Abstract
Mitophagy mediates clearance of dysfunctional mitochondria, and represents one type of mitochondrial quality control, which is essential for optimal mitochondrial bioenergetics. p32, a chaperone-like protein, is crucial for maintaining mitochondrial membrane potential and oxidative phosphorylation. However, the relationship between p32 and mitochondrial homeostasis has not been addressed. Here, we identified p32 as a key regulator of ULK1 stability by forming complex with ULK1. p32 depletion potentiated K48-linked but impaired K63-linked polyubiquitination of ULK1, leading to proteasome-mediated degradation of ULK1. As a result, silencing p32 profoundly impaired starvation-induced autophagic flux and the clearance of damaged mitochondria caused by mitochondrial uncoupler. Importantly, restoring ULK1 expression in p32-depleted cells rescued autophagy and mitophagy defects. Our findings highlight a cytoprotective role of p32 under starvation conditions by regulating ULK1 stability, and uncover a crucial role of the p32–ULK1-autophagy axis in coordinating stress response, cell survival and mitochondrial homeostasis.
Mitophagy is a selective form of autophagy by which mitochondria are degraded in autolysosomes. p32 is a critical regulator of mitochondrial bioenergetics.1 It primarily localizes to the mitochondrial matrix, but has also been reported to be present in other subcellular locations.2, 3, 4, 5 Many human tumors exhibit higher p32 expression levels than their nonmalignant counterpart tissues.6, 7, 8, 9 Depleting p32 in human cancer cells strongly shifts their metabolism from oxidative phosphorylation to glycolysis.1 Consistently, p32 knockout causes mid-gestation lethality of knockout embryos and defects in oxidative phosphorylation. Mouse embryonic fibroblasts (MEFs) generated from p32 knockout embryos exhibited impaired ATP production and reduced mitochondrial membrane potential, which is in agreement with the observation that p32 silencing leads to increased mitochondrial fragmentation.10, 11 Notably, p32 was found to form protein complex with a variety of molecules7, 12, 13 and has been suggested that it may act as a multifunctional chaperone protein.12, 13, 14
ULK1 has a crucial role in mitophagy induction.15 Despite the pivotal role of ULK1 in mitochondrial clearance, little is known as how ULK1 itself is regulated. ULK1 is a relatively stable protein and is subject to proteasome-mediated degradation. Post-translational modifications including K63-linked ubiquitylation16, 17 and phosphorylation18, 19, 20 have been reported to modulate the rates of ULK1 turnover and kinase activity in different cellular contexts. Hsp90 and Cdc37 have been shown to regulate ULK1 stability and activity by forming complex with ULK1, which subsequently influences Atg13-mediated mitophagy.21 Here, we found p32 regulates ULK1 stability by forming protein complex with ULK1. The interaction between ULK1 and p32 is crucial for maintaining the steady-state levels and activity of ULK1. We further show that p32 ablation results in a defect in autophagy in EBSS-starved cells, and impairs clearance of dysfunctional mitochondria in cells exposed to mitochondrial uncoupler. Importantly, these autophagy and mitophagy defects can be restored by re-introducing ULK1 into p32-deficient cells, demonstrating ULK1 functions as a crucial downstream effector of p32.
Results
p32 interacts with ULK1
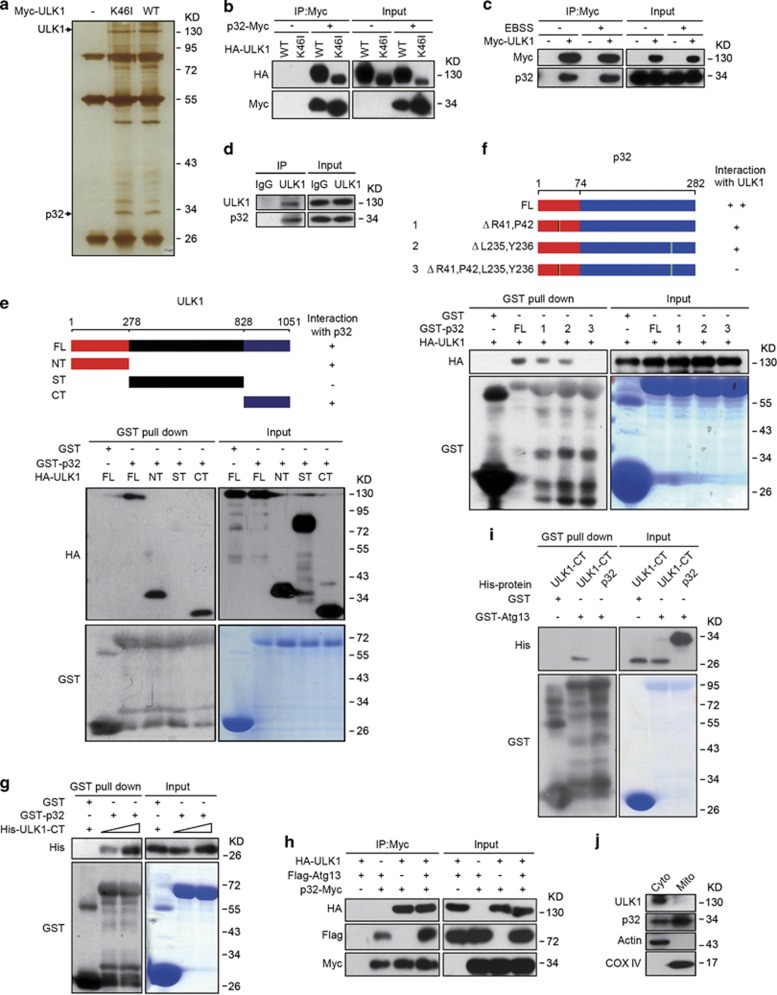
ULK1 is an essential regulator in the autophagy-mediated clearance of mitochondria. To gain insights into ULK1 regulation, we transfected wild-type ULK1 and the dominant negative form of ULK1 (K46I) into HEK293T cells and isolated ULK1-associated proteins by immunoprecipitation approach (Figure 1a). ULK1-binding proteins were analyzed by LC-MS/MS. Candidate binding partners were further validated through immunoprecipitation with ectopically expressed proteins. p32 was identified as ULK1 binding protein. p32-Myc was co-immunoprecipitated with ectopically expressed wild-type ULK1 and mutant ULK1 (K46I), indicating ULK1 kinase activity is dispensable for their interaction (Figure 1b). The interaction between ULK1 and p32 was not affected by nutrient conditions, as endogenous p32 and ectopically expressed ULK1 formed protein complex under normal conditions and upon Earles' Balance Salt Solution (EBSS)-induced starvation (Figure 1c). Furthermore, we were able to show the ULK1–p32 association in Hela cells, which express endogenous ULK1 and p32 (Figure 1d).
Figure 1.
p32 interacts with ULK1. (a) HEK293T cells were transiently transfected with the indicated expression constructs. The anti-Myc immunoprecipitates were resolved by SDS-PAGE, and the proteins were visualized by silver staining, and indicated bands were analyzed by mass spectrometry. (b) Western blotting analysis of input and anti-Myc IP derived from HEK293T cells that were transiently transfected with WT or mutant ULK1 (K46I) and p32-Myc. (c) Hela cells expressing Myc-ULK1 were grown either in normal or in EBSS medium for 6 h. Cell lysates were immunoprecipitated with anti-Myc antibody followed by immunoblotting with anti-p32 antibody. (d) The interaction of endogenous ULK1 and p32 was detected in Hela cells. Normal rabbit IgG was used as a negative control for the immunoprecipitation procedure. (e) Purified GST-p32 was incubated with cell lysates derived from HEK293T cells transfected with the indicated HA-ULK1 constructs. Proteins retained on Sepharose were then blotted with the indicated antibodies. (f) Extracts from HEK293T cells transfected with HA-ULK1 were incubated with recombinant full-length (FL) GST-p32 or GST-p32 mutants coupled to GSH-Sepharose. Proteins retained on Sepharose were then blotted with the indicated antibodies. (g) Purified recombinant His-ULK1-CT was incubated with GST-p32. Proteins retained on Sepharose were then blotted with the indicated antibodies. (h) HEK293T cells were transfected with vectors encoding HA-ULK1, Flag-Atg13 and p32-Myc, as indicated. Cell lysates were immunoprecipitated with anti-Myc antibody. Western blotting was performed using the indicated antibodies. (i) Purified His-ULK1-CT or His-p32 was incubated with purified GST-Atg13. Proteins retained on Sepharose were then blotted with the indicated antibodies. (j) Hela cells were fractionated and analyzed by immunoblotting using antibodies against the indicated proteins. Cyto, cytosol; Mito, mitochondria
We next examined the ULK1 domain responsible for p32 interaction. Data from GST pull-down experiments suggest that both the N-terminal (amino acids 1–278) and the C-terminal regions (amino acids 828–1051) of ULK1 mediates p32-ULK1 association (Figure 1e). To identify key amino acid residues of p32 required for ULK1 interaction, we constructed mutant p32 expression vectors bearing point mutations within the N-terminal and the C-terminal regions of p32. Deletion of R41/P42 or L235/Y236 of p32 impaired its interaction with ULK1, while combined deletion of these four amino acids (denoted as p32Δ) completely disrupted p32–ULK1 complex formation (Figure 1f). Furthermore, purified His-ULK1-CT was able to interact with GST-p32 under cell-free conditions, suggesting a direct interaction between ULK1 and p32 (Figure 1g). To explore if p32 also associates with other ULK1-interacting proteins, we checked Atg13, which is also a ULK1 substrate. Interaction between p32 and Atg13 was readily detectable (Figure 1h), indicating p32, ULK1 and Atg13 associate together as a complex. However, GST-Atg13 failed to interact with His-p32 in vitro (Figure 1i), suggesting p32 may indirectly bind to Atg13.
p32 can be recruited to several subcellular compartments, whereas ULK1 is usually in the cytoplasm. To determine which subcellular compartment that p32 and ULK1 might interact with, we fractionated cell lysates into cytoplasmic (Cyto) and mitochondrial (Mito) fractions and examined the relative abundance of ULK1 and p32 in each of the fractions. A subset of total p32 protein was found in the cytoplasm fractions, in addition to majority p32 in the mitochondrial fractions in Hela cells (Figure 1j). On the other hand, ULK1 was found exclusively in the cytoplasm, as previously reported.21, 22 Immunofluorescent staining further revealed that p32 majorly localized in the mitochondrion, whereas ULK1 predominantly localized in the cytoplasm (Supplementary Figure S1a). However, a very small proportion of cytosolic p32 was found to co-localize with ULK1. These results indicate that ULK1 and p32 may form complex in the cytoplasm. p32 has been shown to relocalize its interacting protein to mitochondria via protein complex formation. However, we failed to detect any endogenous mitochondrial ULK1 signal in 293 T cells overexpressing p32 by western blotting, which is likely attributed to low cellular abundance of ULK1 protein (data not shown). By contrast, a subset of ectopic ULK1 can be distributed to mitochondria in cells expressing WT p32, but not p32Δ (Supplementary Figure S1b). These data suggest that upon binding to p32, a subset of ULK1 protein may translocate to mitochondria.
p32 regulates ULK1 stability and kinase activity
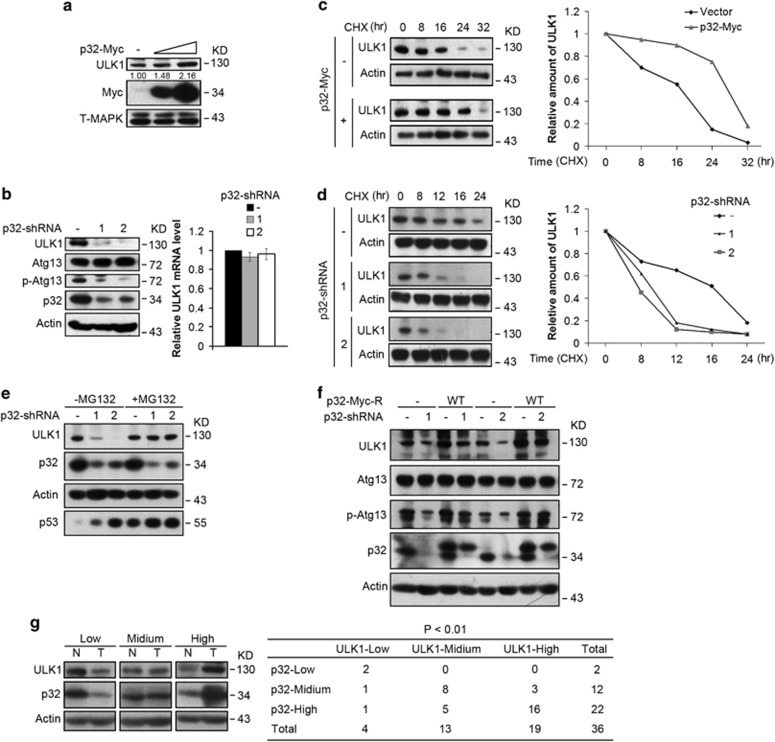
We next explored the biochemical consequence of p32–ULK1 interaction. Ectopically expressed p32 induced ULK1 protein expression (Figure 2a), while p32 ablation led to downregulation of ULK1 (Figure 2b). Notably, ULK1 mRNA levels remain unchanged in p32-depleted cells. We tested three additional human cancer cell lines and observed deduction of ULK1 expression levels upon p32 knockdown (Supplementary Figure S1c). We also found Atg13 phosphorylation was markedly decreased in p32-ablated cells (Figure 2b), likely due to reduced ULK1 expression levels. To test whether p32 regulates the steady-state levels of ULK1, we measured ULK1 protein half-life. In cells expressing ectopic p32, the half-life of ULK1 was extended from 16 h to around 28 h (Figure 2c). Conversely, silencing p32 shortened ULK1 turnover rate from 16 h to less than 10 h (Figure 2d). ULK1 has been suggested to undergo proteasome-mediated degradation.17, 21 We, therefore, tested whether ULK1 reduction in p32-depleted cells could be attributed to increased proteolysis of ULK1. Indeed, proteasomal inhibitor MG132 treatment completely blocked downregulation of ULK1 in p32-ablated cells (Figure 2e), demonstrating that depletion of p32 leads to increased proteasomal degradation of ULK1. To ensure the specificity of p32-mediated ULK1 stabilization, we performed a rescue experiment by introducing a shRNA-resistant form of p32 into Hela cells stably depleted of p32. Reconstitution with shRNA-resistant p32 not only restored ULK1 expression levels, but also largely restored its kinase activity towards Atg13 (Figure 2f), suggesting p32 is essential for maintaining ULK1 stability and activity.
Figure 2.
p32 regulates ULK1 stability and kinase activity. (a) Hela cells were infected with lentiviral vectors expressing p32-Myc. Cell extracts were then prepared and analyzed by western blotting with the indicated antibodies. (b) Protein extracts from Hela cells infected with the indicated lentiviral shRNA constructs were analyzed by western blotting. Total RNA was isolated, and qRT-PCR was performed. (c) Hela cells were infected with the indicated lentiviral vector encoding p32-Myc. Cells were then treated with CHX (25 μg/ml) for the indicated time course. Cell lysates were harvested and analyzed by western blotting. Semiquantification with Actin as a loading control and relative ULK1 levels at time 0 set as 1. (d) Hela cells expressing p32-shRNAs were treated with CHX for the indicated time course. The levels of ULK1 were determined by immunoblotting the total cell lysates and quantification with Actin as a loading control. Results plotted are the amounts of ULK1 at each time point relative to the level at time 0. (e) Hela cells infected with lentivirus encoding the indicated shRNAs were treated with DMSO or MG132 (20 μM) for 6 h. Cell extracts were analyzed by western blotting using the indicated antibodies. p53 immunoblotting was shown as a positive control for inhibited proteasomal degradation by MG132. (f) Hela cells stably expressing shRNA-resistant forms of p32 were infected with the indicated lentiviral shRNA constructs. Cells were then collected and lysates were subjected to immunoblotting with the indicated antibodies. (g) Immunoblotting analysis of p32 and ULK1 levels from tumor tissues (T) and their matched surrounding normal mucosal tissues (N). Actin was used as a loading control. A total of 36 samples of human colorectal cancer were classified into nine groups based on the protein levels of p32 and ULK1 in tumor tissues compared with surrounding normal tissues. The correlation between p32 and ULK1 expression was analyzed by Fisher's exact test
p32 was reported to be overexpressed in human cancers. We assessed the correlation between p32 and ULK1 in human colon cancers by analyzing 36 human colon cancer tissues. Strikingly, we found 22 tumor samples bearing p32 overexpression when compared with nonmalignant tissues. Notably, in these colon cancer samples harboring elevated p32 levels, 16 tumor samples showed significantly increased ULK1 levels (Figure 2g). These data suggest that overexpression of p32 may lead to elevated ULK1 expression in human colon cancers.
p32 interferes with the polyubiquitination of ULK1
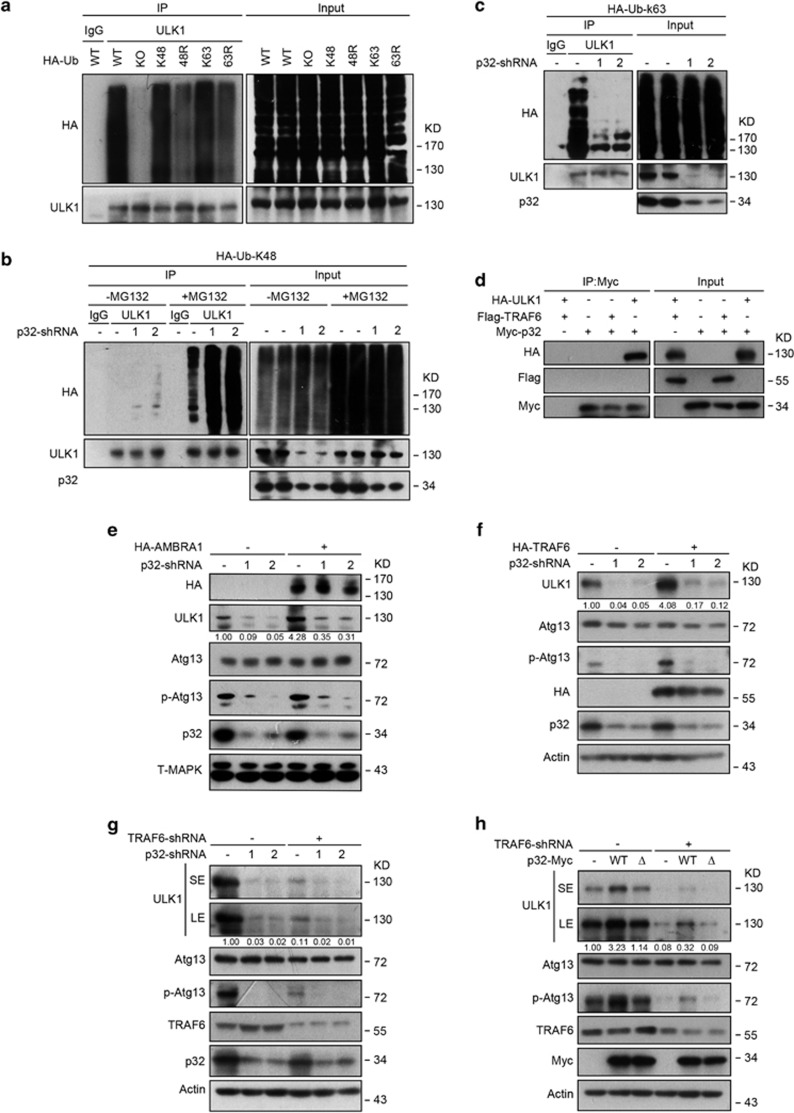
Previous studies reported that ULK1 is a K63-linked but not K48-linked ubiquitylated protein.16, 17 We investigated whether p32 regulates ULK1 stability via modulating its ubiquitylation. Unlike previous studies, when we used ubiquitin mutants Ub-K48 and Ub-K63, which contain arginine substitutions on all their lysine residues except the one at position 48 and 63, respectively, we found ULK1 is subject to both K48-linked and K63-linked ubiquitylation in vivo (Figure 3a). To confirm this finding, we used K48R and K63R ubiquitin mutants that contain a single lysine to arginine mutation at position 48 and 63, respectively. As a control, we also included a polymerization-defective mutant of ubiquitin (Ub-KO). As shown in Figure 3a, overexpression of Ub-K63R or Ub-K48R mutant partially blocked the polyubiquitination of ULK1, whereas polyubiquitination of ULK1 was completely abolished by replacement of Ub-WT with Ub-KO. Taken together, these data suggest that the polyubiquitination we observed on ULK1 depends on both K48-linked and K63-linked ubiquitin. We next assessed the ubiquitylation status of ULK1 in cells depleted of p32. Interestingly, p32-depleted cells exhibited an increase of K48-linked (Figure 3b) and a decrease of K63-linked ubiquitylation in the ULK1 immunoprecipitates (Figure 3c), suggesting p32 may prevent K48-linked but facilitate K63-linked polyubiquitination of ULK1.
Figure 3.
p32 interferes with the polyubiquitination of ULK1. (a) HeLa cells were transfected with HA-tagged WT or mutant ubiquitin constructs. Protein extracts were immunoprecipitated using anti-ULK1 antibody or rabbit IgG as a negative control (IgG). Immunoprecipitates were analyzed by western blotting using anti-ULK1 and anti-HA antibodies. (b) Hela cells expressing HA-Ubiquitin-k48 (HA-Ub-k48) were infected with the indicated p32-shRNAs. Cells were then treated with DMSO or MG132 (20 μM) for 6 h before being collected. Protein extracts were immunoprecipitated using anti-ULK1 antibody. The immunoprecipitates were subjected to western blotting with the indicated antibodies. (c) HeLa cells expressing HA-Ubiquitin-k63 (HA-Ub-K63) were infected with p32 shRNA lentiviral vector. Endogenous ULK1 protein was immunoprecipitated and immunoblotted with the indicated antibodies. Of note, ULK1 levels in the immunoprecipitation experiment do not change in this case in the p32 shRNA lane, owing to saturation of the anti-ULK1 antibody. (d) The indicated expression constructs were transiently transfected into HEK293T cells. Protein extracts were immunoprecipitated using anti-Myc antibody. The immunoprecipitates were subjected to western blotting with the indicated antibodies. (e) Hela cells stably expressing HA-AMBRA1 were infected with p32 shRNA lentiviral vector. Cell lysates were collected and analyzed by western blotting with the indicated antibodies. (f) Hela cells stably expressing HA-TRAF6 were infected with p32 shRNA lentiviral vector as in e. Cell lysates were collected and analyzed by western blotting with the indicated antibodies. (g) Hela cells infected with lentivirus bearing TRAF6-shRNA and p32-shRNA were collected. Protein extracts were subjected to western blotting with the indicated antibodies
TRAF6 is the only known E3 ligase that mediates ULK1 K63 ubiquitylation and its subsequent stabilization.17 We, therefore, tested whether there is an interplay between TRAF6 and p32 in regulating the steady state of ULK1. As both TRAF6 and p32 could bind to ULK1, we wondered whether these two molecules may form protein complex. To address this, we co-transfected Hela cells with ectopic p32 and TRAF6 and then performed immunoprecipitation assay. However, we failed to detect TRAF6 in p32 immunoprecipitants (Figure 3d), suggesting that p32 and TRAF6 may not coexist in the same complex. Given that p32-depletion abrogated K63-linked polyubiquitination of ULK1, which phenocopied TRAF6 silencing, to further explore a possible epistatic relationship between TRAF6 and p32 in regulating ULK1, we asked whether ectopically expressed AMBRA1 or TRAF6 could at least partially rescue ULK1 defect in p32-depleted cells. However, ectopic AMBRA1 or TRAF6 failed to restore ULK1 stability and activity in p32-ablated cells (Figures 3e and f), suggesting p32 did not act through AMBRA1/TRAF6 regulating ULK1.
We next investigated whether TRAF6 and p32 could exert additive or synergistic effects on ULK1 stabilization. To address this, we silenced p32 in Hela cells depleted of TRAF6. Co-depleting p32 and TRAF6 resulted in a reproducible, albeit modest, further reduction of ULK1 protein levels compared with p32 knockdown alone (Figure 3g), indicating p32 may act on distinct mechanisms regulating K63-linked ubiquitylation of ULK1. To ensure this possibility, we conducted the rescue experiment by overexpressing p32-Myc in cells depleted of TRAF6. Ectopically expressed p32 failed to rescue ULK1 degradation upon TRAF6 knockdown, ruling out the possibility that p32 may function downstream of TRAF6 signaling regulating ULK1. Taken together, these data demonstrate that by interfering with K48- and K63-linked polyubiquitination of ULK1, p32 has an essential role in maintaining the steady-state levels of ULK1.
p32 regulates starvation-induced autophagy
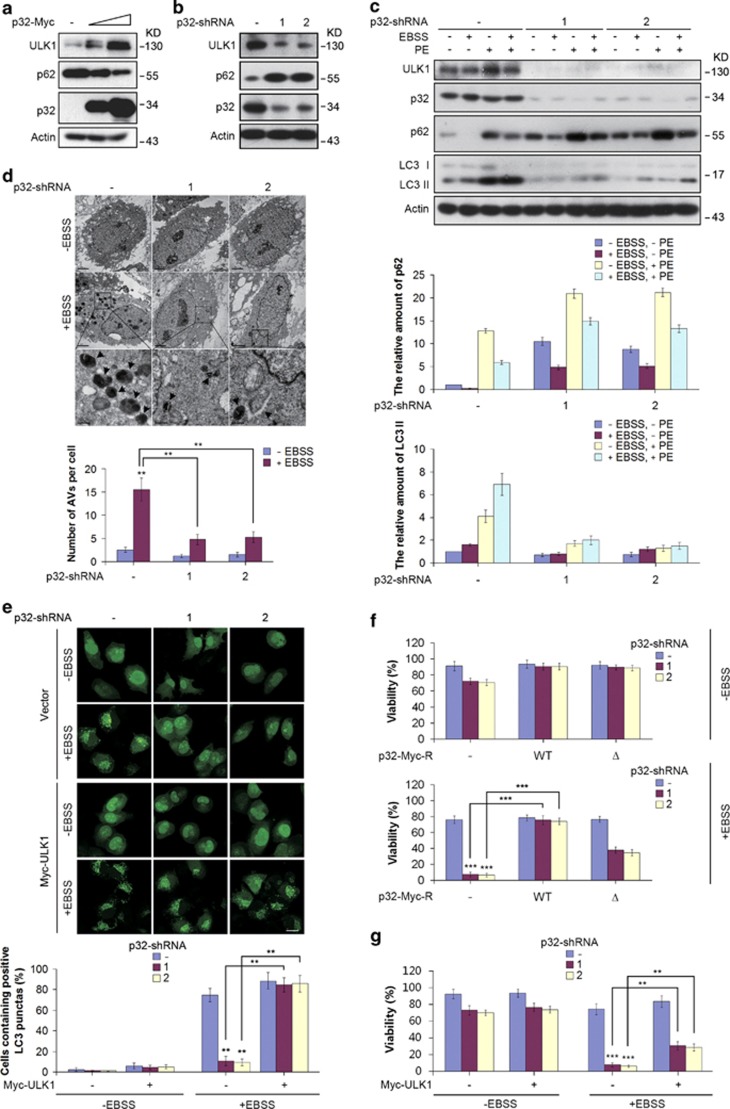
As ULK1 is essential for starvation-induced autophagy,23 we investigated the effects of p32 ablation on autophagy induction. We first monitored the amounts of p62, a protein that is degraded by autophagy. Immunoblot analysis revealed reduction of p62 levels in cells expressing ectopic p32, and elevation of the p62 protein in p32-depleted cells, respectively (Figures 4a and b). This indicates that p32 deficiency may cause autophagy defect. We next introduced ULK1 into cells knocked down for p32 and examined the extent of autophagy upon EBSS treatment. p32-ablated cells reconstituted with ULK1 had reduced p62 levels in the absence or presence of EBSS treatment (Supplementary Figure S2a), suggesting ULK1 acts as a crucial downstream effector of p32 regulating starvation-induced autophagy.
Figure 4.
p32 regulates starvation-induced autophagy. (a) Protein extracts from Hela cells infected with p32-Myc lentiviral construct were analyzed by western blotting with the indicated antibodies. (b) Hela cells infected with the indicated lentiviral p32 shRNAs were collected, and cell lysates were subjected to western blotting with the indicated antibodies. (c) Hela cells stably expressing the indicated shRNAs were placed in normal or in EBSS starvation medium in the presence or absence of lysosomal protease inhibitors PE for 6 h. Lysates were immunoblotted as indicated (upper panel). p62 and LC3-II levels were quantified by densitometry and plotted as mean±S.D. of three independent experiments (middle and lower panel). (d) p32 knockdown Hela cells were untreated or treated with EBSS medium for 6 h. Cells were then fixed and analyzed by transmission electron microscopy (TEM). The bottom panels show enlarged views of the boxed areas. Quantification of the autophagic vacuoles (AVs) was shown as mean±S.D. of 15 random areas. **P<0.01. (e) H1299 cells stably expressing GFP-LC3 were infected with p32-shRNA and stably reconstituted with lentiviral Myc-ULK1 vector. Cells were then placed in EBSS medium for 6 h. GFP-LC3 positive autophagosomes were determined by confocal microscopy. The scale bar represents 20 μM (upper panel). Histogram represents the percentage of cells containing positive LC3 punctas. The error bars represent the mean±S.D. of five random areas (lower panel). **P<0.01. (f) Hela cells stably expressing the indicated shRNA-resistant forms of p32 were infected with p32-shRNA lentiviruses. Histogram represents the percentage of viable cells scored by Annexin V staining and FACS analysis after being cultured in EBSS medium for 6 h. The error bars represent the mean±S.D. of three independent experiments. ***P<0.001. (g) Hela cells were infected with p32-shRNA and stably reconstituted with lentiviral Myc-ULK1 vector. Histogram represents the percentage of viable cells scored by Annexin V staining and FACS analysis after being cultured in EBSS medium for 6 h. The error bars represent the mean±S.D. of three independent experiments. **P<0.01, ***P<0.001
To further verify p32 has a role in autophagy, we conducted the autophagy flux assay by comparing the levels of LC3-II (the lipid modified form of LC3) in p32-depleted cells. When control cells were cultured in EBSS medium containing lysosomal protease inhibitors E64D and pepstatin A (PepA), LC3-II levels markedly increased, indicating an autophagic flux. By contrast, the level of LC3-II in p32-depleted cells failed to increase upon serum starvation and/or E64D plus PepA treatment (Figure 4c). It should be noted that the levels of LC3-II were profoundly decreased in p32-ablated cells under normal conditions, suggesting that low-level autophagy constitutively occurs in p32-depleted cells or that LC3 conversion occurs independently of autophagy (Figure 4c). To provide additional evidence supporting autophagy defect in p32 knockdown cells, we treated Hela cells with Rapamycin in combination with the lysosomal inhibitor Bafilomycin A (Baf A). In control cells, Rapamycin treatment induced a sharp increase of LC3-II levels in the presence of Baf A, as well as a profound induction of p62 degradation. By contrast, the accumulation of LC3-II protein and p62 degradation were significantly impaired in p32-ablated cells (Supplementary Figure S2b). These results further support the notion that p32 inhibition leads to autophagy defect. Consistently, electron microscopic analysis revealed the presence of numerous autophagic vacuoles upon EBSS treatment, which were significantly reduced in cells depleted of p32 (Figure 4d). Taken together, we conclude that autophagy flux is severely impaired in the absence of p32.
To visualize autophagy, H1299 cells stably expressing green fluorescent protein (GFP)-LC3 were cultured in EBSS medium for 6 h. The control cells displayed the conversion of a diffuse cytosolic LC3 pattern to a punctate pattern upon EBSS treatment, indicating the formation of autophagic GFP-LC3 puncta. In contrast, p32 depletion failed to induce GFP-LC3 puncta upon EBSS treatment, consistent with the impaired autophagic flux in p32 knockdown cells. Importantly, p32-depleted cells reconstituted with ULK1 exhibited significantly increased GFP-LC3 puncta and decreased p62 levels when cultured in EBSS medium (Figure 4e; Supplementary Figures S2a and S2e), suggesting p32 acts on ULK1 regulating autophagy.
Autophagy is generally considered as a pro-survival mechanism. We therefore analyzed the survival rate of p32-depleted cells under starvation conditions. It has been proposed that loss of mitochondrial function may underlie the enhanced cell death in p32-depleted cells under non-stress conditions. Indeed, p32 ablation largely impaired cell survival in the absence of starvation treatment. In the presence of EBSS, a significant portion of p32-depleted cells underwent cell death, only 10% cells remained viable (Figure 4f). To determine whether p32–ULK1-autophagy axis has a cytoprotective role upon starvation, we first determined the rescue effect on cell viability by shRNA-resistant form of p32. Notably, when cells were cultured in complete medium, the reduced cell viability in p32-depleted cells were completely restored by both WT p32 and p32Δ, indicating ULK1 is dispensable for p32 depletion-induced cell death under normal conditions. However, when p32-ablated cells were cultured in EBSS medium, reconstitution with WT p32 restored cell viability to a level comparable to that of control cells, whereas p32Δ only exhibited partial, yet significant rescue effect on cell death. This suggests that p32–ULK1-autophagy axis contributes to cell survival under starvation conditions. To assess the contribution of ULK1-dependent autophagy induction in mediating the survival function of p32, we reconstituted ULK1 in p32-depleted cells and found ectopic ULK1 profoundly, albeit partially restored viable cell populations upon EBSS treatment (Figure 4g), demonstrating ULK1 is a pivotal downstream effector mediating the survival function of p32 under starvation conditions. Collectively, p32 can protect cells from starvation-induced cell death partially via ULK1-dependent autophagy induction.
Depletion of p32 leads to mitophagy defects
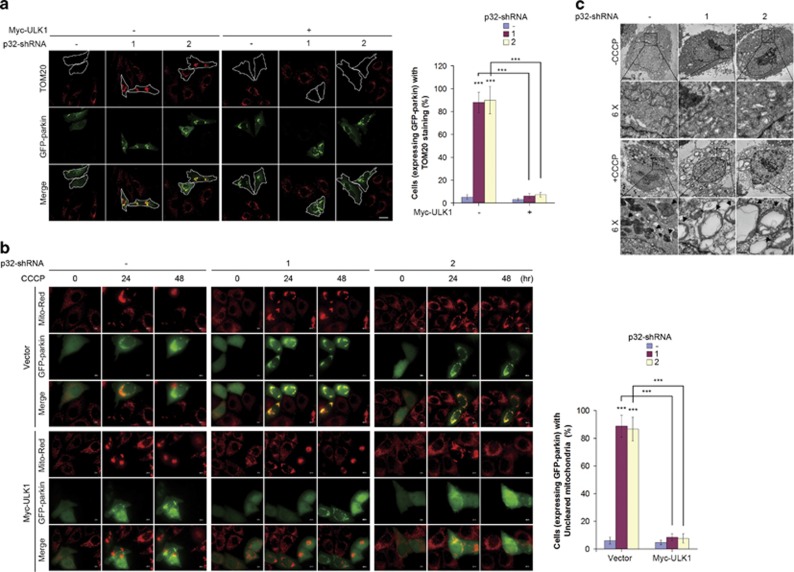
To maintain proper mitochondrial homeostasis under stress conditions, damaged mitochondria must be degraded through autophagy. To determine whether p32 has a role in mitophagy, we performed a well-established mitochondrial degradation assay in cultured cells overexpressing parkin, mitochondrial depolarization induced by the protonophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP) results in loss of mitochondria (measured by immunostaining for mitochondrial outer membrane protein TOM20).24 In control cells, CCCP caused a robust disappearance of TOM20 staining in more than 90% of cells transfected with GFP-conjugated parkin. Strikingly, p32 depletion robustly blocked this loss of TOM20 staining. A significant proportion of the p32-depleted cells contained mitochondria in small clusters that colocalized with parkin, indicating the primary defect in clearance occurs after parkin recruitment to depolarized mitochondria (Supplementary Figure S2c). Reconstitution with ULK1 profoundly restored mitochondrial clearance in p32 shRNA expressing cells, indicated by diminished TOM20 staining (Figure 5a).
Figure 5.
p32 depletion leads to mitophagy defects. (a) Hela cells stably expressing Myc-ULK1, GFP-Parkin and p32-shRNA were treated with CCCP (10 μM) for 48 h. Cells were then immunostained with anti-TOM20 antibody (left) and scored for the percentage of cells expressing GFP-parkin with TOM20 staining, which was shown as mean±S.D. of five random areas (right). The scale bar represents 10 μM. ***P<0.001. (b) Hela cells stably expressing Mito-Red were infected with the indicated lentiviral vectors. Cells were then transiently transfected with GFP-Parkin followed by CCCP treatment (10 μM) for the indicated time course. The process of removing damaged mitochondria in living cells was monitored by live-cell imaging (left panel). Histogram represents the percentage of cells expressing GFP-parkin with uncleared mitochondria after 48 h of CCCP treatment (right panel). The error bars represent the mean±S.D. of five random areas. ***P<0.001. (c) Hela cells stably expressing Myc-parkin were infected with lentivirus encoding control shRNA or p32-shRNA. Cells were then placed in medium containing DMSO or CCCP (10 μM) for 24 h, followed by TEM analysis
Next, we utilized live cell image to examine the influence of p32 on the dynamics of parkin-mediated mitophagy. To this end, we introduced parkin into Hela cells stably expressing Mito-Red. Most parkin-expressing control cells exhibited complete clearance of mitochondria following treatment with CCCP in a time-dependent manner. However, p32 depletion significantly retained parkin-targeted mitochondria even after 48 h of CCCP exposure. These data further demonstrate that p32 is crucial for mitochondrial depolarization-induced mitophagy in parkin-expressing cells. Importantly, mitochondrial clearance defect in p32-deficient cells following CCCP treatment was restored when cells were reconstituted with ULK1 (Figure 5b), suggesting ULK1 is a crucial downstream effector of p32 to mediate mitochondrial clearance by mitophagy. Notably, electron microscopy analysis of CCCP-treated cells revealed typical autophagosomes containing damaged mitochondria, whereas p32 knockdown significantly reduced the number of autophagic vacuoles. Instead, these cells exhibited swelling of all mitochondria and the number of defective mitochondria per cell was significantly increased as compared with that of control cells (Figure 5c; Supplementary Figure S2d). In line with the latter observation, we found COX IV and TOM20 were profoundly elevated in p32-deficient cells (Supplementary Figure S2e), which can be reverted by ectopic ULK1. Taken all together, our data indicate that p32 acts on ULK1 to regulate mitophagy.
p32 regulates autophagy and mitophagy via interacting with ULK1
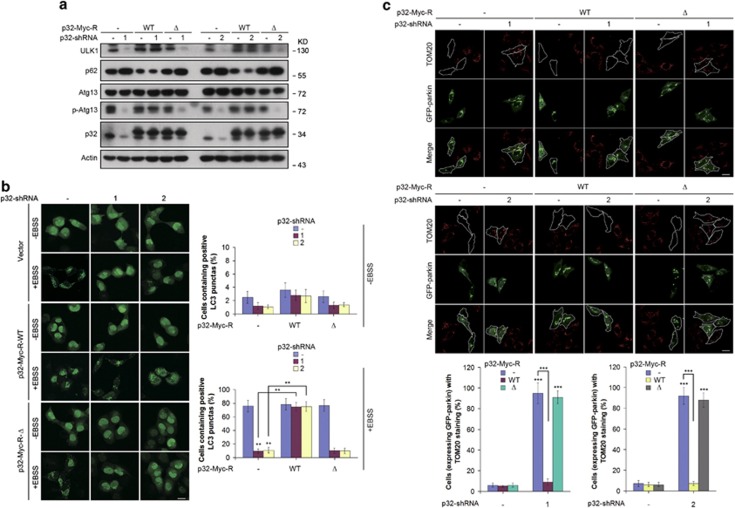
To ensure p32 exerts a specific effect on starvation-induced autophagy and autophagic degradation of damaged mitochondria, we performed rescue experiment by co-transfection of shRNA-resistant human p32 cDNA. Stable lenti-viral reconstitution of a Myc-tagged WT p32 cDNA, but not p32Δ in p32 knockdown cells, restored both ULK1 protein expression levels and Atg13 phosphorylation (Figure 6a). As expected, WT p32 reconstitution also reduced basal p62 protein levels in the absence of EBSS and restored GFP-LC3 puncta upon EBSS treatment (Figure 6b). Collectively, these data indicate p32 has a crucial role in autophagy through binding to ULK1.
Figure 6.
p32 regulates autophagy and mitophagy via interacting with ULK1. (a) Hela cells stably reconstituted with lentivirus bearing the indicated p32 cDNA were infected with p32-shRNA lentiviruses. Lysates were immunoblotted with the indicated antibodies. (b) H1299-GFP-LC3 cells stably expressing the indicated shRNA-resistant forms of p32 were infected with p32-shRNA lentiviruses. Cells were then treated with EBSS medium for 6 h. GFP-LC3 positive autophagosomes were examined by confocal microscopy. The scale bar represents 20 μM (left panel). Histogram represents the percentage of cells containing positive LC3 punctas (right panel). The error bars represent the mean±S.D. of five random areas. **P<0.01. (c) Hela cells stably expressing GFP-Parkin and the indicated shRNA-resistant forms of p32 were infected with p32-shRNA lentiviruses. Cells then were treated with CCCP (10 μM) for 48 h. Cells were immunostained with anti-TOM20 antibody (upper) and scored for the percentage of cells expressing GFP-parkin with TOM20 staining, which was shown as mean±S.D. of five random areas (lower). The scale bar represents 10 μM. ***P<0.001
We next compared the rescue effect between WT p32 and p32Δ on mitochondrial clearance in mitochondrial degradation assay. p32-ablated cells reconstituted with WT p32, but not p32Δ restored their ability to efficiently clear mitochondria following CCCP treatment (Figure 6c). Therefore, p32 is essential for parkin-dependent clearance of depolarized mitochondria via interacting with ULK1.
Discussion
Investigating how ULK1 is maintained under physiological conditions will provide mechanistic insights for understanding the regulation of mitophagy. Previous studies showed that p32 regulates mitochondrial energy status and cell survival. In this report, we identified p32 as a binding partner of ULK1. In the absence of p32, ULK1 turnover rate is profoundly increased, which in turn compromised its kinase activity towards Atg13. Proteasome inhibitor MG132 completely blocked the destabilization of ULK1 in p32-depleted cells, indicating p32 ablation promotes ULK1 proteolytic destruction. Furthermore, we found p32 depletion potentiated K48-linked but impaired K63-linked polyubiquitination of ULK1. These data indicate p32 may prevent the proteasome degradation of ULK1 by interfering with its polyubiquitination. We excluded the possibility that p32 works in concert with TRAF6 signaling regulating K63-linked ubiquitylation of ULK1. Currently, the E3 ubiquitin ligase for ULK1, which is capable of generating K48-linked ubiquitin chains remains unknown. We propose that p32 may exert a positive effect on ULK1 stability and activity via two possible mechanisms. One is that p32 may crosstalk with the unidentified ubiquitination/deubiquitination processes targeting ULK1. Alternatively, p32 has been proposed to act as a multifunctional chaperone protein as evidenced by its participation in different biological processes.1, 13, 14, 25 Therefore, it is likely that the chaperone-like activity of p32 is essential for ULK1 regulation, a process similar to Hsp90–Cdc37 complex-dependent regulation of ULK1.
ULK1 is essential for starvation-induced autophagy and mitochondrial uncoupler-induced mitophagy. In line with a crucial role of p32 in regulating ULK1 steady-state levels and activity, p32 silencing resulted in accumulation of p62 and reduced LC3-II protein levels in EBSS-treated cells, indicative of autophagy defect. Autophagy is a pro-survival mechanism engaged by metabolic stress. p32 ablation augmented cell death following EBSS treatment, which can be partially but significantly rescued by ULK1 reconstitution. This suggests that p32 exerts a protective effect under starvation conditions partially by inducing ULK1-dependent autophagy. p32 is often found aberrantly over-expressed in human tumor tissues. Consistent with this, we found p32 protein levels were upregulated in human colon cancer tissues, and ULK1 proteins levels were significantly correlated with p32 status. On the basis of our data, it is reasonable to speculate that the p32–ULK1-autophagy axis may provide a survival advantage when nutrients are scarce, which ultimately promotes tumorigenesis.
Previous studies showed that p32 is an essential regulator in maintaining mitochondrial oxidative phosphorylation. However, the molecular mechanism underlying this p32-mediated metabolic regulation remains completely unknown. Our findings that upon CCCP treatment, damaged mitochondria failed to be cleared in p32-deficient Hela cells stably expressing parkin, unveiled an unexpected role of p32 in mitophagy. Reconstitution with WT p32, but not the ULK1-binding defective mutant p32Δ, rescued mitochondrial degradation defect by p32 knockdown, indicating the ability of p32 to regulate CCCP-induced mitophagy is through its interaction with ULK1. Consistent with this notion, introducing ectopic ULK1 effectively restored mitochondrial clearance in p32-ablated cells treated with CCCP. Thus, p32 could act upstream of ULK1 participating mitochondrial homeostasis. It has been reported that the stimulation of mitochondrial oxidative phosphorylation enhances mitochondrial renewal by increasing its degradation rate.26 This indicates that mitochondrial homeostasis and oxidative phosphorylation could be mutually beneficial. Mitochondrial quality control may serve as an important mechanism maintaining cellular energy homeostasis. Thus, our results likely provide a plausible explanation for previously reported p32-mediated regulation of mitochondrial bioenergetics. Collectively, our findings underscore the importance of p32 in fine-tuning the mitochondrial quality control and mitochondrial metabolism via ULK1 under physiological conditions.
Materials and Methods
Antibodies
Rabbit anti-Myc (cat. #2272), anti-LC3-II (D11), anti-Total-MAPK (cat. #9102), mouse anti-Myc (9B11) and anti-COX IV (#4844) antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). Rabbit anti-ULK1 (H-240), anti-p32 (FL-282), anti-TOM20 (FL-145), anti-TRAF6 (D-10) and mouse anti-p62 (D-3) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rat anti-HA (3F10) antibodies was purchased from Roche Life Science (Indianapolis, IN, USA). Rabbit anti-Atg13 (cat. SAB4200100), anti-ULK1 (cat. A7481), anti-Flag (cat. F7425), mouse anti-His (cat. SAB1305538), anti-Actin (cat. A5441) and anti-Flag (cat. F3165) antibodies (Gilbertsville, PA, USA) were purchased from Sigma (St. Louis, MO, USA). Rabbit anti-phosphorylated Atg13 (600-401-C49) antibody was purchased from Rockland (Gilbertsville, PA, USA).
Cell culture and reagents
HEK293T, H1299, RKO and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Life Technologies, Carlsbad, CA, USA), and HCT116 cells were grown in McCoy's 5A supplemented with 10% fetal bovine serum (FBS, Hyclone, Logan, UT, USA), all supplemented with 10% (v/v) fetal bovine serum (FBS, Hyclone) and 100 U/ml streptomycin and penicillin (Millipore, Bedford, MA, USA), at 37 °C in 5% CO2. Earle's balanced salt solution (EBSS, Hyclone), Protein G sepharose (GE Healthcare, Uppsala, Sweden), GSH Sepharose (GE Healthcare), MG132 (Calbiochem, San Diego, CA, USA), Rapamycin (Cell Signaling Technology), pepstatin A (PepA, Sigma), E64D (Sigma), Bafilomycin A (Sigma), carbonyl cyanide 3-chlorophenyl-hydrazone (CCCP, Sigma), cycloheximide (Sigma), Annexin V-PE Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA).
DNA constructs and RNA interference
ULK1 expression constructs were generous gifts from Dr S Lin. p32 expression constructs were generated via PCR-based approaches.
shRNAs sequences: human p32: 5′-GGATGAGGTTGGACAAGAAGA-3′ and 5′-GCCTTATATGACCACCTAATG-3′. TRAF6: 5′-CCACGAAGAGATAATG GATGCCAAA-3′.
Immunoprecipitation, immunoblotting and ubiquitination assay
HEK293T cell lysates prepared using ice-cold Triton cell lysis buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 0.5% Triton X-100, containing 1 × complete EDTA-free protease inhibitor cocktails (Roche Life Science) and 1 mM PMSF] were applied to IP or immunoblotting assays with appropriate antibodies. H1299, RKO and HeLa cells were harvested by RIPA lysis buffer (50mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% SDS, 1% sodium deoxycholine acid and 1% Triton X-100, containing 1 × complete EDTA-free protease inhibitor cocktails (Roche Life Science) and 1 mM PMSF).
For immunoprecipitation, the cell lysates were incubated with antibody overnight at 4 °C and precipitated using Protein G sepharose beads (GE Healthcare). The beads were washed at least seven times with cold Triton cell lysis buffer. For ubiquitination assay in cells, cell lysates were first subjected to IP with indicated antibodies, eluted by 5 min boiling in 1% SDS to dissociate protein–protein interactions, then diluted 10-fold with the same lysis buffer and reimmunoprecipitated with indicated antibodies as previously described (2 × IP). The ubiquitin-conjugated proteins were detected by immunoblotting.
Quantitative real-time PCR
Total RNA was isolated with TRIzol (Invitrogen), according to the manufacturer's instructions. Real-time quantitative PCR was performed using SYBR Green PCR Mix (Roche Life Science) according to the manufacturer's protocol. Real-time quantification was performed by using a Fast-Real Time PCR System (Applied Biosystems, Life Technologies). Data were normalized to TBP.
Flow cytometry
For Annexin V staining, cells were washed once with PBS and then were washed in 1 × Annexin V buffer and treated as described by the Annexin V staining protocol (BD Biosciences). Cells were then analyzed using an Epics XL flow cytometer (Beckman Coulter, Mississauga, ON, Canada).
Immunofluorescence and live cell imaging assay
Cells grown on glass coverslips were washed in PBS, fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. Coverslips were blocked in 5% normal serum for 1h at room temperature and then were incubated with primary antibody diluted in blocking buffer and after washed secondary antibodies were applied. Images were acquired by using Zeiss LSM 780 laser-scanning confocal microscope and ZEN software (Carl Zeiss, Oberkochen, Germany).
For live cell imaging assay, Hela cells stably expressing Mito-Red were transiently transfected with GFP-parkin, and then treated as indicated. The movie was monitored by live-cell imaging confocal microscope equipped with a sensitive digital video camera (OLYMPUS, Tokyo, Japan).
Statistical analysis
Values are the mean±S.D. from at least three independent experiments. The data significance was evaluated using Student's t-test unless stated otherwise. P-values <0.05 was considered statistically significant (*P<0.05, **P<0.01 and ***P<0.001).
Acknowledgments
We thank Huihui Liu for technical assistance, Drs Shengcai Lin, Jiahuai Han for critical reading of the manuscript. This work was supported by National Basic Research Program of China 973 Program Grant 2015CB553800 (to HY), Fundamental Research Funds for the Central Universities Grant 2012121043 (to HY), National Natural Science Foundation of China Grant 31170718 (to HY), 31300627 (to LZ), National Science Foundation for Fostering Talents in Basic Research of the National Natural Science Foundation of China (Grant No. J1310027/J0106) and the 111 Project of Ministry of Education of China (B06016 and B12001).
Glossary
- EBSS
Earles' Balance Salt Solution
- PE
Pepstatin A and E64D
- TRAF6
tumor necrosis factor (TNF) receptor-associated factor 6
- CCCP
carbonyl cyanide 3-chlorophenylhydrazone
The authors declare no conflict of interests.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by T Mak
Supplementary Material
References
- 1Fogal V, Richardson AD, Karmali PP, Scheffler IE, Smith JW, Ruoslahti E. Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol Cell Biol 2010; 30: 1303–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Matthews DA, Russell WC. Adenovirus core protein V interacts with p32—a protein which is associated with both the mitochondria and the nucleus. J Gen Virol 1998; 79: 1677–1685. [DOI] [PubMed] [Google Scholar]
- 3Jiang J, Zhang Y, Krainer AR, Xu RM. Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc Natl Acad Sci USA 1999; 96: 3572–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Soltys BJ, Kang D, Gupta RS. Localization of P32 protein (gC1q-R) in mitochondria and at specific extramitochondrial locations in normal tissues. Histochem Cell Biol 2000; 114: 245–255. [DOI] [PubMed] [Google Scholar]
- 5van Leeuwen HC, O'Hare P. Retargeting of the mitochondrial protein p32/gC1Qr to a cytoplasmic compartment and the cell surface. J Cell Sci 2001; 114: 2115–2123. [DOI] [PubMed] [Google Scholar]
- 6Parle-McDermott A, McWilliam P, Tighe O, Dunican D, Croke DT. Serial analysis of gene expression identifies putative metastasis-associated transcripts in colon tumour cell lines. Br J Cancer 2000; 83: 725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Rubinstein DB, Stortchevoi A, Boosalis M, Ashfaq R, Ghebrehiwet B, Peerschke EI et al. Receptor for the globular heads of C1q (gC1q-R, p33, hyaluronan-binding protein) is preferentially expressed by adenocarcinoma cells. Int J Cancer 2004; 110: 741–750. [DOI] [PubMed] [Google Scholar]
- 8Fogal V, Zhang L, Krajewski S, Ruoslahti E. Mitochondrial/cell-surface protein p32/gC1qR as a molecular target in tumor cells and tumor stroma. Cancer Res 2008; 68: 7210–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Chen YB, Jiang CT, Zhang GQ, Wang JS, Pang D. Increased expression of hyaluronic acid binding protein 1 is correlated with poor prognosis in patients with breast cancer. J Surg Oncol 2009; 100: 382–386. [DOI] [PubMed] [Google Scholar]
- 10Yagi M, Uchiumi T, Takazaki S, Okuno B, Nomura M, Yoshida S et al. p32/gC1qR is indispensable for fetal development and mitochondrial translation: importance of its RNA-binding ability. Nucleic Acids Res 2012; 40: 9717–9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Hu M, Crawford SA, Henstridge DC, Ng IH, Boey EJ, Xu Y et al. p32 protein levels are integral to mitochondrial and endoplasmic reticulum morphology, cell metabolism and survival. Biochem J 2013; 453: 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Sunayama J, Ando Y, Itoh N, Tomiyama A, Sakurada K, Sugiyama A et al. Physical and functional interaction between BH3-only protein Hrk and mitochondrial pore-forming protein p32. Cell Death Differ 2004; 11: 771–781. [DOI] [PubMed] [Google Scholar]
- 13Itahana K, Zhang Y. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell 2008; 13: 542–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Storz P, Hausser A, Link G, Dedio J, Ghebrehiwet B, Pfizenmaier K et al. Protein kinase C [micro] is regulated by the multifunctional chaperon protein p32. J Biol Chem 2000; 275: 24601–24607. [DOI] [PubMed] [Google Scholar]
- 15Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 2008; 112: 1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Zhou X, Babu JR, da Silva S, Shu Q, Graef IA, Oliver T et al. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc Natl Acad Sci USA 2007; 104: 5842–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15: 406–416. [DOI] [PubMed] [Google Scholar]
- 18Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011; 331: 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Mao K, Klionsky DJ. AMPK activates autophagy by phosphorylating ULK1. Circ Res 2011; 108: 787–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Joo JH, Dorsey FC, Joshi A, Hennessy-Walters KM, Rose KL, McCastlain K et al. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol Cell 2011; 43: 572–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Wu W, Tian W, Hu Z, Chen G, Huang L, Li W et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep 2014; 15: 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem 2007; 282: 25464–25474. [DOI] [PubMed] [Google Scholar]
- 24Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 2010; 8: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Rozanov DV, Ghebrehiwet B, Ratnikov B, Monosov EZ, Deryugina EI, Strongin AY. The cytoplasmic tail peptide sequence of membrane type-1 matrix metalloproteinase (MT1-MMP) directly binds to gC1qR, a compartment-specific chaperone-like regulatory protein. FEBS Lett 2002; 527: 51–57. [DOI] [PubMed] [Google Scholar]
- 26Melser S, Chatelain EH, Lavie J, Mahfouf W, Jose C, Obre E et al. Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metab 2013; 17: 719–730. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.