Abstract
MicroRNA (miRNA) regulates gene expression by binding to specific sites in the 3′untranslated regions of its target genes. Machine learning based miRNA target prediction algorithms first extract a set of features from potential binding sites (PBSs) in the mRNA and then train a classifier to distinguish targets from non-targets. However, they do not consider whether the PBSs are functional or not, and consequently result in high false positive rates. This substantially affects the follow up functional validation by experiments. We present a novel machine learning based approach, MBSTAR (Multiple instance learning of Binding Sites of miRNA TARgets), for accurate prediction of true or functional miRNA binding sites. Multiple instance learning framework is adopted to handle the lack of information about the actual binding sites in the target mRNAs. Biologically validated 9531 interacting and 973 non-interacting miRNA-mRNA pairs are identified from Tarbase 6.0 and confirmed with PAR-CLIP dataset. It is found that MBSTAR achieves the highest number of binding sites overlapping with PAR-CLIP with maximum F-Score of 0.337. Compared to the other methods, MBSTAR also predicts target mRNAs with highest accuracy. The tool and genome wide predictions are available at http://www.isical.ac.in/~bioinfo_miu/MBStar30.htm.
MicroRNAs (miRNAs) are short, non-coding RNA (~22 nucleotide long) molecules1,2 encoded in both plant and animal genomes. miRNAs are associated with RNA induced silencing complex (RISC) to perform post-transcriptional gene regulation by binding to the 3′-untranslated (3′UTRs) or sometimes to the 5′UTRs of specific mRNAs3. Gene silencing is caused by the translational repression or degradation2,4 of mRNAs by miRNAs. Approximately 30,000 mature miRNAs have been identified over all the species in recent years and now it is known that they regulate diverse biological processes like cell differentiation, development and genomic stability in eukaryotes (miRBase Sequence Database, Release 20)5. They are also involved in many diseases including cancer. Experimental and computational evidence indicates that the expressions of most of the mammalian genes are fine-tuned by miRNAs6. Complex regulatory network which may exist between transcription factors (TFs), genes and miRNAs is also explored7. Studying miRNAs and their targets is an important area of research because of their role in gene expression regulation.
As already mentioned, miRNAs usually regulate gene expression by binding to specific sites in the 3′UTR of its target. Based on miRNA sequence complementarity, several potential binding sites (PBSs) may be identified. However, due to limited knowledge of miRNA biology, identification of the functional binding sites (FBSs) from the list of PBSs remains an unsolved problem. Experimental procedures for detecting the actual binding sites of miRNAs are both costly and time-consuming. These constraints give rise to algorithmic and machine learning challenges to predict FBSs of miRNAs in the target mRNAs. This information will facilitate further wet lab experiments.
Several machine learning based miRNA target prediction algorithms were developed in the last decade4,8,9,10,11,12,13,14,15,16,17,48,49,50. The general flow has been as follows:
For each miRNA, identify PBSs from the validated target (positive) and non-target (negative) mRNAs based on seed site complementarity.
Extract features from these PBSs (irrespective of whether they are functional or not).
Train a classifier to distinguish between target and non-targets.
For an unknown miRNA-mRNA pair, use the classifier to label it as positive (target) or negative (non-target).
An important concern in steps (i) and (ii) is that some of the PBSs of a real target could be inaccessible for miRNA binding because they are occluded by miRNA secondary structure or RNA binding proteins9,19. These sites are therefore not functional. Hence combining the signals from all the PBSs may be erroneous. The classifier trained on such data is likely to have high false positive rates because of the incorrect model built. In this article, we provide an alternate multiple instance learning framework18 for predicting specific functional binding sites of miRNAs. The multiple instance learning (MIL) framework considers PBSs as instances, and the miRNA-mRNA pair as a bag. Note that a bag will have multiple instances (binding sites). For a given problem, a bag may be labeled as positive (target) or negative (non-target). Positive bag indicates that at least one of its instances is positive, while a negative bag means that all its instances are negative. Using a random forests classifier in the MIL framework, we develop MBSTAR, which is able to predict the FBSs in a potential target mRNA. Moreover, the mRNA where MBSTAR predicts an FBS may, in turn, be predicted to be a target of the corresponding miRNA. Transcriptome-wide crosslinking method for RNA-binding proteins (RBPs) and microRNA-containing ribonucleoprotein complexes (PAR-CLIP) has been shown20 to identify the genome wide miRNA binding regions. These results can be used to verify the predicted binding sites generated by MBSTAR.
In MBSTAR, a set of 31 structural and 340 sequence features are extracted and unsupervised feature selection procedure is used to select 40 most relevant features to build the classifier model. Six different MIL techniques are considered and 5-fold cross validation is used to evaluate their performance. Random forests MIL framework is shown to achieve the highest accuracy within the training set. Biologically validated 9531 miRNA-mRNA interactions and 973 non-target miRNA-mRNA pairs are identified from Tarbase 6.0 and the target pairs are confirmed with PAR-CLIP dataset and considered for further evaluation of the model. In addition to MBSTAR, miRanda31,49, TargetScan11,50 with four different score cut-offs, MirTarget215 and SVMicrO17 are considered for comparison. It is found that MBSTAR achieves the highest number of overlapping binding sites with PAR-CLIP with maximum F-Score of 0.337. Compared to the other methods, MBSTAR also predicts target mRNAs with highest accuracy of 78.24% for the validated positive interactions. Another analysis on biological complexity and number of T → C conversion shows that MBSTAR is able to predict many more relevant binding sites compared to other methods.
Results
In this section, we discuss about the feature extraction and training processes of MBSTAR. The proposed method uses miRNA-mRNA pairs, represented by mature miRNA sequences and 3′UTRs of mRNAs, respectively, as the training data. For each pair, the binding sites in the 3′UTR are first identified as described in section “Selection of potential binding sites (PBSs)”. All these binding sites are considered to be instances of a bag. The bag is positive or negative depending on whether the corresponding miRNA-mRNA pair is a validated target or not.
The performance of MBSTAR is tested on a set of independent biologically validated positive and negative examples. Finally, the predicted binding sites are compared with PAR-CLIP dataset for further validation.
Datasets
3′-UTR dataset
The 3′-UTR sequence of human assembly hg19 is extracted from UCSC Genome Browser37.
miRNA dataset
From the miRBase database38,51, 2042 mature human miRNA sequences are extracted.
Negative examples for training MBSTAR
To train MBSTAR, 286 negative (non-target) examples are taken as described in8.
Two data sets containing expressions of both mRNA and miRNA in the same tissue are considered. A two-stage filtering approach, using the two data sets, is carried out such that only those miRNA-mRNA pairs both of which are over expressed or under-expressed in the same tissue are extracted as potential negative examples as these examples do not support the biology of miRNA-mediated target repression event. This set is further reduced by removing all those miRNA-mRNA pairs that show poor interactions in terms of interaction energy score (> 0 K cal/mol). Finally, all those pairs that have a high conservation score (> = 0.5) are also removed. These four stages of filtering result in a set of miRNA-mRNA pairs that are very likely to be non-targets. In fact a further experiment using data at the protein level was carried out in8 to further verify this.
Experimentally validated positive examples for training MBSTAR
We extracted 286 miRNA and validated transcript pairs from the miRecords database36. We use the same number of positive and negative examples to build a balanced classifier model.
Selection of potential binding sites (PBSs)
For each miRNA-mRNA pair, the complementary matching sites in the 3′-UTR of the mRNA corresponding to the seed site of the miRNA are first identified. These are referred to as the PBSs.
There are four possible categories of seed matching sites, namely 6-mer, 7-mer-A1, 7-mer-M8 and 8-mer21. The 6-mer seed region perfectly matches with 6 nucleotide miRNA seed, 7-mer-A1 is actually a 6-mer seed with an additional adenine at target position 1, 7-mer-M8 is a 6-mer site with an extra nucleotide match of miRNA at position 8 and lastly, 8-mer consists of an extra nucleotide match at position 8 as well as the extra adenine at position 1.
Wobble base pair is a non-Watson-Crick base pair model. Four main types of wobble base pairings are found in RNA molecules, namely G-U, I-U, I-A, and I-C. G-U wobble pair is of special mention due to its unique physical, dynamic and ligand binding capacity and acceptable thermodynamic stability. This is almost isomorphic to Watson-Crick base pairs, and thus plays an essential role in a variety of biological processes22. Recent studies have shown that most of the target prediction algorithms fail to obtain good prediction accuracies as they do not consider non-Watson-Crick seed pairing47. We thus consider single G-U wobble pair while finding the PBSs in our proposed method. Further, as suggested in8, we consider only those PBSs that are positioned neither too close (≤ 15 nt) to the stop codon nor near the middle of the 3′-UTR. These PBSs are taken as instances in MBSTAR. Feature extraction is then carried out from the PBSs and their neighboring regions.
Feature extraction
Regions surrounding a PBS play a significant role in determining binding site accessibility of miRNA9. It has also been pointed out in9 that intra-mRNA base pairing probability is significantly low if bases are separated by more than 70 nucleotides. Based on this observation, we consider ±30 nucleotides flanking regions around a PBS to extract 371 features as listed in Supplementary Table S1.
We use both sequence as well as structural features. The sequence features comprise single, di-, tri- and quad-nucleotide frequencies from the flanking regions of the PBS. We use Vienna RNA package version 2.0.723 to calculate the duplex structure and estimate other features, namely (i) internal-loops or interior loops which are found in RNA if non Watson-Crick base pairing between the nucleotides separates the double stranded RNA, (ii) bulge loop which is a single stranded region connecting two adjacent base-paired segments in shape of a “bubble” in the middle of a double helix on one side, (iii) hairpin loop which is a structure with two ends of a single-stranded region (loop) connecting a base-paired region (stem), (iv) multibranch loop which is a loop that brings three or more base-paired segments in close vicinity forming a multi-furcated structure. In addition, the minimum free energy is another feature that is calculated using RNAcofold program for the entire flanking region including the seed matching site. For details about the features see supplementary Table S1.
Feature selection
A total of 371 features are extracted for each PBS in both target and non-targets. Feature selection is used to remove noisy and redundant features from the extracted feature set for better classification accuracy24,25. As the PBSs are not biologically validated, usage of any supervised feature selection algorithm is not possible. We have used Laplacian score based feature selection (LSFS)26, unsupervised discriminative feature selection (UDFS)52 and multi class feature selection (MCFS)53 techniques to find the best set of features within 5 fold cross validation. All the MIL algorithms are trained on selected features in each fold and the method providing the most robust accuracy is selected. LSFS outperformed the other two methods and we have used Laplacian score to select top 40 features from training data. Laplacian score depends on the observation that data belonging to the same class are often close to each other. Thus importance of a feature can be determined by its locality preserving power. Let Lr denote the Laplacian score of the rth feature and xri be the rth feature of the ith sample, where i = 1,...,m, where m is the total number of samples. Let the rth feature over all samples be denoted by fr = [xr1,xr2,...,xrm]T. The nearest neighbor graph for a set of m samples in a metric space is a graph with the samples as nodes and an edge from p to q indicating that q is the nearest neighbor of p with the edge weight being equal to the distance between the two. Consider S as the weight matrix of the nearest neighborhood graph with m sample nodes. Then we define
D = diag(S1m), where 1m = [1,..., m times]T, and Graph Laplacian as L = D-S. Let,
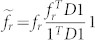 . Finally, the Laplacian score is calculated by26,
. Finally, the Laplacian score is calculated by26,
 |
We have used the top 40 features according to the Laplacian scores out of the 371 initial features to train the classifier. Details of these features with their corresponding Laplacian scores are described in supplementary Table S1.
Training an MIL random forests classifier for MBSTAR
Using these selected features we train an MIL random forests (MIL-RF) classifier with 50 trees in the forest. The cooling parameter of deterministic annealing (equation 7) is set to -0.25 as suggested in27. Hinge loss function and bagging with refine sampling are found to give the most balanced prediction result with sensitivity 0.755 and specificity 0.685 using 5-fold cross validation. We also train Diverse Density (DD), Expectation-Maximization DD (EM-DD), Citation kNN and two variations of multiple instance SVM (MI-SVM) classifiers with the same dataset and perform a 5-fold cross validation. We have used 10 different sets seed points for DD and EM-DD algorithm and then aggregated the results. For Citation kNN, both Euclidean and cosine distance are measured with varying the values of reference and citers from 1 to 10. It is found that the Euclidean distance with reference 2 and citation 4 gives the best result. For both the SVM variants, we used linear polynomial and Radial Basis Function (RBF) kernels with varying the respective parameters. RBF kernel with gamma value 0.05 gives the best result here. The results are reported in Table 1. Since, not all the algorithms can provide instance level predictions and proper instance labels are not known to us, only the bag level predictions are compared here. It is observed that the MIL-RF provides the highest accuracy among all MIL classifiers for the given dataset. Citation kNN achieves second highest accuracy and is able to beat SVM based approaches with a high margin.
Table 1. Comparative study of 5-fold cross validation accuracies for different MIL frameworks.
| Method | Bag level accuracy |
|---|---|
| MIL-RF | 0.7202 |
| Citation kNN | 0.6854 |
| Expectation-Maximization Diverse Density | 0.6644 |
| MI-SVM | 0.5871 |
| mi-SVM | 0.5316 |
| Diverse Density | 0.4861 |
Genome wide biologically verified dataset for independent testing
To evaluate the performance of our model, we have derived all biologically verified positive interactions from TarBase 6.0 database28. After converting genes to corresponding reference sequence identifiers for NCBI standard, we obtain a total of 31,456 unique positive interactions. These interactions are verified by different experimental methodologies such as reporter gene assay, western blotting, northern blotting, microarray analysis, proteomics (such as pSILAC), sequencing (RNA-Seq, HITS-CLIP, PAR-CLIP), qPCR and others (ELISA, RACE, immunohistochemistry, etc.). These unique interactions contain a total of 145 miRNAs and 16,944 mRNAs.
We compare this dataset with PAR-CLIP cluster data of the human genome downloaded from starBase29,30. Biological complexity (BC) of an experiment is a measure of reproducibility between biological experiments. This analysis is carried out with BC greater than or equal to one i.e., at least in one experiment the PAR-CLIP cluster is targeted by miRNAs. We hypothesize that for canonical seed matching to occur, at least a 6-mer site including a possible single G-U wobble pair should be present in the PAR-CLIP cluster. So we first isolate all clusters corresponding to TarBase 6.0 positive dataset. Then we find those clusters which contain at least one 6-mer site corresponding to high confidence miRNA-mRNA pairs obtained from TarBase 6.0 database.
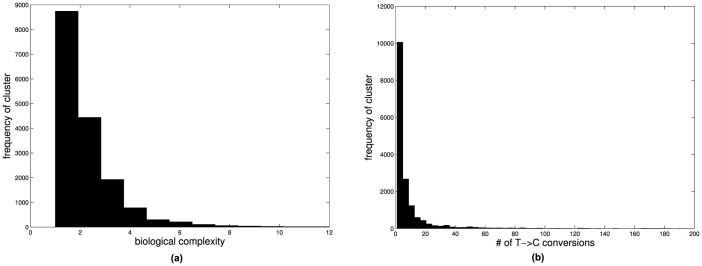
We come up with 16,824 clusters corresponding to 121 miRNAs and 5120 mRNAs. These clusters can be mapped to 9582 miRNA-mRNA interactions. Then we filter the data to remove any common interactions used in building the model and obtained 9531 interactions and 16,681 clusters. These data can be further categorized by biological complexity and the number of T → C mutations in target clusters. The distribution is showed in Figure 1. These 9531 positive miRNA-mRNA interactions (which have no overlap with the training data) are used as 9531 bags for independent testing. Average number of instances per bag is approximately 6 with values ranging from 1 to 59.
Figure 1. Distribution of PAR-CLIP gold standard clusters according to (a) biological complexity and (b) read numbers (number of T→C conversion).
We also extracted 973 non-target pairs of miRNA-mRNA from TarBase 6.0 database.
Comparative performance of MBSTAR at target level
In addition to MBSTAR, we use four popular target prediction algorithms, namely TargetScan11, miRanda31, MirTarget215 and SVMicrO17 to predict targets on the 9531 positive and 973 negative miRNA-mRNA interactions (at the bag level). The latest versions of miRanda executable is downloaded from32 and TargetScan's genome wide result is downloaded from33. MirTarget2 targets are extracted from miRDB16 database and SVMicrO genome wide results are obtained from17. miRanda is executed with its default parameters as described by the package. For TargetScan, it is recommended that users should choose the cut-off value of predicted scores as required. Hence, first quartile (TargetScan25p), second quartile (TargetScan50p), 3rd quartile (TargetScan75p) and whole prediction results (TargetScan100p) are considered according to context score.
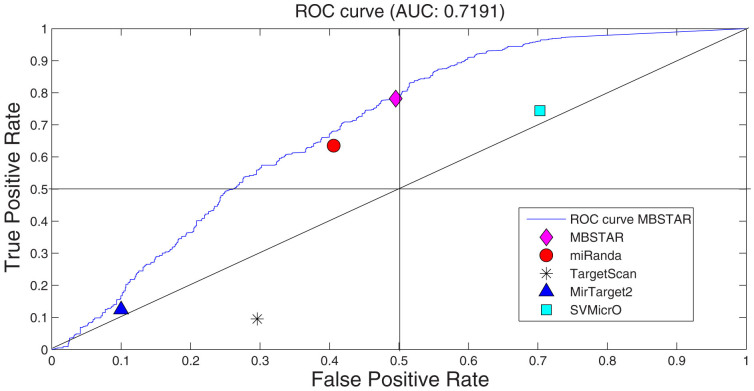
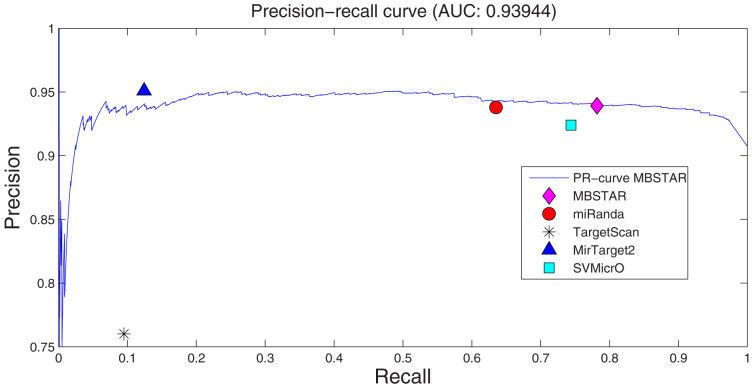
Figure 2 shows the scatter plot of different algorithms compared with ROC plot of MBSTAR, which achieves an AUC (area under curve) of 0.71. From Figure 2, it can be easily verified that MBSTAR comprehensively outperforms all the four approaches. Specifically, at any fixed true positive rate (TPR), it provides the lowest false positive rate (FPR). At the same time, for any fixed FPR, the TPR of MBSTAR is higher than those of all the four methods. A precision recall analysis is also carried out on the prediction results. This can be found in Figure 3.
Figure 2. Scatter plot of false positive rate and true positive rate of MBSTAR and other algorithms (miRanda, TargetScan100p, MirTarget2 and SVMicrO) for verified positive and non-target interactions.
The plot also contains ROC of MBSTAR with AUC (area under curve) of 0.7.
Figure 3. Scatter plot of precision and recall of MBSTAR and other algorithms (miRanda, TargetScan100p, MirTarget2 and SVMicrO) for verified positive and non-target interactions.
The plot also contains Precision-recall curve of MBSTAR with AUC (area under curve) of 0.93.
Comparative performance of MBSTAR at binding site level
As mentioned earlier, the main strength of MBSTAR lies in its ability to predict the functional miRNA binding sites (FBS) within a target mRNA. To demonstrate this, we use the aforementioned 9531 positive miRNA-mRNA pairs for testing and 16,681 clusters to validate the predicted FBSs within these pairs. The following measures are used for comparison: sensitivity 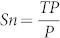 and positive predictive value
and positive predictive value 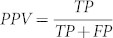 and
and 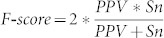 , which is the harmonic mean of PPV and Sn. Here TP = positive predictions overlapping with the 16,681 clusters, P = 16,681 clusters and FP = positive prediction with no overlap with the validated clusters. F-Score provides a comprehensive evaluation of the performance of an algorithm. Results comparing the performance of MBSTAR with the four other techniques are reported in Table 2. The second column of the table shows the number of predicted sites that overlap with the validated 16,681 clusters. The following two columns provide the F-scores and the target level accuracies obtained by the methods.
, which is the harmonic mean of PPV and Sn. Here TP = positive predictions overlapping with the 16,681 clusters, P = 16,681 clusters and FP = positive prediction with no overlap with the validated clusters. F-Score provides a comprehensive evaluation of the performance of an algorithm. Results comparing the performance of MBSTAR with the four other techniques are reported in Table 2. The second column of the table shows the number of predicted sites that overlap with the validated 16,681 clusters. The following two columns provide the F-scores and the target level accuracies obtained by the methods.
Table 2. Comparative study of overlapping results of predicted binding sites using MBSTAR and four other algorithms on extracted positive dataset.
| Algorithm | Total overlapping sites | F-Score of binding site prediction | Target level accuracy |
|---|---|---|---|
| MBSTAR | 7156 | 0.337 | 78.24 |
| miRanda | 3692 | 0.274 | 57.77 |
| TargetScan100p | 3565 | 0.267 | 60.39 |
| TargetScan75p | 2092 | 0.194 | 42.63 |
| SVMicrO* | 1574 | 0.132 | 74.43 |
| TargetScan50p | 1201 | 0.108 | 29.04 |
| MirTarget2 | 770 | 0.082 | 15.8 |
| TargetScan25p | 497 | 0.049 | 16.3 |
* Binding sites information is not provided for all predictions of SVMicrO. The result reported in the table pertains to only those cases where binding site information is available.
As can be seen from the table, among the five algorithms, F-score of MBSTAR is the highest, while TargetScan25p performs the poorest. For SVMicrO, binding site information is not reported for all the genome-wide predictions. The result for SVMicrO reported in Table 2 pertains to only those cases where the binding sites have been reported. From this result, SVMicrO appears to perform poorly.TargetScan100p and miRanda can predict overlapping binding sites with almost the same F-score, but for lower score quartile, the performance of TargetScan degrades. MirTarget2 also shows poor binding site recognition ability. This is expected since even at the target level, its performance is poor. Finally, even with respect to predicting miRNA-mRNA targets from the validated set of 9531 targets, MBSTAR (78.24%) outperforms the others by a large margin.
To further show the superiority of MBSTAR, we compare its performance vis-à-vis random guessing, performed 10000 times. It is found that MBSTAR can outperform the random approach with p-value < 0.0219. Details of this experiment can be found in methods section of supplementary file.
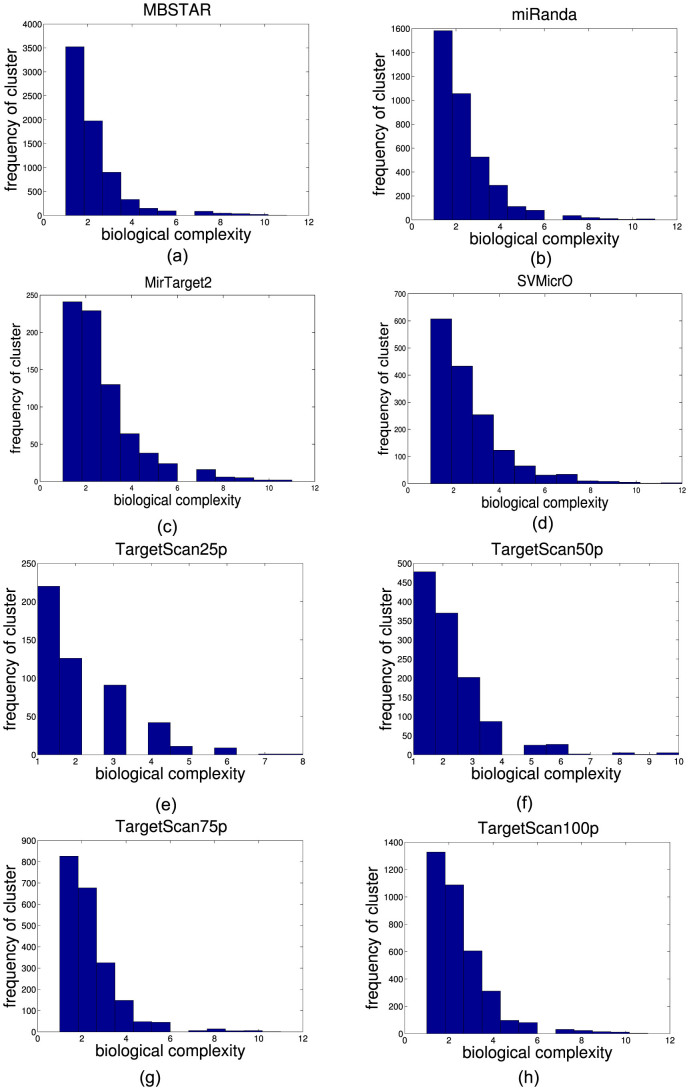
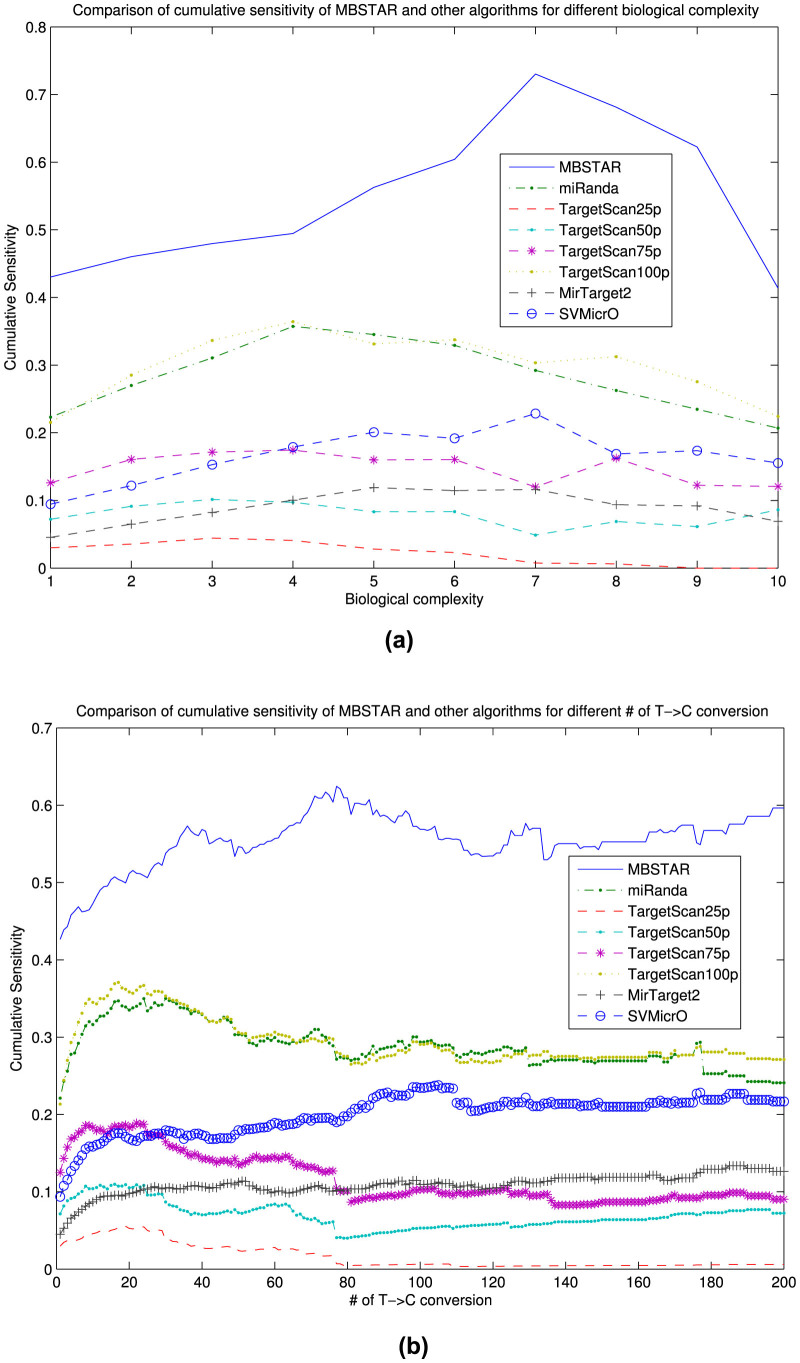
Another analysis has been carried out to find out whether biological complexity and the number of T→C conversion have any effect on prediction of binding sites. Figure 4 shows the distribution of the number of predicted FBSs overlapping with 16,681 PAR-CLIP clusters for different algorithms with varying BC values. As can be seen, MBSTAR provides more than 3500 overlapping FBSs, as compared to TargetScan and miRanda. The distribution pattern for the number of FBSs with the number of T→C conversion is given in supplementary Figure S1. Figure 5(a) shows the variation in cumulative sensitivity of the different algorithms for varying BC. As can be seen, MBSTAR clearly outperforms the other approaches for all BC values. In particular, even for high values of BC ( = 7), MBSTAR can predict clusters with nearly 80% accuracy while the closest competitor, TargetScan100p can attain only ~30%. Figure 5(b) shows the cumulative sensitivities of the different methods while varying the number of T→C conversions. Again, MBSTAR achieves more than 60% accuracy while both TargetScan100p and miRanda provide about 30%. SVMicro achieves around 25% accuracy while MirTarget2 performs poorly with around 10% accuracy.
Figure 4. Distribution of predicted PAR-CLIP gold standard clusters according to (a) biological complexity by MBSTAR, (b) biological complexity by miRanda, (c) biological complexity by MirTarget2, (d) biological complexity by SVMicrO, (e) biological complexity by TargetScan25p, (f) biological complexity by TargetScan50p, (g) biological complexity by TargetScan75p and (h) biological complexity by TargetScan100p.
Figure 5. Cumulative distribution of sensitivity for MBSTAR, miRanda, TargetScan, MirTarget2 and SVMicrO according to (a) biological complexity, (b) number of T→C conversion.
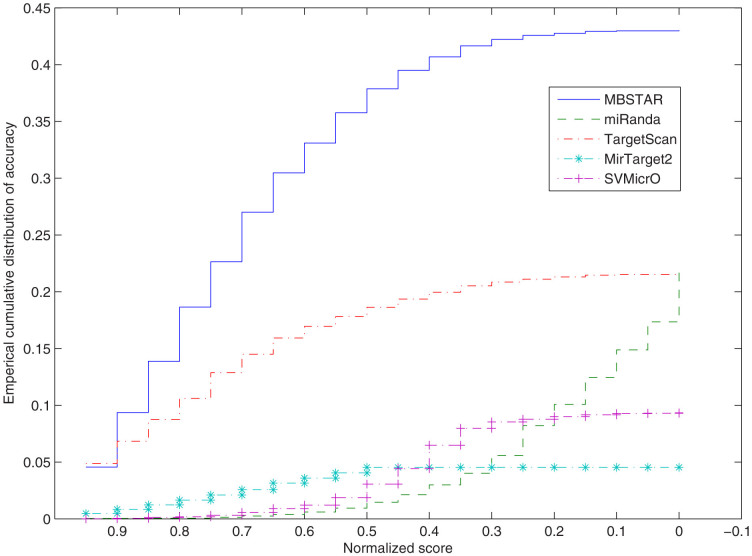
All the algorithms considered in this article provide a score associated with each of its predicted FBSs. We next aim to study the variation of accuracies of the different methods for different values of the scores normalized to the range [0, 1]. Figure 6 shows the variation. It is found that in general MBSTAR provides much superior accuracies irrespective of the cut-off score. In particular, with normalized score cut-off of 0.5, MBSTAR predicts with accuracy of nearly 38%, while miRanda, MirTarget2, SVMicrO and TargetScan can achieve only 1%, 4%, 3% and 18% of accuracies, respectively (Figure 6). We perform Wilcoxon rank sum test (one tailed) to observe the difference between these distributions. It is clear from the obtained p-values that the distributions are significantly different and MBSTAR contains larger median score ranks than miRanda (p = 3.44 × 10−7), TargetScan100p (p = 9.97 × 10−5), MirTarget2 (p = 2.47 × 10−8) and SVMicrO (p = 1.27 × 10−7).
Figure 6. Empirical cumulative distribution of sensitivity for MBSTAR, miRanda, MirTarget2, SVMicrO and TargetScan according to normalized scores.
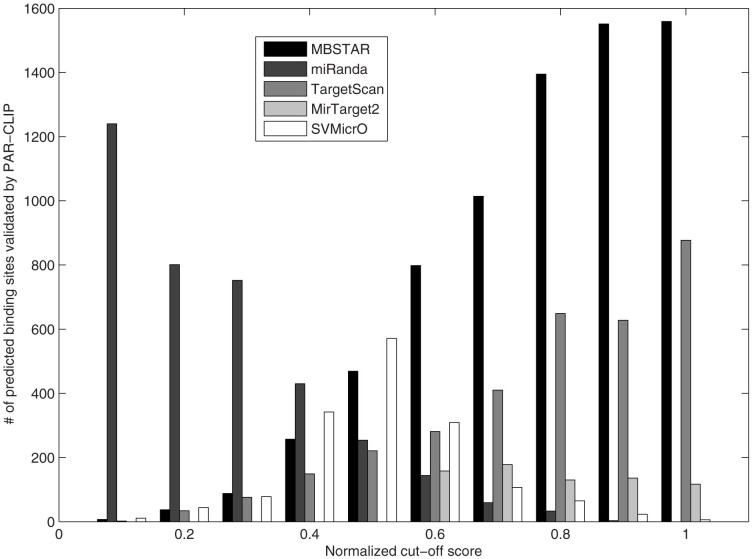
In Figure 7, we show the number of overlapping sites for different cut-off scores obtained from the algorithms. It is clearly seen that in the case of MBSTAR, the number of predicted FBSs increases with the increase in cut-off score. This indicates that MBSTAR predicts most of the binding sites with high confidence. The same trend is observed for TargetScan but the numbers are much smaller compared to MBSTAR. In contrast, miRanda shows an opposite trend where most of the predictions are associated with lower confidence scores. In fact for scores > = 0.9, miRanda is not able to make any significant number of predictions. MirTarget2 and SVMicrO achieve the highest number of binding sites with scores around the median value. As an example, in the cut-off interval (0.9-1), MBSTAR predicts around 1600 overlapping binding sites whereas TargetScan and MirTarget2 predict around 900 and 100 sites respectively. miRanda and SVMicrO fail to provide any significant number of overlapping site within this cut-off range.
Figure 7. Number of predicted sites verified by PAR-CLIP for MBSTAR, miRanda, MirTarget2, SVMicrO and TargetScan according to normalized cut-off score intervals.
Discussion
In this article, for the first time, we have proposed a multiple instance learning based approach to distinguish between functional and non-functional miRNA binding sites. This is based on the fact that not all binding sites predicted by target prediction algorithms are functionally active. Current computational approaches for target predictions do not take into consideration this fact. The proposed method treats each PBS for miRNA-mRNA interaction as an individual instance and uses multiple instance learning approach to find out the functional sites. Both sequence specific and structural features are extracted from the 3′-UTR for training the MIL model. Laplacian feature selection method is used to obtain the top 40 relevant features. The resultant dataset is used to train 6 MIL algorithms and their performance is compared. In terms of cross validation accuracy, the random forests based MBSTAR comprehensively outperforms the other methods viz., citation kNN, EM-DD, two variants of MI-SVM and DD. The performance of MBSTAR is studied on an independent dataset consisting of 9531 positive interactions, derived from intersection of PAR-CLIP clusters and Tarbase 6.0, as well as 973 negative interactions. The proposed method is able to achieve high prediction accuracy with F-score 0.337 while identifying 7156 FBSs which overlap with PAR-CLIP clusters. It also outperforms other algorithms (viz., TargetScan, miRanda, MirTarget2 and SVMicrO) in identifying targets and non-targets at the mRNA level. For different values of biological complexity and T→C conversion of predicted overlapping clusters, MBSTAR comprehensively outperforms other algorithms with a high margin. Wilcoxon rank sum test shows that MBSTAR has larger median score ranks than those of the other algorithms. Also with normalized score cut-off of 0.5 MBSTAR predicts FBSs with accuracy 38% while TargetScan achieves only 18%.
Based on its performance, it can be concluded that MBSTAR will make a valuable impact on future laboratory experiments for finding out functional miRNA binding sites. In this study we included only binding sites in 3′-UTRs and canonical binding sites. Recent studies have demonstrated that the binding sites in the coding regions also have important regulating effects. This can be investigated in future. Learning from PAR-CLIP dataset within the MIL framework can also increase the accuracy of prediction result significantly. However, the PAR-CLIP data is still far from accurate and the clusters fail to identify all of the experimentally verified results. Moreover, this data only indicates genomic regions where some miRNAs bind. The exact set of miRNAs binding to a particular cluster is not known. Research in this direction is ongoing34,35.
Methods
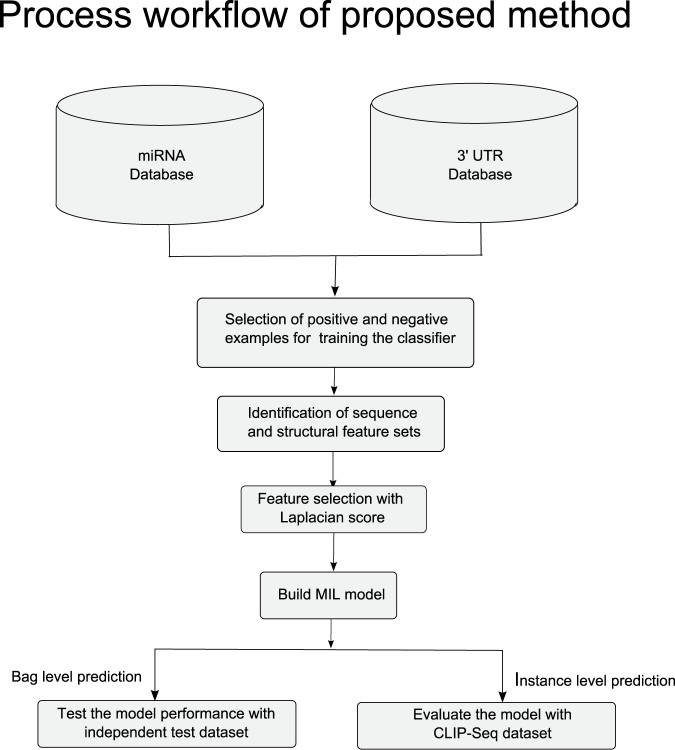
In this section we explain the methods used in this article. The multiple instance learning random forests classifier used for predicting the FBSs, is also explained in detail here. The process flowchart of the complete method is described in Figure 8. At first, human 3′-UTR genome sequence and 2042 mature miRNA sequences are extracted from available database. Biologically verified positive examples of miRNA-mRNA pairs are collected from miRecords database36, while non-target examples are taken from our previous work8. Next, sequence and structural features are extracted from PBSs of miRNA and transcript pairs. Laplacian unsupervised feature selection is used to rank the features on their importance and top 40 features are taken to train the classifier.
Figure 8. Process flowchart of the proposed MBSTAR.
Multiple instance learning
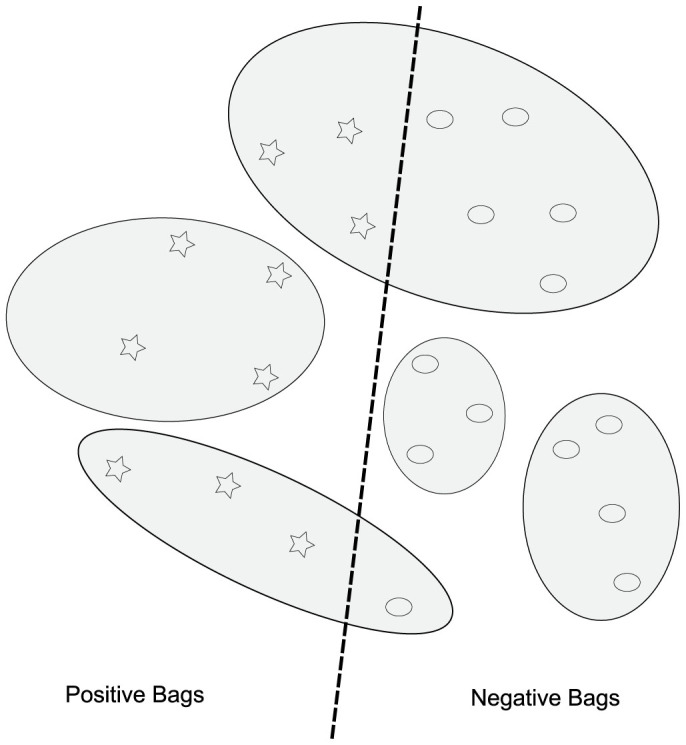
Multiple instance learning is considered to be the fourth learning paradigm after supervised, unsupervised and reinforcement learning in the machine learning community. A graphical example of MIL problem can be found in Figure 9. Here the ellipsoids denote the individual bags and the star and the small ellipsoids denote positive and negative instances respectively. The dotted line represents the hyperplane which separates the instances.
Figure 9. Classification of positive and negative instances by multiple instance learning methodology when only the bag label is known.
In supervised learning the training data consists of instance examples such as {x1,x2, …,xn} and corresponding labels {y1,y2, …,yn} where xi ∈χ and yi∈γ. In a binary classification problem normally χ is a d-dimensional Euclidean space and γ∈{0,1}. A supervised learning algorithm trains a classifier h(χ) : χ→γ that is used to predict the label of a query sample x. In MIL the training samples are in bags {B1,B2, …,Bn} and bag labels {y1,y2,...,yn} are given. Each bag consists of several training samples such that Bi = {xi1,xi2,...,xim} where xij∈χ and γ∈{0,1}. A negative bag consists of only negative examples, where as a positive bag consists of at least one positive example (called a witness)18. The learning ambiguity arises from the fact that, at least one instance of a positive bag is positive; the others can be positive or negative. The MIL assumption is summarized as follows:
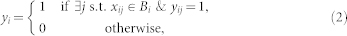 |
Where yij is the label of instance xij in bag Bi.
The task of MIL is to train a bag classifier H(χ):B→{0,1} that is used to classify bags, or an instance classifier h(χ) :χ → {0,1} based on the above information. The bag classifier can be obtained from an instance classifier with max operator: H(χi) = maxj(h(xij)). Below we discuss some common frameworks which are used to solve MIL problems.
Diverse density (DD)
Diverse density39 searches through the feature space for a concept point which lies close to at least one instance of every positive bag. Another condition is that, the concept point should be far away from instances of the negative bags. DD is the measure of distance of instances of positive and negative bags. The maximum DD defines the optimum concept point. Then a distance threshold is computed and a bag is labeled positive if the weighted distance of any of its instances from the optimum concept point is below the threshold.
Citation kNN
Citation kNN40 uses minimum Hausdorff distance which is defined as the shortest distance between any two instances from two different bags. This distance can be used to measure the distance between bags. Using this bag-level distance metric, the k Nearest Neighbor (kNN) approach is used to solve the MIL problem. The minimum Hausdorff distance is defined as,
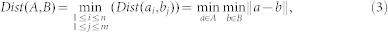 |
where A and B denote two bags, ai and bj are instances from each bag. Using this bag-level distance, label of an unlabeled bag can be predicted using the kNN algorithm.
Support Vector Machine (SVM) for multiple instance learning
Support Vector Machine (SVM) is modified41 to solve MIL problem. The authors proposed an instance-level (mi-SVM) and a bag-level (MI-SVM) classification technique. The goal of mi-SVM algorithm is to maximize the instance margin over kernelized discriminant function and unknown instance labels and can be defined as,
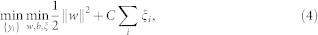 |
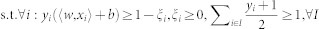 |
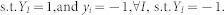 |
On the other hand, MI-SVM maximizes the bag margin, which is the margin of the most positive instance in positive bags, or the margin of the least negative instance of negative bags. This is defined as,
 |
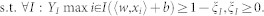 |
Random forests (RF)
RF is an ensemble classifier that consists of many decision trees42,43.The term Random Forests is introduced with the concept of random decision forests44. Each independent decision tree classifier can be represented as fm(x):χ →γ = {1,2, …,K}. A forest of M decision trees F = {f1,f2,....fM} has the regression predictor 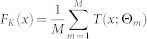 , where Θm defines the mth tree in terms of split variables, cutpoints and terminal node values. For classification problems the predictor is defined as
, where Θm defines the mth tree in terms of split variables, cutpoints and terminal node values. For classification problems the predictor is defined as 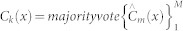 where
where 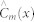 is the class prediction of the mth tree. The complexity of building a random forest model is Θ(M(pq log q)) where p and q are number of instances and attributes of the decision trees respectively.
is the class prediction of the mth tree. The complexity of building a random forest model is Θ(M(pq log q)) where p and q are number of instances and attributes of the decision trees respectively.
MIL RF
The most naive way to implement MIL for decision trees is to use single trees45. MIL for RF is developed as an optimization problem where instance labels are optimization variables and can be solved in an iterative manner27. Once the labels are retrieved, it is trivial to run a supervised RF to classify both bags and instances. Given Bi as the ith bag with label yi and content of each bag as  , the objective function is given as,
, the objective function is given as,
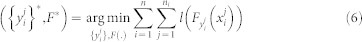 |
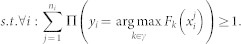 |
Here, loss function l(.) is minimized considering the condition that each bag consists of at least one positive instance of the target class. Fk(x) is the classifier confidence and Pi(.) is an indicator function. Eq. (6) is a non-convex optimization problem and can be solved by deterministic annealing (DA). Non-convex optimization problems can be solved by adding a convex entropy term and minimizing the entropy46.
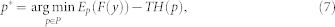 |
Where H is the entropy of distribution p, F(y) is the objective function and T is the cooling parameter, where T0>T1>...>T∞. Deterministic annealing first minimizes the entropy and then solves hidden label of instances and trains an instance classifier. From the RF, the optimal distribution is found by the equation,
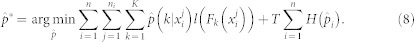 |
Setting derivative with respect to p and equating it to zero we can get the optimal distribution.
MBSTAR: MIL based prediction of functional binding sites
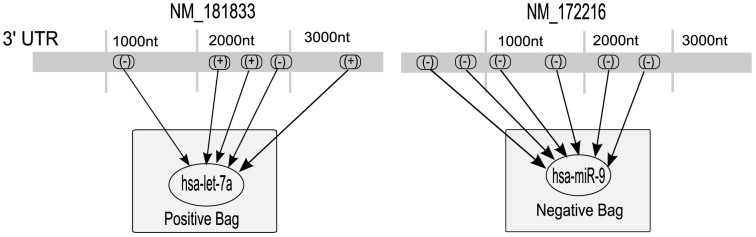
Here, we explain how the miRNA binding site prediction problem has been solved within the MIL framework. Each miRNA can bind to multiple sites on the target mRNAs (see Figure 10). Computational predictions find multiple sites, but not all of these are biologically functional. Hence, we can define each miRNA-mRNA interaction pair as a bag and all PBSs corresponding to the bag as instances. Initially we do not know the proper labeling of the instances (whether they are FBSs or not), but we know whether a given mRNA is targeted by a particular miRNA from our initial training set. So the bags are labeled but the instances are not. By training the MIL classifier with these bags and instances we can predict the nature of both unknown interaction between miRNA and mRNA (bags) and their target binding sites (instances). Figure 10 explains this concept with a toy example. Let us consider a miRNA hsa-let-7a which is biologically known to target the gene NM_181833. Now also consider that there are five PBSs for this miRNA-mRNA pair. Then we can take this pair as a positive bag with the only information available being that at least one of the five PBSs is an FBS. On the other hand miRNA hsa-miR-9 and gene NM_172216 are known to be non-target pair. So we can take this pair as negative example and treat all of their PBSs as negative instances.
Figure 10. Interaction prediction of one positive bag (hsa-let-7a/NM_181833) and a negative bag (hsa-miR-9/NM_172216) with MIL technique where (+) denotes the true binding site of miRNA targets and (-) denotes the negative instance of binding sites.
Supplementary Material
MBSTAR -supplementary
Acknowledgments
This work was partially supported by National Institutes of Health (USA) grants (R01LM011177 and R03CA167695).
Footnotes
Author Contributions S.B. and R.M. conceived the study. S.B., R.M., D.G. and Z.Z. designed the experiments. D.G. and R.M. collected the data, and performed the experiments. All the authors conducted data analysis, corrected and wrote the manuscript.
References
- Ambros V. The functions of animal microRNAs. Nature 431, 350–355 (2004). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Lytle J. R., Yario T. A. & Steitz J. A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. USA. 104, 9667–9672 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright Anton J. et al. MicroRNA targets in Drosophila. Genome Biol. 5, R1–R1 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A. & Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta D. & Bandyopadhyay S. Topological patterns in microRNA-gene regulatory network: studies in colorectal and breast cancer. Mol. BioSyst. 9, 1360–1371 (2013). [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S. & Bhattacharyya M. Analyzing miRNA co-expression networks to explore TF-miRNA regulation. BMC Bioinformatics. 10, 163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S. & Mitra R. TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics. 25, 2625–2631 (2009). [DOI] [PubMed] [Google Scholar]
- Kertesz M., Iovino N., Unnerstall U., Gaul U. & Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 39, 1278–1284 (2007). [DOI] [PubMed] [Google Scholar]
- Kim S., Nam J., Rhee J., Lee W. & Zhang B. miTarget: microRNA target gene prediction using a support vector machine. BMC Bioinformatics. 7, 411 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B. & Bartel D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120, 15–20 (2005). [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Hochsmann M. & Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 10, 1507–1517 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm M., Hackenberg M., Langenberger D. & Frishman D. TargetSpy: a supervised machine learning approach for microRNA target prediction. BMC Bioinformatics. 11, 292 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef M., Jung S., Kossenkov A. V., Showe L. C. & Showe M. K. Naïve Bayes for microRNA target predictions machine learning for microRNA targets. Bioinformatics. 23, 2987–2992 (2007). [DOI] [PubMed] [Google Scholar]
- Wang X. & El Naqa I. M. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 24, 325–332 (2008). [DOI] [PubMed] [Google Scholar]
- Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 14, 1012–1017 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yue D., Chen Y., Gao S. J. & Huang Y. Improving performance of mammalian microRNA target prediction. BMC Bioinformatics. 11, 476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietterich T. G., Lathrop R. H. & Lozano-Pérez T. Solving the multiple instance problem with axis-parallel rectangles. Artificial Intelligence. 89, 31–71 (1997). [Google Scholar]
- Denzler R., Agarwal V., Stefano J., Bartel D. P. & Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 10.1016/j.molcel.2014.03.045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 141, 129–141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A. et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 27, 91–105 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R. B. & Cohen S. M. Principles of microRNA-target recognition. PLoS Biol. 3, e85 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R. et al. ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S., Maulik U. & Wang J. T. Analysis of biological data: a soft computing approach. vol. 3, (World Scientific, Singapore 2007). [Google Scholar]
- Maulik U., Bandyopadhyay S. & Wang J. T. Computational Intelligence and Pattern Analysis in Biology Informatics. vol. 20, (Wiley Interscience, USA 2011). [Google Scholar]
- He X., Cai D. & Niyogi P. Laplacian score for feature selection. In: Adv. Neural Inf. Process. Syst. 507–514; 10.1.1.71.3712 (2005). [Google Scholar]
- Leistner C., Saffari A. & Bischof H. MIForests: Multiple-instance learning with randomized trees. In: Computer Vision-ECCV. 29–42; 10.1007/978-3-642-15567-3_3 (Springer, 2010). [DOI] [Google Scholar]
- Vergoulis T. et al. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 40, D222–D229 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jianhua Y. starBase. (2010) Available at: http://starbase.sysu.edu.cn/download.php. (Accessed 15th March 2013). [Google Scholar]
- Yang J. et al. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 39, D202–D209 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B. et al. Human microRNA targets. PLoS Biol., 2, e363 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memorial Sloan-Kettering Cancer Center, miRanda. (2010) Available at: http://www.microrna.org/microrna/getDownloads.do. (Accessed: 1st May 2013).
- Whitehead Institute for Biomedical Research. TargetScan Human. (2012). Available at: http://www.targetscan.org/cgi-bin/targetscan/data_download.cgi?db=vert_61 (Accessed 1st May 2013). [Google Scholar]
- Khorshid M., Hausser J., Zavolan M. & van Nimwegen E. A biophysical miRNA-mRNA interaction model infers canonical and noncanonical targets. Nat. Methods. 10, 253–255 (2013). [DOI] [PubMed] [Google Scholar]
- Majoros W. H. et al. MicroRNA target site identification by integrating sequence and binding information. Nat. Methods. 10, 630–633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F. et al. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 37, D105–D110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- University of California Santa Cruz. UCSC Genome Browser. Available at: http://genome.ucsc.edu/. (Accessed 1st February 2013). [Google Scholar]
- University of Manchester. miRBase. (2006) Available at: http://www.mirbase.org. (Accessed 30th January 2013). [Google Scholar]
- Maron O. & Lozano-Perez T. A framework for multiple-instance learning. In: Adv. Neural Inf. Process. Syst. 570–576 (Morgan Kaufmann, 1998). [Google Scholar]
- Wang J. & Zucker J. D. Solving the Multiple-Instance Problem: A Lazy Learning Approach. In: Proc. of the Seventeenth Int. Conf. on Machine Learning, 1119–1126 (Morgan Kaufmann Publishers Inc., 2000). [Google Scholar]
- Andrews S., Tsochantaridis I. & Hofmann T. Support vector machines for multiple-instance learning. In: Advances in neural information processing systems 561–568 (2002). [Google Scholar]
- Ho T. K. Random decision forests. In: Proc. of the Third Int. Conf. on Document Analysis and Recognition 1, 278–282 (IEEE 1995). [Google Scholar]
- Amit Y. & Geman D. Shape quantization and recognition with randomized trees. Neural computation 9, 1545–1588 (1997). [Google Scholar]
- Breiman L. Random forests. Machine learning 45, 5–32 (2001). [Google Scholar]
- Blockeel H., Page D. & Srinivasan A. Multi-instance tree learning. In: Proc. of the 22nd int. conf. on Machine learning 57–64; 10.1145/1102351.1102359 (ACM, 2005). [DOI] [Google Scholar]
- Rose K., Gurewitz E. & Fox G. C. Deterministic annealing, constrained clustering and optimization. In: IEEE Int. Joint Conf. on Neural Networks 2515–2520; 10.1109/IJCNN.1991.170767 (IEEE, 1991). [DOI] [Google Scholar]
- Peter M. E. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene, 29, 2161–2164 (2010) [DOI] [PubMed] [Google Scholar]
- Mitra R. & Bandyopadhyay S. MultiMiTar: A novel multi objective optimization based miRNA-target prediction method. PloS one 6, e24583 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D., Koppal A., Agius P., Sander C. & Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 11, R90; 10.1186/gb-2010-11-8-r90 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R. C., Farh K. K. H., Burge C. B. & Bartel D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A. & Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Shen H. T., Ma Z., Huang Z. & Zhou X. l2, 1-Norm Regularized Discriminative Feature Selection for Unsupervised Learning. In: IJCAI Proc.-Int. Joint Conf. on Artificial Intelligence. 22, 1589–1594 (2011). [Google Scholar]
- Cai D., Zhang C. & He X. Unsupervised feature selection for multi-cluster data. In: Proc. of the 16th ACM SIGKDD int. conf. on Knowledge discovery and data mining. 333–342; 10.1145/1835804.1835848 (2010). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MBSTAR -supplementary