actin (Sigma-Aldrich).
Gene Expression Analysis
Total RNA was isolated using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer's instructions. For RNA-Seq, total RNA was submitted to the Sequencing and Microarray Facility at MD Anderson Cancer Center for next-generation sequencing. The raw reads were aligned to the human reference genome build HG19, and HTSeq software was utilized to count the number of reads mapped to each gene. Differential gene expression tests were conducted utilizing DESeq, based on the reads and counts data. For the investigation of apoptosis signaling pathways affected, we used the Human Apoptosis RT2 Profiler PCR Array according to the manufacturer's instructions (Qiagen/SABiosciences). For validation of array results, cDNA was generated using the iScript cDNA Synthesis Kit (BioRad), and qPCR was performed using iTaq UniverSYBR master mix (BioRad). All data were normalized to B-actin, and fold change was determined by comparison to vehicle-treated controls for each cell line.
p53 and p73 Protein Analysis
After treatment, cells were harvested, and total cell lysates were prepared by lysis with Triton X-100 lysis buffer (PBS, 25 mM Tris pH 7.5, 150 mM NaCl, 1% Triton X-100) containing protease inhibitors for 1 hour at 4°C followed by centrifugation. Protein (50 µg) was separated by SDS-PAGE, transferred to PVDF, and immunoblotted with specific antibodies as indicated. Immunoreactive bands were detected by chemiluminescence (GE Healthcare). Densitometry was performed using Image J (NIH).
Determination of Cell Death
DNA fragmentation was used as a surrogate marker for apoptosis in GBM cells as previously described.2 Trypan blue staining, followed by cell counting via hemacytometer, was used to determine cell viability.
In Vivo Xenograft Experiments and RNA Analysis
All experimental procedures were approved by the Institution Animal Care and Uses Committee (IACUC) at the University of Texas MD Anderson Cancer Center (Protocol: 030402934). For all experiments, guide screws were implanted in 6-week-old, female NCR (nu/nu) mice (NCI), and the mice were inoculated 7 days later with 5 × 105 U87MG or luciferase-labeled U87MG GBM cells as previously described.26 Three distinct experiments were performed as follows. (i) Seven days post inoculation of cells, mice were treated with 50 mg/kg vorinostat 5 days/week, 1 mg/mouse tranylcypromine 7 days/week or the combination via i.p. injection for a 2-week period. Dosing was based upon previously reported single agent use of vorinostat27 and tranylcypromine.28 Mice were monitored until signs of morbidity and euthanized to determine the effects of the combination on overall survival. (ii) Mice inoculated with luciferase-labeled U87-MG cells were monitored noninvasively prior to treatment and at the end of treatment by injecting 3.75 mg D-luciferin 10 minutes prior to imaging using an IVIS 100 platform. Quantification of luciferase signal was calculated using Xenogen Living Systems software (Perkin Elmer). (iii) For analysis of gene expression, mice inoculated with U87-MG cells were euthanized one day following the end of the treatment schedule, and brain tissue was excised and flash frozen. Total RNA was isolated from flash-frozen tissue using the RNeasy Plus Universal kit (Qiagen), and qRT-PCR was performed as described above.
Results
Combined Inhibition of KDM1A and HDACs Decreases the Viability of Glioma Stem Cells
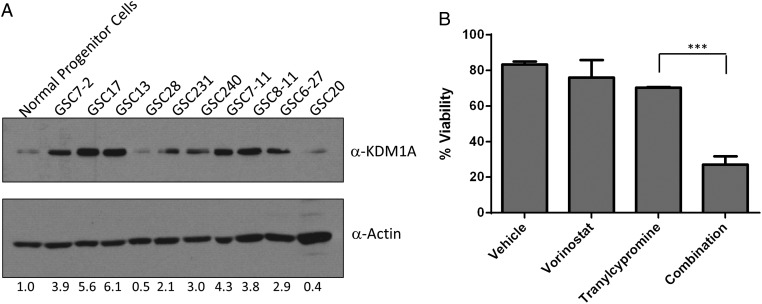
Our previous studies demonstrated that KDM1A inhibition, through knockdown and chemical means, combined with inhibitors of HDACs enhances cell death in GBM cell lines.2 KDM1A plays a role in stem cell propagation and self-renewal,29 and the KDM1A-RCOR2 chromatin complex is essential for GBM stem-like tumor propagating cells.30 Additionally, KDM1A is elevated in GBM cell lines relative to normal human astrocytes.2 Therefore, we examined expression of KDM1A in an expanded panel of patient-derived GSCs compared with normal neural progenitor cells. Eight of the 10 GSC lines displayed elevated KDM1A protein compared with normal counterparts (Fig. 1A). We next wanted to determine whether the combination of HDAC and KDM1A inhibition increased cell death in this population of cells using the FDA-approved inhibitors vorinostat (a pan-HDACi) and tranylcypromine (a small molecule inhibitor known to target KDM1A). Treatment of GSCs with the combination of vorinostat and tranylcypromine (with doses previously identified to be the most synergistic in GBM cell lines2) reduced viability to approximately 30% (Fig. 1B). A similar trend was seen for the combination of entinostat, (a class I HDAC inhibitor) with tranylcypromine (data not shown). These data indicate that the GSC population frequently overexpress KDM1A and is susceptible to cell death when treated with HDACis and tranylcypromine.
Fig. 1.
Combined targeting of HDACs and KDM1A leads to decreased viability of glioma stem cells (GSCs). (A) Expression of KDM1A protein expression in GSCs compared with normal neural progenitor cells. Actin was used as a loading control. (B) Viability of GSCs was measured 5 days after single dosing with 5 uM vorinostat, 1 mM tranylcypromine or the combination. n = 3, mean ± SEM. ***P ≤ .001.
Inhibition of KDM1A and HDACs Leads to Changes in Apoptotic Gene Expression
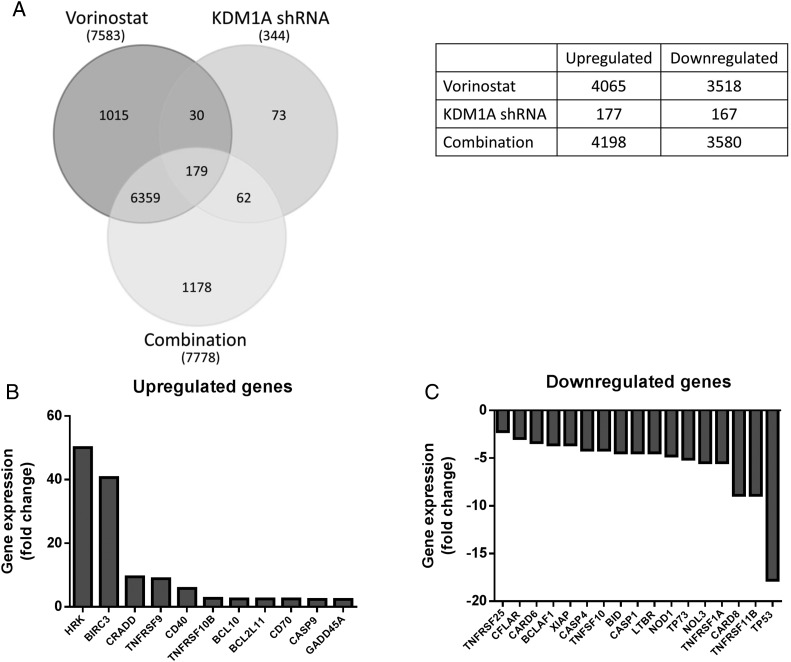
In order to further understand the molecular mechanisms by which inhibition of KDM1A and HDACs cooperate to enhance cell death, we evaluated global gene expression changes in KDM1A knockdown LN-18 cells with or without vorinostat treatment. We found that there were an equal number of genes upregulated as downregulated (Fig. 2A), which is consistent with other studies evaluating gene control by KDM1A that demonstrate context dependence of gene expression (reviewed in31). Further comparison of mRNAs changed by each of the treatment groups reveals 179 genes that are commonly changed by all 3 treatments. Interestingly, there are 1178 genes that are uniquely altered in cells where both KDM1A and HDACs are inhibited, some of which are apoptosis-related genes (Fig. 2A).
Fig. 2.
Inhibition of HDACs and KDM1A alter gene expression in glioblastoma cell lines. (A) Venn diagram depicting the total number of genes changed in LN-18 cells by vorinostat, KDM1A knockdown, and the combination. The table illustrates the number of genes up- and downregulated by each of the treatment groups. (B) LN-18 cells transfected with KDM1A shRNA were treated with 5 µM vorinostat for 24 hours, and gene expression was measured using a RT-qPCR array. A waterfall plot shows the expression profiles of genes that displayed >2-fold upregulation and (C) >2-fold downregulation.
To further interrogate apoptosis-related genes, we performed a focused RT-qPCR array that evaluated 84 apoptosis-specific genes. Results from these analyses revealed that 38 genes were either directly or indirectly changed ≥2-fold after combined KDM1A knockdown and treatment with vorinostat. Of these, 11 genes were upregulated (Fig. 2B), and 11 genes were downregulated (Fig. 2C) in both the RT-qPCR and RNA-Seq datasets. Interestingly, both TP53 and family member TP73, are highly downregulated by the combination (Fig. 2C). Analysis of TCGA project data for GBM revealed that p53 signaling was altered in 87% of the samples collected.8 Disruption of the p53 pathway is known to contribute to tumorigenesis through inactivation of p53-target genes such as Puma, Noxa, Bad and Bid,32 providing the rationale for further study.
KDM1A and HDACs Regulate p53 Expression in Glioblastoma Cells
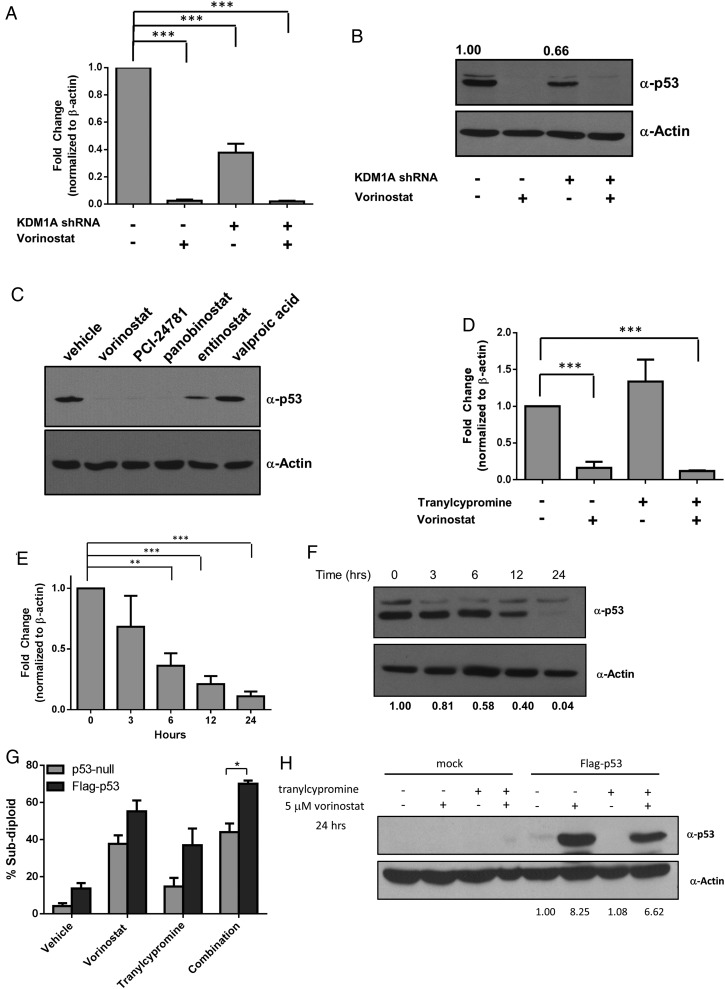
Surprisingly, our focused array identified TP53 as the most highly downregulated gene (>17-fold) in KDM1A knockdown cells treated with vorinostat (Fig. 2C). qPCR validation of TP53 expression in LN-18 cells revealed that KDM1A knockdown alone decreases TP53 expression by 2-fold (Fig. 3A). Additionally, vorinostat alone or the combination of KDM1A knockdown and vorinostat decreased TP53 mRNA (Fig. 3A) and protein (Fig. 3B) to almost undetectable levels. HDACis are classified based on inhibition of specific HDAC family members and by their chemical structure. We found that PCI-24781 and panobinostat, pan-HDACis that share structural similarity to vorinostat, decreased the expression of p53 protein, whereas entinostat (a specific class I/II HDACi) only partially decreased p53, and valproic acid (an aliphatic acid) had no effect at equimolar doses (Fig. 3C). These data suggest that certain classes of HDACis may be more effective at causing loss of p53, which may be due to the HDACs that they inhibit.
Fig. 3.
p53 mRNA and protein expression is rapidly regulated by HDAC and KDM1A inhibition. LN-18 cells transfected with control or KDM1A shRNA were treated with 5 µM vorinostat for 24 hours, and (A) TP53 gene expression or (B) p53 protein expression was measured. (C) LN-18 cells were treated with 5 µM of the HDACi indicated. p53 protein expression was evaluated 24 hours after treatment. (D) LN-18 cells were treated with 5 µM vorinostat, 1 mM tranylcypromine, or the combination for 24 hours, and p53 mRNA expression was evaluated. (E) p53 mRNA was measured in LN-18 cells treated with 5 µM vorinostat at the time points indicated. (F) p53 protein was assessed after treatment with 5 µM vorinostat by Western blot at the indicated time points. (G) LNZ308 (p53-null) cells were transfected with empty vector or vector-expressing wild-type p53 (Flag-p53). DNA fragmentation was measured 72 hours after treatment with 5 µM vorinostat, 1 mM tranylcypromine, or the combination. (H) Western blots demonstrating lack of p53 protein in LNZ308 cells and ectopic expression of wild-type p53 protein under conditions stated in part G. All Western blots are representative of 3 independent experiments. Actin was used as a loading control. n = 3, mean ± SEM. *P ≤ .05, ** P ≤ .01, ***P ≤ .001.
To determine whether chemical inhibition of KDM1A causes a similar decrease in TP53 expression, we treated LN-18 cells with previously determined synergistic doses of vorinostat and tranylcypromine2 and measured levels of p53 mRNA by qPCR (Fig. 3D). Similar to control and KDM1A knockdown cells, vorinostat alone or the combination of vorinostat with tranylcypromine causes a 6–8-fold decrease in p53 mRNA. These results suggest that HDACs may be responsible for the majority of the effect on p53, but KDM1A is also a contributing factor especially when KDM1A protein is knocked down. This is consistent with a report by Yan et al showing that HDACis suppress TP53 transcription via HDAC8 control of HoxA5.33
Since vorinostat caused the greatest change in p53 mRNA and protein expression, we next wanted to understand the mechanism by which inhibition of HDACs leads to decreased TP53 expression in GBM cells. It is well known that p53 is regulated by HDACs at both the transcriptional and posttranslational levels.34 To determine whether vorinostat decreases p53 mRNA prior to loss of protein, we evaluated p53 mRNA (Fig. 3E) and protein (Fig. 3F) loss at several time points following exposure to vorinostat. We found that p53 mRNA decreased as early as 6 hours after the addition of vorinostat (Fig. 3E). p53 protein expression decreased with similar kinetics (Fig. 3F). These data suggest that the effects of vorinostat primarily alter levels of mRNA expression. Importantly, GBM cells that lack p53 are insensitive to the combination of vorinostat and tranylcypromine (Fig. 3G). However, reintroduction of p53 sensitizes cells to the combination (Fig. 3G), suggesting that p53 plays an important role in promoting apoptosis following simultaneous inhibition of HDACs and KDM1A. Western blot analysis confirming expression of p53 protein in LNZ308 cells is shown in Fig. 3H.
The p53 Family Member TP73 Is Also Regulated by HDACs and KDM1A
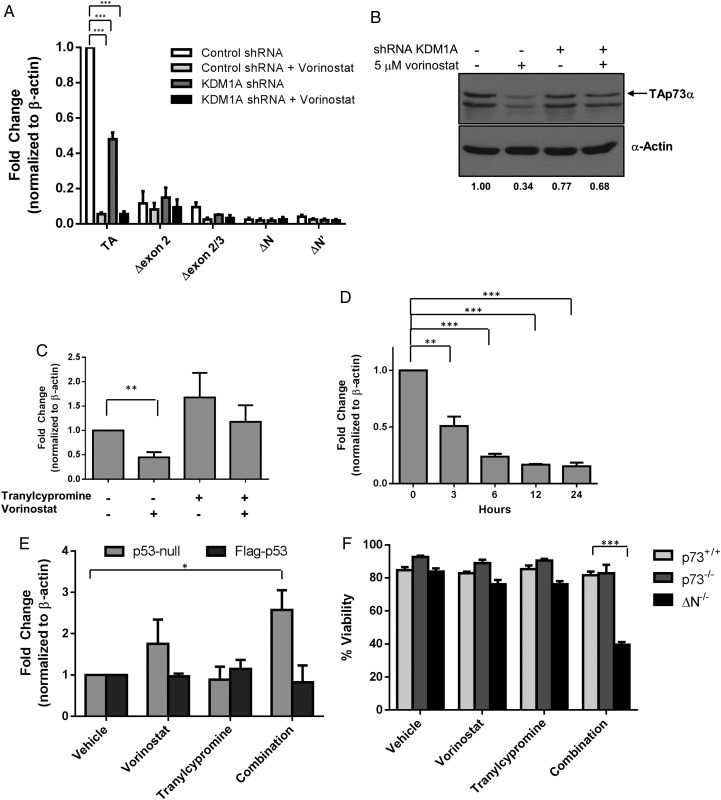
p53 family members p63 and p73 also play a role in human malignancies, albeit in different capacities. Interestingly, p73, but not p63, was also largely downregulated by combined KDM1A and HDAC inhibition in the focused apoptosis array (Fig. 2C). The TP73 gene has 2 distinct promoters that result in transactivating (TA) or ΔN isoforms.35 TA-p73 is able to induce the expression of several p53 target genes rendering it proapoptotic.19–21 In contrast, ΔN-p73 isoforms serve as antiapoptotic proteins because they lack the transactivation domain and act as dominant-negative inhibitors of TA-p73 and p53.36 There are also several alternatively spliced variants that are found in human cancers.37–39 To determine which of the p73 isoforms was expressed and changed by knockdown of KDM1A, vorinostat or the combination, we performed qPCR using primers specific for TA, Δexon 2, Δ exon 2/3, ΔN, and ΔN′. We observed that TAp73 was the most abundant isoform of p73 in LN-18 GBM cells and was the only isoform altered by inhibition of HDACs and KDM1A (Fig. 4A). Additionally, TAp73 protein expression mirrored the mRNA expression (Fig. 4B). Similar to changes in p53 mRNA (Fig. 3D), vorinostat caused downregulation of p73 mRNA expression, whereas the inhibition of KDM1A with tranylcypromine had little effect (Fig. 4C). Since the regulation of TAp73 was similar to that of p53, we measured the kinetics of TAp73 mRNA expression in response to vorinostat (Fig. 4D) and found it to be similar to the kinetics of p53 (Fig. 3E). Taken together, these data suggest that p53 and TAp73 are regulated similarly by HDACs and KDM1A.
Fig. 4.
HDAC and KDM1A inhibition alter TAp73 expression. LN-18 cells transfected with control or KDM1A shRNA were treated with 5 µM vorinostat for 24 hours: (A) TA, Δexon 2 (Δex2), Δexon 2 and 3 (Δex2/3), ΔN or ΔN′ isoforms of p73 were measured by RT-qPCR., or (B) p73 protein expression was evaluated by Western blot. (C) LN-18 cells were treated with 5 µM vorinostat, 1 mM tranylcypromine, or the combination and TAp73 mRNA expression was evaluated by qPCR. (D) LN-18 cells were treated with 5 µM vorinostat, and TAp73 mRNA expression was measured at the indicated time points. (E) TAp73 expression was measured in p53-null glioblastoma cells (LNZ308) were transfected with vector or Flag-tagged wild-type p53 followed by treatment with 5 µM vorinostat, 1 mM tranylcypromine, or the combination. (F) Viability was measured in wild-type (p73+/+), p73-deficient (p73−/−), or ΔN-p73-deficient (ΔN−/−) mouse embryonic fibroblasts treated with 5 µM vorinostat, 1 mM tranylcypromine, or the combination for 24 hours. n = 3, mean ± SEM. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
The similar kinetics of p53 and TAp73 expression changes suggest that p53 may regulate TAp73 after inhibition of HDACs and KDM1A. Several studies link p53 and p73; p53 directly regulates p73 at the transcriptional level40,41 as well as indirectly by influencing p73 RNA stability.42 To investigate the specific role of p53 in the regulation of TAp73, we transfected p53-null GBM cells with wild-type p53 and measured changes in TAp73 mRNA expression after each treatment (Fig. 4E). We observed little-to-no difference in the expression of TAp73 mRNA in response to vorinostat or tranylcypromine alone in either p53-expressing or p53-null cells. However, when cells were treated with the combination of vorinostat and tranylcypromine, there was a significant (P ≤ .05) upregulation of TAp73 expression (Fig. 4E). Interestingly, overexpression of p53 followed by combined treatment with vorinostat and tranylcypromine largely blocks this upregulation, suggesting that p53 negatively regulates the expression of TAp73. It is important to note that mouse embryonic fibroblasts lacking ΔN isoforms (ΔN−/−) of p73 had a dramatic loss in cell viability (P < .001) following combination treatment, which suggests that TAp73 is also required to promote cell death by the combination of vorinostat and tranylcypromine (Fig. 4F).
Combination of Vorinostat and Tranylcypromine Reduces p53 and TAp73 Expression in an Orthotopic Glioblastoma Xenograft Tumor Model
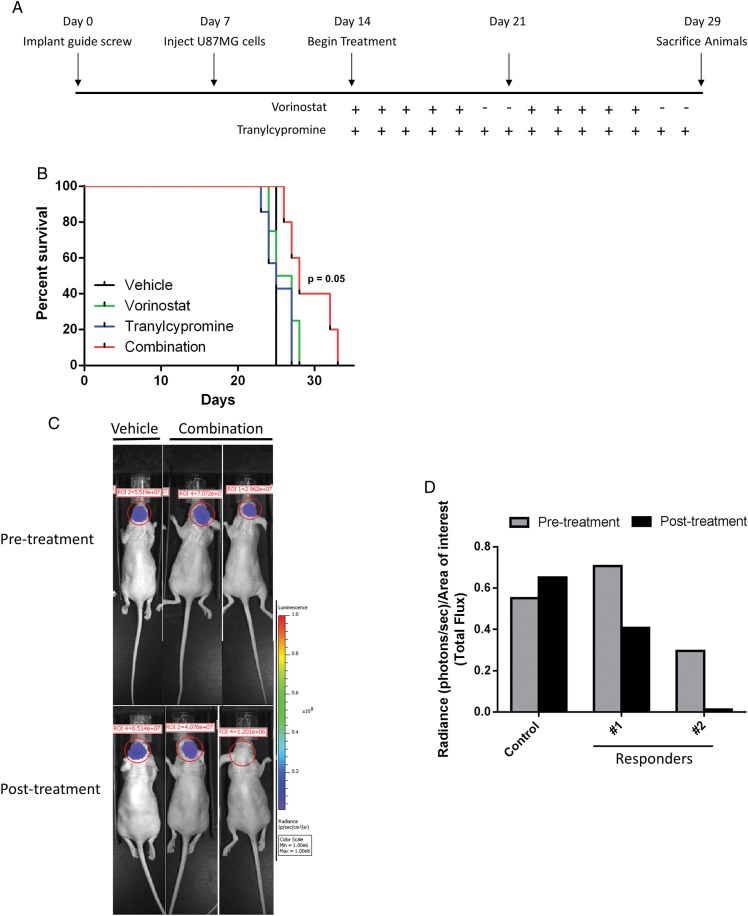
Given our promising in vitro data in GBM cell lines,2 we next wanted to evaluate the effects of vorinostat and tranylcypromine (2 FDA-approved drugs that inhibit HDACs and KDM1A), respectively, in in vivo models of GBM. We utilized the orthotopic GBM xenograft model using U87 cells developed by Lal et al.26 The overall dosing scheme is depicted in Fig. 5A. To evaluate the effect of the combination on overall survival, we randomized mice into 4 treatment groups 7 days post U87 cell implantation. Non–luciferase-labeled U87-MG cells were used for overall survival studies to eliminate possible differences in sensitivity to vorinostat and tranylcypromine conferred by gene manipulation. While individual treatment with vorinostat or tranylcypromine provided no improvement in overall survival compared with control, the combination-treated group showed a trend towards a survival advantage (P = .05) (Fig. 5B). To determine whether the improvement in overall survival correlated with reduced tumor size, we utilized luciferase-labeled U87 cells in the orthotopic xenograft model in a separate experiment. Seven days after tumor cell implantation, mice were randomized into 2 groups: control (n = 2) and combination (n = 8). Fig. 5C and D show examples of combination-treated mice with reduced tumor burden that correlates with the overall survival data in Fig. 5B. Taken together, these data show promise for the utilization of vorinostat and tranylcypromine as a novel therapeutic combination strategy.
Fig. 5.
Combination of vorinostat and tranylcypromine extends survival of glioblastoma xenograft mouse model. (A) Schematic diagram illustrating the dosing schedule for animal experiments. (B) Mice were treated 7 days after glioma cell implantation with the following regimens: 50 mg/kg vorinostat, 5 days/week (n = 4); 1 mg/mouse tranylcypromine, 7 days/week (n = 7), combination of vorinostat and tranylcypromine (n = 5), or vehicle as a control (n = 2). Kaplan-Meyer curves depict time to moribund characteristics (hunched posture, significant weight loss, or hemiparesis). (C) Luciferase-labeled U87 was injected into athymic nu/nu mice and imaged 2 weeks after implantation (pretreatment) and at the conclusion of the 2-week combination regimen (post treatment). (D) Luminescence signal was quantified and presented graphically.
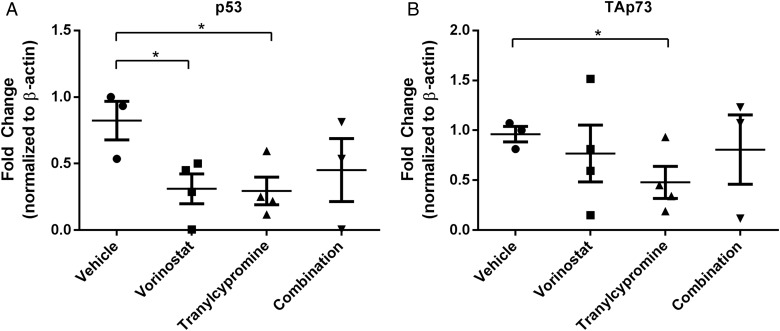
We next wanted to determine whether the gene expression changes identified by RNA-Seq are also altered in the xenograft tumors. To do this, we euthanized 4 mice from each treatment group after the 2-week treatment schedule and isolated total RNA from tumor tissue. RT-qPCR was performed to evaluate changes in p53 (Fig. 6A) and TAp73 (Fig. 6B) mRNA. We observed that p53 and TAp73 mRNA was downregulated by either treatment with vorinostat or tranylcypromine as a single agent (Fig. 6A and B). Additionally, combined treatment with both agents yielded results similar to individual agent treatment and reflected our observations in vitro (Fig. 3D and 4C). These data suggest that the effects of vorinostat and tranylcypromine on gene expression can be extended to in vivo models of GBM.
Fig. 6.
Vorinostat affects p53 and TAp73 expression in an in vivo mouse model for glioblastoma. RNA from brain tissue was isolated from xenograft mice, and qPCR performed as previously described for (A) p53 and (B) TAp73. n = 3 or 4 depending on RNA quality, mean ± SEM. *P ≤ .05.
Discussion
Epigenetic enyzmes have become popular targets for cancer therapeutics. We previously demonstrated that combined inhibition of KDM1A and HDACs leads to increased apoptosis in GBM cells.2 Our current research investigated the molecular mechanism underlying the increase in apoptosis following HDAC and KDM1A inhibition downstream of these complexes. We observed several gene expression changes associated with apoptosis in KDM1A-knockdown cells treated with vorinostat (Fig. 2B and C). We also found that p53 and its family member, p73, were both highly affected by this combination. Interestingly, we found that KDM1A knockdown alone decreased the expression of p53 and p73 mRNA by at least 2-fold, a result that is among the first to link KDM1A to the expression of these genes. In addition, we demonstrated that the combination of vorinostat and tranylcypromine shows efficacy in an orthotopic GBM mouse model and that TP53 and TP73 gene expression changes observed in vitro are reflected in the tumors of these animals.
Mutation of the p53 gene is the most common genetic mutation in cancer, making p53 an attractive target for cancer therapeutics. HDAC inhibitors have been shown to affect p53 in a variety of ways. Acetylation of wild-type p53 by p300/CBP leads to increased stability and transcriptional activity for apoptotic targets.43,44 However, HDACis disrupt the HDAC6-HSP90 chaperone pathway, which leads to the degradation of mutant p53 and increased cytotoxicity to these agents.45
Our data suggest that p53 is regulated in GBM cells (more so at the transcriptional level) (Fig. 3). Consistent with our findings, Yan et al demonstrated that inhibition of HDACs decreases p53 transcription.33 We also demonstrated changes in TP53 in tumors of xenograft mice (Fig. 6), providing evidence for the use of HDACi to target p53.
There are multiple interactions and cross talk between the p53 family members. We observed that p53 altered expression of TAp73 in response to combined inhibition of HDACs and KDM1A. Expression of wild-type p53 blocked the upregulation of TAp73 mRNA upon treatment with the combination of vorinostat and tranylcypromine (Fig. 4E). These data are supported by knockdown and knockout studies demonstrating that loss of p53 increases p73 expression at the transcriptional level through the E2F-1 transcription factor.41 p53 can also directly induce the expression of p73 in response to DNA-damaging agents through a p53-binding site in the p73 promoter.40
In addition to transcriptional control of p73, p53 also indirectly affects the stability of p73 mRNA through the regulation of RNPC142 as well as protein stability through direct interactions with the p53 core domain.46 It is possible that inhibition of KDM1A and/or HDACs influences not only the transcriptional control of p73 in cell lines and xenograft tumors but may also affect other factors to control p73 protein expression.
Our previous study demonstrated that simultaneous treatment of GBM cells with HDAC and KDM1A inhibitors increased cell death.2 Here we show that this combination can be extended to the GSC (Fig. 1B). GSCs, or tumor-propagating cells (TPCs), are thought to drive tumor progression, recurrence, and resistance to current therapeutic strategies. Recent studies by Suvá et al have demonstrated the requirement of KDM1A for survival and sphere-forming capability of TPCs as well as tumor-propagating potential of these cells in in vivo mouse models of GBM.30
We showed that KDM1A is upregulated in GSCs (Fig. 1A), which provides one possible explanation for the sensitivity of these cells to KDM1A inhibition. The mechanism by which KDM1A is overexpressed in GSCs is currently unknown. However, there is evidence of an alternatively spliced variant of KDM1A in neuronal cells that alters enzyme function when phosphorylated.47 Alternative splicing of KDM1A in GBM remains to be determined.
We also showed efficacy of the combination in an orthotopic xenograft GBM mouse model (Fig. 5). These data are consistent with reports of combined HDAC and KDM1A inhibition in in vivo AML mouse models. In this study, the authors demonstrate that treatment of mice with the combination of SP2509, a more specific KDM1A inhibitor, with panobinostat, an HDACi with structural similarity to vorinostat, significantly increased overall survival.7 Further studies evaluating specific KDM1A inhibitors for their ability to cross the blood-brain barrier and affect tumor biology in Phase I trials are already underway and will improve specificity of treatment for GBM.
Funding
This work was supported by the NIH/NCI under award numbers (i) P30CA016672 and used the Characterized Cell Line Core and the Research Animal Support Facility; (ii) Brain Tumor SPORE Award Number P50 CA127001 (Developmental Project Grant to J.C.); and (iii) F32CA150610 (to M.M.S). We also gratefully acknowledge funding support by the Center for Cancer Epigenetics at the University of Texas MD Anderson Cancer Center (Solexa Core Allowance to J.C.).
Acknowledgments
We would like to thank Dr. Oliver Bogler for the p53-null LNZ308 cells.
Conflict of interest statement. The authors declare no conflict of interest.
References
- 1.Miller CP, Singh MM, Rivera-Del Valle N, et al. Therapeutic strategies to enhance the anticancer efficacy of histone deacetylase inhibitors. J Biomed Biotechnol. 2011;2011:514261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber TG, Poschinger T, Galban S, et al. Noninvasive monitoring of pharmacodynamics and kinetics of a death receptor 5 antibody and its enhanced apoptosis induction in sequential application with doxorubicin. Neoplasia. 2013;15(8):863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahl P, Gullotti L, Heukamp LC, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66(23):11341–11347. [DOI] [PubMed] [Google Scholar]
- 4.Galban S, Martindale JL, Mazan-Mamczarz K, et al. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol Cell Biol. 2003;23(20):7083–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding J, Zhang ZM, Xia Y, et al. LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer. 2013;109(4):994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasilatos SN, Katz TA, Oesterreich S, et al. Crosstalk between lysine-specific demethylase 1 (LSD1) and histone deacetylases mediates antineoplastic efficacy of HDAC inhibitors in human breast cancer cells. Carcinogenesis. 2013;34(6):1196–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiskus W, Sharma S, Shah B, et al. Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia. 2014;28(11):2155–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blough MD, Beauchamp DC, Westgate MR, et al. Effect of aberrant p53 function on temozolomide sensitivity of glioma cell lines and brain tumor initiating cells from glioblastoma. J Neurooncol. 2011;102(1):1–7. [DOI] [PubMed] [Google Scholar]
- 10.Gjerset RA, Turla ST, Sobol RE, et al. Use of wild-type p53 to achieve complete treatment sensitization of tumor cells expressing endogenous mutant p53. Mol Carcinog. 1995;14(4):275–285. [DOI] [PubMed] [Google Scholar]
- 11.Hirose Y, Berger MS, Pieper RO. p53 effects both the duration of G2/M arrest and the fate of temozolomide-treated human glioblastoma cells. Cancer Res. 2001;61(5):1957–1963. [PubMed] [Google Scholar]
- 12.Squatrito M, Brennan CW, Helmy K, et al. Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell. 2010;18(6):619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ichimiya S, Nimura Y, Kageyama H, et al. p73 at chromosome 1p36.3 is lost in advanced stage neuroblastoma but its mutation is infrequent. Oncogene. 1999;18(4):1061–1066. [DOI] [PubMed] [Google Scholar]
- 14.Flores ER, Sengupta S, Miller JB, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7(4):363–373. [DOI] [PubMed] [Google Scholar]
- 15.Tomasini R, Tsuchihara K, Wilhelm M, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22(19):2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casciano I, Mazzocco K, Boni L, et al. Expression of DeltaNp73 is a molecular marker for adverse outcome in neuroblastoma patients. Cell Death Differ. 2002;9(3):246–251. [DOI] [PubMed] [Google Scholar]
- 17.Concin N, Becker K, Slade N, et al. Transdominant DeltaTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004;64(7):2449–2460. [DOI] [PubMed] [Google Scholar]
- 18.Yang A, Walker N, Bronson R, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404(6773):99–103. [DOI] [PubMed] [Google Scholar]
- 19.Zhu J, Jiang J, Zhou W, et al. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 1998;58(22):5061–5065. [PubMed] [Google Scholar]
- 20.De Laurenzi V, Costanzo A, Barcaroli D, et al. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med. 1998;188(9):1763–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueda Y, Hijikata M, Takagi S, et al. New p73 variants with altered C-terminal structures have varied transcriptional activities. Oncogene. 1999;18(35):4993–4998. [DOI] [PubMed] [Google Scholar]
- 22.Tannapfel A, Wasner M, Krause K, et al. Expression of p73 and its relation to histopathology and prognosis in hepatocellular carcinoma. J Natl Cancer Inst. 1999;91(13):1154–1158. [DOI] [PubMed] [Google Scholar]
- 23.Zaika AI, Slade N, Erster SH, et al. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med. 2002;196(6):765–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shingu T, Chumbalkar VC, Gwak HS, et al. The polynuclear platinum BBR3610 induces G2/M arrest and autophagy early and apoptosis late in glioma cells. Neuro Oncol. 2010;12(12):1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhat KP, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lal S, Lacroix M, Tofilon P, et al. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg. 2000;92(2):326–333. [DOI] [PubMed] [Google Scholar]
- 27.Yin D, Ong JM, Hu J, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor: effects on gene expression and growth of glioma cells in vitro and in vivo. Clin Cancer Res. 2007;13(3):1045–1052. [DOI] [PubMed] [Google Scholar]
- 28.Schulte JH, Lim S, Schramm A, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69(5):2065–2071. [DOI] [PubMed] [Google Scholar]
- 29.Adamo A, Sese B, Boue S, et al. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13(6):652–659. [DOI] [PubMed] [Google Scholar]
- 30.Suva ML, Rheinbay E, Gillespie SM, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157(3):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forneris F, Binda C, Battaglioli E, et al. LSD1: oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem Sci. 2008;33(4):181–189. [DOI] [PubMed] [Google Scholar]
- 32.Kuribayashi K, El-Deiry WS. Regulation of programmed cell death by the p53 pathway. Adv Exp Med Biol. 2008;615:201–221. [DOI] [PubMed] [Google Scholar]
- 33.Yan W, Liu S, Xu E, et al. Histone deacetylase inhibitors suppress mutant p53 transcription via histone deacetylase 8. Oncogene. 2013;32(5):599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oren M. Regulation of the p53 tumor suppressor protein. J Biol Chem. 1999;274(51):36031–36034. [DOI] [PubMed] [Google Scholar]
- 35.Grob TJ, Novak U, Maisse C, et al. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 2001;8(12):1213–1223. [DOI] [PubMed] [Google Scholar]
- 36.Pozniak CD, Radinovic S, Yang A, et al. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289(5477):304–306. [DOI] [PubMed] [Google Scholar]
- 37.Stiewe T, Theseling CC, Putzer BM. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J Biol Chem. 2002;277(16):14177–14185. [DOI] [PubMed] [Google Scholar]
- 38.Ng SW, Yiu GK, Liu Y, et al. Analysis of p73 in human borderline and invasive ovarian tumor. Oncogene. 2000;19(15):1885–1890. [DOI] [PubMed] [Google Scholar]
- 39.Fillippovich I, Sorokina N, Gatei M, et al. Transactivation-deficient p73alpha (p73Deltaexon2) inhibits apoptosis and competes with p53. Oncogene. 2001;20(4):514–522. [DOI] [PubMed] [Google Scholar]
- 40.Chen X, Zheng Y, Zhu J, et al. p73 is transcriptionally regulated by DNA damage, p53, and p73. Oncogene. 2001;20(6):769–774. [DOI] [PubMed] [Google Scholar]
- 41.Tophkhane C, Yang SH, Jiang Y, et al. p53 inactivation upregulates p73 expression through E2F-1 mediated transcription. PLoS One. 2012;7(8):e43564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan W, Zhang J, Zhang Y, et al. p73 expression is regulated by RNPC1, a target of the p53 family, via mRNA stability. Mol Cell Biol. 2012;32(13):2336–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268(10):2764–2772. [DOI] [PubMed] [Google Scholar]
- 44.Prives C, Manley JL. Why is p53 acetylated? Cell. 2001;107(7):815–818. [DOI] [PubMed] [Google Scholar]
- 45.Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011;18(12):1904–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaiddon C, Lokshin M, Ahn J, et al. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21(5):1874–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toffolo E, Rusconi F, Paganini L, et al. Phosphorylation of neuronal Lysine-Specific Demethylase 1LSD1/KDM1A impairs transcriptional repression by regulating interaction with CoREST and histone deacetylases HDAC1/2. J Neurochem. 2014;128(5):603–616. [DOI] [PubMed] [Google Scholar]