Abstract
Unstable Repeat Diseases (URDs) share a common mutational phenomenon of changes in the copy number of short, tandemly repeated DNA sequences. More than 20 human neurological diseases are caused by instability, predominantly expansion, of microsatellite sequences. Changes in the repeat size initiate a cascade of pathological processes, frequently characteristic of a unique disease or a small subgroup of the URDs. Understanding of both the mechanism of repeat instability and molecular consequences of the repeat expansions is critical to developing successful therapies for these diseases. Recent technological breakthroughs in whole genome, transcriptome and proteome analyses will almost certainly lead to new discoveries regarding the mechanisms of repeat instability, the pathogenesis of URDs, and will facilitate development of novel therapeutic approaches. The aim of this review is to give a general overview of unstable repeats diseases, highlight the complexities of these diseases, and feature the emerging discoveries in the field.
Keywords: Unstable repeat diseases, repeat instability, repeat expansions and contractions, induced pluripotent stem cells
Introduction
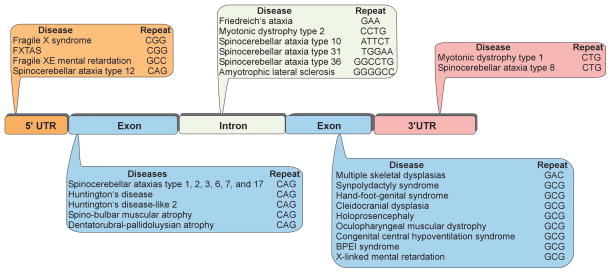
The phenomenon of dynamic mutations occurring in simple repetitive sequences was discovered over 20 years ago (1). Since the first report of the CAG•CTG repeat expansion (where the center dot indicates two complementary sequences of a duplex DNA) leading to spino-bulbar muscular atrophy (SBMA), more than 20 other human diseases caused by a mutation in unstable repeat sequences have been identified (2–4). This group includes common neurological and neuromuscular inherited disorders such as fragile X syndrome (FRAXA) or myotonic dystrophy type 1 (DM1), as well as rare diseases such as spinocerebellar ataxias (SCAs). While the vast majority of these diseases are caused by expansions of trinucleotide repeat sequences, expansions of other repeat tracts, including tetra-, penta- and hexanucleotide repeats (5–8), have also been identified (Figure 1). Therefore, to encompass all of the repeat associated disorders, we will refer to the entire group as Unstable Repeat Diseases (URDs). CG-rich trinucleotide repeat sequences such as CAG•CTG and CGG•CCG are the most common unstable DNA repeats. In the unaffected population, repeat tracts are short and stable, whereas in affected individuals the tracts become longer and frequently become somatically unstable. In some cases, individuals carrying premutation alleles can be identified, as the premutation alleles are longer than the repeat tracts in the unaffected population. Although premutation alleles are non-pathogenic, they frequently become unstable and subsequently expand to the full mutation range in subsequent generations (4).
Figure 1. Unstable repeat diseases.
Unstable repeats can be located in both coding and non-coding regions of genes. Due to the recent discoveries of widespread bi-directional transcription and RAN translation, classification of URDs based upon repeat location within a gene, or mechanism leading to disease is arbitrary and represents our current understanding of the molecular mechanism of a particular disease.
Pathogenic repeat sequences can be found in any region of a gene, including coding sequences, 5′ and 3′ UTRs, as well as introns (Figure 1). A correlation can be observed between the location and the size of the expansions, with a greater propensity for expansion associated with non-coding repeat tracts. The largest observed expansions can span several thousand repeats (6,7). In the majority of cases, the repeats elongate from one generation to the next, which is associated with increasing severity of disease symptoms and decreasing age of onset - a phenomenon that has been termed anticipation (9). Interestingly in the case of the FMR1 gene mutation, the extent of the CGG•CCG repeat expansion determines entirely distinct clinical syndromes: fragile X mental retardation (FRAXA, >200 CGG•CCG repeats) and fragile X-associated tremor/ataxia syndrome (FXTAS, 55 – 200 CGG•CCG repeats) (10). In addition to germ line instability, somatic repeat expansions can also be detected in the tissues of the affected individuals. Tissue-specific increase in the repeat size may accelerate disease progression and aggravate the severity of symptoms (11,12).
Although repeat expansions are the primary cause of most of the pathological changes associated with URDs, in rare cases repeat contractions can also be associated with disease (13,14). Repeat contractions are the predominant events observed in the majority of model systems used to study the mechanisms of repeat instability. While in most cases expansion is the underlying cause of a disease, analyzing the mechanisms of contraction is highly relevant from the perspective of possible therapeutic interventions (15).
Countless cis elements and trans factors affect repeat instability in different model systems that range from in vitro investigations to mammalian model organisms (4,16). While several new discoveries reported over the past two decades have changed our comprehension of expansions and contractions of repeating sequences, all of these studies have confirmed the initial hypothesis that, in the majority of cases, non-B DNA conformations play a central role in mediating repeat instability (17). However, proving the existence of these conformations in vivo turned out to be quite a challenging task. The notable exceptions are polyalanine expansion diseases (18,19) most likely resulting from gene conversion-mediated repeat instability. In the case of small expansions or contractions (e.g. 1 or 2 repeat units), a slippage event, without formation of stable structural intermediates, can be responsible for change in the repeat length.
The initial search for a unified mechanism of repeat instability and the search to identify similarities in disease pathogenesis have been gradually replaced by the notion that every URD has unique characteristics that are not shared by other repeat instability syndromes (4,20). Although change in repeat length is an underlying cause of all URDs, the mechanism of molecular pathogenesis that these disorders employ may vary from the effects caused by silencing of the mutated genes to the toxic effects exerted by highly expressed RNAs or proteins. The similarities and differences between the URDs suggest that both disease-specific as well as universally applicable URD therapeutic strategies may be developed.
It is also noteworthy that pathologies associated with changes in the number of short tandem repeats are not limited to humans and have been identified in both animals and plants as well (21,22).
The purpose of this review is to highlight recent progress in our understanding of the molecular pathogenesis of the unstable repeat diseases, including novel aspects of repeat instability, as well as new emerging cellular models of these disorders. Detailed characteristics of the repeat-associated diseases can be found in recent review articles (3,4).
DNA structures in vivo and repeat instability
Simple repeat sequences are an important component of the human genome. While they represent a potential source of genetic variability to aid in evolution, elongation of these sequences beyond a certain threshold is associated with instability and an increasing propensity for further, pathological expansions.
It has been shown that numerous processes affect repeat instability, but the precise mechanisms and exact contributions from each involved pathway are difficult to ascertain using cellular model systems. As such, the most progress made in understanding detailed mechanisms has been made in vitro. Precisely designed oligonucleotide substrates were employed in the biochemical dissection of various DNA processing events. This work primarily focused on hairpin-forming CTG•CAG sequences (23–27).
The in vitro evidence that repeat sequences have a high propensity to form hairpins and slipped structures indicates that these non-canonical DNA structures are likely to form in vivo as a result of any of a variety of DNA processing events. Efficient repair systems may thus be present at non-pathological length repeats and be critical for maintaining their stability. Defects in these repair systems would very likely lead to a much greater rate of mutations that result in normal repeat lengths expanding to pathological lengths. Results of recent in vitro studies utilizing CTG and CAG oligonucleotides and nuclear extracts indeed demonstrate that human cells possess a DNA hairpin repair system (HPR) that is responsible for error-free removal of these hairpins (25,27).
The HPR system appears to be similar to the nucleotide excision repair pathway (NER), however XPG (xeroderma pigmentosum, complementation group G) is not necessary for repair activity (26). The newly replicated nicked DNA strand is the one targeted for repair, and the hairpins are removed either by dual incisions or by a combination of incision and flap endonucleolytic cleavage (27). Proliferating cell nuclear antigen (PCNA) is also required for efficient repair (27). The HPR process is stimulated by components of NER as well as Werner syndrome protein (WRN helicase) which is involved in unwinding of hairpins hence facilitating repair-associated DNA synthesis (24). Surprisingly, mismatch repair (MMR) proteins do not play important roles in HPR as shown in vitro by using MMR-deficient cell extracts (27).
It is still not clear exactly which proteins are required for HPR activity. More studies are needed to test whether a similar system exists for other non-hairpin forming expandable repeats (e.g. GAA•TTC or ATTCT•AGAAT). From the therapeutic perspective, stimulation of the activity of the error-free hairpin repair system would reduce somatic instability and possibly alter disease progression.
DNA or RNA oligonucleotides are not only used to dissect mechanisms of repeat instability in vitro. Recently antisense oligonucleotides and short RNA duplexes have been of great interest as potential very specific and effective therapeutic strategies. Several reports have indicated that antisense oligonucleotides (ASOs) and duplex RNAs can induce silencing of URD genes that are responsible for producing toxic RNA or protein products (reviewed in (28)). Selective silencing of the “toxic” allele via translation inhibition or RNA cleavage is the ultimate therapeutic goal in the case of several dominantly inherited diseases including Huntington’s Disease (HD) and SCAs. This strategy was recently applied successfully in a myotonic dystrophy type 1 mouse model. Systemic administration of ASOs effectively silenced transcription of expanded CUG repeats, with sustained therapeutic benefit lasting up to 1 year (29). Another class of therapeutically interesting compounds capable of sequence specific binding to DNA repeats are pyrrole-imidazole polyamides (PAs), which were demonstrated to bind to GAA•TTC tracts (30). Interestingly, opposite to ASOs, binding of PAs lead to upregulation of expression of the frataxin gene in human cells (30).
In addition to oligonucleotide, dsRNA, and polyamide approaches, small molecules targeting RNAs containing expanded CUG, CAG, and CGG repeats are being implemented to improve molecular abnormalities associated with myotonic dystrophy, FAXTAS and other RNA gain-of-function diseases (31–33).
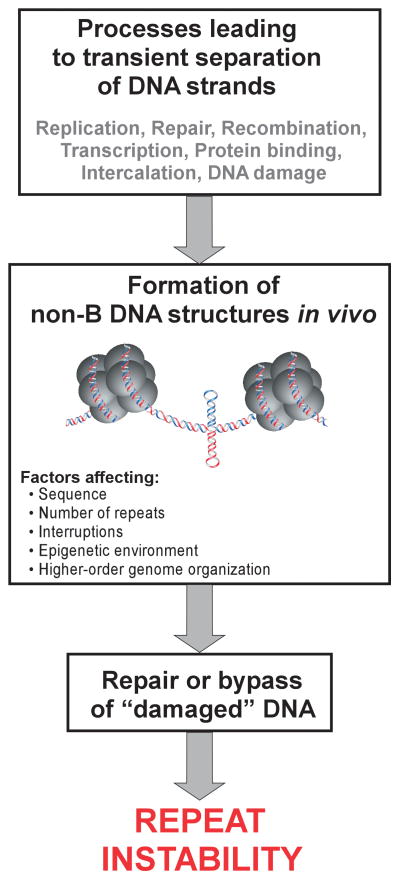
Practically any process acting on DNA has the potential to either stimulate or prevent repeat expansions and/or contractions (Figure 2) (4,16,34–36). It has been hypothesized since the earliest studies on the instability of di- and trinucleotide repeats that the propensity of these sequences to transiently adopt non-canonical DNA structures is tightly linked to instability (Figure 2) (17,37). Numerous studies using a variety of biochemical and biophysical approaches have demonstrated the formation of stable hairpins, cruciforms, triplexes, tetraplexes, and other non-canonical DNA conformations by repeat sequences associated with URDs (36–38). Moreover, the stability of these structures depends on the length of the repeat tracts, frequently with the threshold of instability at the border between long-normal and short-pathogenic alleles. Indirect evidence, such as, the stabilizing effect of interruptions disturbing the homogeneity of repeat tracts, the effect of superhelical density on frequency of expansions or contractions, and the relative stability of sequences not prone to structure formation (e.g. CAA•TTG repeats) indicates that non-B DNA structures are formed in vivo (34,39–42).
Figure 2. Complex mechanisms leading to repeat instability.
The crucial step in all models of repeat instability is a transient formation of stable non-B DNA conformations (hairpins, slipped structures, cruciforms, triplexes, tetraplexes and etc.). The extent of repeat instability is affected by the activity of processes leading to formation of stable secondary structures, capacity of the repeat tracts to adopt these unusual structures, and the efficiency of a variety of DNA repair activities functioning to restore normal B-DNA conformations.
The first direct evidence that unstable repeat sequences can adopt non-B DNA structures in vivo came from recent studies on replication-dependent instability of CTG•CAG repeats (43). Liu et al. used engineered zinc finger nucleases (ZFNs) capable of specific recognition of the CAG or CTG repeat tracts (43). Each ZFN consisted of two functional domains: DNA binding and DNA cleaving (encoded by FokI nuclease). Because the FokI enzyme acts as a dimer to induce DNA cleavage, a functional nuclease must contain a pair of proteins that typically recognize specific 18 – 30 bp sequences of DNA (44–46).
Concurrent delivery of a CTG specific ZFN with a CAG specific nuclease leads to a functional ZFN pair that recognizes and efficiently cleaves double stranded B-DNA CTG•CAG tracts. The ZFNs used by Liu et al. were designed to cleave CTG•CTG duplex (ZFNCTG) and CAG•CAG DNA (ZFNCAG). Interestingly, when a single ZFN was expressed in the cells (targeting either CTG•CTG or CAG•CAG repeats), repeat contractions could readily be detected, indicating that CTG•CTG and CAG•CAG duplexes were formed in the cells (43). Such sequences can only be present if individual strands of the CTG•CAG duplex adopt a hairpin conformation. Moreover, the instability induced by ZFNCTG alone or ZFNCAG alone was dependent on active replication and could only be detected in cell lines harboring 45 or 102 repeats. No contractions were found in cells containing only 12 CTG•CAG repeats, in agreement with the prediction that any of the ZFN dimers require a stretch of 8 repeat units to bind to, and a tract of 12 CTG or CAG repeats cannot adopt hairpins with a stem spanning 8 repeat units. Rationally, modifying the ability of these sequences to adopt non-B DNA structures could be therapeutically important for controlling both germline and somatic instability and perhaps even be used as a tool to drive the contraction of pathological expansions (4,15).
Experiments conducted using repeat-specific ZFNs demonstrated that in vivo formation of hairpins during replication is critical for repeat instability. Replication is an important process driving formation of non-B DNA structures, thus in the past several years many laboratories uncovered various cis elements and trans factors connected to DNA replication which interfered with repeat stability (for excellent reviews see (16,47,48)). Due to space constraints, we will predominantly focus on the role of repeat-containing RNAs and transcription processes on the instability and pathogenesis of URDs.
RNA, R-loops, and RAN
As mentioned before, practically any process that acts on DNA molecules, which leads to transient separation of the complementary strands or the exposure of a single stranded DNA, will affect the stability of tandem repeats. Initially, DNA replication was thought to be the primary source of instability (17,49,50), however, frequent reports of repeat expansions and contractions in terminally differentiated, non-dividing cells indicated that other processes must play a critical role in triggering instability (51). One of the processes recurrently leading to the separation of the DNA duplex is transcription (39,52–57). RNA synthesis occurs continuously in cells, irrespective of cell cycle progression. Additionally, a large portion of the human genome, including the majority of URD loci, is transcribed bidirectionally using both sense and antisense DNA strands as templates (58,59). In fact, genes harboring expandable repeats are likely to be transcribed more frequently than they are replicated.
Transcription was first demonstrated to stimulate CA•TG and CTG•CAG repeat instability in yeast and bacteria (56,57,60). Progression of the transcription machinery leads to the separation of complementary strands over relatively long stretches of DNA. Moreover, it creates a wave of negative supercoiling, which facilitates separation of DNA strands and promotes formation of non-B DNA structures (61,62). Recent studies, conducted independently in three laboratories, demonstrated that the movement of the transcriptional machinery along the DNA is not the only mechanism involved in transcriptionally-induced repeat instability. Transcribed trinucleotide and tetranucleotide repeats associated with URDs have the propensity to form stable DNA•RNA hybrids between the DNA template and nascent RNA strands (39,63,64). Short 8 – 9 nt hybrids are normally present in the transcriptional bubble and they are progressively dissociated during the progression of RNA synthesis. In the case of tandem repeats, much longer, stable DNA•RNA hybrids (R-loops) have been described. Data obtained using transcribed plasmid models (39,64) indicate that repeat-containing DNA•RNA hybrids can span the entire repeat tract and even extend into flanking regions. On the other hand, bisulfite conversion experiments in mammalian cells demonstrated uninterrupted modification of up to 8 repeats, suggesting formation of much shorter hybrids (39). However, these results may be influenced by extended isolation and purification protocols. Moreover, the rate of bisulfite conversion detected on the non-template DNA strand may be biased because the repeat-containing single stranded DNA regions are likely to be involved in formation of secondary structures, hence decreasing the apparent length of the bisulfite-accessible DNA strand. In vitro these hybrid structures are resistant to RNase A cleavage and depend on both the length of the repeat and supercoiling. Lowering the activity of RNase HI in bacteria or RNase H1 and H2 expression in mammalian cells significantly increases the instability of expanded trinucleotide repeat sequences in a transcription-dependent manner, which suggests the existence of stable DNA•RNA hybrids (39).
Formation of R-loops renders non-template DNA strands persistently unpaired and increases the likelihood of the formation of intrastrand non-canonical DNA structures within them. Additionally, the formation of these structures decreases the probability that the complementary DNA strands will correctly re-associate. Lastly, formation of R-loops has been shown to interfere with DNA synthesis, causing stalling of the replication fork (65,66). Impediment of DNA synthesis can contribute to repeat instability and is frequently observed at various repeat sequences (67,68).
During transcription, non-canonical structures formed by the non-template strand are likely to further facilitate formation of stable R-loops that will persist until DNA replication. It would be logical to assume that these structures would subsequently be bypassed by replication machinery, resulting in preferential contractions of repeat tracts. However, R-loop formation was shown to stimulate both expansions and contractions, indicating that cells utilize additional mechanisms to cope with these structures instead of simply bypassing them. Accordingly, there are several mechanisms that may be involved in promoting repeat instability at R-loops (63). Like other non-canonical structures, R-loops may be recognized by transcription-coupled nucleotide excision repair machinery (TC-NER) (69). Hairpins or other non-canonical DNA structures can also be recognized by components of the mismatch repair complex (MMR) (70,71). Binding of these complexes to R-loops or DNA structures formed as a result of R-loop presence may lead to error-prone repair and consequently repeat instability. Additionally, persistent R-loops can potentially induce DNA double-strand breaks (DSBs) (66), which may also facilitate repeat expansions or contractions. It is tempting to speculate that tissue specific factors could determine the choice of the pathway involved in the processing of R-loops, leading to distinct outcomes including faithful repair, expansions, or contractions.
All of the above-mentioned effects are elicited by unidirectional transcription. Recent work has indicated that simultaneous RNA synthesis on both complementary DNA strands can have a significantly greater impact on repeat instability than unidirectional transcription (72–74). Bidirectional transcription throughout trinucleotide repeats has also been shown to facilitate the formation of double R-loops (63). Such structures may be, at least in part, responsible for enhanced instability of convergently transcribed repeat sequences and may also potentially induce apoptotic effects via the DNA damage response pathways. Using a genetic assay that detects contractions of the (CTG•CAG)95 tract, Lin and colleagues demonstrated that repeat contractions are stimulated ~3 fold during bidirectional transcription as compared to unidirectional RNA synthesis (72,74). Additionally, a rather surprising consequence of convergent transcription throughout the CTG•CAG tract was rapid cell cycle arrest and induction of massive apoptotic cell death (see next section).
The studies examining the effect of bidirectional transcription on repeat instability were recently extended into much larger repeat tracts. Nakamori et al. demonstrated, using a genomic reporter harboring an 800 CTG•CAG tract, that convergent transcription increased instability approximately 30-fold when compared to non-transcribed control and approximately 3 to 6-fold relative to unidirectional expression (73).
Bidirectional transcription through the repeats certainly increases the potential of DNA to form hairpins, slipped structures and persistent DNA•RNA hybrids on both DNA strands. Consequently, processing of these non-canonical structures by different repair mechanisms increases the likelihood of instability at repeat tracts. Convergent transcription might also cause topological problems due to the local accumulation of positive waves of supercoiling in front of the transcriptional machinery. Although the effect of such topological contributions to repeat instability has never been analyzed, changes in superhelical tension were previously identified as a factor contributing to the stability of repetitive sequences (40). In yeast, convergent transcription led to the inhibition of the progression of the RNA polymerase II (RNAPII) machinery (75). Transcriptional stalling may affect instability of DNA sequences by increasing the potential of non-canonical structure formation and impeding progression of the replication fork. Accessibility of DNA to protein factors as well as chromatin status, including nucleosomal positioning, can also be affected at the bidirectionally transcribed loci (75). The interplay between bidirectional transcription and replication fork progression has never been analyzed. Taking into the account potential deleterious effects arising from collision between transcription and replication, the effect of convergent transcription combined with replication needs to be addressed in future studies.
In addition to the direct effect of repeat-containing RNAs on DNA instability via the preferential formation of DNA•RNA hybrids, accumulation of expanded CUG, CGG, CAG and CCUG RNAs in the form of foci is considered the instigator of molecular pathogenesis in toxic RNA gain of function diseases such as DM1, DM2 and FAXTAS (76,77). In the case of SCA8, both RNA and protein mechanisms are likely co-contributors, as two antisense transcripts harboring CAG and CUG repeats are synthesized (ATXN8 and AXN8OS, respectively) (78,79). Accumulation of the expanded CUG RNAs and polyglutamine-containing proteins has been detected in SCA8 cells. Recent discoveries of non-ATG initiated translation triggered by RNAs containing expanded repeats introduced yet another level of complexity into the molecular pathogenesis of URDs (80). In studying the mechanism of SCA8 pathogenesis, Zu et al. showed that constructs containing long CAG•CTG repeat tracts expressed polyGln protein even without the ATG initiation codon. More surprisingly, the RAN translation (repeat-associated non-ATG translation) was not limited to a single frame because polyAla (GCA frame) and polySer (AGC frame) proteins were readily expressed. Analysis of CAG constructs of differing lengths ranging from 15 – 107 repeats revealed that RAN translation is length dependent and that polyGln, polyAla and polySer proteins can only be detected for the longer repeat lengths (80).
These facts taken together with the observation that constructs expressing non-hairpin forming CAA repeats do not exhibit ATG independent translation, indicated that the formation of stable hairpin structures by repeat-containing RNAs may be essential to initiate RAN translation. Zu et al. suggested that a mechanism similar to internal ribosome entry site (IRES) translation may be involved in RAN synthesis. As RNA hairpins are formed by several repeat sequences associated with URDs, the RAN mechanism could be a common contributor to the repeat pathogenesis. In fact, analyses of SCA8 and DM1 mice tissues revealed than not only CAG repeats, but also CUG tracts initiate RAN translation in vivo. Homopolymeric proteins corresponding to RAN products were also detected in various tissues from DM1 patients (80).
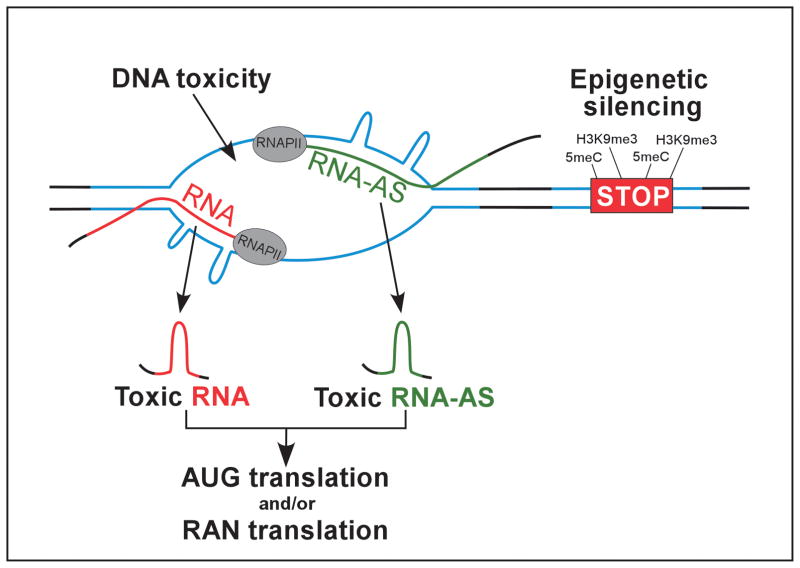
The discovery of RAN translation may have a tremendous impact on the understanding of molecular pathogenesis of the URDs. After establishing the existence of homo aminoacid RAN products, a critical task will be to determine whether these proteins contribute to disease pathology. Considering toxic RNA (both CUG and CAG strand), AUG-initiated translation of CAG repeats (polyGln toxicity), and RAN initiated translation (both strands in all 3 reading frames), repeat expansion in a single locus may lead to the generation of up to nine potentially toxic entities affecting the phenotype of the disease ((80,81) Figure 3).
Figure 3. Molecular mechanisms of pathogenesis induced by expanded repeats.
Pathological consequences elicited by long repeat sequences can be classified into two categories (i) loss of gene expression due to the changes in the epigenetic landscape and (ii) consequences of RNA and/or protein expression, which includes RNA-toxic-gain of function induced by sense and/or antisense transcripts, as well as protein gain of function encompassing AUG-translated and RAN-translated proteins. DNA toxicity resulting from convergent transcription through the repeats may also contribute to the pathogenesis of URDs.
Interplay between repeat expansion, bidirectional transcription, and epigenetic regulation
Antisense transcripts have been detected in at least 16 genes associated with URDs (58,59). The rate of antisense transcription varies between loci from robust expression that is greater than the sense transcription, to almost undetectable levels. Frequently, antisense transcripts can be detected only in specific cell lines or tissues, complicating the task of dissecting their role in regulating gene expression or affecting disease pathogenesis. Therefore, more analyses conducted by strand-specific high throughput RNA sequencing confirming the existence of these antisense transcripts and studies defining their functions are required.
As described in the previous section, antisense transcription may stimulate instability of the repeats and directly influence pathology of the disease by contributing potentially toxic RNAs and protein products. Strong evidence from studies on SCA8 and Huntington’s disease-like 2 (HDL2) demonstrate that both RNA and protein gain of function pathways can co-exist and contribute to pathogenesis (78,82,83). In addition to a direct contribution of toxic RNAs and/or proteins to pathogenesis, new studies on the mechanism of SCA7 demonstrated an interplay between bidirectional transcription, chromatin landscape at the SCA7 locus, and molecular pathogenesis (84).
SCA7 is caused by CTG•CAG/polyGln expansion in the N-terminal portion of the protein encoded by the SCA7 sense transcript. The antisense, non-coding RNA – SCAANT1 is synthesized from the alternative promoter located upstream of the repeats (downstream considering the SCA7 sense promoter). Experiments conducted in mouse models demonstrated that CTG•CAG repeats in the SCA7 gene are flanked at the 3′, as well as the 5′ side by binding sites for the CTCF protein (CCCTC binding factor) (84). The CTCF protein is widely distributed throughout the genome and plays a role in chromatin organization and insulation, transcriptional regulation, and genomic imprinting (85). CTCF was initially demonstrated to regulate cis repeat instability of CTG•CAG repeats in the SCA7 locus (86). A more recent study has revealed that CTCF also regulates expression of ataxin 7 influencing pathogenesis of this disease. Upon CTCF binding, the antisense SCAANT1 transcript is upregulated leading to the downregulation of SCA7 transcription accompanied by the epigenetic silencing of the sense promoter (84). On the other hand, expansion of the CTG•CAG repeats diminishes binding of CTCF in the vicinity of the repeat tract, resulting in reduced activity of the antisense promoter and a high rate of transcription of the sense SCA7 mRNA and an increased synthesis of the mutant ataxin 7. Hence, in SCA7 patients, the precise balance between sense and antisense is disturbed.
Additionally, interplay between CTCF binding and antisense transcription affects pathogenesis of DM1 and FRDA (87,88). In contrast to SCA7, depletion of CTCF caused by expansion of CTG•CAG or GAA•TTC repeats may allow transcription of the antisense RNA leading to heterochromatin formation and epigenetic silencing of these loci.
The effect of the chromatin context, especially distinct posttranslational histone modifications and the presence of the appropriate protein complexes responsible for writing and reading the histone code, on the repeat stability and disease pathogenesis is only beginning to be recognized and elucidated. Results of recent studies on the role of histone deacetylases (HDACs) and histone acetyltransferases (HATs) indicate that posttranslational modifications of histone and non-histone proteins can affect instability of repeating tracts in yeast and human cells (89–91). Future work will reveal the role of the epigenetic environment in the transcriptional regulation of genes associated with URDs, as well as on repeat instability. Several therapeutic strategies are currently being investigated that interfere with the epigenetic status of the affected cells underscoring the importance of this research area (92,93).
Transcription-mediated DNA toxicity
RNAs and proteins encoded by expanded repeats can be toxic to the cell (81,94,95). In contrast, DNA is considered an instigator (via repeat expansions) that initiates a cascade of events that lead to cell death, but is not considered a direct contributor to cellular toxicity. The mutagenic potential of repeats, or more precisely the structures adopted by them, has been demonstrated in different organisms using various repeat sequences (14,40,96). Some global effects on cell proliferation (increased doubling time and reduced fitness) resulting from repeats at the DNA level were reported in bacteria harboring plasmids with expanded CTG•CAG tracts (97).
New results generated from studies addressing the effect of transcription on CTG•CAG repeat instability revealed a surprising consequence of convergent transcription throughout these sequences. Induction of bidirectional transcription through the (CTG•CAG)95 sequence stimulated rapid cell cycle arrest and induced massive apoptotic cell death (74). Neither of these processes were observed during unidirectional sense or antisense transcription through the repeats or during bidirectional transcription of non-repeating DNA fragments. Additionally, apoptosis was not induced when transcription was initiated using both DNA strands simultaneously in spatially separated regions of the genome, eliminating the possibility of a role for double-stranded RNA in triggering cell death or toxic effects associated with RAN translation of two repeating sequences. The phenomenon of DNA initiating cell death while undergoing convergent transcription was termed “DNA toxicity” (72,74). In non-proliferating cells, apoptotic death occurred more rapidly than in the dividing cells, suggesting that DNA toxicity does not result from interaction between transcription and replication machineries. The extent of cell death was directly correlated to the rate of sense or antisense transcription.
Given that convergent transcription through a non-repeating template elicited no ill effects, but triggered significant cell death with a repeating template suggests that the collision of sense and antisense transcription bubbles at expanded repeats creates an abnormal “double-bubble” that is necessary for the induction of apoptosis. Formation of this “double-bubble” at CTG•CAG repeats may lead to a conglomeration of DNA, RNA, and proteins at these sites that are then inefficiently processed and repaired, ultimately resulting in increased cell death (72).
Biochemical analyses demonstrated that convergent transcription of the (CTG•CAG)95 tract was associated with increased levels of active caspase 3 and active ATR (ataxia telangiectasia and Rad3 related), indicating stimulation of the apoptotic response. Components of the ATR pathway were shown to be recruited to CAG repeats upon convergent transcription, most likely to stimulate repair of DNA•RNA structures that initiate apoptosis. Although the exact signal initiating this response is not known, several possible scenarios have been proposed (72,74), including RNAPII stalling by hairpins formed during convergent transcription of both DNA strands, accumulation of single-stranded DNA binding protein replication protein A (RPA), or simultaneous formation of RNA•DNA hybrids on both template strands.
It has been demonstrated in the past that global impediment of transcription by physical or chemical mutations can lead to apoptosis. Remarkably, in the case of expanded CTG•CAG repeats, convergent transcription of a single locus in the entire genome (which undergoes frequent global bidirectional transcription) can trigger a robust cellular response. This raises the possibility that other pathways contribute to the apoptotic response. Perhaps rapid accumulation of repeat-containing dsRNAs resulting from a spatial proximity of transcribed CTG and CAG strands triggers toxic RNA effects via pathways different than canonical processing of dsRNAs and miRNAs. Additionally, we cannot rule out that yet unknown factors exist that are capable of recognizing the double-bubble repeat structures and initiating an apoptotic response. Possibly, a combination of two or more mechanisms leads to this catastrophic effect.
In contrast, no induction of apoptosis was reported in bidirectional transcription experiments conducted using mammalian cells harboring a very long (CTG•CAG)800 tract (73). It is probable that a significant difference in the repeat length and/or rate of transcription affected the globally toxic DNA effect observed in the (CTG•CAG)95 constructs. The rate of transcription may be a critical factor, influencing not only the cellular toxicity of bidirectional transcription, but also repeat instability. It has been shown using GAA•TTC repeat-containing models that the rate of transcription can affect the balance between repeat contractions and expansions. High levels of transcription throughout the GAA•TTC region stimulate contractions, while low to moderate transcription leads to expansions (98).
It is possible that convergent transcription of long repeats results in the induction of apoptosis in patients’ cells. However, a possible toxic effect of transcription through repeats may be difficult to distinguish from toxic RNA/protein effects. Several lines of evidence suggest that convergent transcription of long repeat DNA induces local stress at the cellular level leading to the destruction of intracellular homeostasis and shifts cellular metabolism towards apoptosis (72,74). Recent work conducted in yeast using CTG•CAG repeat sequences showed that expanded repeats could induce the DNA damage checkpoint response (99). Analyses revealed that yeast cells harboring the repeats demonstrated prolonged cell division cycles with a delay of S phase and a G2/M arrest. Intriguingly, induction of the DNA damage checkpoint and lengthening of the cell cycle facilitated repeat expansions. Although low-level transcription was observed at the repeat locus by RT-PCR, the data was more consistent with the DNA damage sensing that occurs during DNA replication. Considering the potential of DNA toxicity to affect both repeat instability and pathogenesis, its mechanism and contribution needs to be further investigated.
Induced pluripotent stem cells - new disease models with therapeutic potential
Appropriate cell culture models of neurological diseases are extremely important for studies to uncover the molecular mechanisms of these diseases, as well as testing of novel therapeutic approaches. The majority of the cellular studies of URD pathogenesis were conducted using patient-derived lymphoblasts, primary fibroblasts and/or myoblasts, as these cell types are readily available. In an attempt to generate more relevant disease models, immortalized neuronal-like cell lines have been generated to study the neuronal aspects of these diseases. Although patient-derived lymphoblasts or fibroblasts can recapitulate some molecular features of URDs, they are inadequate for modeling the physiological state of cells predominantly affected by the disease. Additionally, studies with tissues and cells derived directly from patients are limited by the accessibility as well as quantity of the material and are frequently conducted using material that represents the end-stage disease. Hence, development of the appropriate, disease-relevant cellular models of unstable repeat diseases is as important as generating adequate animal models.
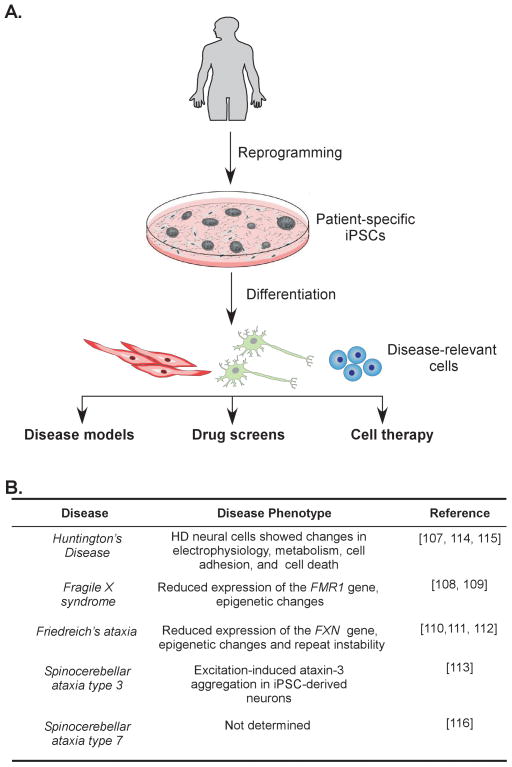
Recent advances in stem cell biology and the development of somatic cell reprogramming technology enabled the generation of patient-specific induced pluripotent stem cells (iPSCs) (100–102). These embryonic stem cell-like cells can be differentiated in vitro into cell types derived from all three germ layers, including neurons, cardiomyocytes, and pancreatic cells (Figure 4). The iPSCs can be derived directly from easily accessible patient material (e.g. skin fibroblasts, keratinocytes, blood samples) and efficiently reprogrammed into iPSCs via chemical modifiers and expression of pluripotency factors. Recently, several reprogramming methods have been developed that impose minimal or no alterations on the genome of reprogrammed cells (103–106). The iPSCs can recapitulate the step-by-step development of disease and changes of the molecular phenotype during progression of differentiation. They can model early stages of the diseases, which are impossible to analyze using patient material. The use of iPSCs and their differentiated derivatives allow for the study of the molecular mechanisms of repeat expansion, details of pathogenesis, and testing of potential therapeutic strategies all in appropriate disease-relevant cell types (Figure 4). Additionally, human iPSCs convey the unique and critical advantage of a platform upon which to study different aspects of URDs in cells undergoing natural processes such as early development, lineage commitment, differentiation, and senescence.
Figure 4. Induced pluripotent stem cells in disease modeling and therapy.
(A). Induced pluripotent stem cell technology combined with lineage specific differentiation methods have great potential in generating disease-relevant models critical for studying mechanisms of URDs. Generation of patient-specific iPSCs will stimulate development of therapeutic approaches including high throughput screens for novel drugs and cell therapy. (B) Current iPSC models of URDs.
To date, iPSC lines have been generated and characterized for Huntington’s disease, fragile X syndrome, Friedreich’s ataxia and spinocerebellar ataxias type 3 and 7 (Figure 4) (107–116). Reprogramming and establishing iPSCs is associated with dramatic changes in chromatin status (117,118). Therefore, modeling human diseases caused by epigenetic abnormalities such as FRDA or FRAXA is of particular importance. First, iPSC models of Friedreich’s ataxia have clearly demonstrated epigenetic silencing of the FXN locus, similar to that observed in somatic cells from FRDA patients (111,112). Additionally, reprogramming led to a significant increase of GAA•TTC repeat instability with a strong bias towards repeat expansions (111). Large GAA•TTC repeat expansions, spanning in some cases more than a hundred repeats, were observed in all reprogrammed FRDA iPSCs, as well as in the mutation carriers. Instability was not found in the short GAA•TTC alleles and in other non FXN GAA•TTC-containing loci in the human genome. Expansions were progressive and accumulated with increasing passages of the iPSC lines. Analyses of the effect of mismatch repair (MMR) on GAA•TTC repeat stability in FRDA iPSCs demonstrated that high expression of MMR in these cells (characteristic of pluripotent cells (119)) stimulated the expansions, while depletion of MSH2 (a major component of MMR) led to the stabilization of the repeat sequences (111). Chromatin immunoprecipitation analyses indicated that MMR proteins were enriched in the vicinity of the expanded GAA•TTC tracts. Similar findings related to the role of MMR in the CTG•CAG repeat instability were also described for human DM1 embryonic stem cells (hESCs). Differentiation of these cells coincided with the stabilization of the expanded CTG•CAG tracts and downregulation of the MMR system (120). Although MMR has been recurrently demonstrated in various model systems to affect repeat stability, the iPSCs showed a unique propensity for expansion of the GAA•TTC repeats (121,122).
Similarly to FRDA, fragile X syndrome patient-derived iPSCs preserved the epigenetic defect observed in the FRAXA fibroblasts harboring expanded CGG•CCG repeats (108,109). Interestingly, the epigenetic marks characteristic of silenced chromatin (H3K9me3, CpG methylation) were absent in the hESCs derived from FRAXA embryos, clearly demonstrating that differences exist between ES cells and their induced counterparts (109). It is also likely that remodeling of the epigenetic landscape during reprogramming can be modulated during transition from a somatic to a pluripotent state by culture conditions, perhaps allowing for a reversal of epigenetic silencing of the FRAXA and FXN loci. In this case, somatic cell reprogramming could be used to elucidate the exact mechanism of the epigenetic silencing caused by expanded triplets and to uncover new strategies of therapeutic intervention.
FRAXA iPSCs not only recapitulated epigenetic defects observed in patient cells but also demonstrated a clear neuronal phenotype, characterized by aberrant neuronal differentiation with significantly shorter and fewer neural processes (108). Such obvious phenotypic differences between iPSC-derived control and FRAXA neurons can be used as a readout in high throughput screens to discover novel therapeutic approaches for FRAXA.
Although time-consuming, the reprogramming of somatic cells and characterization of the iPSC lines can be routinely performed, while the methodology of differentiating pluripotent cells into specific cell types is much less developed. This, along with phenotypic characterization of the patient specific cell lines, will be an important step towards the establishment of iPSCs and their derivatives as invaluable resources in modeling diseases and uncovering new therapeutic strategies (102,123).
Recent progress in genome editing technologies brings the possibility of sequence specific removal of pathogenic repeats using ZFNs or TALENs (transcription activator like effector nucleases) and the development of cell based therapy (44,46,124). In conjunction with safe methods of reprogramming and efficient differentiation of pluripotent cells into terminally defined cell types, it opens the possibility of a future regenerative therapy to treat some of the unstable repeat diseases.
Concluding remarks
Twenty years of research on the mechanisms of pathogenesis of the URDs has significantly broadened our knowledge of these diseases to the point where rational therapeutic approaches are being designed and tested using a plethora of excellent model systems. It is impossible to review all the therapeutic approaches currently being developed or being tested in clinical trials that are aimed at transforming the currently available symptom-management strategies towards targeted disease modulation. In the past two years, approximately 250 clinical trials have been initiated, are active, or have been completed worldwide, and are aimed at uncovering novel treatments for HD, FRAXA, FRDA, DM1 and SCAs (ClinicalTrial.gov). Additionally, several unbiased drug screens have been conducted to uncover novel, potential therapeutics. These efforts combined with the alluring prospects of genome editing and regenerative medicine may in the near future result in the development of successful therapies for these URDs.
Acknowledgments
This work was partially supported by grants from the Friedreich’s Ataxia Research Alliance and a pilot grant from the Arnold Family Foundation and The Center for Stem Cells and Developmental Biology at the MD Anderson Cancer Center to MN. We thank Hilary Graham, MA for editorial assistance.
References
- 1.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 2.Wells RD, Ashizawa T. Genetic Instabilities and Neurological Diseases. 2. Elsevier-Academic Press; San Diego, CA: 2006. [Google Scholar]
- 3.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 5.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 7.Matsuura T, Yamagata T, Burgess DL, Rasmussen A, Grewal RP, et al. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat Genet. 2000;26:191–194. doi: 10.1038/79911. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi H, Abe K, Matsuura T, Ikeda Y, Hitomi T, et al. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am J Hum Genet. 2011;89:121–130. doi: 10.1016/j.ajhg.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter NJ. Genetic anticipation. Expanding tandem repeats. Neurol Clin. 1994;12:683–697. [PubMed] [Google Scholar]
- 10.Willemsen R, Levenga J, Oostra BA. CGG repeat in the FMR1 gene: size matters. Clin Genet. 2011;80:214–225. doi: 10.1111/j.1399-0004.2011.01723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma R, Bhatti S, Gomez M, Clark RM, Murray C, et al. The GAA triplet-repeat sequence in Friedreich ataxia shows a high level of somatic instability in vivo, with a significant predilection for large contractions. Hum Mol Genet. 2002;11:2175–2187. doi: 10.1093/hmg/11.18.2175. [DOI] [PubMed] [Google Scholar]
- 13.Delot E, King LM, Briggs MD, Wilcox WR, Cohn DH. Trinucleotide expansion mutations in the cartilage oligomeric matrix protein (COMP) gene. Hum Mol Genet. 1999;8:123–128. doi: 10.1093/hmg/8.1.123. [DOI] [PubMed] [Google Scholar]
- 14.Wells RD. Discovery of the role of non-B DNA structures in mutagenesis and human genomic disorders. J Biol Chem. 2009;284:8997–9009. doi: 10.1074/jbc.X800010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomes-Pereira M, Monckton DG. Chemical modifiers of unstable expanded simple sequence repeats: what goes up, could come down. Mutat Res. 2006;598:15–34. doi: 10.1016/j.mrfmmm.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 16.Pearson CE, Edamura KN, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 17.Kang S, Jaworski A, Ohshima K, Wells RD. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 18.Warren ST. Polyalanine expansion in synpolydactyly might result from unequal crossing-over of HOXD13. Science. 1997;275:408–409. doi: 10.1126/science.275.5298.408. [DOI] [PubMed] [Google Scholar]
- 19.Amiel J, Trochet D, Clement-Ziza M, Munnich A, Lyonnet S. Polyalanine expansions in human. Hum Mol Genet. 2004;13:235–243. doi: 10.1093/hmg/ddh251. [DOI] [PubMed] [Google Scholar]
- 20.Mirkin SM. Toward a unified theory for repeat expansions. Nat Struct Mol Biol. 2005;12:635–637. doi: 10.1038/nsmb0805-635. [DOI] [PubMed] [Google Scholar]
- 21.Lohi H, Young EJ, Fitzmaurice SN, Rusbridge C, Chan EM, et al. Expanded repeat in canine epilepsy. Science. 2005;307:81. doi: 10.1126/science.1102832. [DOI] [PubMed] [Google Scholar]
- 22.Sureshkumar S, Todesco M, Schneeberger K, Harilal R, Balasubramanian S, et al. A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science. 2009;323:1060–1063. doi: 10.1126/science.1164014. [DOI] [PubMed] [Google Scholar]
- 23.Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat Struct Mol Biol. 2005;12:654–662. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- 24.Chan NL, Hou C, Zhang T, Yuan F, Machwe A, et al. The Werner Syndrome Protein Promotes CAG/CTG Repeat Stability by Resolving Large (CAG)n/(CTG)n Hairpins. J Biol Chem. 2012;287:30151–30156. doi: 10.1074/jbc.M112.389791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang T, Huang J, Gu L, Li GM. In vitro repair of DNA hairpins containing various numbers of CAG/CTG trinucleotide repeats. DNA Repair (Amst) 2012;11:201–209. doi: 10.1016/j.dnarep.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou C, Zhang T, Tian L, Huang J, Gu L, et al. The Role of XPG in Processing (CAG)n/(CTG)n DNA Hairpins. Cell Biosci. 2012;1:11. doi: 10.1186/2045-3701-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou C, Chan NL, Gu L, Li GM. Incision-dependent and error-free repair of (CAG)(n)/(CTG)(n) hairpins in human cell extracts. Nat Struct Mol Biol. 2009;16:869–875. doi: 10.1038/nsmb.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsui M, Corey DR. Allele-selective inhibition of trinucleotide repeat genes. Drug Discov Today. 2012;17:443–450. doi: 10.1016/j.drudis.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burnett R, Melander C, Puckett JW, Son LS, Wells RD, et al. DNA sequence-specific polyamides alleviate transcription inhibition associated with long GAA.TTC repeats in Friedreich’s ataxia. Proc Natl Acad Sci U S A. 2006;103:11497–11502. doi: 10.1073/pnas.0604939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Disney MD, Liu B, Yang WY, Sellier C, Tran T, et al. A Small Molecule That Targets r(CGG)(exp) and Improves Defects in Fragile X-Associated Tremor Ataxia Syndrome. ACS Chem Biol. 2012 doi: 10.1021/cb300135h. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Childs-Disney JL, Hoskins J, Rzuczek SG, Thornton CA, Disney MD. Rationally designed small molecules targeting the RNA that causes myotonic dystrophy type 1 are potently bioactive. ACS Chem Biol. 2012;7:856–862. doi: 10.1021/cb200408a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, et al. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. J Am Chem Soc. 2012;134:4731–4742. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cleary JD, Nichol K, Wang YH, Pearson CE. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 35.McIvor EI, Polak U, Napierala M. New insights into repeat instability: role of RNA*DNA hybrids. RNA Biol. 2010;7:551–558. doi: 10.4161/rna.7.5.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wells RD, Dere R, Hebert ML, Napierala M, Son LS. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson CE, Sinden RR. Trinucleotide repeat DNA structures: dynamic mutations from dynamic DNA. Curr Opin Struct Biol. 1998;8:321–330. doi: 10.1016/s0959-440x(98)80065-1. [DOI] [PubMed] [Google Scholar]
- 38.Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr Opin Struct Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci U S A. 2010;107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Napierala M, Bacolla A, Wells RD. Increased negative superhelical density in vivo enhances the genetic instability of triplet repeat sequences. J Biol Chem. 2005;280:37366–37376. doi: 10.1074/jbc.M508065200. [DOI] [PubMed] [Google Scholar]
- 41.Napierala M, Dere R, Vetcher A, Wells RD. Structure-dependent recombination hot spot activity of GAA.TTC sequences from intron 1 of the Friedreich’s ataxia gene. J Biol Chem. 2004;279:6444–6454. doi: 10.1074/jbc.M309596200. [DOI] [PubMed] [Google Scholar]
- 42.Pearson CE, Eichler EE, Lorenzetti D, Kramer SF, Zoghbi HY, et al. Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry. 1998;37:2701–2708. doi: 10.1021/bi972546c. [DOI] [PubMed] [Google Scholar]
- 43.Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG) x (CAG) repeat hairpins in human cells. Nat Chem Biol. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JS, Lee HJ, Carroll D. Genome editing with modularly assembled zinc-finger nucleases. Nat Methods. 2010;7:91–92. doi: 10.1038/nmeth0210-91b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 47.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 48.Lenzmeier BA, Freudenreich CH. Trinucleotide repeat instability: a hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet Genome Res. 2003;100:7–24. doi: 10.1159/000072836. [DOI] [PubMed] [Google Scholar]
- 49.Schumacher S, Fuchs RP, Bichara M. Expansion of CTG repeats from human disease genes is dependent upon replication mechanisms in Escherichia coli: the effect of long patch mismatch repair revisited. J Mol Biol. 1998;279:1101–1110. doi: 10.1006/jmbi.1998.1827. [DOI] [PubMed] [Google Scholar]
- 50.Freudenreich CH, Stavenhagen JB, Zakian VA. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol Cell Biol. 1997;17:2090–2098. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cleary JD, Pearson CE. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet Genome Res. 2003;100:25–55. doi: 10.1159/000072837. [DOI] [PubMed] [Google Scholar]
- 52.Mangiarini L, Sathasivam K, Mahal A, Mott R, Seller M, et al. Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nat Genet. 1997;15:197–200. doi: 10.1038/ng0297-197. [DOI] [PubMed] [Google Scholar]
- 53.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 54.Lin Y, Hubert L, Jr, Wilson JH. Transcription destabilizes triplet repeats. Mol Carcinog. 2009;48:350–361. doi: 10.1002/mc.20488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowater RP, Jaworski A, Larson JE, Parniewski P, Wells RD. Transcription increases the deletion frequency of long CTG.CAG triplet repeats from plasmids in Escherichia coli. Nucleic Acids Res. 1997;25:2861–2868. doi: 10.1093/nar/25.14.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schumacher S, Pinet I, Bichara M. Modulation of transcription reveals a new mechanism of triplet repeat instability in Escherichia coli. J Mol Biol. 2001;307:39–49. doi: 10.1006/jmbi.2000.4489. [DOI] [PubMed] [Google Scholar]
- 58.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dion V, Wilson JH. Instability and chromatin structure of expanded trinucleotide repeats. Trends Genet. 2009;25:288–297. doi: 10.1016/j.tig.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wierdl M, Greene CN, Datta A, Jinks-Robertson S, Petes TD. Destabilization of simple repetitive DNA sequences by transcription in yeast. Genetics. 1996;143:713–721. doi: 10.1093/genetics/143.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinden RR. DNA Structure and Function. Academic Press; San Diego, California: 1994. [Google Scholar]
- 62.Cozzarelli NR, Wang JC. DNA topology and its biological effects. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- 63.Reddy K, Tam M, Bowater RP, Barber M, Tomlinson M, et al. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2011;39:1749–1762. doi: 10.1093/nar/gkq935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grabczyk E, Mancuso M, Sammarco MC. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007;35:5351–5359. doi: 10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gan W, Guan Z, Liu J, Gui T, Shen K, et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes Dev. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- 67.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol Cell Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samadashwily GM, Raca G, Mirkin SM. Trinucleotide Repeats Affect Dna Replication In Vivo. Nat Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 69.Tian M, Alt FW. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J Biol Chem. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- 70.Pearson CE, Ewel A, Acharya S, Fishel RA, Sinden RR. Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Human Molecular Genetics. 1997;6:1117–1123. doi: 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]
- 71.Owen BA, Yang Z, Lai M, Gajec M, Badger JD, 2nd, et al. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 72.Lin Y, Wilson JH. Transcription-induced DNA toxicity at trinucleotide repeats: double bubble is trouble. Cell Cycle. 2011;10:611–618. doi: 10.4161/cc.10.4.14729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakamori M, Pearson CE, Thornton CA. Bidirectional transcription stimulates expansion and contraction of expanded (CTG)*(CAG) repeats. Hum Mol Genet. 2011;20:580–588. doi: 10.1093/hmg/ddq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin Y, Leng M, Wan M, Wilson JH. Convergent transcription through a long CAG tract destabilizes repeats and induces apoptosis. Mol Cell Biol. 2010;30:4435–4451. doi: 10.1128/MCB.00332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Puig S, Perez-Ortin JE, Matallana E. Transcriptional and structural study of a region of two convergent overlapping yeast genes. Curr Microbiol. 1999;39:369–0373. doi: 10.1007/s002849900474. [DOI] [PubMed] [Google Scholar]
- 76.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 77.Napierala D, Napierala M. Toxic RNA in Pathogenesis of Human Neuromuscular Disorders. Springer Verlag; Berlin: 2008. [Google Scholar]
- 78.Ikeda Y, Daughters RS, Ranum LP. Bidirectional expression of the SCA8 expansion mutation: one mutation, two genes. Cerebellum. 2008;7:150–158. doi: 10.1007/s12311-008-0010-7. [DOI] [PubMed] [Google Scholar]
- 79.Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pearson CE. Repeat associated non-ATG translation initiation: one DNA, two transcripts, seven reading frames, potentially nine toxic entities! PLoS Genet. 2011;7:e1002018. doi: 10.1371/journal.pgen.1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nemes JP, Benzow KA, Moseley ML, Ranum LP, Koob MD. The SCA8 transcript is an antisense RNA to a brain-specific transcript encoding a novel actin-binding protein (KLHL1) Hum Mol Genet. 2000;9:1543–1551. doi: 10.1093/hmg/9.10.1543. [DOI] [PubMed] [Google Scholar]
- 83.Wilburn B, Rudnicki DD, Zhao J, Weitz TM, Cheng Y, et al. An antisense CAG repeat transcript at JPH3 locus mediates expanded polyglutamine protein toxicity in Huntington’s disease-like 2 mice. Neuron. 2011;70:427–440. doi: 10.1016/j.neuron.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sopher BL, Ladd PD, Pineda VV, Libby RT, Sunkin SM, et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron. 2011;70:1071–1084. doi: 10.1016/j.neuron.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Libby RT, Hagerman KA, Pineda VV, Lau R, Cho DH, et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 2008;4:e1000257. doi: 10.1371/journal.pgen.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De Biase I, Chutake YK, Rindler PM, Bidichandani SI. Epigenetic silencing in Friedreich ataxia is associated with depletion of CTCF (CCCTC-binding factor) and antisense transcription. PLoS ONE. 2009;4:e7914. doi: 10.1371/journal.pone.0007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 89.Debacker K, Frizzell A, Gleeson O, Kirkham-McCarthy L, Mertz T, et al. Histone deacetylase complexes promote trinucleotide repeat expansions. PLoS Biol. 2012;10:e1001257. doi: 10.1371/journal.pbio.1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gannon AM, Frizzell A, Healy E, Lahue RS. MutSbeta and histone deacetylase complexes promote expansions of trinucleotide repeats in human cells. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks810. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lahue RS, Frizzell A. Histone deacetylase complexes as caretakers of genome stability. Epigenetics. 2012;7:806–810. doi: 10.4161/epi.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gottesfeld JM, Pandolfo M. Development of histone deacetylase inhibitors as therapeutics for neurological disease. Future Neurol. 2009;4:775–784. doi: 10.2217/fnl.09.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sippl W, Jung M. Epigenetic targets in drug discovery. Wiley-VCH; Weinheim: 2009. [Google Scholar]
- 94.Wojciechowska M, Krzyzosiak WJ. CAG repeat RNA as an auxiliary toxic agent in polyglutamine disorders. RNA Biol. 2011;8:565–571. doi: 10.4161/rna.8.4.15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Mezer M, Wojciechowska M, Napierala M, Sobczak K, Krzyzosiak WJ. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011;39:3852–3863. doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wojciechowska M, Napierala M, Larson JE, Wells RD. Non-B DNA Conformations Formed by Long Repeating Tracts of Myotonic Dystrophy Type 1, Myotonic Dystrophy Type 2, and Friedreich’s Ataxia Genes, Not the Sequences per se, Promote Mutagenesis in Flanking Regions. J Biol Chem. 2006;281:24531–24543. doi: 10.1074/jbc.M603888200. [DOI] [PubMed] [Google Scholar]
- 97.Bowater RP, Rosche WA, Jaworski A, Sinden RR, Wells RD. Relationship between Escherichia coli growth and deletions of CTG.CAG triplet repeats in plasmids. J Mol Biol. 1996;264:82–96. doi: 10.1006/jmbi.1996.0625. [DOI] [PubMed] [Google Scholar]
- 98.Ditch S, Sammarco MC, Banerjee A, Grabczyk E. Progressive GAA.TTC repeat expansion in human cell lines. PLoS Genet. 2009;5:e1000704. doi: 10.1371/journal.pgen.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sundararajan R, Freudenreich CH. Expanded CAG/CTG repeat DNA induces a checkpoint response that impacts cell proliferation in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1001339. doi: 10.1371/journal.pgen.1001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 101.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 102.Tiscornia G, Vivas EL, Belmonte JC. Diseases in a dish: modeling human genetic disorders using induced pluripotent cells. Nat Med. 2011;17:1570–1576. doi: 10.1038/nm.2504. [DOI] [PubMed] [Google Scholar]
- 103.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2011;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou H, Wu S, Joo JY, Zhu S, Han DW, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS ONE. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Polak U, Hirsch C, Ku S, Gottesfeld JM, Dent SYR, et al. Selecting and isolating colonies of human induced pluripotent stem cells reprogrammed from adult fibroblasts. J Vis Exp. 2012;60:e3416. doi: 10.3791/3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ku S, Soragni E, Campau E, Thomas EA, Altun G, et al. Friedreich’s ataxia induced pluripotent stem cells model intergenerational GAATTC triplet repeat instability. Cell Stem Cell. 2010;7:631–637. doi: 10.1016/j.stem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu J, Verma PJ, Evans-Galea MV, Delatycki MB, Michalska A, et al. Generation of induced pluripotent stem cell lines from Friedreich ataxia patients. Stem Cell Rev. 2010;7:703–713. doi: 10.1007/s12015-010-9210-x. [DOI] [PubMed] [Google Scholar]
- 113.Koch P, Breuer P, Peitz M, Jungverdorben J, Kesavan J, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480:543–546. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 114.Zhang N, An MC, Montoro D, Ellerby LM. Characterization of Human Huntington’s Disease Cell Model from Induced Pluripotent Stem Cells. PLoS Curr. 2011;2:RRN1193. doi: 10.1371/currents.RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.An MC, Zhang N, Scott G, Montoro D, Wittkop T, et al. Genetic Correction of Huntington’s Disease Phenotypes in Induced Pluripotent Stem Cells. Cell Stem Cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Luo Y, Fan Y, Zhou B, Xu Z, Chen Y, et al. Generation of induced pluripotent stem cells from skin fibroblasts of a patient with olivopontocerebellar atrophy. Tohoku J Exp Med. 2012;226:151–159. doi: 10.1620/tjem.226.151. [DOI] [PubMed] [Google Scholar]
- 117.Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mattout A, Biran A, Meshorer E. Global epigenetic changes during somatic cell reprogramming to iPS cells. J Mol Cell Biol. 2011;3:341–350. doi: 10.1093/jmcb/mjr028. [DOI] [PubMed] [Google Scholar]
- 119.Maynard S, Swistowska AM, Lee JW, Liu Y, Liu ST, et al. Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cells. 2008;26:2266–2274. doi: 10.1634/stemcells.2007-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Seriola A, Spits C, Simard JP, Hilven P, Haentjens P, et al. Huntington’s and myotonic dystrophy hESCs: down-regulated trinucleotide repeat instability and mismatch repair machinery expression upon differentiation. Hum Mol Genet. 2011;20:176–185. doi: 10.1093/hmg/ddq456. [DOI] [PubMed] [Google Scholar]
- 121.Jaworski A, Rosche WA, Gellibolian R, Kang S, Shimizu M, et al. Mismatch repair in Escherichia coli enhances instability of (CTG)n triplet repeats from human hereditary diseases. Proc Natl Acad Sci U S A. 1995;92:11019–11023. doi: 10.1073/pnas.92.24.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ezzatizadeh V, Pinto RM, Sandi C, Sandi M, Al-Mahdawi S, et al. The mismatch repair system protects against intergenerational GAA repeat instability in a Friedreich ataxia mouse model. Neurobiol Dis. 2012;46:165–171. doi: 10.1016/j.nbd.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]