Abstract
The aims of this study were to evaluate the microbial diversity of different lignocellulosic biomasses during degradation under natural conditions and to isolate, select, characterise new well-adapted bacterial strains to detect potentially improved enzyme-producing bacteria. The microbiota of biomass piles of Arundo donax, Eucalyptus camaldulensis and Populus nigra were evaluated by high-throughput sequencing. A highly complex bacterial community was found, composed of ubiquitous bacteria, with the highest representation by the Actinobacteria, Proteobacteria, Bacteroidetes and Firmicutes phyla. The abundances of the major and minor taxa retrieved during the process were determined by the selective pressure produced by the lignocellulosic plant species and degradation conditions. Moreover, cellulolytic bacteria were isolated using differential substrates and screened for cellulase, cellobiase, xylanase, pectinase and ligninase activities. Forty strains that showed multienzymatic activity were selected and identified. The highest endo-cellulase activity was seen in Promicromonospora sukumoe CE86 and Isoptericola variabilis CA84, which were able to degrade cellulose, cellobiose and xylan. Sixty-two percent of bacterial strains tested exhibited high extracellular endo-1,4-ß-glucanase activity in liquid media. These approaches show that the microbiota of lignocellulosic biomasses can be considered an important source of bacterial strains to upgrade the feasibility of lignocellulose conversion for the ‘greener' technology of second-generation biofuels.
Lignocellulosic biomass includes dedicated energy crops, such as miscanthus, switchgrass, Arundo donax, Populus nigra and Eucalyptus camaldulensis. These plants can easily grow in farmland not suitable for food crops or in soils subjected to accelerated erosion, which affects most of the Mediterranean hilly croplands1,2,3. Vegetable biomasses represent an inexpensive alternative to fossil sources of fermentable sugars that can be utilised in several industrial applications, including second-generation biofuels and biopolymer production4.
The enzymatic hydrolysis of plant carbohydrates has emerged as the most prominent eco-technology for the degradation of such biomasses. From the microbiological point of view, lignocellulosic biomass represents a complex ecosystem in which environmental conditions influence living organisms. In particular, geochemical (pH and salinity) and physical (temperature, pressure and radiation) factors can have a selective pressure on the biodiversity of microorganisms5. As a consequence, autochthonous microbial communities may prevail over other microorganisms because they possess enzymes that are able to degrade complex molecules such as cellulose and hemicellulose, forming the lignocellulosic biomasses that are the most abundant renewable energy source on Earth6.
Generally, at an industrial level, the bioconversion of pretreated cellulose-based materials into fermentable sugars is performed by using a reaction mixture composed of multiple enzymes for complete hydrolysis. However, because the biorefining process is still economically unfeasible, novel biocatalysts from bacteria could help overcome costly hurdles due to the operative steps of cooling, oxygen pumping, stirring and neutralisation, as well as the intrinsically high cost of hydrolytic enzyme production7. Different microorganisms producing hemicellulolytic enzymes that are potentially usable as new biocatalysts for hemicellulose hydrolysis have been isolated from different natural environments such as compost8,9. They belong to specific groups of microorganisms that are able to synthesise cellulase, xylanases and other biocatalysts necessary to allow a complete hydrolysis of the recalcitrant components of the lignocellulosic biomass10. Cellulolytic microorganisms can synthesise distinct enzymes such as endoglucanases, exoglucanases, including d-cellodextrinases, cellobiohydrolases and ß-glycosidase, which cooperate in cellulose degradation. Indeed, the hemicellulolytic microorganisms produce xylanases for degrading xylan into xylose that include endo-β-1,4-xylanases and β-xylosidases as well as auxiliary enzymes such as α-glucuronidases, α-arabinofuranosidases, acetylesterases and acetyl xylan esterases11.
The use of culture-independent high-throughput sequencing can potentially reveal uncultivable microbiota and enables the study of the microbial ecology and taxonomic diversity at a high resolution. A thorough determination of the microbial diversity in biomass degradation can be fundamental to evaluating potential sources of novel enzymes and activities12,13.
In the present work, the changes in the microbiota during the natural biodegradation of lignocellulosic biomasses of A. donax, E. camaldulensis and P. nigra were studied. In addition, new well-adapted bacterial strains from the three lignocellulose biomasses were isolated, identified and characterised. This study shows that the microbiota of lignocellulosic biomasses can be considered an important source of bacterial strains to upgrade the feasibility of lignocellulose conversion for the ‘greener' technology of second-generation biofuels.
Results
Physicochemical measurement
The temperature values observed in the piles were approximately 24°C during the first 45 days. This value increased up to about 29°C after 135 days of biodegradation, before declining up to 25°C at the end of the experiment (180 days). The values of aw ranged from to 0.91 to 0.99. The environmental temperature increased from April (20.5°C on the average) to August (34.2°C on the average) and declined in September (29.2°C).
Microbial Diversity of Lignocellulosic Biomasses by High-Throughput Sequencing
The microbiota of three different lignocellulosic biomasses (A. donax, E. camaldulensis and P. nigra) was characterised by partial 16S rRNA gene sequencing obtained from DNA directly extracted from environmental samples. A total of 238,450 number of reads were obtained by high-throughput sequencing. However, following the removal of short, ambiguous and/or low-quality pyrotag reads, the final data set consisted of 138,336 high-quality reads with an average sequence length of 456 bp. The alpha-diversity was determined by calculating the Shannon diversity index and the Chao1 richness index based on OTUs of 97% identity (Table 1). The results showed that the highest diversity indices were observed in the E. camaldulensis and P. nigra biomasses after 180 days of biodegradation in the underwood condition (P < 0.05) but were quite variable in the A. donax biomass (Table 1). Good's coverage indicated that more than 90% of the microbial diversity was described in most of the samples.
Table 1. Number of sequences analysed, observed diversity and estimated sample coverage for 16S rRNA amplification from DNA extracted from the chipped lingo-cellulosic biomasses.
| Sample | No. reads | No. OTUs | Chao1 | Shannon indexa | Good's coverage (%) |
|---|---|---|---|---|---|
| At0 | 12,021 | 322 | 432.68 | 4.28AB | 99.24 |
| At1OF | 6,235 | 477 | 682.92 | 6.59DE | 97.45 |
| At1UW | 9,713 | 506 | 713.56 | 5.51BC | 98.38 |
| At2OF | 4,158 | 592 | 955.73 | 7.09DEF | 93.70 |
| At2UW | 2,564 | 286 | 443.81 | 6.09C | 95.44 |
| At3OF | 1,699 | 404 | 618.70 | 7.33F | 88.99 |
| At3UW | 5,451 | 441 | 561.42 | 6.36CDE | 97.54 |
| At4OF | 5,160 | 620 | 1111.02 | 6.94DEF | 94.46 |
| At4UW | 4,293 | 501 | 671.50 | 6.61DE | 95.64 |
| Et0 | 5,582 | 340 | 555.28 | 6.05C | 97.76 |
| Et1OF | 5,271 | 322 | 485.11 | 3.92A | 97.67 |
| Et1UW | 3,207 | 286 | 430.38 | 4.65B | 96.35 |
| Et2OF | 3,983 | 453 | 656.84 | 6.65DE | 95.81 |
| Et2UW | 1,069 | 228 | 405.60 | 6.52DE | 89.5 |
| Et3OF | 4,520 | 470 | 671.11 | 6.52DE | 96.00 |
| Et3UW | 7,091 | 623 | 978.51 | 6.46CDE | 96.63 |
| Et4OF | 4,271 | 571 | 882.78 | 7.49FG | 94.66 |
| Et4UW | 5,309 | 887 | 1367.25 | 8.37GH | 93.60 |
| Pt0 | 10,116 | 471 | 623.72 | 5.94C | 98.65 |
| Pt1OF | 4,292 | 501 | 865.63 | 6.18CD | 94.43 |
| Pt1UW | 10,117 | 845 | 1128.45 | 8.24G | 97.68 |
| Pt2OF | 3,524 | 728 | 1097.07 | 7.89FG | 91.06 |
| Pt2UW | 1,226 | 367 | 727.57 | 7.58FG | 84.01 |
| Pt3OF | 5,031 | 743 | 1086.64 | 6.63DE | 93.76 |
| Pt3UW | 2,188 | 583 | 1169.58 | 8.08FG | 86.15 |
| Pt4OF | 2,576 | 654 | 975.19 | 7.82FG | 88.94 |
| Pt4UW | 7,669 | 1,629 | 3122.43 | 9.48H | 89.79 |
aDifferent letters after Shannon index values indicate significant differences (P < 0.05).
Abbreviations. A: A. donax; E: E. camaldulensis; P: P. nigra; T0: 0 days of degradation; t1: 45 days of degradation; t2: 90 days of degradation; t3: 135 days of degradation; t4: 180 days of degradation; OF: open field degradation condition; UW: underwood degradation condition.
The relative abundances of bacterial taxa were examined at the level of phyla and class to determine whether there were any significant shifts in the composition of the bacterial communities according to the plant species, degradation conditions and sampling time.
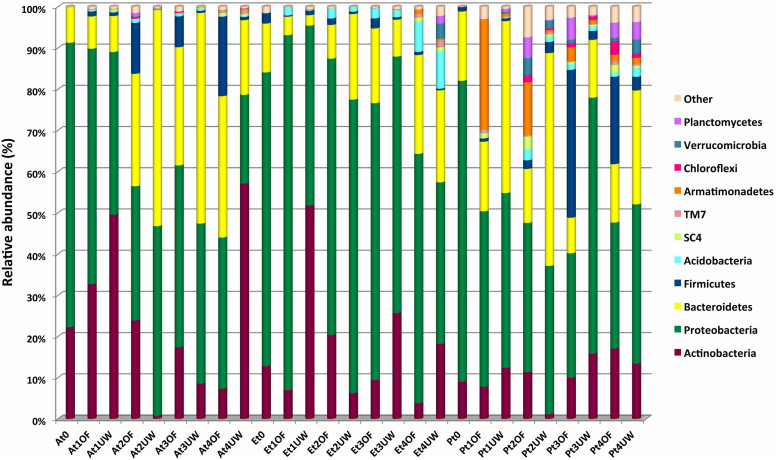
In total, twenty-six different phyla were detected in the biomass samples, but only Actinobacteria, Proteobacteria, Bacteroidetes and Firmicutes were detected in all samples (Fig. 1). These phyla together accounted for approximately 98%, 99% and 91% of the total biodiversity in A. donax, E. camaldulensis and P. nigra, respectively.
Figure 1. Abundance of bacterial phyla in lignocellulosic biomasses during the biodegradation process.
Only OTUs with an incidence >1% in at least two samples are shown. Abbreviations. A: A. donax; E: E. camaldulensis; P: P. nigra; T0: 0 days of degradation; t1: 45 days of degradation; t2: 90 days of degradation; t3: 135 days of degradation; t4: 180 days of degradation; OF: open field degradation condition; UW: underwood degradation condition.
The native composition of the microbial community in the A. donax biomass was strongly dominated by Proteobacteria (69.03%), followed by Actinobacteria (22.23%) and Bacteroidetes (8.61%). During the biodegradation process, the microbial composition remained the same in terms of diversity but varied in terms of abundance. This pattern was primarily observed in the piles processed under the open field condition (Fig. 1). Interestingly, in this environment, an inverse correlation was observed between Actinobacteria and Bacteroidetes. In particular, Actinobacteria decreased from 32.70% after 45 days of degradation under the open field condition to 7.31% after 180 days (P < 0.05), whereas Bacteroidetes increased from 7.84% after 45 days to 34.30% after 180 days (P < 0.05). Firmicutes was very low at the initial time (0.02%) and remained roughly constant in the underwood pile (0.77% after 180 days of biodegradation). By contrast, in the open field pile, the relative abundance of this taxon gradually increased from 1.20% after 45 days of biodegradation to 19.28% after 180 days (P < 0.05). Minor phyla included Acidobacteria and candidate phyla SC4 and TM7 were recovered in all A. donax biomass samples, although at an incidence ≤ 1% (Fig. 1).
As in the A. donax biomass, Proteobacteria was the taxa that heavily dominated the native microbial community in E. camaldulensis and P. nigra (71.26% and 73.01%, respectively) (P < 0.05), remaining high during all degradation processes, followed by Actinobacteria, Bacteroidetes, Firmicutes and Acidobacteria (Fig. 1). In the E. camaldulensis pile, Acidobacteria increased to 7.35% and 9.02% after 180 days of degradation under the open field and underwood conditions, respectively, whereas a slight variation was detected in the P. nigra samples. By contrast, the abundance of Firmicutes increased in the P. nigra pile after 135 (35.88%, P < 0.05) and 180 days (21.20%, P < 0.05) of degradation under the open field condition and was relatively stable in the E. camaldulensis pile (Fig. 1). Other taxa, including Armatimonadetes, Chloroflexi, Planctomycetes, Verrucomicrobia, SC4 and TM7, were also detected. Interestingly, Armatimonadetes increased in the P. nigra pile after 45 days under the open field condition (26.70%, P < 0.05) and then gradually decreased during the degradation process to 1.63% (Fig. 1).
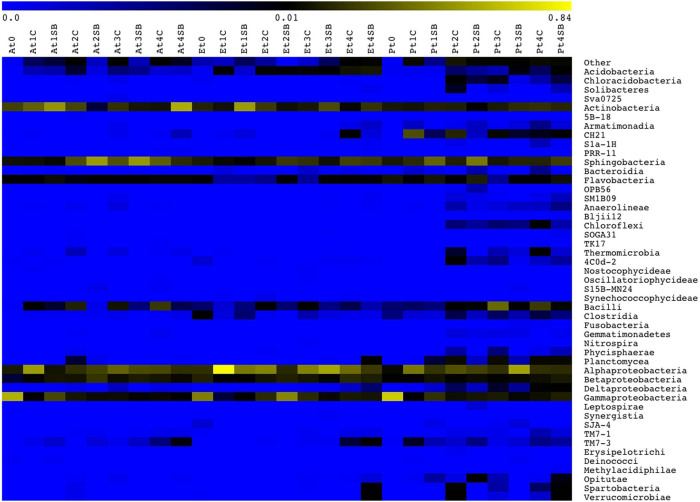
The microbial diversity was also analysed at a deeper taxonomic level. The identification of OTUs at the class level is reported in the heatmap shown in Fig. 2. Actinobacteria, γ-Proteobacteria, β-Proteobacteria, α-Proteobacteria, Acidobacteria, Sphingobacteria, Flavobacteria and Bacilli were recovered in all samples, with the exception of Acidobacteria, which was not detected in the A. donax biomass at the initial time. The abundance of α-Proteobacteria was high and exhibited the same variations during the biodegradation experiment in all three lignocellulosic biomasses. Interestingly, γ-Proteobacteria was the dominant taxa at the beginning of the experiment, with an abundance of 58.32%, 42.64% and 67.48% in the A. donax, E. camaldulensis and P. nigra samples, respectively; however, a dramatic reduction was recorded in all plant biomasses after 180 days (1.07%, 8.12% and 11.49%, respectively) (Fig. 2). A steady reduction in class Actinobacteria during degradation of the A. donax biomass under the open field condition was observed. By contrast, Bacilli showed an opposite trend, increasing from 0.02% at time zero to 19.26% after 180 days. Under the underwood condition, the middle phase of the biodegradation process was primarily dominated by Sphingobacteria, which showed an abundance of approximately 50%. Ever-increasing Sphingobacteria and β-Proteobacteria taxa abundance was recorded in the E. camaldulensis pile during the degradation process under the open field experiment. Acidobacteria showed a similar trend under both the open field and underwood conditions. The P. nigra biomass was characterised by an increase in the incidence of Bacilli and β-Proteobacteria during the biodegradation process under the open field conditions (Fig. 2). Other taxa were present at a very low incidence and with a great variability in the different samples. However, as shown in Fig. 2, the highest bacterial diversity was recorded in the P. nigra biomass. An interesting finding was the high incidence of the uncultured bacterium CH21 (26.45%) in the P. nigra pile during the first phase (after 45 days) and its constant decrease during the other phases of the degradation process in the open field. Chloroflexi was detected only in the P. nigra biomass and from 90 days to 180 days of biodegradation, showing an abundance ranging from 0.33% to 1.55%.
Figure 2. Distribution of bacterial classes in lignocellulosic biomasses during the biodegradation process.
Colour scale indicates the relative abundance of each OTU within the samples. Abbreviations. A: A. donax; E: E. camaldulensis; P: P. nigra; T0: 0 days of degradation; t1: 45 days of degradation; t2: 90 days of degradation; t3: 135 days of degradation; t4: 180 days of degradation; OF: open field degradation condition; UW: underwood degradation condition.
However, the PCoA of the weighted UniFrac community distances showed a marked difference between the native microbiota of the lignocellulosic biomasses and the microbiota in the analysed samples during the biodegradation process. The three samples of the chipped biomasses of A. donax, E. camaldulensis and P. nigra at harvest time clustered separately (Fig. 3). However, the microbial diversity seemed to be influenced by a correlation between the degradation condition and the biomass type. In fact, no difference was observed among the samples during the biodegradation process under the underwood condition (Fig. 3 panel a). By contrast, under the open field condition, P. nigra samples at different time points clustered separately from the other biomasses (Fig. 3, panel b). A similar trend was observed in the A. donax biomass, except for the samples collected after 45 days of degradation (Fig. 3, panel b). Moreover, the statistical ADONIS test showed that the composition of bacterial community in the different piles during biodegradation process was significantly influenced by lignocellulosic plant species (P < 0.001), by the degradation conditions (P < 0.05) and by sampling time (P < 0.01). This difference increased combining the three factors. In fact, the analysis performed with ADONIS found significant difference combining lignocellulosic plant species x sampling time (P < 0.001), lignocellulosic plant species x degradation condition (P < 0.001) and degradation condition x sampling time (P < 0.01).
Figure 3. Principal Coordinates Analysis of weighted UniFrac distances for 16S rRNA gene sequence data of lignocellulosic biomasses during the biodegradation process under the underwood (panel a) and open field (panel b) conditions.
Colour label. red: A. donax at harvest time; blue: E. camaldulensis at harvest time; orange: P. nigra at harvest time; green: A. donax; violet: E. camaldulensis; yellow: P. nigra. Abbreviations. A: A. donax; E: E. camaldulensis; P: P. nigra; T0: 0 days of degradation; t1: 45 days of degradation; t2: 90 days of degradation; t3: 135 days of degradation; t4: 180 days of degradation; OF: open field degradation condition; UW: underwood degradation condition.
Multienzymatic Screening and Identification of Isolated Cellulolytic Bacterial Strains
A culture-dependent approach enables the isolation of putative cellulolytic strains. A total of 366 aerobic endo- and exo-cellulolytic bacteria were isolated from samples of the A. donax, E. camaldulensis, and P. nigra biomasses. In particular, more than one hundred isolates were obtained from each pile, although the P. nigra biomass produced the highest number (127 isolates). In total, 95.9% of the isolates that were obtained from Avicel agar plates, exhibited exo-cellulase activity, whereas isolates with endo-cellulase activity on the CMC agar plates were detected at values less than 50%. To establish the number of putative multifunctional-degrading bacteria, all isolates were assayed for different enzymatic activities using the methods described below. Forty aerobic endo- and exo-cellulolytic strains that showed higher ICMC values (> 10) were chosen for further characterisation. The polyphasic approach of identification resulted in bacterial isolates with different shapes, dimensions and, in some cases, spore presence (data not shown) and great biodiversity, as twenty genera and twenty-seven different species were found (Table 2). Curtobacterium spp. and Bacillus spp. were the most representative genera with the species Curtobacterium flaccumfaciens and Curtobacterium citreum as well as Bacillus amyloliquefaciens, Bacillus licheniformis and Bacillus subtilis, respectively. The bacterial strains showed activities for three to six different enzymes. All strains showed endo- and exo-cellulolytic activities and, at same time, expressed cellobiase, xylanase and pectinase (Table 2). The highest endo-cellulase activity was observed in the CE86 strain, identified as Promicromonospora sukumoe (ICMC = 34, P < 0.05), in Isoptericola variabilis CA84b (ICMC = 32, P < 0.05) and Staphylococcus warneri CE83 (ICMC = 32, P < 0.05). The first two strains were also able to degrade exo-cellulose, cellobiose and xylan. Moreover, Isoptericola variabilis CA84b produced the highest pectinase activity (IPEC = 22, P < 0.05). Curtobacterium citreum CE711 and Bacillus amyloliquefaciens CA81 represented the most versatile strains because they possessed six different enzymatic activities. Thirty-two bacterial strains showed, in addition to endo- and exo-cellulases, cellobiase and xylanase activities that were missed in only 10% of the screened strains (Staphylococcus warneri CE83, Lysobacter enzymogenes CE710, Lysobacter gummosus CP72, Mycobacterium frederiksbergense CP710b, Promicromonospora citrea CE73, Novosphingobium resinovorum CE77, Bacillus subtilis subsp. subtilis CA816 and Aurantimonas altamirensis SBP73). All the strains belonging to the different Bacillus spp. and Curtobacterium spp. had the highest cellobiase activities. In particular, the genus Curtobacterium, represented by the species Curtobacterium citreum and Curtobacterium flaccumfaciens, was able to grow at least on four/five different substrates and, in one case, also showed peroxidase activity. In total, 87.5% of the bacterial strains produced xylanase (Table 2). Similar behaviour was observed for pectinase activity production as only 15% of the strains possessed an IPEC from 12 to 22. Laccase and peroxidase activities were detected in five strains belonging to the species Curtobacterium citreum CE711, Novosphingobium resinovorum CE77 and CE84, Lysobacter enzymogenes CE710 and Bacillus amyloliquefaciens CA81. Lignin hydrolysis was not detected in any of the strains tested.
Table 2. Identification and enzymatic activities of bacterial strains isolated from different lignocellulosic biomasses.
| Strain | Source | Ca | Ab | CEc | Xc | Pa | AZc | ABc | Lc | ADc | Identification (% identity) | Accession Number |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CE86 | E. camaldulensis | 34 ± 1.0N | + | ++ | + | 0.0 ± 0.0A | - | - | - | - | Promicromonospora sukumoe (99%) | KF057947 |
| CA84b | A. donax | 32 ± 0.0N | + | ++ | + | 22 ± 0.5G | - | - | - | - | Isoptericola variabilis (98%) | KF057948 |
| CE83 | E. camaldulensis | 32 ± 1.0N | + | + | - | 0.0 ± 0.0A | - | - | - | - | Staphylococcus warneri (100%) | KF057949 |
| CP72 | P. nigra | 28 ± 1.0M | + | - | ++ | 0.0 ± 0.0A | - | - | - | - | Lysobacter gummosus (99%) | KF040972 |
| CP710b | P. nigra | 28 ± 0.5M | + | - | + | 12 ± 0.5E | - | - | - | - | Mycobacterium frederiksbergense (99%) | KF057950 |
| CA817 | A. donax | 27 ± 0.5M | + | + | + | 2 ± 0.2B | - | - | - | - | Cellulosimicrobium cellulans (99%) | KF040973 |
| CA812 | A. donax | 27 ± 1.0M | + | +++ | +++ | 0.0 ± 0.0A | - | - | - | - | Bacillus amyloliquefaciens (99%) | KF040974 |
| SBP79 | P. nigra | 26 ± 0.5LM | + | ++ | + | 12 ± 0.0E | - | - | - | - | Promicromonospora citrea (99%) | KF057951 |
| SBA88 | A. donax | 26 ± 0.3LM | + | + | + | 0.0 ± 0.0A | - | - | - | - | Microbacterium lacus (99%) | KF057952 |
| SBE74 | E. camaldulensis | 24 ± 1.0IL | + | ± | + | 0.0 ± 0.0A | - | - | - | - | Enterobacter aerogenes/Kluyvera cryocrescens (98%) | KF057960 |
| CE77 | E. camaldulensis | 22 ± 1.0HI | + | - | - | 0.0 ± 0.0A | ++ | - | - | - | Novosphingobium resinovorum (100%) | KF040976 |
| CP81 | P. nigra | 22 ± 0.5HI | + | + | + | 3 ± 0.0C | - | - | - | - | Xanthomonas campestris (100%) | KF040971 |
| CP77 | P. nigra | 22 ± 0.9HI | + | + | + | 4 ± 0.0D | - | - | - | - | Xanthomonas orizae (98%) | KF040975 |
| CE75b | E. camaldulensis | 22 ± 0.2HI | + | ++ | + | 0.0 ± 0.0A | - | - | - | - | Pediococcus acidilactici (99%) | KF057953 |
| CP77b | P. nigra | 22 ± 0.5HI | + | ++ | ± | 0.0 ± 0.0A | - | - | - | - | Curtobacterium flaccumfaciens (99%) | KF057954 |
| SBA76 | A. donax | 22 ± 1.0HI | + | + | + | 0.0 ± 0.0A | - | - | - | - | Pantoea ananatis (99%) | KF057955 |
| SBP71 | P. nigra | 22 ± 1.0HI | + | + | + | 0.0 ± 0.0A | - | - | - | - | Schumannella luteola (97%) | KF057956 |
| CP78b | P. nigra | 22 ± 0.0HI | + | ± | + | 12 ± 0.0E | - | - | - | - | Mycobacterium frederiksbergense (99%) | KF057957 |
| CA816 | A. donax | 21 ± 0.5H | + | +++ | - | 20 ± 1.0F | - | - | - | - | Bacillus subtilis subsp. subtilis (99%) | KF040977 |
| CA81 | A. donax | 20 ± 1.0GH | + | +++ | +++ | 0.0 ± 0.0A | + | + | - | - | Bacillus amyloliquefaciens (99%) | KF040978 |
| CA82 | A. donax | 20 ± 0.0GH | + | +++ | ++ | 4 ± 0.2D | - | - | - | - | Bacillus licheniformis (99%) | KF040979 |
| CE84 | E. camaldulensis | 20 ± 0.5GH | + | + | +++ | 0.0 ± 0.0A | ++ | - | - | - | Novosphingobium resinovorum (99%) | KF040980 |
| CA81b | A. donax | 20 ± 1.0GH | + | +++ | ++ | 4 ± 0.1D | - | - | - | - | Bacillus licheniformis (99%) | KF040981 |
| CE73b | E. camaldulensis | 20 ± 0.9GH | + | ++ | + | 0.0 ± 0.0A | - | - | - | - | Pediococcus acidilactici (99%) | KF057958 |
| SBP73 | P. nigra | 20 ± 0.0GH | + | - | ± | 0.0 ± 0.0A | - | - | - | - | Aurantimonas altamirensis (99%) | KF057959 |
| CE85 | E. camaldulensis | 18 ± 1.0FG | + | ++ | ++ | 2 ± 0.0B | - | - | - | - | Isoptericola variabilis (98%) | KF040982 |
| CE710 | E. camaldulensis | 18 ± 0.5FG | + | - | ++ | 2 ± 0.1B | - | ++ | - | - | Lysobacter enzymogenes (99%) | KF040983 |
| CP81b | P. nigra | 18 ± 1.0FG | + | ++ | ++ | 0.0 ± 0.0A | - | - | - | - | Promicromonospora sukumoe (99%) | KF040984 |
| CA83 | A. donax | 17 ± 1.0EF | + | ++ | + | 2 ± 0.1B | - | - | - | - | Curtobacterium flaccumfaciens (99%) | KF040985 |
| CA77 | A. donax | 16 ± 0.5DEF | + | + | +++ | 2 ± 0.0B | - | - | - | - | Sphingobacterium multivorum (99%) | KF040986 |
| CE75 | E. camaldulensis | 16 ± 1.0DEF | + | ++ | + | 2 ± 0.1B | - | - | - | - | Curtobacterium flaccumfaciens (99%) | KF040987 |
| CE73 | E. camaldulensis | 16 ± 0.5DEF | + | + | - | 0.0 ± 0.0A | - | - | - | - | Promicromonospora citrea (99%) | KF040988 |
| CA83b | A. donax | 15 ± 0.3CDE | + | ++ | ++ | 0.0 ± 0.0A | - | - | - | - | Curtobacterium flaccumfaciens (99%) | KF040989 |
| CA84 | A. donax | 14 ± 0.5BCD | + | +++ | + | 0.0 ± 0.0A | - | - | - | - | Curtobacterium citreum (99%) | KF040990 |
| CP713 | P. nigra | 14 ± 0.3BCD | + | +++ | ++ | 20 ± 0.3F | - | - | - | - | Cellulomonas flavigena (98%) | KF040991 |
| CA818 | A. donax | 13 ± 1.0BC | + | ++ | + | 2 ± 0.0B | - | - | - | - | Microbacterium testaceum (99%) | KF040992 |
| CE78 | E. camaldulensis | 12 ± 0.0AB | + | ++ | + | 2 ± 0.3B | - | - | - | - | Curtobacterium citreum (99%) | KF040993 |
| CP710 | P. nigra | 12 ± 0.5AB | + | + | + | 0.0 ± 0.0A | - | - | - | - | Labedella gwakjiensis (99%) | KF040994 |
| CE711 | E. camaldulensis | 10 ± 0.9A | + | ++ | + | 2 ± 0.0B | + | - | - | - | Curtobacterium citreum (99%) | KF040995 |
| CP78 | P. nigra | 10 ± 0.5A | + | +++ | + | 0.0 ± 0.0A | - | - | - | - | Raoultella terrigena (99%) | KF040996 |
Enzymatic activities: C = endo-cellulase; A = eso-cellulase; CE = cellobiase; X = xylanase; P = pectinase; AZ = peroxidase; AB = laccase; L = ligninase with guaiacol and lignin alkali; AD = ligninase with guaiacol and Arundo donax; aICMC or IPEC index, values represent the means ± SD of three replicates. Different letters after values indicate significant differences (P<0.05); bgrowth; c – negative; + low intensity; ++ middle intensity; +++ high intensity.
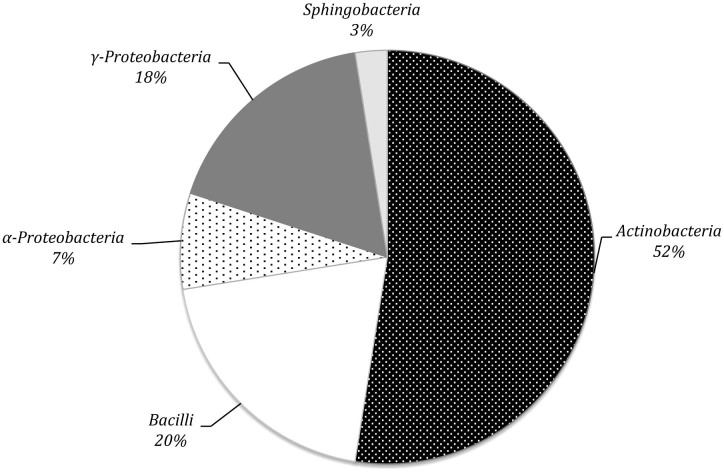
Phylogenetic Analysis of Selected Cellulolytic Bacteria
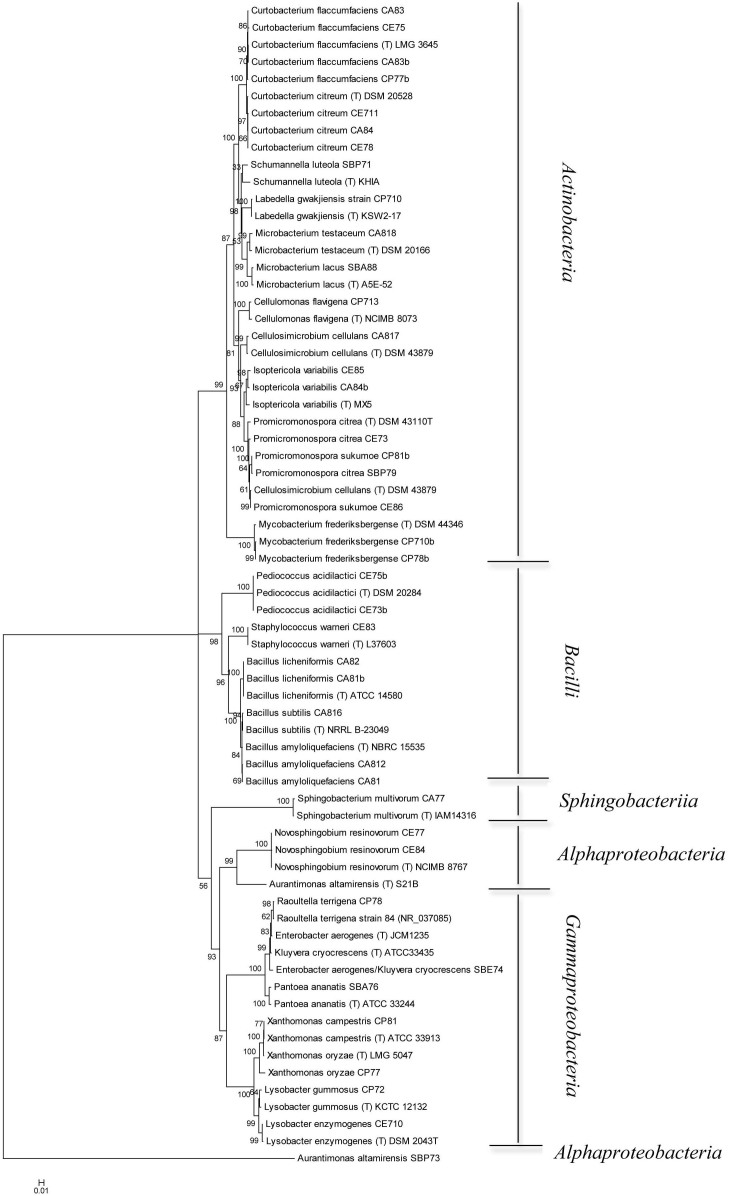
16S rRNA gene sequences of the forty cellulolytic bacteria identified as described below were grouped by phylogenetic analysis into five different clusters, generating a consensus tree. The clusters were Actinobacteria (n = 21; 52.5%), Bacilli (n = 8; 20.0%), α-Proteobacteria (n = 3; 7.5%), γ-Proteobacteria (n = 7; 17.5%), Sphingobacteria (n = 1; 4%) (Fig. 4). A phylogenetic tree was generated from the distance data using the Neighbour-Joining method with the Maximum Composite Likelihood model in a MEGA4 Program (Fig. 5). The nucleotide sequences of related type strains of different genera were included in the data set. High bootstrap values were observed and indicated significant branching points in the phylogenetic tree. Strains representative of the dominant class of Actinobacteria were placed in a cluster with bootstrap values higher than 52% and could be differentiated into nine subclusters of different genera. The bacterial strains primarily belonged to Curtobacterium (33.3%) and Promicromonospora (19.0%), whereas other genera (Schumannella, Labedella, Cellulomonas, Cellulosimicrobium, Microbacterium, Isoptericola, and Mycobacterium) were represented by one or two strains. The strains Curtobacterium citreum CE711, Curtobacterium citreum CA84 and Curtobacterium citreum CE78 showed a sequence similarity of 100%. Representatives of the Bacilli class were placed in three clusters of the genera Bacillus (62.5%), Staphylococcus (12.5%) and Pediococcus (25.0%). The strains belonging to Bacillus licheniformis CA82 and CA81b showed a similarity level of 100%, as did Bacillus amyloliquefaciens CA812 and CA81 and Pediococcus acidilactici CE75b and CE73b (Fig. 5). The α-Proteobacteria strains were placed in two different genera, in which two strains of Novosphingobium resivorum (CE77 and CE84) exhibited a sequence similarity of 100% with 100% of the bootstrap analysis. The other strain, belonging to Aurantimonas altamirensis (SBP73), formed a phylogenetically distinct cluster with a low 16S rRNA gene sequence similarity (60%) (Fig. 5). The representative strains of the γ-Proteobacteria cluster belonged to the genera Xanthomonas and Lysobacter and included the species Xanthomonas campestris, Xanthomonas oryzae, Lysobacter gummosus, and Lysobacter enzymogenes. The other genera, identified as the Enterobacter aerogenes/Klebsiella cryocrescens group, Raoultella and Pantoea, were represented by one strain. The strain Sphingobacterium multivorum CA77 formed a phylogenetically distinct cluster with the type strain of the respective species with 100% of the bootstrap analysis (Fig. 5).
Figure 4. Percentage composition of different phyla of eso- and endo-cellulolytic bacteria isolated from lignocellulosic biomasses on the basis of 16S rRNA gene sequence similarity.
Figure 5. Neighbour-Joining tree based on the comparison of 16S rRNA gene sequences showing the relationships among cellulolytic strains.
Bootstrap values (expressed as percentages of 1,000 replications) greater than 50% are given at the nodes. Strains marked with “(T)” represent type strains. The scale bar estimates the number of substitutions per site.
Screening of Cellulolytic Microorganisms in Liquid Medium
Sixteen bacterial strains were selected based on at least one of these characteristics: higher halo dimension on CMC agar, multienzymatic activities, high pectinase activity or high peroxidase activity, and no pathogenicity for humans; these strains were submitted to a quantitative CMCase assay. All the analysed strains achieving the maximum AZO-CMCase level of production between the 8th and 26th hours, corresponding to the exponential and stationary phases of growth, respectively (data not shown). In Table 3, the values of the maximum AZO-CMCase activity measured for each strain and the corresponding time of production are reported. Schumannella luteola SBP71 showed the highest AZO-CMCase activity, at 0.33 ± 0.09 U mL−1, although no significant differences were found with other strains. In particular, high activity levels ranging from 0.20 ± 0.06 to 0.32 ± 0.08 U mL−1 were also found for Pediococcus acidilactici CE75b, Curtobacterium citreum CA84, Curtobacterium flaccumfaciens CA83, Cellulomonas flavigena CP713 and Pantoea ananatis SBA76a (Table 3). Mycobacterium frederiksbergense CP78b, Curtobacterium flaccumfaciens CP77b and Promicromonospora citrea SBP79 (with a maximum value of 0.18 ± 0.02 U mL−1 and 0.16 ± 0.02 U mL−1) exhibited less AZO-CMCase activity than the microorganisms mentioned above but more than Cellulosimicrobium cellulans CA817 (0.15 ± 0.01 U mL−1) and Isoptericola variabilis CA84b (0.13 ± 0.01 U mL−1). All other bacterial strains tested showed values lower than 0.10 U mL−1.
Table 3. Maximum value of Azo-CMCase activity measured for each strain and the corresponding time of production.
| Bacterial strains | Maximum value of AZO-CMCase activitya (U mL−1) | Time (h) |
|---|---|---|
| Schumannella luteola SBP71 | 0.33 ± 0.09F | 12 |
| Pediococcus acidilactici CE75b | 0.32 ± 0.10EF | 12 |
| Curtobacterium citreum CA84 | 0.29 ± 0.01D-F | 17 |
| Curtobacterium flaccumfaciens CA83 | 0.24 ± 0.03C-F | 15 |
| Cellulomonas flavigena CP713 | 0.22 ± 0.10B-F | 20 |
| Pantoea ananatis SBA76a | 0.20 ± 0.06A-F | 8 |
| Mycobacterium frederiksbergense CP78b | 0.18 ± 0.02A-E | 26 |
| Curtobacterium flaccumfaciens CP77b | 0.16 ± 0.02A-D | 20 |
| Promicromonospora citrea SBP79 | 0.16 ± 0.02A-D | 12 |
| Cellulosimicrobium cellulans CA817 | 0.15 ± 0.01A-D | 20 |
| Isoptericola variabilis CA84b | 0.13 ± 0.01A-C | 12 |
| Bacillus subtilis subsp. subtilis CA816 | 0.09 ± 0.00AB | 12 |
| Microbacterium testaceum CA818 | 0.09 ± 0.01AB | 17 |
| Curtobacterium flaccumfaciens CA83b | 0.08 ± 0.03AB | 12 |
| Promicromonospora sukumoe CE86 | 0.07 ± 0.02A | 14 |
| Bacillus licheniformis CA82 | 0.07 ± 0.00A | 15 |
aThe values represent the means ± SD of three replicates of three independent experiments. Different letters after the values indicate significant differences (P < 0.05).
Discussion
In recent years, the competitive production of alternative renewable biofuels has stimulated research into new bacteria as a source of highly active and specific cellulases. They exhibit several advantages such as a fast growth rate, production of enzymes that are often more effective catalysts due to less feedback inhibition, and secretion of a complete multi-enzyme system for an efficient conversion of lignocelluloses into fermentable sugars11,14. In this context, particular attention must be given to exploring the biodiversity of natural niches so that cellulase-producing bacteria can be isolated and characterised. For these reasons, in this work, the microbial diversity of natural ecosystems, represented by lignocellulosic biomasses of A. donax, E. camaldulensis and P. nigra, was evaluated by culture-independent and culture-dependent approaches.
A highly complex bacterial community was found, in which the most frequently occurring bacteria were those belonging to the Actinobacteria, Proteobacteria, Bacteroidetes and Firmicutes phyla. Proteobacteria was the most abundant taxa recovered in the E. camaldulensis and P. nigra piles, followed by Actinobacteria, Bacteroidetes, Firmicutes and Acidobacteria. These taxa are related to microorganisms previously characterised as biomass degraders. The biodiversity of the microbial community in our study corresponded well with a previous study in which Proteobacteria, Firmicutes and Bacteroidetes, along with members of the class Proteobacteria, comprised 83% of the microbial richness and heavily dominated switchgrass-adapted communities15. Moreover, bacterial species belonging to Proteobacteria and Acidobacteria, together with Firmicutes (Clostridium and Bacillus genera) and followed by Bacteroidetes, Chlamydiae/Verrucomicrobia and Actinobacteria (mainly Streptomyces), are known as the major plant biomass-degrading microbes in peat swamp forests16. During the natural biodegradation process of plant substrates, the indigenous bacterial community would initially have grown by utilising the more accessible cellulose and hemicellulose and only later would use the more resilient lignin component17. With regard to lignin decomposition, Actinobacteria, Firmicutes and Acidobacteria are the major taxa involved in this process18. Acidobacteria was recovered in the late stage of our experiment and its abundance increased during the biodegradation process, especially in the E. camaldulensis piles. Firmicutes showed a similar trend in the A. donax and P. nigra biomasses, with the relative abundance of this taxon gradually increasing in the piles processed under the open field condition. Wu and He19 reported that Firmicutes could be the main microbes for lignin depolymerisation since a dominance of this phylum was recovered in enriched microbial consortia using a medium with lignin. Moreover, this phylum is common in natural processes such as rice straw compost20 and decaying wood21, suggesting its importance in the degradation of lignocellulolytic materials.
Analysing the microbial diversity more deeply, Actinobacteria, γ-Proteobacteria, β-Proteobacteria, α-Proteobacteria, Acidobacteria, Sphingobacteria, Flavobacteria, Bacilli and Acidobacteria were recovered in all samples. In particular, Actinobacteria, α-Proteobacteria, Sphingobacteria and β-Proteobacteria, all potent plant polysaccharide-degrading microbes that play an important role in plant biomass degradation in the tropical peat swamp forest ecosystem16, were the most abundant taxa during the biodegradation process in all lignocellulosic piles.
Different bacteria belonging to the Actinobacteria class are involved in complex glycoside degradation such as chitin and cellulose and are fundamental in lignin and polyphenol degradation22. Martins and co-workers23 reported that biomass degradation in the composting process, including the deconstruction of recalcitrant lignocellulose, is fully performed by bacterial enzymes, most likely by members of the Clostridiales and Actinomycetales orders. β-Proteobacteria and α-Proteobacteria were also recovered in our study. According to Castillo et al.22, the dominance of members of the phylum Proteobacteria such as γ-Proteobacteria is observed only at the beginning of the biodegradation process, and its strong reduction during the experiment could be due to its involvement in lignocellulosic waste declining during the early stages of the process. In the open field experiment, Bacilli increased in both the A. donax and P. nigra biomasses. Members belonging to this taxon are known to have specific genes encoding enzymes involved in cellulose and hemicellulose degradation24,25.
The constant increase in the relative abundance of Sphingobacteria and Acidobacteria recorded in the E. camaldulensis pile during the degradation process in the open field experiment suggests that these species play a role in the decomposition of lignocellulosic material. Kanokratana et al.26, using complementary shotgun pyrosequencing, identified different genes encoding glycosyl hydrolases targeting cellulose and hemicellulose degradation in a bagasse pile, most of which were found in orders Clostridiales, Bacteroidales, Sphingobacteriales and Cytophagales. Moreover, the Acidobactria taxon is able to a use a diversity of carbon sources, from simple sugars to complex plant biomass substrates26.
Another adapted-lignocellulosic taxon was β-Proteobacteria, which increased in the E. camaldulensis and P. nigra piles. In recent work, Stursova et al.27 identified β-Proteobacteria, Bacteroidetes and Acidobacteria as the primary cellulose decomposers in forest litter.
An interesting finding was the high incidence of the uncultured bacterium CH21 in the P. nigra pile during the first phase of the degradation process in the open field. CH21 is a member of the phylum Armatimonadetes, formerly called candidate division OP1028. The phylum Armatimonadetes is very poorly studied and its phylogeny is still poorly defined29. Members of this phylum are detected in different ecosystems and they are phylogenetically different. This phylum includes species with a wide variety of metabolic potentials30. Wang and co-workers31 reported that their prevalence in plant-fed anaerobic bioreactors indicates a role in degradation of plant material. In our research, CH21 is the only recovered member of this phylum. Since it's relative abundance increased in the Populus nigra pile after 45 days and decreased during the other phases of the degradation process in the open field, it is possible its involving in the biodegradation of this specific lignocellulosic biomass during the first phase of the process.
The differing trends observed in this study in terms of taxa abundance during the biodegradation process and between the vegetable species used demonstrated a local selective pressure in the lignocellulosic ecosystems.
The culture-dependent methodology used here, which was based on a functional approach of detection and isolation to find new lignocellulose-degrading bacterial strains, provides us with key insight. Special attention to the methodology was required to determine the optimal culture and assay conditions. A comparison with another study11 revealed that differential substrates containing CMC and Avicel are effective for the enumeration and isolation of putative colonies of cellulolytic microorganisms32. According to Soares et al.6, exo- and endo-cellulolytic bacterial isolates are found at different frequencies, and the number of microorganisms that were able to grow on Avicel as the sole carbon source was high. The cellulolytic strains isolated from the biomasses showed multienzymatic activities useful to perform the hydrolysis of a complex substrate such as lignocellulose, an important initial step in many technological applications33 that require the action of different specific enzymes. All forty bacterial strains submitted to the multienzymatic screening showed both endo- and exo-glucanase activities, confirming that these enzymes act synergistically during the saccharification of celluloses. These observations were reinforced by the fact that many of these bacterial strains also possessed β-cellobiase as well as cellulolytic activity. Xylanase activity was also commonly observed, which is unsurprising because a close correlation between cellulase and xylanase activities has been demonstrated and is due to their coexpression in the same operon.
The Neighbour-Joining phylogenetic method generated a consensus tree that grouped all 16S rRNA gene sequences of the isolated strains into five different clusters at the class level. Culture-dependent data showed similar predominant bacterial classes detected by high-throughput sequencing in the lignocellulosic biomasses, with an abundance of Actinobacteria, Bacilli, α-Proteobacteria, γ-Proteobacteria and Sphingobacteria. Moreover, the bacteria isolated are dominant players since these microbial strains were isolated from a high serial decimal dilutions (10−6–10−7). According to phylogenetic research on cellulose-decomposing bacteria isolated from soil carried out by Ulrich et al.34, Actinobacteria are the most prevalent bacterial group based on 16S rRNA gene sequences. This cluster included the species Curtobacterium citreum, which is able to use up to six different lignocellulose components and that is phylogenetically related to Microbacterium testaceum and Microbacterium lacus. In particular, previous studies reported the production of enzymes involved in cellulose and xylan degradation by Microbacterium species35. The Actinobacteria cluster included other genera involved in lignocellulosic biomass degradation, such as Cellulomonas. Akasaka et al.36 reported that more than 60% of isolates from rice plant residues was closely related to Cellulomonas and involved in their degradation. Our study revealed the production of cellobiase and pectinase activities in Actinobacteria members, such as Cellulosimicrobium cellulans, Isoptericola variabilis, Promicromonospora sukumoe and Promicromonospora citrea, which belong to the suborder Micrococcineae. Many of the representatives of the Promicrosporaceae and Corinebacteriaceae families can degrade polysaccharides such as cellulose and xylan37,38. The Bacilli cluster was primarily represented by the Pediococcus and Bacillus genera on the basis of 16S rRNA gene sequence analysis. Interesting, Zhao and co-workers39 reported the use of a strain of Pediococcus acidilactici with high tolerance to temperature and a lignocellulose-derived inhibitor in simultaneous saccharification and fermentation (SSF) for high lignocellulosic lactic acid production. The strains CA812, CA81, CA816, CA82 and CA81b, isolated from the A. donax biomass and phylogenetically correlated to the Bacillus genus, showed high endo-cellulolytic and multi-enzymatic activities. The capacity of Bacillus strains to produce large quantities of extracellular enzymes has placed them among the most important industrial enzyme producers isolated from compost, soil and several other natural habitats40. In particular, Bacillus amyloliquefaciens, Bacillus subtilis and Bacillus licheniformis, isolated from soil and compost, are able to hydrolyse cellulosic waste-material8 by both cellulolytic activities41 and multi-enzyme complexes42. The selected bacterial strains could have cross-specificity facilitated by specific or non-specific active sites and also distinct catalytic domains binding to different substrates33. Xanthomonas campestris, known as a phytopathogenic bacterium, and Lysobacter gummosus and Lysobacter enzymogenes, potent biocontrol agents that release cellulolytic enzymes such as glucanase43, were identified in the phylogenetic group of γ-Proteobacteria. Other strains of this class, such as Enterobacter sp. and Pantoea sp., although are known as insect-associated bacteria that are able to produce bioactive compounds and digestive enzymes that are responsible for lignocellulose degradation44, they are less attractive for possible biotechnological application since human disease has been reported to be caused by these bacteria as well as Raoultella terrigena and Sphingobacterium multivorum.
The two strains CE77 and CE84, included in the α-Proteobacteria cluster and characterised for their peroxidase activity, were closely related to the species Novosphingobium resinovorum, previously isolated from soil and studied for its capacity to degrade oil resins45. Interestingly, the strain Aurantimonas altamirensis SBP73 showed multienzymatic activity. To our knowledge, this study is the first to report the multienzymatic activity of this specie.
The bacterial strains screened in liquid medium showed enzymatic activity levels similar to the values reported in previous studies46. For example, Ekperigin47 reported that the maximum activity of the cellulose-degrading enzyme determined for CMC in the culture supernatant of Branhamella spp. was 0.34 U mL−1. Even though comparing cellulase production across studies is difficult because there are too many differences in the production of extracellular enzymes (e.g., media composition, fermentation conditions and raw materials), the results demonstrated that the bacterial strains isolated and characterised in this study could represent a very interesting biological source for the conversion of lignocellulose carbohydrates into products of commercial significance. Moreover, their biotechnological performance could be enhanced by modifying the biotechnological parameters of the fermentation process such as the temperature33.
In conclusion, in this work, pyrosequencing-based technology increased our knowledge of lignocellulosic-adapted microbiota and the microbial dynamic during the degradation of three different biomasses under natural conditions. The dominant taxa found during the biodegradation process were members of classes Actinobacteria, γ-Proteobacteria, α-Proteobacteria and Sphingobacteria. However, the abundance of the major and minor taxa retrieved during the process was determined by selective pressure employed by the lignocellulosic plant species and the degradation conditions.
In addition, new multifunctional degrading bacteria have been selected and identified that are potential producers of multiple enzymes that have synergistic actions on cellulose and hemicellulose. This step is fundamental for biotechnological applications of interest to industry because they constitute a microbial source of new multifunctional enzymes that can increase the efficiency of the hydrolysis of lignocellulosic biomass into fermentable sugars for biofuel eco-technology.
Methods
Lignocellulosic Biomasses and Sampling
Chipped wood from A. donax, E. camaldulensis and P. nigra was processed for biodegradation under natural conditions for 180 days. The chipped wood was used to form two piles of approximately 30 kg (length 0.85 m, width 0.85 m and height 0.70 m) for each plant species; on March 22, 2012, the piles were placed under two different environmental conditions in order to increase microbial biodiversity: open field conditions, in which the biomass piles were established on agronomic soil without any coverage shady trees, in the experimental station of Department of Agriculture (Naples, Italy; 40°48′50.1″N, 14°20′48.2″E); the underwood conditions performed reproducing the natural environment that usually arise in a forest since the piles were placed under oak trees in the woodland at the Department of Agriculture (Naples, Italy; 40°48′47.8″N, 14°20′50.4″E). Samples of 0.5 kg were collected from the external part (right and left side of the pile) and the internal central part of the biomass immediately after preparation (T0) and at 45, 90, 135 and 180 days (T1, T2, T3 and T4, respectively) of biodegradation. During the process, temperature (°C) was measured by using specific temperature sensors (VWR International PBI, Milan, Italy) placed directly in the core of the piles at a depth of 30–40 cm. The water activity (aw, water readily available for microbial metabolic activities) was also monitored using a HygroPalm23-AW (Rotronic AG, Basserdorf, Germany).
Analysis of the Microbiota by High-Throughput Sequencing of the 16S rRNA Gene
Total DNA recovery from microorganisms adherent to the plant biomass was performed as previously described48. The bacterial diversity was evaluated by pyrosequencing using the primers Gray28F (5′-TTTGATCNTGGCTCAG-3′) and Gray519r (5′-GTNTTACNGCGGCKGCTG-3′) spanning the V1-V3 region of the 16S rDNA of E. coli49. 454-adaptors were included in the forward primer followed by a 10-bp sample-specific Multiplex Identifier (MID). The PCR mixture (50-µl total volume) included 20 ng of target DNA, 1 x Taq DNA polymerase buffer (Invitrogen, Milano, Italy), 2.5 mM MgCl2, 0.5 mM of each dNTP, 0.4 µM of each primer and 2.5 U of Taq DNA polymerase (Invitrogen). PCR amplification was performed as previously reported50. After agarose gel electrophoresis, PCR products were purified twice by Agencourt AMPure kit (Beckman Coulter, Milano, Italy) and quantified using the QuantiFluorTM (Promega, Milano, Italy), and an equimolar pool was obtained prior to further processing. The amplicon pool was used for pyrosequencing on a GS Junior platform (454 Life Sciences, Roche, Italy) according to the manufacturer's instructions using Titanium chemistry.
Bioinformatics and Data Analysis
The pyrosequencing data were filtered using the following quality check parameters using the QIIME software package51: a minimum sequence length of 200 bp, maximum homopolymer number of 5, minimum quality score of 25, and maximum number of ambiguous bases and primer mismatches of 0. The QIIME software was also used to denoise, split sequences into the proper samples, and pick operational taxonomy units (OTUs) at 97% sequence identity using UCLUST52. The representative sequences were submitted to the RDPII classifier53 to obtain a taxonomy assignment using the Greengenes 16S rRNA gene database54.
Alpha diversity was evaluated by rarefaction curves, Good's coverage, Chao1 richness55 and Shannon diversity indices56. To test for significant differences in alpha diversity, QIIME's compare_alpha_diversity.py script was used to run nonparametric two-sample t-tests. The default number of Monte Carlo permutations (999) was used to calculate P-values in the nonparametric t-tests, and a significance threshold of P < 0.05 was used. Statistical tests for calculating the influence of sample variables on microbial ecology were applied using QIIME's group_significance.py (ANOVA) and compare_categories.py (ADONIS) scripts. The OTU taxonomy tables generated by QIIME were used to produce heatmaps using the software TMeV v 4.857. Beta diversity was also evaluated by UniFrac58, and principal coordinate analysis (PCoA) was generated by QIIME. Unweighted UniFrac distances were also calculated, but we only report weighted distances here because sample clustering was more informative.
Isolation of Cellulolytic Functional Groups in the Lignocellulosic Biomasses
Different samples were collected from the chipped vegetable biomass piles of A. donax, E. camaldulensis and P. nigra for the isolation of aerobic cellulolytic bacteria. Serial dilutions (0.1 mL) of the samples were spread on the surface of plates containing basal medium (1 g L−1 [NH4]NO3, 1 g L−1 yeast extract, 50 mL L−1 standard salt solution, 1 mL L−1 trace element solution and 15 g L−1 agar bacteriological, pH 7.0) with 5 g L−1Avicel (Sigma-Aldrich, Milan, Italy) or 5 g L−1 carboxymethylcellulose (CMC) with 0.1% Congo red (Sigma-Aldrich) for exo-cellulolytic or endo-cellulolytic differential isolation, respectively. After 4 days under aerobic conditions at 28°C, the endo-cellulolytic colonies were differentially visualised by a clear halo on CMC. All colonies grown on the substrate Avicel as only carbon source were considered as exo-cellulolytic bacteria. Morphologically different exo- and endo-cellulolytic colonies were isolated, purified again on the same isolation media and stored at 4°C until their characterisation.
Screening for Functional Activities on Solid Media
The endo-cellulolytic activity of all isolates was measured on CMC by a semi-quantitative agar spot method. After adjusting the turbidity of the bacterial suspensions (0.5 of McFarland Turbidity Standard corresponding to approximately 1.5 × 108 CFU mL−1), the cells were spotted on CMC agar medium in triplicate. After a 4-day incubation at 28°C, the plates were stained with 0.1% Congo red solution for 30 min followed by washing with 5 M NaCl8,14. Cellulase activities on screening media were recorded as the “Indices of Relative Enzyme Activity, ICMC = diameter of clearing or halo zone/colony diameter”11. The exo-cellulase activity was similarly estimated in a semi-quantitative way by inoculating all the isolates by spotting on the Avicel agar and observing the development of bacterial colonies after incubation at 28°C for 10 days.
All isolates were also assayed for cellobiase, xylanase, pectinase and ligninase activities. In particular, cellobiase activity was assessed with cellobiose agar (10 g L−1 bacteriological peptone, 15 g L−1 agar bacteriological [Oxoid, Milan, Italy], 7.5 g L−1 cellobiose and 0.06 g L−1 bromothymol blue [Sigma-Aldrich]). After incubation at 28°C for 3 days, the cellobiase activity was detected by observing yellow haloes around the colonies. For xylanase detection, the method described by Ko et al.59 was applied, adding 0.05% Remazol brilliant blue-R and a 0.5% solution of sonicated (FALC Instruments, HK3300) xylan (Sigma-Aldrich) to the LB Agar medium (Oxoid). After 3–5 days of incubation at 28°C, the hydrolysis of xylan was detected by observing a clear zone around the colonies. Pectinase activity was measured following the method described by Pepe et al.60 and the results were recorded as the “Indices of Relative Enzyme Activity, (IPEC) = diameter of clearing or halo zone/colony diameter”.
Ligninase activity was detected by using the CGA culture medium described by Okino et al.61, modified by replacing the sugarcane bagasse lignin with vegetable biomass powder from A. donax or 2 g L−1 lignin alkali (Sigma-Aldrich). A reddish-brown halo around a colony indicates the successful oxidation of guaiacol by ligninolytic enzymes. Laccase activity was determined for all the isolated strains by growth on agar medium composed of liquid basal medium (LBM) supplemented with 0.1% 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid [ABTS], Sigma-Aldrich) as described by Toutella et al.62. After 10 days of incubation at 25°C in darkness, a green colour in the medium indicated the presence of laccase activity. Azure-B (Sigma-Aldrich) agar was used to evaluate the production of lignin peroxidase and manganese-dependent peroxidase by the formation of a clear halo around a colony after 10–30 days of incubation at 25°C in darkness63.
Identification and Phylogenetic Analysis of Cellulolytic Strains
Bacterial isolates were identified by molecular method (16S rRNA gene sequencing). Total genomic DNA was extracted and purified by InstaGeneTM Matrix (Bio-Rad Laboratories, Hercules, CA, USA) according to the supplier's recommendations. Approximately 50 ng of DNA was used as the template for PCR assays. Synthetic oligonucleotide primers fD1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and rD1 (5′-AAGGAGGTGATCCAGCC-3′) were used to amplify the 16S rRNA gene. The PCR mixture was prepared as reported by Alfonzo et al.64. The PCR conditions were performed as described by Pepe et al.65. The PCR products, after visualisation by agarose (1.5% wt/vol) gel electrophoresis at 100 V for 1 h, were purified by using a QIAquick gel extraction kit (Qiagen S.p.A., Milan, Italy) and sequenced. The DNA sequences were analysed as previously reported61 and compared with the GenBank nucleotide data library using Blast software at the National Centre of Biotechnology Information website (http://www.ncbi.nlm.nih.gov/Blast.cgi).
Multiple nucleotide alignments of nearly full-length 16S rRNA sequences of isolated strains and type strains within each of the defined species were performed using the Clustal W program66 from MEGA version 4.067. The nucleotide sequences of the type strains were retrieved from the Ribosomal Database Project (RDP - http://rdp.cme.msu.edu/). The phylogenetic tree was inferred by using the Neighbour-Joining method with the Maximum Composite Likelihood model in MEGA4 program with bootstrap values based on 1,000 replications.
Screening of Cellulolytic Activity in Liquid Media
The bacterial strains were pre-inoculated by dissolving a single colony in 3 mL of liquid medium containing 0.5% CMC, 0.7% yeast extract, 4 g L−1 KH2PO4, 4 g L−1 Na2HPO4, 0.2 g L−1 MgSO4·7H2O, 0.001 g L−1 CaCl2·2H2O and 0.004 g L−1 FeSO4·7H2O. After overnight incubation at 37°C, a volume of the broth culture corresponding to 0.1 O.D. was used to inoculate 100-mL plugged Erlenmeyer flasks, each containing 20 mL of the same medium. During incubation at 37°C on a rotary shaker at 225 rpm, samples of the liquid culture were withdrawn and used to measure the optical density (O.D.600nm) and the extracellular endo-1,4-ß-glucanase activity by AZO-CMCase assay (Megazyme, Ireland), following the supplier's instructions. The analytical determinations correspond to the mean value of three replicates.
Accession Numbers
The 16S rRNA gene sequences obtained were deposited in the GenBank nucleotide database under accession numbers from KF040971 to KF040996 and from KF057947 to KF057960 (http://www.ncbi.nlm.nih.gov/Blast.cgi).
The pyrosequencing data are available in the Sequence Read Archive database of the National Center of Biotechnology Information (PRJNA248067).
Statistical Analyses
One-way ANOVA followed by Tukey's HSD post hoc for pair-wise comparison of means (at P < 0.05) was used to assess the difference in the enzymatic activities of isolated strains such as ICMC, IPEC and Azo-CMCase. Statistical analyses were performed using SPSS 13.0 statistical software package (SPSS Inc., Cary, NC, USA).
Acknowledgments
This work was supported by grant from the Ministero dell'Università e della Ricerca Scientifica Industrial Research Project “Integrated agro-industrial chains with high energy efficiency for the development of eco-compatible processes of energy and biochemical production from renewable sources and for the land valorization (EnerbioChem)” PON01_01966, funded in the frame of Operative National Programme Research and Competitiveness 2007–2013 D. D. Prot. n. 01/Ric. 18.1.2010.
Footnotes
Author Contributions V.V. wrote the main manuscript text and, in particular, microbial diversity of lignocellulosic biomasses by High-Throughput Sequencing and Phylogenetic analysis of selected cellulolytic bacteria. She prepared Figure 1, 2, 3, 4, 5 and table 1. A.Aliberti and A.R. wrote the isolation and identification of cellulolytic bacterial strains by culture-dependent approach and selection by multienzymatic screening. They prepared Table 2. V.F., S.G. and A.Amore wrote the screening of cellulolytic microorganisms in liquid medium and prepared Table 3. D.E. wrote the microbial diversity of lignocellulosic biomasses by High-Throughput Sequencing. M.F. wrote lignocellulosic biomasses and sampling. O.P. is the corresponding author. All authors reviewed the manuscript.
References
- Diodato N., Fagnano M. & Alberico I. CliFEM – Climate Forcing and Erosion Response Modelling at Long-Term Sele River Research Basin (Southern Italy). Nat. Hazard Earth Syst. Sci. 9, 1693–1702 (2009). [Google Scholar]
- Fiorentino N. et al. Biomass accumulation and heavy metal uptake of giant reed on polluted soil in southern Italy. J. Biotechnol. 150, S1261 (2010). [Google Scholar]
- Fiorentino N. et al. Assisted phytoextraction of heavy metals: compost and Trichoderma effects on giant reed (Arundo donax L) uptake and soil N-cycle microflora. Ital. J. Agron. 8, 244–254 (2013). [Google Scholar]
- Pirozzi D. et al. Lipids production by yeast grown on giant reed biomass. J. Biotechnol. 150, S167–168 (2010). [Google Scholar]
- Van Den Burg B. Extremophiles as a source for novel enzymes. Curr. Opin. Microbiol. 6, 213–218 (2003). [DOI] [PubMed] [Google Scholar]
- Soares F. L. Jr, Melo I. S., Dias A. C. & Andreote F. D. Cellulolytic bacteria from soils in harsh environments. World J. Microbiol. Biotechnol. 28, 2195–2203 (2012). [DOI] [PubMed] [Google Scholar]
- Maki M., Leung K. T. & Qin W. S. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Biol. Sci. 5, 500–516 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amore A. et al. Industrial waste based compost as a source of novel cellulolytic strains and enzymes. FEMS Microbiol. Lett. 339, 93–101 (2013). [DOI] [PubMed] [Google Scholar]
- Sizova M. V., Izquierdo J. A., Panikov N. S. & Lynd L. R. Cellulose- and xylan-degrading thermophilic anaerobic bacteria from biocompost. Appl. Environ. Microbiol. 77, 2282–2291 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taherzadeh M. J. & Karimi K. Enzyme-based hydrolysis processes for ethanol from lignocellulosic materials: a review. Bioresources 2, 707–738 (2007). [Google Scholar]
- Saini J. K. & Tewari A. L. Simultaneous isolation and screening of cellulolytic bacteria: selection of efficient medium. J. Pure Appl. Microbiol. 3, 1339–1344 (2012). [Google Scholar]
- Duan C. J. & Feng J. X. Mining metagenomes for novel cellulose genes. Biotechnol. Lett. 32, 1765–1775 (2010). [DOI] [PubMed] [Google Scholar]
- Thompson C. E. et al. A potential source for cellulolytic enzyme discovery and environmental aspects revealed through metagenomics of Brazilian mangroves. AMB Express 3, 65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amore A. et al. Cloning and recombinant expression of a cellulase from the cellulolytic strain Streptomyces sp. G12 isolated from compost. Microb. Cell Fact. 11, 164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis K. M. et al. Strategies for enhancing the effectiveness of metagenomic-based enzyme discovery in lignocellulolytic microbial communities. Bioenerg. Res. 3, 146–158 (2010). [Google Scholar]
- Kanokratana P. et al. Insights into the phylogeny and metabolic potential of a primary tropical peat swamp forest microbial community by metagenomic analysis. Microb. Ecol. 61, 518–528 (2011). [DOI] [PubMed] [Google Scholar]
- Van der Heijden M. G. A., Bardgett R. D. & van Straalen N. M. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310 (2008). [DOI] [PubMed] [Google Scholar]
- DeAngelis K. M. et al. Characterization of trapped lignin-degrading microbes in tropical forest soil. PLoS ONE 6, e19306 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. R. & He J. Characterization of anaerobic consortia coupled lignin depolymerization with biomethane generation. Bioresour. Technol. 139C, 5–12 (2013). [DOI] [PubMed] [Google Scholar]
- Matsuyama T. et al. Bacterial community in plant residues in a Japanese paddy field estimated by RFLP and DGGE analyses. Soil Biol. Biochem. 39, 463–472 (2007). [Google Scholar]
- Zhang H. B., Yang M. X. & Tu R. Unexpectedly high bacterial diversity in decaying wood of a conifer as revealed by a molecular method. Int. Biodeterior. Biodegrad. 62, 471–474 (2008). [Google Scholar]
- Castillo J. M., Romero E. & Nogales R. Dynamics of microbial communities related to biochemical parameters during vermicomposting and maturation of agroindustrial lignocellulose wastes. Bioresour. Technol. 146, 345–354 (2013). [DOI] [PubMed] [Google Scholar]
- Martins L. F. et al. Metagenomic analysis of a tropical composting operation at the São Paulo Zoo Park reveals diversity of biomass degradation functions and organisms. PLoS ONE 8, e61928 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. J. et al. Purification and characterization of cellulase produced by Bacillus amyoliquefaciens DL-3 utilizing rice hull. Bioresour. Technol. 99, 378–386 (2009). [DOI] [PubMed] [Google Scholar]
- Di Pasqua R. et al. Influence of different lignocellulose sources on endo-1,4-ß-glucanase gene expression and enzymatic activity of Bacillus amyloliquefaciens B31C. Bioresources 9, 1303–1310 (2014). [Google Scholar]
- Kanokratana P. et al. Phylogenetic analysis and metabolic potential of microbial communities in an industrial bagasse collection site. Microb. Ecol. 66, 322–334 (2013). [DOI] [PubMed] [Google Scholar]
- Stursova M., Zifcakova L., Leigh M. B., Burgess R. & Baldrian P. Cellulose utilization in forest litters and soils: identification of bacterial and fungal decomposers. FEMS Microbiol. Ecol. 80, 735–746 (2012). [DOI] [PubMed] [Google Scholar]
- Dalevi D., Hugenholtz P. & Blackall L. L. A multiple-outgroup approach to resolving division-level phylogenetic relationships using16S rDNA data. Int. J. Syst. Evol. Microbiol. 51, 385–391 (2001). [DOI] [PubMed] [Google Scholar]
- Lee K. C. Y. et al. Phylogenetic Delineation of the Novel Phylum Armatimonadetes (Former Candidate Division OP10) and Definition of Two Novel Candidate Divisions. Appl. Environ. Microbiol. 79, 2484–2487 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunfield P. F. et al. Electing a candidate: a speculative history of the bacterial phylum OP10. Environ. Microbiol. 14, 3069–3080 (2012). [DOI] [PubMed] [Google Scholar]
- Wang H. et al. Development of microbial populations in the anaerobic hydrolysis of grass silage for methane production. FEMS Microbiol. Ecol. 72, 496–506 (2010). [DOI] [PubMed] [Google Scholar]
- Ventorino V., Amore A., Faraco V., Blaiotta G. & Pepe O. Selection of cellulolytic bacteria for processing of cellulosic biomass. J. Biotechnol. 150, S181 (2010). [Google Scholar]
- Van Dyk J. S. & Pletschke B. I. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes - Factors affecting enzymes, conversion and synergy. Biotechnol. Adv. 30, 1458–1480 (2012). [DOI] [PubMed] [Google Scholar]
- Ulrich K., Ulrich A. & Ewald D. Diversity of endophytic bacterial communities in poplar grown under field conditions. FEMS Microbiol. Ecol. 63, 169–180 (2008). [DOI] [PubMed] [Google Scholar]
- Okeke B. C. & Lu J. Characterization of a defined cellulolytic and xylanolytic bacterial consortium for bioprocessing of cellulose and hemicelluloses. Appl. Biochem. Biotechnol. 163, 869–881 (2011). [DOI] [PubMed] [Google Scholar]
- Akasaka H., Izawa T., Ueki K. & Ueki A. Phylogeny of numerically abudant culturable anaerobic bacteria associated with degradation of rice plant residue in Japanese paddy field soil. FEMS Microbiol. Ecol. 43, 149–161 (2003). [DOI] [PubMed] [Google Scholar]
- Lo Y. C., Saratale G. D., Chen W. M., Bai M. D. & Chang J. S. Isolation of cellulose-hydrolytic bacteria and applications of the cellulolytic enzymes for cellulosic biohydrogen production. Enzyme Microb. Technol. 44, 417–425 (2009). [Google Scholar]
- Song J. M. & Wei D. Z. Production and characterization of cellulases and xylanases of Cellulosimicrobium cellulans grown in pretreated and extracted bagasse and minimal nutrient medium M9. Biomass Bioenerg. 34, 1930–1934 (2010). [Google Scholar]
- Zhao K. et al. Simultaneous saccharification and high titer lactic acid fermentation of corn stover using a newly isolated lactic acid bacterium Pediococcus acidilactici DQ2. Bioresour. Technol. 135, 481–489 (2013). [DOI] [PubMed] [Google Scholar]
- Amore A., Pepe O., Ventorino V., Aliberti A. & Faraco V. Cellulolytic Bacillus strains from natural habitats - A review. Chimica Oggi/Chemistry Today 31, 49–52 (2013). [Google Scholar]
- Kim Y. K., Lee S. C., Cho Y. Y., Oh H. J. & Ko Y. H. Isolation of cellulolytic Bacillus subtilis strains from agricultural environments. ISRN Microbiol 0.5402/2012/650563 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. M., Van Dyk J. S. & Pletschke B. I. Bacillus subtilis SJ01 produces hemicellulose degrading multi-enzyme complexes. Bioresources 7, 1294–1309 (2012). [Google Scholar]
- Palumbo J. D., Yuen G. Y., Jochum C. C., Tatum K. & Kobayashi D. Y. Mutagenesis of β-1,3-glucanase genes in Lysobacter enzymogenes strain C3 results in reduced biological control activity toward bipolaris leaf spot of tall fescue and Pythium damping-off of sugar beet. Phytopathology 95, 701–707 (2005). [DOI] [PubMed] [Google Scholar]
- Anand A. A. et al. Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J. Insect Sci. 10, 107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y. W., Moon E. Y. & Chun J. Reclassification of Flavobacterium resinovorum Delaporte and Daste 1956 as Novosphingobium resinovorum comb. nov., with Novosphingobium subarcticum (Nohynek et al. 1996) Takeuchi et al. 2001 as a later heterotypic synonym. Int. J. Syst. Evol. Microbiol. 57, 1906–1908 (2007). [DOI] [PubMed] [Google Scholar]
- Gupta P., Samant K. & Sahu A. Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int. J. Microbiol. 10.1155/2012/578925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekperigin M. M. Preliminary studies of cellulase production by Acinetobacter anitratus and Branhamella sp. Afr. J. Biotechnol. 6, 28–33 (2007). [Google Scholar]
- Ventorino V., Parillo R., Testa A., Aliberti A. & Pepe O. Chestnut biomass biodegradation for sustainable agriculture. Bioresources 8, 4647–4658 (2013). [Google Scholar]
- Andreotti R. et al. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol. 11, 6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercolini D., De Filippis F., La Storia A. & Iacono M. “Remake” by high-throughput sequencing of the microbiota involved in the production of water buffalo mozzarella cheese. Appl. Environ. Microbiol. 78, 8142–8145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naïve Bayesan classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D. et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archea. ISME J. 6, 610–618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. & Bunge J. Estimating the number of species in a stochastic abundance model. Biometrics 58, 531–539 (2002). [DOI] [PubMed] [Google Scholar]
- Shannon C. E. & Weaver W. The mathematical theory of communication. Urbana: University of Illinois Press. 125 p. (1949). [Google Scholar]
- Saeed A. I. et al. TM4: a free, opensource system for microarray data management and analysis. Biotechniques 34, 374–378 (2003). [DOI] [PubMed] [Google Scholar]
- Lozupone C., Hamady M. & Knight R. UniFrac e an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7, 371 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko K. C., Han Y., Shin B. S., Choi J. H. & Song J. J. A rapid and simple method for preparing an insoluble substrate for screening of microbial xylanase. Appl. Biochem. Biotechnol. 167, 677–84 (2012). [DOI] [PubMed] [Google Scholar]
- Pepe O., Ventorino V. & Blaiotta G. Dynamic of functional microbial groups during mesophilic composting of agro-industrial wastes and free-living (N2)-fixing bacteria application. Waste Manage. 33, 1616–1625 (2013). [DOI] [PubMed] [Google Scholar]
- Okino L. K., Machado K. G. M., Fabric C. & Bonomi V. L. R. Ligninolytic activity of tropical rainforest basidiomycetes. World J. Microbiol. Biotechnol. 16, 889–893 (2000). [Google Scholar]
- Toutella G. R., Rubilar O., Gianfreda L., Valenzuela E. & Diez M. C. Enzymatic characterization of Chilean native wood-rotting fungi for potential use in the bioremediation of polluted environment with chlorophenols. World J. Microbiol. Biotechnol. 24, 2805–2818 (2008). [Google Scholar]
- Pointing S. B. Qualitative methods for the determination of lignocellulolytic enzyme production by tropical fungi. Fungal Divers. 2, 17–33 (1999). [Google Scholar]
- Alfonzo A. et al. Antifungal peptides produced by Bacillus amyloliquefaciens AG1 active against grapevine fungal pathogens. Ann. Microbiol. 62, 1593–1599 (2012). [Google Scholar]
- Pepe O. et al. Characterization in the archaeological excavation site of heterotrophic bacteria and fungi of deteriorated wall painting of Herculaneum in Italy. J. Environ. Biol. 32, 241–250 (2011). [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M. & Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007). [DOI] [PubMed] [Google Scholar]