Abstract
Neurological disorders such as Alzheimer's disease, stroke and epilepsy are currently marred by the lack of effective treatments to prevent neuronal death. Erroneous cell cycle reentry (CCR) is hypothesized to have a causative role in neurodegeneration. We show that forcing S-phase reentry in cultured hippocampal neurons is sufficient to induce neurodegeneration. We found that kainic-acid treatment in vivo induces erroneous CCR and neuronal death through a Notch-dependent mechanism. Ablating Notch signaling in neurons provides neuroprotection against kainic acid-induced neuronal death. We further show that kainic-acid treatment activates Notch signaling, which increases the bioavailability of CyclinD1 through Akt/GSK3β pathway, leading to aberrant CCR via activation of CyclinD1-Rb-E2F1 axis. In addition, pharmacological blockade of this pathway at critical steps is sufficient to confer resistance to kainic acid-induced neurotoxicity in mice. Taken together, our results demonstrate that excitotoxicity leads to neuronal death in a Notch-dependent manner through erroneous CCR.
Neuronal atrophy and eventual neuronal demise is the leading cause of irreversible behavioral deficits associated with several neurological disorders such as Alzheimer's disease (AD), ischemic stroke, Parkinson's disease (PD) and so on. As neuronal loss usually happens in the later stages of the progressive disorders,1 cell signaling events that take place before cell death during the early phases of disease progression are of particular therapeutic importance.
Aberrant cell cycle reentry (CCR) has been proposed to be one of the important causative steps before neuronal loss.2 Several studies have identified changes in the expression of cell cycle-related genes in neuronal populations vulnerable to progressive neurodegenerative disorders.3, 4, 5, 6 Recent studies have proposed some important molecular players involved in CCR in neurons.7, 8, 9, 10 Yet, a precise molecular pathway triggered by diverse neurodegenerative stimuli that leads to CCR in neurons remains unidentified.
Notch1 is a transmembrane receptor that is activated through sequential enzymatic cleavages, through ADAM-17 and γ-secretase, to generate the Notch1 intracellular domain (NICD1) with transcriptional activity through RBPJK. Notch/RBPJK canonical signaling, is a highly conserved signaling cascade, which has an important role in nervous system development and in stem cell proliferation and maintenance.11 Notch signaling also has crucial roles in the adult brain, in neurons12 and neurogenic niches.13 Notch1 appears to be an important signaling receptor in neurons regulating synaptic plasticity, and learning and memory.12 On the other hand, overactivation of Notch signaling, in response to hypoxic–ischemic or epileptic injury appears to contribute to neuronal demise.14, 15
Here we show that excitotoxic kainic acid (KA) treatment results in S-phase reentry with concomitant nuclear localization of Notch1. Notch signaling upon KA regulates Akt/Cyclind1 axis activation. Blockade of the canonical Notch signaling, Akt or Cyclind1 (Ccnd1) activity provides significant protection against neuronal death. Our results unravel an important role for Notch1 in Akt/Ccnd1-dependent CCR in neurons and identify this signaling cascade as a possible therapeutic target for treating neurodegenerative disorders.
Results
Excitotoxic KA treatment triggers S-phase reentry and is associated with Notch signaling
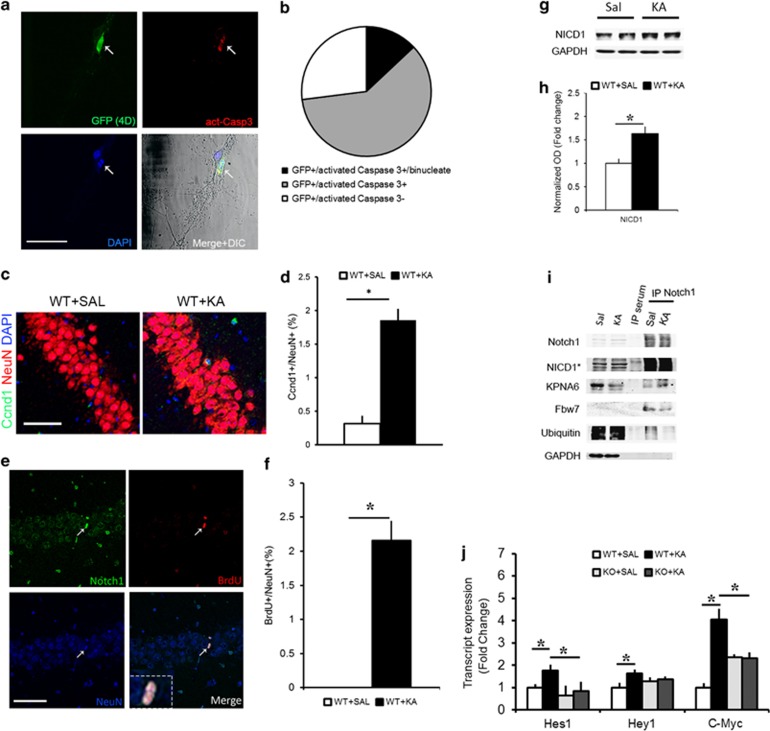
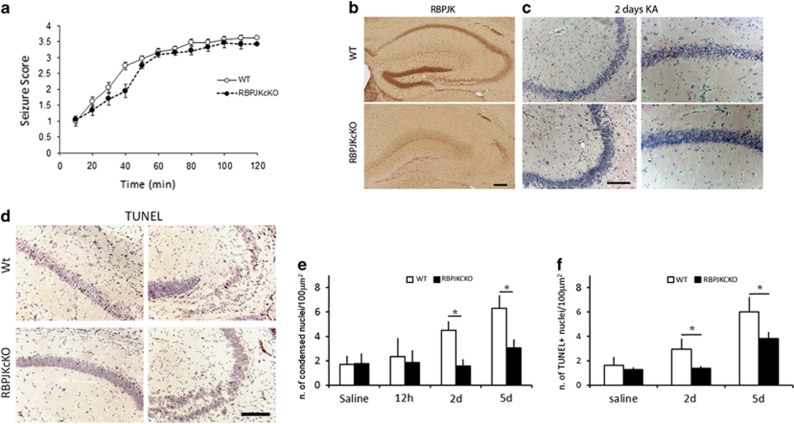
To address whether CCR in neurons affects cell survival, we transfected primary hippocampal cultures with a plasmid expressing Ccnd1 and CDK4 along with a GFP tag and a nuclear localization signal (Ccnd1-CDK4-GFP-Nls, 4D), which was shown to force G1–S phase transition in neuronal progenitors.16 We tested whether overexpression of the 4D protein complex was sufficient to induce cell death in hippocampal neurons in vitro (Figure 1a). We observed that about 73% of the neurons transfected with the 4D-GFP construct were act-Caspase 3 (act-Casp3) positive. Interestingly, about 13% of the GFP/act-Casp3 positive neurons also displayed binucleate morphology, as indicated by DAPI staining. About 27% of the transfected neurons did not display any sign of cell death (n=50 transfected cells counted; Figure 1b). The nontransfected neurons and the neurons transfected with GFP control plasmid showed little act-Casp3 staining (Figure 1a and data not shown). To further investigate whether CCR occurs in response to excitotoxicity, we injected wild-type (WT) mice with 25 mg/kg dose of KA given intraperitoneally (i.p.) and analyzed the nuclear localization of Ccnd1 in the hippocampus as compared with Saline controls (Figure 1c). Two days following KA, we observed a sixfold increase in the percentage of neurons showing Ccnd1 immunolabeling in cornu ammonis (CA) fields (n=1592 neurons from n=3 mice for Saline and n=3032 neurons from n=4 mice for KA; P<0.0001, Student's t-test; Figure 1d). We next assessed whether BrdU (5-bromo-2'-deoxyuridine) incorporation occurs in neurons in response to KA. We applied one single injection of BrdU (100 mg/kg) 12 h before sacrifice at 2 days after KA injection. Incorporation of BrdU in principal neurons of the hippocampal CA fields (Figure 1e) was visible within 12 h of BrdU injection. Interestingly, all of the BrdU-positive neurons showed nuclear localization of the Notch1 protein, along with a condensed nuclear morphology suggesting an impending cell death (Figure 1e). Cell quantitation indicated that upon KA, about 2% of CA field neurons are BrdU positive (Figure 1f), whereas in controls, no BrdU positive neurons were visible (n=951 cells, n=4 mice per condition; P<0.001, t-test). We next investigated whether Notch1 signaling is involved in excitotoxic neuronal death in vitro and in vivo. Using a bath application of 40 μM of NMDA, we observed that NICD1 levels increases within 1 h (Supplementary Figures 1A and B) and persists for over 6 h (Supplementary Figure 1E). Within this time frame, Notch pathway activation, as indicated by the Notch transcriptional reporter TNR–GFP,17 is also significantly increased in neuronal cultures (n=5 experiments; P<0.05, t-test; Supplementary Figures 1C and D). Interestingly, after 3 and 6 h of NMDA application, the progressive rise in NICD1 is associated with increasing amount of Ccnd1 protein (Supplementary Figure 1E). In vivo, we found that NICD1 rose by up to 60% within 12 h of KA treatment (n=8 mice per condition, P<0.05, t-test; Figures 1g and h). By co-immunoprecipitation (co-IP) assay on CA field lysates, we found that KA treatment results in an increased interaction of Notch1 protein with importin7 (Kpna6; Figure 1i) suggesting increased nuclear translocation of Notch1.18 We also found that KA treatment results in a reduction in binding of Notch1 to the ubiquitin ligase fbw7 and in reduced ubiquitination of the Notch1 protein, possibly increasing its bioavailability (n=4 independent IP; Figure 1i). We further determined that the excitotoxic KA treatment results in a significant increase in the transcription of Notch/RBPJK canonical targets: Hes1 (n=5–8 mice per condition; P<0.05, t-test), Hey1 (n=4–5 mice per condition; P<0.05, t-test) and C-myc (n=4–6 mice per condition; P<0.05, t-test; Figure 1j). As expected, the KA-mediated increase in transcription of Hes1, Hey1 and C-myc, was absent in the RBPJKcKO mice (n=4–6 mice per group; hes1: P=0.6; hey1: P=0.7; c-myc, P=0.9, t-test). The responses in Hes1, Hey1 and C-myc expression upon KA treatment were significantly different between WTs and RBPJKcKOs (Hes1: F1,7=6.51, P<0.05; Hey1: F1,7=6.18, P<0.05; C-myc, F1,10=6.79, P<0.05, one-way ANOVA; Figure 1j). The significantly higher baseline transcript levels for C-myc (P<0.01, t-test) and marginally higher levels of Hey1 in RBPJKcKOs can be explained by the role of RBPJK as a constitutive repressor, when not activated by NICD (Figure 1j).19 On the basis of the evidence that Notch signaling is induced upon KA excitotoxicity, we addressed whether nuclear (canonical) Notch signaling has a causative role in cell death upon KA. We crossed the RBPJKflox/flox mice20 with the CamKII::cre (T29-1) driver line.21 The RBPJK deletion from hippocampal CA fields was confirmed at both protein and mRNA levels using immunohistochemistry (Figure 2b) and in situ hybridization (data not shown), respectively. Despite no apparent difference in the seizure behavior (F1,28=1.75, P=0.2, one-way ANOVA; Figure 2a), the RBPJKcKO mice showed a significant protection against KA-induced neuronal death as compared with WTs at 2 (n=4 mice per genotype; P<0.01, t-test; Figures 2c and e) and 5 days (n=5 mice per genotype; P<0.01, t-test) as assessed by hematoxylin and eosin (HE) staining (Figure 2e). The number of condensed cells observed at 12 h following KA administration in both genotypes were comparable to the controls (n=3 mice per genotype; P=0.63, t-test). This indicates that at 12 h, processes of neuronal vulnerability to excitotoxicity can be investigated without significant interference from neuronal demise. Neuronal cell death was also assessed using TUNEL staining (Figure 2d). In the RBPJKcKO mice a comparable neuro-protection was observed at 2 (n=4 mice per genotype; P<0.05, t-test) and 5 days (n=5 mice per genotype; P<0.05, t-test) following KA injection as compared with WT (Figure 2f). These results show that the canonical Notch signaling has a causative role in KA-mediated neurodegeneration and that Notch signaling activation correlates with CCR in neurons following excitotoxicity.
Figure 1.
Cell cycle reentry following KA insult is associated with nuclear Notch signaling. (a) Representative fluorescence immunohistochemistry photomicrographs of primary neuronal cultures transfected with a Ccnd1:CDK4 (4D) construct colabeled with act-Casp3 and DAPI. Arrow indicates a binucleate 4D+ neuron. (b) Pie chart summarizing the proportion of neurons labeled with GFP only, those colabeled with act-Casp3 or those colabeled with act-Casp3 and with binucleate morphology (n=50 cells). (c) Representative immunofluorescence photomicrograph of Ccnd1 expressing neuron from the WT hippocampi treated with KA. (d) Quantitation of the percentage of Ccnd1-positive neurons in Saline and KA hippocampi (n=4 WT mice per condition). (e) Hippocampal CA1 layer, 2 days following KA insult, shows the presence of BrdU+ neurons expressing Notch1 in the nucleus (white arrow and insert). (f) Graph summarizing the proportion of BrdU+ cells as a percentage of NeuN+ neurons in the CA layer following KA as compared with Saline controls (n=951 cells from n=4 mice per condition). (g) Representative immunoblot shows the extent of cleaved Notch1 in hippocampal lysates at 12 h after KA as compared with Saline controls. (h) Optical density quantitation of the NICD1 bands 12 h after Saline and KA injection (n=8 per condition). (i) Representative immunoblot on immunoprecipitated hippocampal lysates using an antibody against the cytoplasmic tail of Notch1 shows the following interactions- Notch1:Kpna6 and Notch1:fbw7, and ubiquitination of Notch1 in Saline and KA conditions. No trace of protein is visible in the control IP sample with serum. GAPDH is used to control for loading of the inputs and contamination in the IP samples (n=4 independent IPs). (j) Graph summarizing the expression of the Notch targets; Hes1, Hey1 and C-myc, upon KA in WT and RBPJKcKO hippocampi as compared with the respective Saline controls (n=4-8 mice per condition). Asterisks indicate significant differences. Error bars represent mean±S.E.M. Scale bars in A and C is 50 μm and E is 100 μm
Figure 2.
RBPJKcKOs display significant neuroprotection in response to KA. (a) Graph summarizing the time course of seizure behavior in WT and RBPJKcKO mice within the 120 min of observation (n=15 mice per genotype). (b) Immunohistochemistry for RBPJK shows targeted ablation of RBPJK expression in CA1, CA3 fields in the RBPJKcKO as compared to WT. (c) Representative photomicrographs of HE staining on hippocampal sections from WT and RBPJKcKO mice in CA1 and CA3 fields. (d) Representative images of TUNEL staining in WT and RBPJkcKO CA1 and CA3 fields. (e) Graph summarizing the counts of condensed nuclei as revealed by HE stainings in WT and RBPJKcKO at 12 h, 2 and 5 days following systemic challenge with KA as compared with Saline controls (n=3–5 mice per genotype per condition). (f) Graph summarizing the counts of TUNEL positive nuclei in WT and RBPJKcKO, 2 and 5 days following KA administration as compared to Saline controls (n=3–5 mice per genotype per condition). Asterisks indicate significant differences. Error bars represent mean±S.E.M. Scale bar in B-D is 200 μm
KA treatment results in Notch-dependent activation of Akt pathway
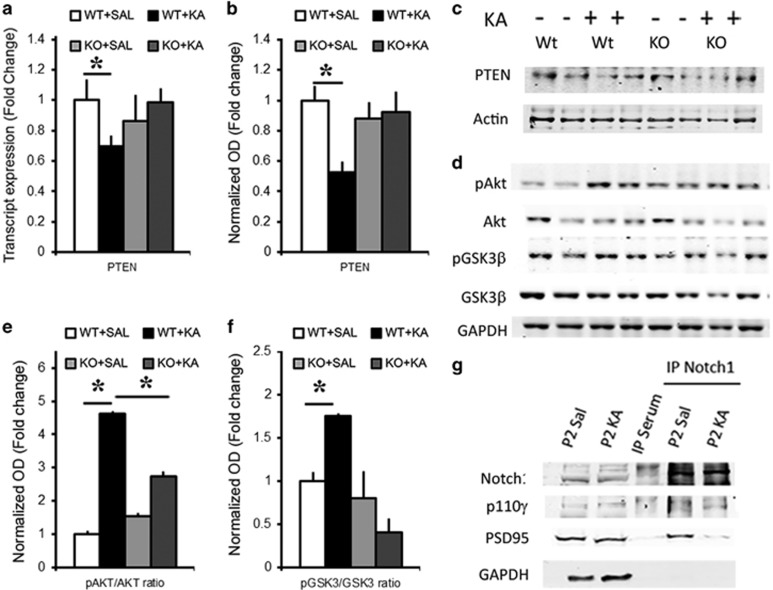
Tumor suppressor phosphatase and tensin homolog (PTEN) is a well-known cell cycle repressor through inhibition of Akt,22 which appears to be downregulated upon ischemic injury.23 We found that KA treatment results in a significant decrease in the transcript expression of the PTEN gene within 12 h in WT (n=3–4 mice per condition; P<0.05, t-test; Figure 3a). Interestingly, no change was observed in RBPJKcKO hippocampi upon KA (n=3–4 mice per condition; P=0.87, t-test). The response to KA in PTEN transcripts (F1,5=23.52, P<0.01, one-way ANOVA) and PTEN protein levels (n=8 mice per condition; P<0.05, t-test; Figures 3b and c) were significantly different between WT and RBPJKcKO. This supports the evidence that PTEN is under Notch signaling regulation through Hes1.24 Both mRNA and protein data showed no change in PTEN protein levels in RBPJKcKO hippocampi upon KA (n=6–8 mice per condition; P=0.96, t-test) and the difference between the two genotypes was significant (F1,6=8.8, P<0.05, one-way ANOVA). As a result of PTEN downregulation upon KA administration, Akt phosphorylation was increased in WT hippocampi (n=3 per condition; P<0.01, t-test) and this change was significantly higher than in the RBPJKcKOs (F1,5=26.15, P<0.01, one-way ANOVA), despite a significant increase in Akt phosphorylation in the KOs (n=3 per condition; P<0.01, t-test; Figures 3d and e). Phosphorylated Akt inactivates GSK3β through phosphorylation.25 As expected, we found that the excitotoxic KA treatment results in an increased phosphorylation of GSK3β in WT hippocampi (n=3–4 mice per condition; P<0.01, t-test), and this increase was absent in the RBPJKcKO mice (n=5 mice per condition; P=0.29) and the difference between the two genotypes was significant (F1,6=22.14, P<0.005, one-way ANOVA; Figures 3d and f). On the basis of the evidence that Notch1 activates Akt pathway by directly interacting with Phospho-inositol 3 Kinase (PI3K),26 and recent work showing that PI3Kγ is induced by excitotoxic NMDA stimulation,27 we investigated a possible interaction of Notch1 protein with PI3Kγ (p110γ). By using synaptosomal fractionation followed by co-IP, we found an increase in the interaction of Notch protein with p110γ following KA challenge (n=3 independent IP; Figure 3g). Interestingly, the anchoring to the postsynaptic density, as indicated by the interaction Notch1:PSD95, is lowered upon KA suggesting that mobilization of the Notch1 receptor takes place upon excitotoxicity. These results indicate that Notch1 may induce Akt signaling through a canonical transcriptional regulation of PTEN as well as a noncanonical interaction with PI3Kγ.
Figure 3.
KA induces Notch-dependent activation of Akt pathway. (a) Graph summarizing PTEN transcript expression: In WTs, KA application reduces the levels of PTEN transcripts as compared with Saline, whereas no change is observed in RBPJKcKOs (n=3–4 per condition). (b) Graph summarizing PTEN protein expression: In WTs, KA application reduces the levels of PTEN protein as compared with Saline, whereas no change is observed in RBPJKcKOs (n=6–8 per condition). (c) Representative immunoblot of hippocampal CA lysates shows that PTEN is decreased upon KA treatment in WT hippocampi but not RBPJKcKO as compared with the respective Saline controls. β-actin is used as loading control. (d) Representative images showing Akt and GSK3β phosphorylation upon KA treatment in WT and RBPJKcKO hippocampi as compared with Saline condition. GAPDH is used as a loading control. The treatments and genotypes are the same as in c. Graphs summarizing the differences in optical density (e) of phosphorylated Akt versus total Akt (n=3 per condition) and (f) of phosphorylated GSK3β versus total GSK3β upon KA as compared with Saline in WT and RBPJKcKO hippocampi (n=3–5 per condition). (g) Representative immunoblot on immunoprecipitated hippocampal crude synaptosomal fractions using an antibody against the cytoplasmic tail of Notch1 shows the interaction between Notch1:p110γ and Notch1:PSD95 in Saline and KA conditions. No trace of protein is visible in the control IP sample with serum. GAPDH is used as a loading control for the inputs and to detect contamination in the IP samples (n=3 independent IPs). Asterisks indicate significant differences. Error bars represent mean±S.E.M.
KA-mediated activation of Akt pathway results in G1–S transition
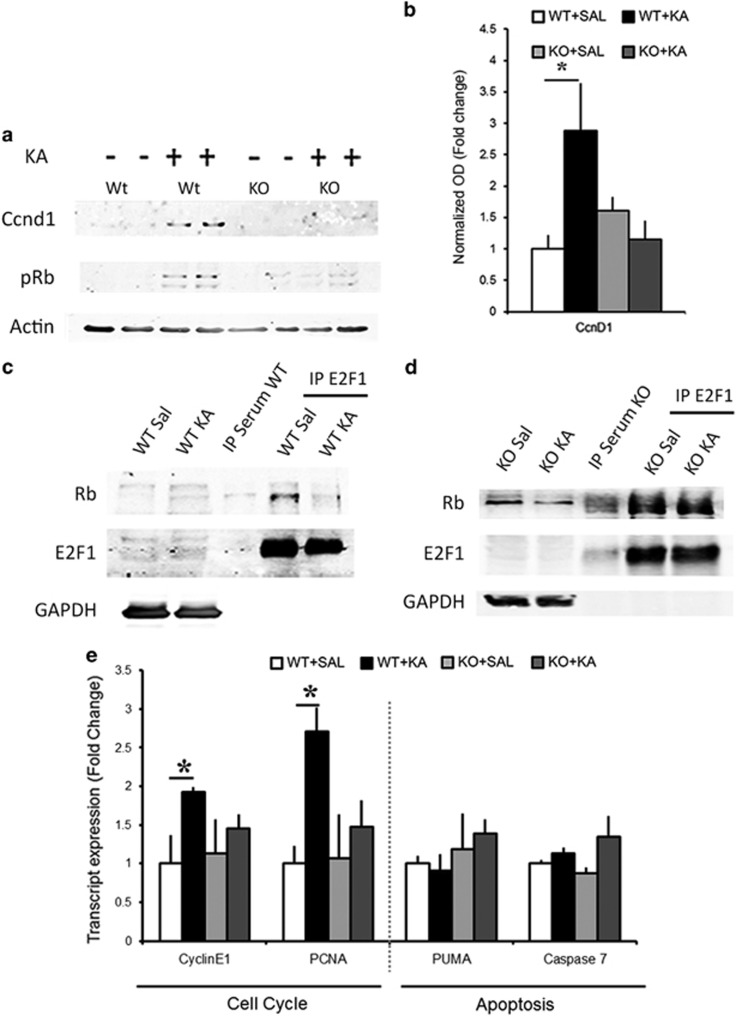
We addressed whether KA-induced inactivation of GSK3β results in greater bioavailability of Ccnd1, a known direct target of GSK3β.28 We observed a significant increase in the Ccnd1 levels in KA treated WT hippocampi as compared to Saline controls (n=6 mice per condition; P<0.05, t-test; Figures 4a and b). Baseline levels of Ccnd1 in neurons appeared low and increased with excitotoxic activity also in vitro (Supplementary Figure 1E). In line with the sustained levels of active GSK3β in the RBPJKcKO hippocampi, Ccnd1 levels remained low following KA (n=6 mice per condition; P=0.24, t-test) and the difference between genotypes was significant (F1,10=7.73, P<0.05, one-way ANOVA; Figures 4a and b). During the G1–S transition, Ccnd1 forms a complex with CDK4/6 and phosphorylates the retinoblastoma protein (Rb).29 We found that KA treatment results in increased phosphorylation of Rb in WT hippocampi, which was much less pronounced in the RBPJKcKOs (n=4-6 per condition; Figure 4a). Increased phosphorylation of Rb results in its detachment from E2F1 and disinhibition of E2F1-mediated transcription.30 We found that KA treatment results in a decreased interaction between Rb and E2F1 as compared with Saline controls (n=3 independent IP; Figure 4c). In line with the failure in Rb phosphorylation in the RBPJKcKOs, binding of the Rb:E2F1 complex persists after KA treatment (Figure 4d; n=3 independent IP). We further observed that transcript expression of E2F1 targets was differentially regulated upon KA. We found a significant increase in the expression of Cyclin E1 (n=5–6 mice per condition; P<0.05, t-test) and PCNA (n=3–4 mice per condition; P<0.05, t-test) in KA treated WT hippocampi as compared with Saline controls (Figure 4e). This increase was not seen in the RBPJKcKO mice upon KA (n=4–6 mice per group; Cyclin E1: P=0.42. PCNA: P=0.52, t-test). The induction in Cyclin E1 in the WTs was significantly different as compared with RBPJKcKOs (F1,10=13.78, P<0.005, one-way ANOVA) and there was a trend toward a differential increase in PCNA in WT as compared with KOs (F1,5=5.67, P=0.07, one-way ANOVA) (Figure 4e). On the other hand, the expression of proapoptotic E2F1 targets, PUMA and Casp7, was unchanged following KA treatment at 12 h in both WT (PUMA: n=4–6 mice per condition; P=0.62; Casp7: n=4–6 mice per condition; P=0.21, t-test) and RBPJKcKO (PUMA: n=4–6 mice per condition; P=0.60; Casp7: n=4–6 mice per condition; P=0.16, t-test; Figure 4e). This suggests that Notch signaling can regulate the expression of the cell cycle-related E2F1 targets in neurons.
Figure 4.
KA treatment induces Ccnd1/E2F1 activation and is Notch signaling dependent. (a) Representative immunoblots of hippocampal CA lysates show the changes in Ccnd1 expression and Rb phosphorylation in WT and RBPJKcKO hippocampi upon KA as compared with Saline controls. (b) Graph summarizing the fold changes in the optical density of Ccnd1 in WT and KO hippocampi (n=6 per condition and genotype). (c) Representative immunoblot shows that the E2F1:Rb interaction is reduced upon KA as compared with Saline in the WT mice. No trace of protein is visible in the control IP sample with serum. GAPDH is used to control for loading of the inputs and contamination in the IP samples (n=3 independent IPs). (d) Representative immunoblot shows that Rb:E2F1 interaction is prominent even after KA treatment in the RBPJKcKO mice as compared with Saline. The GAPDH band serves as in C (n=3 independent IPs). (e) Graph summarizing transcript expression of E2F1 targets in WTs and RBPJKcKOs upon KA as compared with Saline (n=3–6 per condition). Asterisks indicate significant differences. Error bars represent mean±S.E.M.
Neurodegeneration is a result of Notch/pAkt/Ccnd1 axis activation
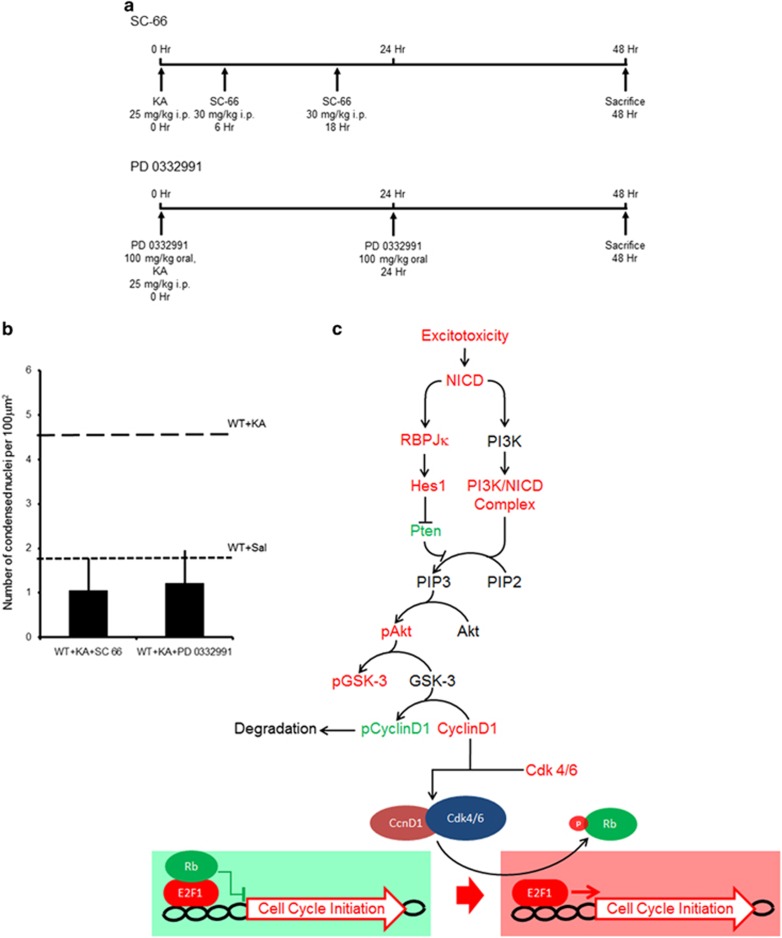
We next determined whether the Notch-mediated activation of the pAkt/Ccnd1 pathway is the cause of neurodegeneration. We combined the in vivo KA model with blockade of the pathway at various steps (Figure 5a). We found that pharmacological blockade of either pAkt (SC-66) or Ccnd1 (PD 0332991) activity was sufficient to provide a significant protection against KA-induced neurodegeneration in WT hippocampi at 2 days following KA as compared with the mice treated with KA only (n=3 mice treated with SC-66; P<0.001; n=4 mice treated with PD 0332991; P<0.001, t-test; Figure 5b). This effect was comparable to the loss of RBPJK (F2,10=2.23, P=0.16, one-way ANOVA; Figure 2e). Taken together, these results suggest that excitotoxic neuronal death, particularly at the early stages, happens as a result of erroneous CCR through Notch/Akt pathway activation (Figure 5c).
Figure 5.
Blockade of either Akt or Ccnd1 activity confers neuroprotection comparable to the RBPJK loss. (a) Scheme depicting the strategy and time course of pharmacological treatment with SC-66 and PD 0332991. (b) Graph summarizing the counts of condensed nuclei in hippocampal CA fields upon application of an Akt (SC-66; n=3 mice per condition) and a Ccnd1 (PD 0332991; n=4 mice per condition) inhibitor following KA challenge. (c) Flow chart summarizing the proposed pathway involving Notch, Akt, Ccnd1 and E2F1 leading to cell cycle reentry in neurons. Error bars represent mean±S.E.M.
Discussion
In this work, we addressed the involvement of Notch signaling in neuronal vulnerability to excitotoxicity. Notch signaling has been hypothesized to have a causative role in brain damage following both acute insults, such as stroke,14, 15 as well as chronic neurodegenerative disorders such as AD.31 Interestingly, excitotoxicity has been strongly implicated in the pathophysiology of both of these categories of disorders.32 Here we show that systemic KA administration as an in vivo model of excitotoxicity can induce Notch pathway activation leading to CCR.
Several studies have identified changes in the expression of cell cycle-related genes in neuronal populations vulnerable to progressive neurodegenerative disorders. Signs of CCR, such as quadruploid neurons and expression of cell cycle genes have been observed before plaque deposition in postmortem brains of preclinical and mild AD patients.33 Furthermore, CCR has also been observed following ischemic stroke,34 traumatic brain injury,35 spinal cord injury36 and temporal lobe epilepsy.37 This suggests that this event is not specific only to the neuronal populations intrinsically vulnerable to heritable- or aging-related neurodegenerative disorders, but may be a direct result of excitotoxicity in neurons. Indeed, we show that administration of KA in vivo leads to incorporation of BrdU in the principal neurons of the hippocampal CA fields and is associated with nuclear localization of Notch1. In addition, for the first time, we show that the overexpression of a Ccnd1:CDK4 complex16 is sufficient to cause neuronal demise in primary hippocampal cultures in absence of any known neurodegenerative stimulus. Only a small percentage of neurons overexpressing Ccnd1:CDK4 in the nucleus show a binucleate morphology (13%). This is consistent with the human data from AD patients indicating that, despite the significant numbers of neurons upregulating cell cycle genes (1 in 15),4 only a minute percentage of those show a binucleate morphology (1 in 20 000).38 We hypothesize that G1–S transition is the cell death triggering event in neurons. Nevertheless, depending on the repertoire of resistance mechanisms, individual neurons can survive for different lengths of time, and hence proceed to different stages of cell cycle at the time of death. Therefore, it is possible that neurons overexpressing cell cycle-related genes make the G1–S transition; however, only very few would survive through anaphase, in order to display binucleate morphology. Also, incorporation of BrdU occurs in 2% of the neurons after KA application. This number is comparable to the number of neurons expressing Ccnd1, a protein that peaks during the transition to the S-phase. The proportion of BrdU and Ccnd1 positive neurons is in line with the number of nuclei showing condensed morphology in HE staining and TUNEL reactivity when the approximate neuronal density and the section thickness is taken into consideration. These results along with the robust neuroprotection conferred by the loss of Ccnd1 activity hints at a strong causative role of cell cycle initiation in neurodegeneration.
Notch functions as a proliferation factor in neuroblasts in response to physical exercise and also KA injury.13 Similar to neuroblasts, we show that upon KA excitotoxicity, Notch cleavage and signaling to the nucleus is sustained in pyramidal CA fields. According to our own and previous studies, Notch pathway activation in physiological condition is very limited and Notch transcriptional targets are strongly induced only after injury.39 This suggests that Notch canonical activity may be instrumental for neuronal demise upon brain insult but does not contribute to age-dependent neurodegeneration.40 Indeed, we observe that conditional loss of RBPJK in the hippocampus provides significant neuroprotection from KA-mediated cell death. Similarly, systemic γ-secretase inhibition can provide significant neuroprotection in a model of stroke in mice.15, 41
We further showed that excitotoxic KA treatment leads to increase in phosphorylation of Akt, through PTEN-mediated disinhibition and partially through the activation of PI3Kγ. Our study indicates that PTEN is negatively regulated by canonical Notch signaling in line with repression of PTEN by Hes1.24 On the other hand, we observe that upon KA the interaction between Notch1 and the catalytic domain of PI3K, p110γ, is stronger. It has been previously shown that excitotoxic stimulation can activate p110γ27 and several studies in cancer have reported a critical cross-talk between Notch1 and PI3K/Akt in cellular proliferation.42 In support of the canonical Notch regulation of PI3K/Akt signaling, we observe that Akt phosphorylation in the RBPJKcKOs is significantly reduced. Nevertheless, the RBPJKcKOs upon KA display a small (1.7-fold) but significant increase in Akt phosphorylation, which may result from the existing interaction between Notch1 and PI3Kγ. In fact, although the canonical Notch pathway is abrogated in the RBPJKcKO mice, the noncanonical interaction of activated Notch with PI3Kγ may account for increased Akt phosphorylation. Once Akt is activated, it inactivates GSK3β through phosphorylation in WTs, but not in RBJKcKOs. As GSK3β activity is required for Notch inactivation,43, 44 it is possible that in absence of canonical Notch signaling, as in the RBPJKcKOs, compensatory steady levels of GSK3β may be adopted to reduce the amount of NICD as transcriptional signaling is shunted.
The active unphosphorylated form of GSK3β has been shown to phosphorylate Ccnd1, leading to its degradation and thus keeping the cells from entering S-phase of cell cycle.28 Indeed, we show that inactivation of GSK3β in hippocampal CA fields in response to excitotoxicity is associated with increased bioavailability of Ccnd1, which does not occur in the RBPJKcKOs. Ccnd1, in association with CDK4, can subsequently phosphorylate Rb releasing the binding between Rb and E2F1 and allowing for E2F1 dependent transcription to take place.29 Activation of E2F1-mediated transcription in neurons upon injury, AD and PD has been reported before and has been associated to neuronal damage.9, 45, 46 In our study, we observe that upon KA, the Ccnd1/Rb/E2F1 axis is positively regulated leading to an increase in transcription of PCNA and Cyclin E1 in neurons. Such regulation was absent in the RBPJKcKOs. Nevertheless, in both genotypes, no change in pro-apoptotic E2F1 target genes could be observed. The differential regulation of proapoptotic versus cell cycle-related targets may be specific to neurons, in line with a previous report.47 On the whole, these data show that Notch signaling positively regulates the Akt/Ccnd1 axis leading to cell cycle-related gene expression (Figure 5c).
On the basis of the role of Akt in the activation of CCR, we observe that the blockade of Akt after KA injection at the time when Akt activity is maximal 48 confers neuroprotection. The effects of blockade of the Akt pathway on neuronal vulnerability appear paradoxical with some studies showing that such blockade is beneficial,7 while others reporting it to be detrimental.49 It was also shown that Rapamycin causes paradoxical effects on KA-induced neurodegeneration through pAkt signaling, depending on the time of administration.48 To further confirm the role of CCR in cell vulnerability, we used a potent CDK4 antagonist PD 0332991 50 to block Ccnd1/CDK4 activity. In line with our hypothesis, Ccnd1 blockade provides neuroprotection comparable to the loss of transcriptional Notch signaling and the Akt blockade. These data show that the activation Notch1/Akt/Ccnd1 pathway has a causative role in excitotoxicity-mediated neurodegeneration.
Taken together, we have shown that the neurons die spontaneously after CCR. Activation of canonical Notch signaling is facilitated upon excitotoxic insult, which in turn leads to the activation of Akt signaling and an increase in the bioavailability of Ccnd1, which results in E2F1-mediated CCR. Our study highlights a novel Notch signaling-dependent pathway leading to erroneous CCR, which we believe has tremendous therapeutic potential. Further studies will be needed to investigate the link between CCR and initiation of neuronal death pathways in neurons.
Materials and Methods
Animals
All mice experiments were performed with permission of the local animal care committee (Canton of Fribourg, Switzerland) and according to the present Swiss law and the European Communities Council Directive of November 24 1986 (86/609/EEC). All animals (2–4 months of age, 30–35 g) were housed on 12 h light–dark cycle with access to food and water ad libitum. RBPJKcKO and WT littermate control (RBPJKflox/flox, and CamKII::Cre) mice were obtained by crossing RBPJKflox/flox mice20 to the CamKII::Cre (T29-1) mouse line21 on a C57BL6/129 background. The TNR–GFP mouse line17 is maintained and distributed by the Jackson Laboratory (Bar Harbor, ME, USA).
Kainate treatment
The choice of drugs and dosage was based on previous studies. To investigate the KA-mediated excitotoxicity, mice were single housed for 30 min before the KA treatment. KA (Abcam, Cambridge, UK) was injected i.p. at 25 mg/kg of the body weight. This dosage was shown to induce seizures and neurodegeneration in the hippocampus.51 Control mice were injected i.p. with an equivalent volume of vehicle (0.9% saline). Mice were observed for 2 h after the KA treatment and seizure behavior was scored every 10 min during the observation period as described previously.51 After the 2 h observation period, mice were returned to their home cage. Mice were then sacrificed at various time points after the KA injections to assess molecular and histological changes.
Pharmacological treatments
All mice undergoing pharmacological treatment were sacrificed 2 days after KA treatment and transcardially perfused with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS). For BrdU analysis studies, mice were injected i.p. with a single dose of 100 mg/kg of BrdU (Abcam), 12 h prior to sacrifice. For studies involving pharmacological blockade of pAkt, 30 mg/kg of SC-66 (Sigma-Aldrich, St. Louis, MO, USA) was injected i.p.52 into WT mice at 6 and 18 h after the KA injection (Figure 5a). Our data and those from earlier reports show that the pAkt levels peak after excitotoxic neuronal damage, only briefly during the 6–12 h period.49 Based on these data and the pharmacokinetics of SC-66, we blocked the activity of Akt pathway 6 h on either sides of the 12 h peak (6 and 18 h). For blockade of Ccnd1 activity, mice were fed with 100 mg/kg of specific inhibitor of CDK4/6 activity, PD 0332991 isethionate (Sigma-Aldrich),50 orally using an oral gavage, just before KA injection and once again 24 h later (Figure 5a).
Cell Cultures and in vitro Manipulations
Neuronal cultures were prepared from the hippocampus of E17 WT or TNR–EGFP embryos and plated on poly-l-lysine-coated 60 mm dishes or 18 mm glass coverslips. Cultures were maintained in P-neuronal media (PAA, Pasching, Austria) supplemented with Neuromix (PAA) and 10% horse serum as previously described.53 After 10 days in vitro (d.i.v), neurons were either transfected with 4D and GFP (gift from F Calegari, Dresden University)16 constructs and fixed 1 day later. At 14 d.i.v., neurons were exposed for different times to bath application of 40 μM NMDA (Tocris, Bristol, UK). Control cultures were remained untreated. Cultures were then fixed using 4% PFA or lysed in ice-cold PBS.
Antibodies
Following antibodies were used for immunohistochemistry in this study: rat anti BrdU (1 : 200; ab6326, Abcam), mouse anti Ccnd1 (1 : 250; MA1-10324, Thermo Fisher Scientific, Waltham, MA, USA), mouse anti NeuN (1 : 250; MAB377, Chemicon/Millipore, Darmstadt, Germany), rabbit anti NeuN (1 : 250; Abcam), rabbit anti-act-Casp3 (1 : 500; ab2302, Abcam) and rabbit anti-RBPJK (1 : 500; #5313, Cell Signaling, Cambridge, MA, USA). Following antibodies were used for immunoblot in this study: rabbit anti Notch1 (1 : 1000; 07-220, Upstate/Millipore, Darmstadt, Germany), mouse anti Notch1 (mN1A; 1 : 250; SAB4700742, Sigma-Aldrich), rabbit anti cleaved N1, NICD1 (1 : 1000; #2421, Cell Signaling), rabbit anti Importin7 (Kpna6; 1 : 1000; ab105350, Abcam), mouse anti Fbw7 (1 : 1000; NBP1-59631, Novus, Cambridge, UK), mouse anti PSD95 (1 : 500; sc-32290, Santa Cruz Biotechnology, Dallas, TX, USA), mouse anti Ubiquitin (1 : 1000; sc-8017, Santa Cruz Biotechnology), rabbit anti PTEN (1 : 500; ab154812, Abcam), mouse anti Akt (1 : 1000; sc-5298, Santa Cruz Biotechnology), rabbit anti phospho-Akt (1 : 1000; #4060, Cell Signaling), mouse anti GSK3β (1 : 1000; NBP1-47470, Novus), rabbit anti phospho-GSK3β (1 : 1000; ab107166, Abcam), rabbit anti-RBPJK (1 : 1000; #5313, Cell Signaling), mouse anti Rb (1 : 100; #MS595P, Thermo Fisher Scientific), rabbit anti phospho-Rb (1 : 1000; orb14852, Biorbyt, Cambridge, UK), mouse anti Ccnd1 (1 : 200; MA5-12702, Thermo Fisher Scientific), rabbit anti E2F1 (1 : 500; ab112580, Abcam), mouse anti GAPDH (1 : 5000; 1D4, Novus) and mouse anti β-Actin (1 : 2000; sc-8432, Santa Cruz Biotechnology).
Following antibodies were used for immunoprecipitation in this study: rabbit anti Notch1 (10 ug/1.2 mg of lysate; 07-220, Upstate/Millipore), rabbit anti E2F1 (10ug/1.2 mg of lysate; 1 : 500; ab112580, Abcam).
The secondary antibodies were used in the study for immunohistochemistry were: directly conjugated to Cy2, Cy3 or Cy5 in donkey-raised secondary antibodies (all 1 : 500; Jackson ImmunoResearch Europe Ltd, Suffolk, UK). For the BrdU staining, biotinylated in donkey raised secondary antibody against rat IgG (1 : 250, Vector Laboratories, Burlingame, CA, USA) followed by Streptavidin-FITC (1 : 1000, Jackson ImmunoResearch Europe Ltd) was used. After the completion of immunofluorescence protocols, the sections were stained with DAPI to visualize nuclear morphology, mounted on Super frost slides (Thermo Fisher Scientific) and coverslipped with a custom made Polyvinyl alcohol and 1,4-diazabicyclo[2.2.2]octane-based mounting media. For chromagen immunohistochemistry, biotinylated in horse-raised secondary antibody against rabbit (1 : 500, Jackson ImmunoResearch Europe Ltd) was developed using Streptavidin-HRP (1 : 1000, Jackson ImmunoResearch Europe Ltd) followed by 3,3'-Diaminobenzidine (Thermo Fisher Scientific). Sections were mounted as previously described, air dried, dehydrated and after immersion in Xylene coverslipped with Entelan (Fluka, Buchs, Switzerland).
Histological Analysis of cell death, immunohistochemistry and immunofluorescence
Mice were sacrificed by transcardial perfusion with 0.9% saline solution followed by a solution of 4% PFA. The brains were post-fixed overnight in 4% PFA and subsequently cryo-protected for two overnights in 30% sucrose solution. 50 μm thick coronal sections were cut on a cryostat (Leica, Wetzlar, Germany) in the anterior–posterior plane: −1.55 to −2.355 from Bregma. The sections were stored in PBS with 0.01 sodium azide (Sigma-Aldrich) at 4 °C until further analysis.
For visualizing the number of dying cells having condensed nuclear morphology, HE staining and TUNEL methods were used. Stained and mounted sections were imaged using a 20 × objective under a light transmission microscope, DMRX (Leica). CA1 and CA3 field were imaged from three to five consecutive sections. Number of condensed cells and TUNEL positive cells were quantified post hoc on the acquired images by a blinded investigator using ImageJ (NIH). Each image was tiled using a grid of 100 μm2 large squares. Cells per square area were counted over the CA fields. The immunohistochemistry stainings using fluorescently tagged secondary antibodies were done as described previously.12
Transcript expression analysis by RT-qPCR
12 h after the KA injection, mice were transcardially perfused with 0.9% saline solution. The brain was dissected out and was transferred into an ice-cold PBS solution. Hippocampus was dissected out and CA region was obtained by gently peeling the DG apart under a dissection microscope (Nikon, Chiyoda, TK, Japan). The tissue was flash-frozen in liquid nitrogen and stored at −80 °C until further use. Total RNA was extracted using peqGOLD TriFast reagent (Peqlab, Erlangen, Germany). Isolated RNA was quantified and the quality was assessed with a Nanodrop (NanoDrop2000, Thermo Scientific, Waltham, MA, USA). 2 μg of RNA per sample was subjected to reverse transcription using M-MLV Reverse Transcriptase (Promega, Madison, WI, USA). Gene expression analysis was done by RT-qPCR (GoTaq qPCR Master Mix, Promega) using gene specific primers (Supplementary Table 1) with a Rotorgene (Qiagen, Duesseldorf, Germany). Quantitative PCR data analysis was performed using the ΔΔCt method as described previously.54 Gene expression analysis data were normalized to the endogenous housekeeping gene, β-actin.
Co-immunoprecipitation assays and western blot analysis
Hippocampal CA fields were dissected as described above and snap frozen. Hippocampal CA tissues were either fractionated to obtain the crude synaptosomal fraction (n=3 bilateral CA fields per fractionation per condition) or homogenized using non-ionic buffer. Protein concentrations from both preparations were determined using the BCA method (Carl Roth, Karlsruhe, Germany). Notch1 immunoprecipitation was performed on the membrane enriched fractions from hippocampal CA fields12 and hippocampal CA lysates, whereas E2F1 IP was performed on hippocampal CA fields lysates only (two bilateral CA fields per IP per condition). The homogenates were incubated for 1.5 h with 10 μg of primary antibodies or with 10 μg of Goat serum (PAA). Protein G Magnetic beads (Thermo Fisher Scientific) were then added to the samples, which were incubated for another 1.5 h. Next, the beads were washed three times (0.1% Triton, 50 mM Tris-HCL pH.7.5, 300 mM NaCl). The fourth wash was performed using the washing buffer containing 0.2% SDS, and the fifth wash with PBS containing 0.1% Triton. The beads were eluted using 50 μl of 2x Laemmli Buffer (Carl Roth). Proteins were separated using standard electrophoresis and western blot techniques. Proteins transferred to a Nitrocellulose membrane (Membrane Solutions, Dallas, TX, USA) and were probed with primary antibodies, and Infrared dye-conjugated secondary antibodies (LiCOR, Bad-Homburg, Germany). An Infrared scanner (LiCOR) was used to visualize the signal. Densitometric analysis to quantitate the intensity of individual protein bands was done using ImageJ software (NIH). Values were then averaged among experiments.
Statistical analysis
Transcript and protein values were normalized to housekeeping genes and the loading controls, β-actin or GAPDH, respectively. Values were averaged among the indicated number of samples. For the cell countings, cells or nuclei per square area (100 μm2) were counted over the CA fields by a blinded investigator (Figures 2e and f). The proportion of activated Casp3 expressing neurons and/or binucleate neurons was calculated as a fraction of the total transfected GFP (4D) positive neurons (Figures 1a and b). Only neurons with supra threshold intensity of BrdU, calibrated on dividing progenitors in subgranular zone of the dentate gyrus, were taken into consideration. Number of GFP+ neurons (Supplementary Figure 1D), CcnD1+ neurons (Figure 1d) and BrdU+ neurons (Figure 1f) were counted as a percentage of the total number of NeuN+ neurons by setting an intensity threshold using the ImageJ cell counter plugin (NIH). All data were compiled and analyzed using Excel and the Real Statistic Add-in (Dr. Charles Zaiontz). Student's t-test was used in all pairwise analysis for statistical comparison. One-way ANOVA with Bonferroni's post hoc test was used when performing multiple comparisons. The seizure score over the 120 min observation period was quantified by calculating the area under the curve for each animal and the comparison between genotypes was carried out by one-way ANOVA with Bonferroni's post hoc test.
Acknowledgments
This work was supported by funds from the Swiss National Foundation, Swiss Heart Association and Synapsis Foundation. We would like to thank Dr. Nicholas Gaiano for his continuous support to our lab. We thank T Honjo for sharing the RBPJKfloxed/floxed mouse line. F Calegari and B Artegiani for providing us the 4D construct. We are thankful to Dr. Solinas for providing the p110γ antibody. We thank P Clarke and J Puyal for their helpful scientific input on the study.
Glossary
- 4D
Ccnd1-CDK4-GFP-Nls
- AD
Alzheimer's disease
- BrdU
5-bromo-2'-deoxyuridine
- CA
cornu ammonis
- CCR
cell cycle reentry
- HE
hematoxylin and eosin
- KA
kainic acid
- PD
Parkinson's disease
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by N Bazan
Supplementary Material
References
- 1Bredesen DE, Rao RV, Mehlen P. Cell death in the nervous system. Nature 2006; 443: 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Herrup K. Post-mitotic role of the cell cycle machinery. Curr Opin Cell Biol 2013; 25: 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Copani A, Caraci F, Hoozemans JJM, Calafiore M, Angela Sortino M, Nicoletti F. The nature of the cell cycle in neurons: Focus on a ‘non-canonical' pathway of DNA replication causally related to death. Biochim Biophys Acta 2007; 1772: 409–412. [DOI] [PubMed] [Google Scholar]
- 4Yang Y, Mufson EJ, Herrup K. Neuronal cell death is preceded by cell cycle events at all stages of Alzheimer's disease. J Neurosci 2003; 23: 2557–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Pelegrí C, Duran-Vilaregut J, del Valle J, Crespo-Biel N, Ferrer I, Pallàs M et al. Cell cycle activation in striatal neurons from Huntington's disease patients and rats treated with 3-nitropropionic acid. Int J Dev Neurosci 2008; 26: 665–671. [DOI] [PubMed] [Google Scholar]
- 6Ranganathan S, Bowser R. Alterations in G1 to S phase cell-cycle regulators during amyotrophic lateral sclerosis. Am J Pathol 2003; 162: 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Bhaskar K, Miller M, Chludzinski A, Herrup K, Zagorski M, Lamb BT. The PI3K-Akt-mTOR pathway regulates Aβ oligomer induced neuronal cell cycle events. Mol Neurodegener 2009; 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Rashidian J, Iyirhiaro G, Aleyasin H, Rios M, Vincent I, Callaghan S et al. Multiple cyclin-dependent kinases signals are critical mediators of ischemia/hypoxic neuronal death in vitro and in vivo. Proc Natl Acad Sci USA 2005; 102: 14080–14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Absalon S, Kochanek DM, Raghavan V, Krichevsky AM. MiR-26b, upregulated in Alzheimer's disease, activates cell cycle entry, Tau-phosphorylation, and apoptosis in postmitotic neurons. J Neurosci 2013; 33: 14645–14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Park DS, Morris EJ, Bremner R, Keramaris E, Padmanabhan J, Rosenbaum M et al. Involvement of retinoblastoma family members and E2F/DP complexes in the death of neurons evoked by DNA damage. J Neurosci 2000; 20: 3104–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 2011; 69: 840–855. [DOI] [PubMed] [Google Scholar]
- 12Alberi L, Liu S, Wang Y, Badie R, Smith-Hicks C, Wu J et al. Activity-induced Notch signaling in neurons requires Arc/Arg3.1 and is essential for synaptic plasticity in hippocampal networks. Neuron 2011; 69: 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Lugert S, Basak O, Knuckles P, Haussler U, Fabel K, Gotz M et al. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell 2010; 6: 445–456. [DOI] [PubMed] [Google Scholar]
- 14Alberi L, Chi Z, Kadam SD, Mulholland JD, Dawson VL, Gaiano N et al. Neonatal stroke in mice causes long-term changes in neuronal Notch-2 expression that may contribute to prolonged injury. Stroke 2010; 41: S64–S71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Arumugam TV, Chan SL, Jo DG, Yilmaz G, Tang SC, Cheng A et al. Gamma secretase-mediated Notch signaling worsens brain damage and functional outcome in ischemic stroke. Nat Med 2006; 12: 621–623. [DOI] [PubMed] [Google Scholar]
- 16Artegiani B, Lindemann D, Calegari F. Overexpression of cdk4 and cyclinD1 triggers greater expansion of neural stem cells in the adult mouse brain. J Exp Med 2011; 208: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 2007; 449: 351–355. [DOI] [PubMed] [Google Scholar]
- 18Huenniger K, Kramer A, Soom M, Chang I, Kohler M, Depping R et al. Notch1 signaling is mediated by importins alpha 3, 4, and 7. Cell Mol Life Sci 2010; 67: 3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 2009; 137: 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol 2002; 14: 637–645. [DOI] [PubMed] [Google Scholar]
- 21Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ et al. Subregion- and cell type–restricted gene knockout in mouse brain. Cell 1996; 87: 1317–1326. [DOI] [PubMed] [Google Scholar]
- 22Ramaswamy S, Nakamura N, Vazquez F, Batt DB, Perera S, Roberts TM et al. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci USA 1999; 96: 2110–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Li W, Huang R, Chen Z, Yan L-J, Simpkins JW, Yang S-H. PTEN degradation after ischemic stroke: a double-edged sword. Neuroscience 2014; 274: 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 2007; 13: 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Cross DAE, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 1995; 378: 785–789. [DOI] [PubMed] [Google Scholar]
- 26Sade H, Krishna S, Sarin A. The anti-apoptotic effect of Notch-1 requires p56lck-dependent, Akt/PKB-mediated signaling in T cells. J Biol Chem 2004; 279: 2937–2944. [DOI] [PubMed] [Google Scholar]
- 27Brennan-Minnella AM, Shen Y, El-Benna J, Swanson RA. Phosphoinositide 3-kinase couples NMDA receptors to superoxide release in excitotoxic neuronal death. Cell Death Dis 2013; 4: e580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev 1998; 12: 3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet 2001; 10: 699–703. [DOI] [PubMed] [Google Scholar]
- 30Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol 2002; 4: 859–864. [DOI] [PubMed] [Google Scholar]
- 31Berezovska O, Xia MQ, Hyman BT. Notch is expressed in adult brain, is coexpressed with presenilin-1, and is altered in Alzheimer disease. J Neuropathol 1998; 57: 738–745. [DOI] [PubMed] [Google Scholar]
- 32Salińska E, Danysz W, Łazarewicz JW. The role of excitotoxicity in neurodegeneration. Folia Neuropathol 2005; 43: 322–339. [PubMed] [Google Scholar]
- 33Arendt T, Brückner MK, Mosch B, Lösche A. Selective cell death of hyperploid neurons in Alzheimer's disease. Am J Pathol 2010; 177: 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Rashidian J, Iyirhiaro GO, Park DS. Cell cycle machinery and stroke. Biochim Biophys Acta 2007; 1772: 484–493. [DOI] [PubMed] [Google Scholar]
- 35Di Giovanni S, Movsesyan V, Ahmed F, Cernak I, Schinelli S, Stoica B et al. Cell cycle inhibition provides neuroprotection and reduces glial proliferation and scar formation after traumatic brain injury. Proc Natl Acad Sci USA 2005; 102: 8333–8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Di Giovanni S, Knoblach SM, Brandoli C, Aden SA, Hoffman EP, Faden AI. Gene profiling in spinal cord injury shows role of cell cycle in neuronal death. Ann Neurol 2003; 53: 454–468. [DOI] [PubMed] [Google Scholar]
- 37Fiala M, Avagyan H, Merino JJ, Bernas M, Valdivia J, Espinosa-Jeffrey A et al. Chemotactic and mitogenic stimuli of neuronal apoptosis in patients with medically intractable temporal lobe epilepsy. Pathophysiology 2013; 20: 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Zhu X, Siedlak SL, Wang Y, Perry G, Castellani RJ, Cohen ML et al. Neuronal binucleation in Alzheimer disease hippocampus. Neuropathol Appl Neurobiol 2008; 34: 457–465. [DOI] [PubMed] [Google Scholar]
- 39Tatsumi K, Okuda H, Makinodan M, Yamauchi T, Makinodan E, Matsuyoshi H et al. Transient activation of Notch signaling in the injured adult brain. J Chem Neuroanat 2010; 39: 15–19. [DOI] [PubMed] [Google Scholar]
- 40Sato C, Turkoz M, Dearborn JT, Wozniak DF, Kopan R, Hass MR. Loss of RBPj in postnatal excitatory neurons does not cause neurodegeneration or memory impairments in aged mice. PLoS One 2012; 7: e48180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Arumugam TV, Cheng YL, Choi Y, Choi YH, Yang S, Yun YK et al. Evidence that gamma-secretase-mediated Notch signaling induces neuronal cell death via the nuclear factor-kappaB-Bcl-2-interacting mediator of cell death pathway in ischemic stroke. Mol Pharmacol 2011; 80: 23–31. [DOI] [PubMed] [Google Scholar]
- 42Gutierrez A, Look AT. NOTCH and PI3K-AKT pathways intertwined. Cancer Cell 2007; 12: 411–413. [DOI] [PubMed] [Google Scholar]
- 43Espinosa L, Inglés-Esteve J, Aguilera C, Bigas A. Phosphorylation by glycogen synthase kinase-3β down-regulates Notch activity, a link for Notch and Wnt pathways. J Biol Chem 2003; 278: 32227–32235. [DOI] [PubMed] [Google Scholar]
- 44Mckenzie G, Ward G, Stallwood Y, Briend E, Papadia S, Lennard A et al. Cellular Notch responsiveness is defined by phosphoinositide 3-kinase-dependent signals. BMC Cell Biol 2006; 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Park DS, Obeidat A, Giovanni A, Greene LA. Cell cycle regulators in neuronal death evoked by excitotoxic stress: implications for neurodegeneration and its treatment. Neurobiol Aging 2000; 21: 771–781. [DOI] [PubMed] [Google Scholar]
- 46Höglinger GU, Breunig JJ, Depboylu C, Rouaux C, Michel PP, Alvarez-Fischer D et al. The pRb/E2F cell-cycle pathway mediates cell death in Parkinson's disease. Proc Natl Acad Sci USA 2007; 104: 3585–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Andrusiak MG, Vandenbosch R, Park DS, Slack RS. The retinoblastoma protein is essential for survival of postmitotic neurons. J Neurosci 2012; 32: 14809–14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Chen L, Hu L, Dong J-Y, Ye Q, Hua N, Wong M et al. Rapamycin has paradoxical effects on S6 phosphorylation in rats with and without seizures. Epilepsia 2012; 53: 2026–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Liu J, Xu Q, Wang H, Wang R, Hou X-Y. Neuroprotection of ischemic postconditioning by downregulating the postsynaptic signaling mediated by kainate receptors. Stroke 2013; 44: 2031–2035. [DOI] [PubMed] [Google Scholar]
- 50Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther 2004; 3: 1427–1438. [PubMed] [Google Scholar]
- 51Koeller HB, Ross ME, Glickstein SB. Cyclin D1 in excitatory neurons of the adult brain enhances kainate-induced neurotoxicity. Neurobiol Dis 2008; 31: 230–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52Jo H, Lo P-K, Li Y, Loison F, Green S, Wang J et al. Deactivation of Akt by a small molecule inhibitor targeting pleckstrin homology domain and facilitating Akt ubiquitination. Proc Natl Acad Sci USA 2011; 108: 6486–6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Banker GA, Cowan WM. Rat hippocampal neurons in dispersed cell culture. Brain Res 1977; 126: 397–425. [DOI] [PubMed] [Google Scholar]
- 54Bookout AL, Mangelsdorf DJ. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal 2003; 1: e01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.