Abstract
Agrobacterium mediated transformation (AMT) has been embraced by biotechnologists as the technology of choice to introduce or alter genetic traits of plants. However, in plants it is virtually impossible to predetermine the integration site of the transferred T-strand unless one is able to generate a double stranded break (DSB) in the DNA at the site of interest. In this study, we used the model organism Saccharomyces cerevisiae to investigate whether the Agrobacterium mediated translocation of site-specific endonucleases via the type IV secretion system (T4SS), concomitantly with T-DNA transfer is possible and whether this can improve the gene targeting efficiency. In addition to that, the effect of different chromatin states on targeted integration, was investigated. It was found that Agrobacterium mediated translocation of the homing endonuclease I-SceI has a positive effect on the integration of T-DNA via the homologous repair (HR) pathway. Furthermore, we obtained evidence that nucleosome removal has a positive effect on I-SceI facilitated T-DNA integration by HR. Reversely; inducing nucleosome formation at the site of integration removes the positive effect of translocated I-SceI on T-DNA integration.
Agrobacterium tumefaciens is a soil born plant pathogen well known for its ability to genetically alter the cells of the dicotyledonous plants that form its main natural host. These genetically modified plant cells are inclined to produce amino acid derivatives, which can be utilized by Agrobacterium as a carbon and nitrogen source. In addition to this, plant tissue infected by Agrobacterium shows excessive cell proliferation resulting in tumorous outgrowths. Although for agriculturists these are the symptoms of crown gall disease, for biotechnologists the Agrobacterium mediated transformation (AMT) of plants has formed the foundation for new technologies leading the way to crop improvement and the production of medicines or biofuels by plants. The precursor of the DNA that is integrated into the plant genome is located on a tumor-inducing (Ti) plasmid flanked by two imperfect direct 25 bp repeats; the left and right border sequences1. This transfer-DNA (T-DNA) is processed by the Agrobacterium virulence protein VirD2 which induces a nick at the lower strand of the right and left border sequence2,3. A single stranded section of the T-DNA with VirD2 covalently attached to the 5′ end, the T-strand, is released and targeted to a type IV secretion system (T4SS) which injects the T-strand into the host cell4. There are two models for the integration of T-DNA in plants, one involves the free 3′-end of the T-strand to find microhomologies in the genomic DNA of the host cells. After the free 3′-end anneals to the genomic DNA, this would then prime the formation of the complementary strand5. Later an alternative route for T-DNA integration was suggested where double stranded DNA is formed from the T-strands prior to opportunistic integration in genomic DSBs6,7,8. Probably host cell primases can synthesize de-novo RNA primers complementary to the T-strand, thereby facilitating the synthesis of a secondary T-strand prior to integration9.
T-DNA tends to integrate randomly into the genome of the recipient cell in respect to predicted transcriptional activity based on the DNA methylation patterns10. Inherent to the randomness of T-DNA integration is the risk of unintended gene disruption at the integration site or deviating expression levels of the introduced transgene caused by different positional effects at integration sites. To address these concerns related to AMT, it would be highly desirable to utilize the homologous recombination (HR) system of plants to replace a predetermined genomic locus with an incoming T-strand that comprises a strong homology to that locus. Unfortunately, HR-mediated integration of T-DNA in plants is a rare event, estimated to occur only once per 104 to 105 integration events11,12.
A breakthrough regarding the targeted integration of T-DNA via HR was the finding that this is facilitated by the formation of DNA double strand breaks (DSBs) at the target locus7,8,13. With the development of highly versatile site specific artificial nucleases, it is now theoretically possible to generate a DSB at any particular genomic position of interest14 and a major bottleneck for site-directed mutagenesis has thus disappeared. However, without directly selectable gain of function mutations after gene targeting, finding the few HR-mediated gene targeting events remains challenging. Negative selection against randomly integrated T-DNA copies can be instrumental for the recovery of rare gene targeting events in rice15, but this seems not to be generically applicable in other plant species. Other studies towards enhancing gene targeting have explored down regulation of enzymes involved in the NHEJ pathway which was very efficient in yeast and fungi16 but also this approach has variable efficiency in plants17,18.
Altogether, despite the abundant use of AMT for plant transformation, reports about its successful application for gene targeting are still scarce and a generally applicable strategy is lacking. Additionally, obvious parameters likely to be important for successful application of AMT for gene targeting have still been insufficiently explored. The first parameter that is subject of the present study concerns the chromatin state of the target locus. It has been demonstrated that the effect of site specific artificial transcription factors very much depends on their access to DNA19,20. As a more open chromatin structure might allow for better access of site specific nucleases as well, our experiments have addressed the possibility of nuclease access of the chromosomes for different states of chromatin condensation. The second parameter that is addressed in this manuscript was related to the delivery of the required site specific nucleases to the recipient cells. In the present study, different approaches were explored to deliver nuclease proteins to the recipient cell via Agrobacterium mediated translocation concomitant with T-strand delivery as will be explained below.
Agrobacterium effector proteins are targeted for translocation to the recipient host cell because of a series of positively charged amino acids at their C terminal end that act as a T4SS translocation signal21. An illustrative example of this is the C terminal end of the virulence protein VirF (VirFCT). By making translational fusions with VirFCT a number of DNA modifying proteins have been shown to translocate to the host cells, like the recombinase Cre21,22. Recently, it was demonstrated that complex fusion proteins consisting of a nuclear localization signal (NLS) for nuclear targeting, the Agrobacterium relaxase VirD2, I-SceI for the induction of double stranded breaks and a VirFCT moiety to allow the translocation of this protein complex to the host cell can be translocated to recipient plant cells23.These experiments were performed by means of assaying simultaneous transfer of T-strands coupled to such fusion proteins. The occurrence of footprints within the I-SceI recognition site in the host genome proved that the I-SceI moiety retained its nuclease activity after Agrobacterium mediated translocation. Hence, T-strands can be equipped with covalently attached modified VirD2 proteins, combining the relaxase activity of VirD2 with enzymatic activities of choice24.
A possibility to achieve gene targeting via AMT by introducing particles consisting of a T-strand with target site homology, covalently linked with a nuclease moiety that creates the required DSB to trigger an HR-mediated gene targeting event is well worth exploring. However, AMT offers opportunities to deliver the T-strand and the nuclease via the T4SS also without covalent linkage of the nuclease to a T-strand. In these cases the T-strand passes to the recipient cell piloted by an unmodified VirD2 protein bound at its 5′end, while the nuclease of interest is produced in the same Agrobacterium cell and is transported via the T4SS by means of a VirFCT fusion.
Previously, it was shown that AMT of Saccharomyces cerevisiae (budding yeast) can be used as a model system to address the roles of HR and NHEJ pathways in T-DNA integration16,25. Readily available mutants, time-efficient transformation protocols for the introduction of genomic target loci, as well as convenient transformant screening and subsequent analysis contributes to making the AMT of yeast a valuable asset. Of special interest to this study are the well characterized promoter regions of yeast genes involved in the galactose pathway that, depending on the carbon source, can have open or closed chromatin states26. These promoter sequences hence should allow for the in vivo modulation of the chromatin state during the AMT of yeast. In the present work, we used the yeast model to assess the possibilities for combined Agrobacterium mediated introduction of T-strands and the site-specific nuclease I-SceI to enhance gene targeting. In addition to this, the effects of the chromatin state for gene targeting via AMT were explored. These experiments were done in experimental set-ups conducive for transfer of T-strands covalently attached to NLS-VirD2-I-SceI-VirFCT fusion proteins or, alternatively, for the transfer of T-strands covalently attached to an unaltered VirD2 molecule with a NLS-I-SceI-VirFCT fusion protein translocated separately, albeit from single Agrobacterium cells.
As is shown in this manuscript, without introduced nucleases, the chromatin state of a target locus was of little importance for gene targeting frequencies. The Agrobacterium mediated transfer of a site specific nuclease protein during AMT resulted in higher transformation frequencies. This elevated efficiency of T-DNA integration, when transferring I-SceI fusion proteins with the T-strands, was however abolished when nucleosome formation was induced at the target locus.
Methods
The construction of the yeast target lines RSY12 pID2GU and RSY12 pIDU
For the amplification of the UAS typed promoter sequences from the yeast galactose pathway, genomic DNA from the Saccharomyces cerevisiae strain YPH25027 was used as a template. The PCR is performed using the primers listed in table 1. The PCR product from UAS7 was digested with BamHI and XhoI and cloned into pSKNSgraI which was also digested with BamHI and XhoI. The resulting vector was named pOPSL3. The PCR product from UAS10 was digested with SpeI and BamHI and cloned into pSKNSgraI also digested with SpeI and BamHI. The resulting vector was named pOPSL5. The cloning vector pSKNSgraI has been described by Neuteboom et al28. After these initial cloning steps pOPSL5 was digested with BamHI and SpeI. The resulting fragment containing the UAS10 fragment was cloned into pOPSL3 digested with BglII and SpeI resulting in a new vector containing both upstream activating sequences; p2Gstart. This vector in turn was digested with SpeI and XhoI. A PCR fragment of URA3 digested with SpeI and XhoI was cloned in between the upstream activating sequences resulting in p2GU. The primers used for the amplification of URA3 are depicted in Table 1.
Table 1. Oligos designed for the amplification of the galactose pathway promoter sequences UAS7 and UAS10 and the URA3 marker gene.
| Oligo name | Nucleotide sequence (5′-3′) |
|---|---|
| UASGAL7TTGAL10 BamHI F | GAGTAGGATCCAATATTCAACTGTTTTTTTTTATCATG |
| UASGAL7TTGAL10 XhoI R | ATCGCTCGAGTTGCCAGCTTACTATCCTTC |
| UASGAL10-GAL1 SpeI F | TGACACTAGTTTGAATTTTCAAAAATTCTTACTTTTTTTTTGG |
| UASGAL10-GAL1 BamHI R | GCATGGATCCTTGACGTTAAAGTATAGAGGT |
| URA Xho FW | TCCTCTCGAGCTTTTCAATTCAATTCASTC |
| URA Spe RV | TGGCACTAGTGGGTAATAACTGATATAATTAAATT |
The shuttle vector pRS31627 was used as template for the PCR. From p2GU a UAS10-URA3-UAS7 BamHI fragment was excised and cloned into pINT29, a vector that integrates into the PDC6 locus30 resulting in pID2GU. The intact PDC6 locus contains a weakly expressed pyruvate decarboxylase (PDC) gene that is not required for full PDC activity in yeast cells30,31. To prepare the pINT vector for this cloning step it was digested with BamHI and SphI and treated with T4 DNA polymerase to create blunt ends recreating the BamHI recognition site which was used to clone the UAS10-URA3-UAS7 BamHI fragment into. As a negative control, another target locus was required without the galactose inducible promoter sequences to rule out any down or upstream effects of the UAS nucleosome status on the neighboring PDC6 locus. For this, the digested PCR product of URA3 with XhoI and SpeI overhangs was cloned directly into pINT digested with SalI and SpeI. The yeast strains RSY12 (MATa leu2-3, 112 his3-11, 15 ura3D::HIS332) and YPH250 (MATα, ura3-52, lys2-801, ade2-101, trp1-Δ1, his3- Δ 200, leu2- Δ 127) were transformed with pID2GU and pIDU by LiAc/ssDNA/PEG transformation33.
The construction of the yeast target lines pID2G2SU and pID2G2LU
The plasmids pID2G2SU (pINT:PDC6:UAS10:I-SceI RS:URA3:I-SceI RS:UAS7:PDC6) and pID2G2LU (pINT:PDC6:UAS10:loxP:URA3:loxP:UAS7:PDC6) were constructed by the successive addition of two DNA fragments containing the enzyme recognition sites, directly upstream and downstream the URA3 autotrophy marker gene of pID2GU (pINT:PDC6:UAS10:URA3:UAS7:PDC6). Pairs of complementary oligos depicted in Table 2 were annealed resulting in dsDNA fragments with ss overhangs compatible with pID2GU, successively digested with XhoI and SpeI.
Table 2. Oligos used for the construction of DNA fragments added to the integrative vector pID2GU (pINT:PDC6:UAS10:URA3:UAS7:PDC6) resulting in pID2G2SU equipped with I SceI recognition sites (pINT:PDC6:UAS10:I-SceI RS:URA3:I-SceI RS:UAS7:PDC6) and pID2G2LU equipped with Lox sites (pINT:PDC6:UAS10:loxP:URA3:loxP:UAS7:PDC6).
| Plasmid | Oligo name | Nucleotide sequence (5′-3′) |
|---|---|---|
| pID2G2SU | scesalxhoFW scesalxhoRV scespexbaFW scespexbaRV | TCGACCTTAAGTTACGCTAGGGATAACAGGGTAATATAAC TCGAGTTATATTACCCTGTTATCCCTAGCGTAACTTAAGG CTAGTACGCTAGGGATAACAGGGTAATATAACGGAATTCT CTAGAGAATTCCGTTATATTACCCTGTTATCCCTAGCGTA |
| pID2G2LU | loxsalxhoFW loxsalxhoRV loxspexbaFW loxspexbaRV | TCGACATAACTTCGTATAGCATACATTATACGAAGTTATC TCGAGATAACTTCGTATAATGTATGCTATACGAAGTTATG CTAGTATAACTTCGTATAGCATACATTATACGAAGTTATT CTAGAATAACTTCGTATAATGTATGCTATACGAAGTTATA |
The yeast strains RSY1232 and YPH25027 were transformed with pID2G2SU and pID2G2LU by LiAc/ssDNA/PEG transformation33.
The construction of the binary and protein translocation vectors for Agrobacterium
Two types of vectors were used; binary vectors for the transfer of T-DNA and a set of vectors involved in protein translocation expressing chimeric DNA modifying proteins equipped with a translocation signal. For the construction of the binary vectors p14-2GKX (allowing for the translocation of T-DNA harbouring UAS10:KanMX:UAS7), and p14-PDC6KX (allowing for the translocation of T-DNA harbouring PDC6:KanMX:PDC6), the following steps were taken. A KanMX marker gene flanked with SpeI and XhoI restriction sites was generated by PCR using pSDM800016 as DNA template. This fragment was cloned into p2GU also digested with SpeI and XhoI resulting in p2GKX. The fragment containing the KanMX marker gene flanked with the UAS7 and UAS10 sequence was cloned into the binary vector pSDM1434 as a KpnI, SalI fragment resulting into p14-2GKX. The PCR primers used for the amplification of KanMX are shown in table 3. To assemble a binary vector with two PDC6 flanks homologous to the PDC6 sequences as are present in pID2GU (pINT:PDC6:UAS10:URA3:UAS7:PDC6) and its derivatives, PCR was performed using the primers depicted in Table 3 using pINT as template. The resulting PCR product was digested with BglII and cloned into BamHI digested pSDM14, resulting in p14-PDC. A KanMX marker gene with TEF1 promoter and terminator was obtained from pSDM8000 as a BamHI fragment and was cloned in p14-PDC digested with BamHI, resulting into p14-PDCKX. The binary vectors p14-2GKX and p14-PDC6KX were transferred to the Agrobacterium strain LBA1100 or LBA2556 by electroporation. LBA1100 has the C58C1 Agrobacterium chromosomal background and carries a disarmed octopine pTiB6 plasmid35. LBA2556 is a virD2 null-deletion mutant, isogenic to LBA110036.
Table 3. Oligos designed for the amplification of the PDC6 locus and the KanMX marker gene. Restriction sites added to the PCR primer sequences are underlined.
| Oligo name | Nucleotide sequence (5′-3′) |
|---|---|
| PDC6FW | TAATAGATCTAGGTAAATAAATGTGCAGATGCA |
| PDC6RV | TAATAGATCTTGAACAAAGGGCGAAACTTCGCA |
| Kan Xho | TAATCTCGAGTTTAGCTTGCCTCGTC |
| Kan Spe | TAATACTAGTTTTCGACACTGGATGG |
For the construction of all the fusion protein translocation vectors describe here, pBFF was used as the vector backbone. This non-transmissable vector was derived from the broad host-range plasmid pRL66237 and was adjusted to enable the translocation of a diversity of fusion proteins22,24. Cloning ORFs as NotI fragments into the pBFF vector puts protein expression under control of the virF promoter sequence from the octopine Ti-plasmid. When properly allowing for translational fusions, produced proteins will contain an N-terminal FLAG-tag for immunodetection, a SV40 nuclear localization signal (NLS) to ensure its nuclear entry and a 37 aa C-terminal end of VirF allowing for T4SS mediated translocation21. The vector pBFF I-SceI and pBFF VirD2 I-SceI were generated by PCR-amplifying the I-SceI coding region (kind gift of Dr. Holger Puchta, University of Karlsruhe, Germany) and cloning it into the pBFF vector, thus supplying it with the features mentioned above.
The construction of the yeast expression vectors
The yeast expression vectors were constructed by first cloning a DNA fragment encoding an ATG translational initiation codon followed by the FLAG::NLS::virFCT sequence into pRS425 ADH38 resulting into pRS425 ADH FLAG::NLS::virFCT. New ORFs were subsequently introduced as NotI fragments within the same site present directly upstream of virFCT. The vector pRS425 FLAG::NLS::virD2::I-SceI::virFCT was thus constructed by insertion of a NotI fragment from pBFF VirD2-I-SceI and pRS425 FLAG::NLS::virD2::Cre::virFCT by insertion of a NotI fragment from pBFF VirD2-CRE. The fusion proteins produced in Agrobacterium and yeast were derived from identical ORFs.
Qualitative assay for enzymatic activity of DNA modifying fusion proteins in yeast
The pRS425ADH expression vectors encoding NLS-VirD2-I-SceI-VirFCT or NLS-VirD2-CRE-VirFCT were transferred to yeast cells carrying the different target loci by LiAc-transformation and transformants were selected for leucine autotrophy. After 3 or 4 days, 50 transformed yeast cells were removed from the selective plates and pooled in 500 μl of sterile 0.9% salt solution. The OD600 of this cell suspension was adjusted to 0.1 and diluted 100-fold. Subsequently, 100 μl of this cell suspension was replated on SD medium selective for leucine and uracil autotrophy, in order to maintain yeast cells carrying an expression vector and an intact target locus. The same aliquot of cells was plated on SD medium selective for leucine autotrophy but containing uracil as well as 5-FOA. The surviving colonies represented the yeast cells that carried the expression vector but lost the uracil selection marker gene.
For the AMT of yeast cells, we followed our earlier published method39 with some modifications. Briefly, Agrobacterium cells were precultured on LB medium with appropriate antibiotic selection. The transition from medium containing glucose to medium containing galactose has no impact on the growth rate of the Agrobacterium strains described here. For reliable results, the yeast strain YPH250 and RSY12 required acclimatization to galactose before and during the preculture step for sufficient growth on IM plates with galactose as the sole carbon source. Before mixing the Agrobacterium cell suspension with the yeast cells, the Agrobacterium cells were washed with induction medium (IM) without any carbon source. All cocultivations were performed for one week at 21°C, without any antibiotic or autotrophic selection in order to avoid any experimental biases. Whenever galactose induction of the yeast cells was applied, the IM used for preculturing the yeast strains as well as the induction plates were prepared with galactose instead of glucose.
Results
Experimental setup
To get a deeper insight into the requirements of T-DNA integration by HR a versatile target locus was developed and introduced into the yeast genome. As can be seen in Figure 1, this target locus had a URA3 marker gene flanked by two 18 bp I-SceI recognition sites. To enable the modulation of the nucleosome occupancy of this target locus, two different DNA sequences, normally present upstream of galactose inducible genes of the yeast galactose metabolism pathway, were cloned adjacent to the I-SceI sites. The same sequences, UAS7 and UAS10 were also introduced into a binary vector as part of the T-DNA sequence flanking an yeast KanMX marker which has no homology to the yeast genome. During Agrobacterium mediated transformation (AMT), a T-strand carrying these sequences will enter recipient yeast cells. With the UAS7 and UAS10 sequences, the T-strand can interact with the homologous chromosomal sequences at the target locus. Regarding the target locus, such an interaction can result in URA3 for KanMX marker exchange by a two-sided HR event. As for the UAS7 and UAS10 sequences, their nucleosome occupancy is known to be reduced in the presence of galactose, and increased in the presence of glucose, resulting in increased and reduced expression of downstream ORFs, respectively. Detailed nuclease protection patterns for the sequences derived from yeast growing on galactose or glucose have become available26. The wide application of UAS sequences for carbon-source dependent gene expression provides compelling evidence that their characteristics, including the nucleosome occupancy, are portable traits.
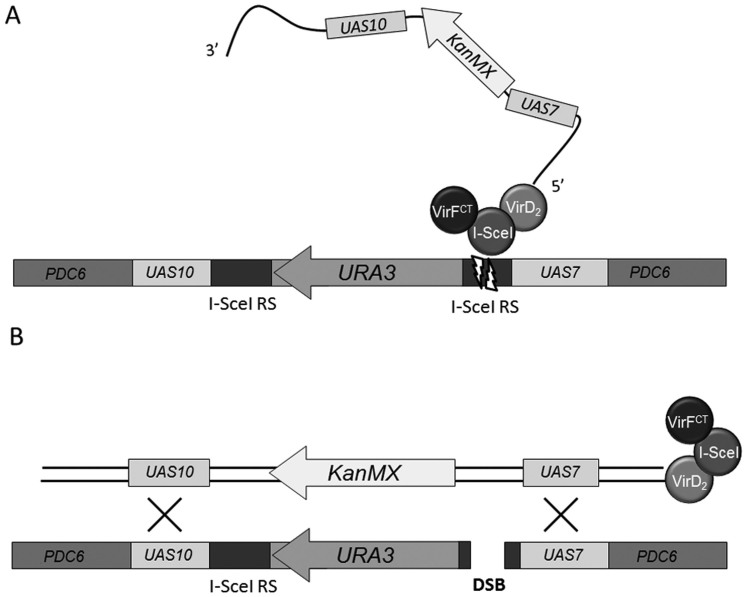
Figure 1. Experimental set-up for the study of T-DNA integration via HR by the I-SceI mediated DSB formation.
(1A) The T-strand entering yeast cells is covalently attached to a fusion protein consisting of an FLAG-NLS module (not depicted in the figure), VirD2, I-SceI, and an Agrobacterium translocation signal derived from the VirF C-terminus. The I-SceI moiety induces the formation of a DSB at one or both the I-SceI recognition sites flanking the URA3 gene, which triggers DNA recombination. (1B) Precise HR of the target locus with the incoming T-strand results in the exchange of the URA3 autotrophy marker with the KanMX marker gene leading to G418 resistance as well as uracil auxotrophy. The regions on the T-strand homologous to the UAS10 and UAS7 sequences in the target locus are approximately 700 bp in length.
In order to increase HR at the target locus, DSBs can be induced by I-SceI at the cognate restriction sites. We tested whether such DSBs can be induced efficiently by I-SceI translocated from Agrobacterium as a VirD2-I-SceI-VirFCT fusion protein that is covalently attached to the translocated T-strand (Figure 1A). This protein contains an N-terminal FLAG tag followed by an SV40 nuclear localization signal (NLS) to ensure its nuclear entry as do the other fusion proteins used in this study. Since the full length VirD2 protein contains an active NLS sequence, an extra NLS is not really required24. For the I-SceI-VirFCT protein, an active NLS is a prerequisite. Expression in Agrobacterium was mediated by the virF promoter.
The I-SceI moiety should induce one or more DSBs at the target locus, thereby enhancing the integration of T-DNA mediated by the HR pathway. Since the Holliday junctions involved in crossing over events during HR pathway require two dsDNA molecules, marker exchange is depicted as occurring between two double stranded DNA molecules (Figure 1B), thus as if the incoming T-strand has formed a complementary strand prior to integration. Incoming T-strands did not contain a nuclease target site.
The target locus depicted in Figure 1, as well as those shown in Figure 2, were integrated in the genomic PDC6 locus. By performing the AMT of these yeast target lines on medium containing galactose or glucose, the respective removal or formation of nucleosomes can be induced and their influence on the integration efficiency can be assessed. For control experiments, alternative T-DNA regions were constructed, aimed to generate T-strands just representing the KanMX marker gene, thus without flanking UAS regions or to possess similarly sized flanking PDC6 sequences instead of UAS regions (see Table 4).
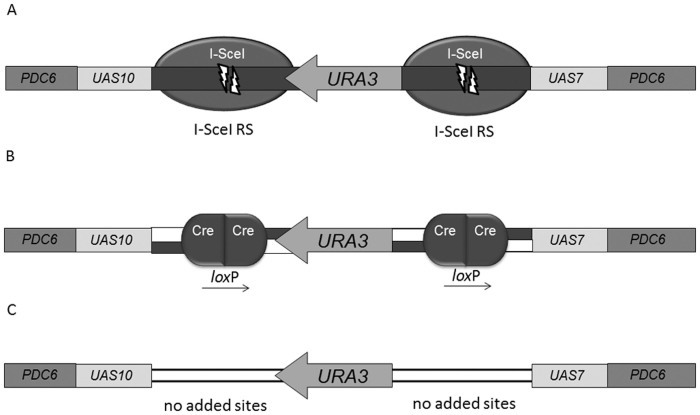
Figure 2. Target loci used to assay the effect of DSBs and nucleosome occupancy on the integration of T-DNA by HR.
These target loci were all integrated at the PDC6 locus, all containing sequence homology to the incoming T-strands by two different UAS type promoters derived from the yeast galactose pathway or by the flanking PDC6 fragments. To remove nucleosomes from these promoter sequences, the yeast cells are grown in a medium with galactose as the sole carbon source. Growth in a glucose medium would induce the formation of nucleosomes on the UAS typed promoters. The central URA3 marker is flanked by: (2A) recognition sites of the homing endonuclease I-SceI that catalyzes the formation of DSBs as a monomer, (2B) recognition sites for the recombinase Cre allowing for the removal of the URA3 marker gene without inducing double stranded break formation, (2C) target locus without any added recognition sites but still provided with homology to the incoming T-strands.
Table 4. Overview of all the constructs mentioned in the experimental setup.
| Abbreviation | Name | Description | Vector type, organism |
|---|---|---|---|
| pIDU | pSDM3585 | pINT:PDC6:URA3:PDC6 | Integrative vector, yeast |
| pID2GU | pSDM3588 | pINT:PDC6:UAS10:URA3:UAS7:PDC6 | Integrative vector, yeast |
| pID2G2LU | pSDM3586 | pINT:PDC6:UAS10:loxP:URA3:loxP:UAS7:PDC6 | Integrative vector, yeast |
| pID2G2SU | pSDM3587 | pINT:PDC6:UAS10:I-SceI RS:URA3:I-SceI RS:UAS7:PDC6 | Integrative vector, yeast |
| pRS425 VirD2-I-SceI | pSDM3592 | pRS425:pADH:NLS::virD2::I-SceI::virFCT | Expression vector, yeast |
| pRS425 VirD2-Cre | pSDM3591 | pRS425:pADH:FLAG::NLS::virD2::Cre::virFCT | Expression vector, yeast |
| pBFF I-SceI | pSDM8017 | pBFF:FLAG::NLS::I-SceI::virFCT | Expression vector, Agrobacterium |
| pBFF VirD2-I-SceI | pSDM3884 | pBFF:FLAG::NLS::virD2::I-SceI::virFCT | Expression vector, Agrobacterium |
| pBFF Cre | pSDM8019 | pBFF:FLAG::NLS::Cre::virFCT | Expression vector, Agrobacterium |
| pBFF VirD2-Cre | pSDM8020 | pBFF:FLAG::NLS::virD2::Cre::virFCT | Expression vector, Agrobacterium |
| p14-PDC6KX | pSDM3590 | pSDM14:rightborder:PDC6:KanMX:PDC6:leftborder | Binary vector, Agrobacterium |
| P14-2GKX | pSDM3589 | pSDM14:rightborder:UAS7: KanMX:UAS10:leftborder | Binary vector, Agrobacterium |
| pSDM8000 | pSDM14:rightborder:KanMX:leftborder16 | Binary vector, Agrobacterium |
For convenience, the constructs were given a short working name. All uninterrupted ORFs in the description are depicted in bold writing. The abbreviation “I-SceI-RS” indicates recognition sites for I-SceI.
AMT experiments with T-strands covalently attached to VirD2 containing fusion proteins, such as FLAG-NLS-VirD2-I-SceI-VirFCT, were performed using Agrobacterium strain LBA255636 containing a Ti plasmid deleted for the virD2 gene. For T-strand transfer combined with transfer of non-VirD2 containing proteins, such as FLAG-NLS-I-SceI-VirFCT, the isogenic strain LBA1100 was used. In this strain, a wild type VirD2 protein is still produced.
In addition to a target locus that is substrate for I-SceI (Figure 2A), similarly designed target loci were prepared harboring recognition sites of the recombinase Cre (Figure 2B) and a negative control locus lacking any particular recognition sites (Figure 2C). A complete list of all gene constructs used for the experiments and their abbreviations is presented in Table 4.
Enzyme activity of I-SceI and Cre fusion proteins
To test if the expressed fusions proteins were able to process the target loci described in Figure 2, an in vivo assay was designed. For this, the ORFs of the VirD2 fusion proteins were transferred to the yeast constitutive expression vector pRS425ADH equipped with a leucine autotrophy marker38. This resulted in pRS425ADH expressing NLS-VirD2-I-SceI-VirFCT (pRS425 VirD2-I-SceI) and NLS-VirD2-Cre-VirFCT (pRS425 VirD2-Cre) to serve as a positive control. These vectors were transferred to yeast cells carrying the corresponding target loci (derived from pID2G2SU and pID2G2LU respectively), or a negative control without any cognate recognition sites (derived from pID2GU). Fifty yeast colonies resulting from transformation with either of these expression vectors were pooled. An aliquot containing at least 1000 suspended yeast cells was plated on selective medium without leucine and uracil and on medium supplemented with 5-Fluoroorotic Acid (5-FOA) and uracil in order to determine the fraction of yeast cells that had lost the URA3 marker, rendering them 5-FOA resistant. Although the described assay did not provide quantitative data on enzyme activity, it did show that the two VirD2 fusion proteins were able to process their cognate target loci but not the negative control target locus without recognition sites. The expression of I-SceI fusion proteins resulted in a complete loss of the URA3 marker gene, Cre fusion proteins removed this marker in 88% of the cells.
The effect of nucleosome occupancy on AMT transformation efficiency
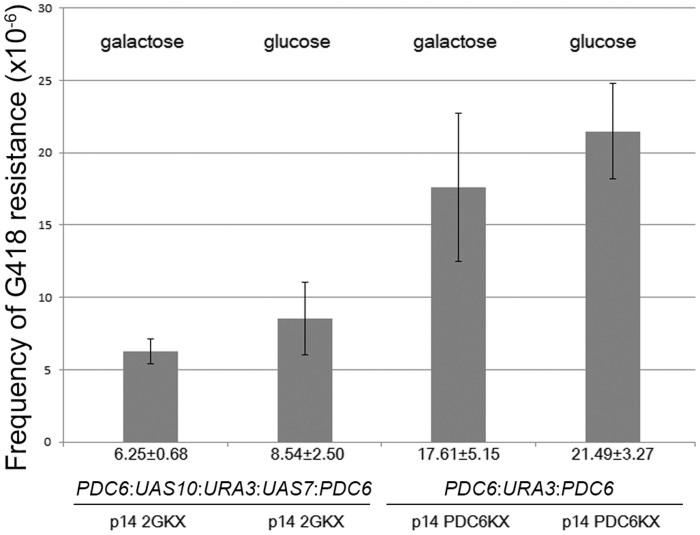
The yeast strain RSY12 harboring a pID2GU target locus (PDC6:UAS10:URA3:UAS7:PDC6) was cocultivated with an Agrobacterium strain translocating T-DNA from the binary vector p14-2GKX (pSDM14:rightborder:UAS7: KanMX:UAS10:leftborder). This T-DNA can integrate via HR at the UAS fragments present in the pID2GU target locus. Cocultivations on medium containing glucose or galactose, that should modulate the nucleosome occupancy of the UAS promoter sequences present at the target locus, resulted in similar transformation efficiencies. From these results it could be concluded that the presence or absence of nucleosomes at the UAS containing area of the target locus had no measurable effect on the efficiency of T-DNA integration (Figure 3, bar one and two). The same was observed for a negative control containing the pIDU reporter locus (PDC6:URA3:PDC6), that lacked the UAS promoter sequences, which was targeted by homologous T-DNAs derived from p14-PDC6KX which enables translocation of T-DNA with PDC6:KanMX:PDC6 (Figure 3, bar three and four). From this result it could be concluded that there was also general effect of the different carbon sources on the transformation efficiencies.
Figure 3. Effects of carbon source on AMT efficiency using yeast strain RSY12.
Bar one and two represent RSY12 with the pID2GU target locus (PDC6:UAS10:URA3:UAS7:PDC6) cocultivated with LBA1100 harboring p14-2GKX. Bar three and four represent yeast cells with the pIDU target locus (PDC6:URA3:PDC6), lacking the UAS promoter sequences, cocultivated with LBA1100 harboring p14-PDC6KX (pSDM14:rightborder:PDC6:KanMX:PDC6:leftborder). Every bar represents the average of 10 independent experiments (n = 10). The transformation efficiency as is shown at the Y-axes should be read as ‘per million’ as is indicated with (×10−6). Error bars indicate the SEM. The hypothesis of equality could not be rejected with α set to 0.05 when performing a two-tailed heteroscedastic Student's T-test comparing “galactose” with “glucose”.
A comparison of two approaches to induce DSBs to affect T-DNA integration via HR
Using the yeast strain RSY12 harboring the target locus depicted in Figure 1, it was assessed whether it would be possible to obtain evidence for a stimulation of gene targeting efficiency by delivery of nuclease activity during AMT. The first approach was based on translocation of T-strands piloted by a chimerical NLS-VirD2-I-SceI-VirFCT protein using an Agrobacterium strain with a virD2 deletion (LBA2556), the second one on T-strand delivery via an isogenic Agrobacterium strain containing a WT VirD2 locus (LBA1100) concomitant with transfer of NLS-I-SceI-VirFCT. Apart from the plasmids expressing the proteins of interest, both strains contained the binary vector p14-2GKX (pSDM14:rightborder:UAS7: KanMX:UAS10:leftborder). T-DNA derived from p14-2GKX can integrate into the yeast genome via the HR pathway at the target locus consisting of UAS7:I-SceI RS:URA3:I-SceI RS:UAS10. The translocated I-SceI fusion proteins might enhance this by inducing the formation of DSBs at the target locus.
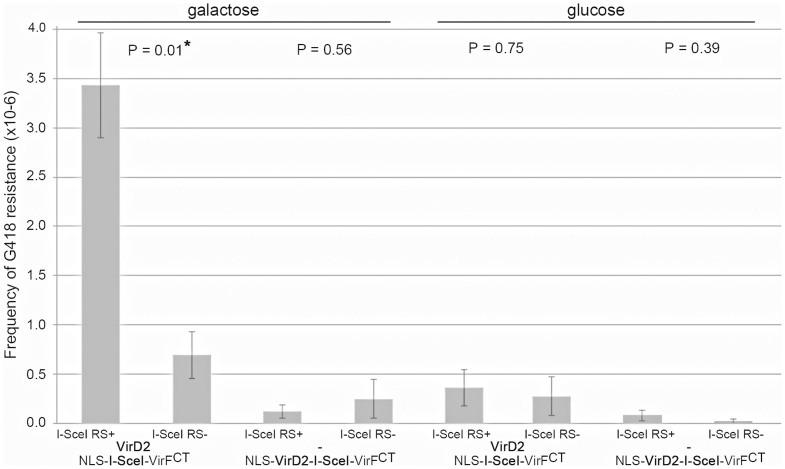
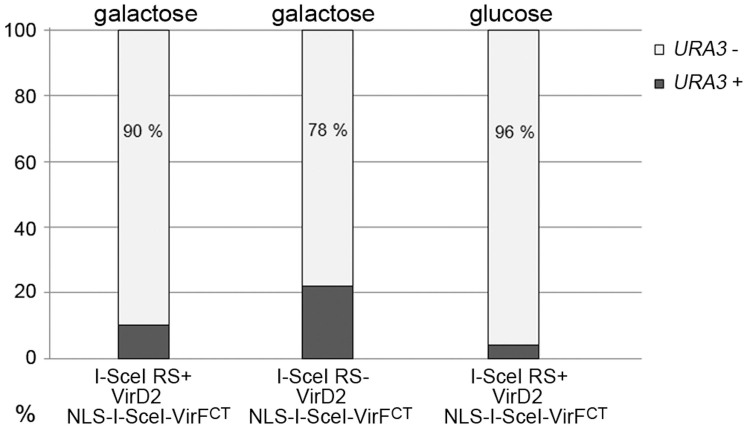
The observed AMT frequencies are depicted in Figure 4. As can be seen in this figure, the usage of a yeast strain harboring a target locus with two I-SceI restriction sites flanking the URA3 marker gene did not lead to an increase in transformation efficiency using LBA2556 harboring p14-2GKX (which delivers T-DNA with UAS7: KanMX:UAS10) and pBFF VirD2-I-SceI. A different result was obtained when these yeast cells were cocultivated with LBA1100 harboring p14-2GKX and pBFF I-SceI. Here, WT VirD2 directed the translocation of the T-strand while NLS-I-SceI-VirFCT was translocated separately to induce DSBs at the target locus. Now a significantly higher transformation efficiency was obtained when I-SceI recognition were present sites at the target locus (Figure 5, first two bars).
Figure 4. Influence of galactose or glucose treatment on the I-SceI assisted integration of T-DNA by HR.
The yeast strain RSY12 was cocultivated with LBA1100 harboring p14-2GKX (pSDM14:rightborder:UAS7: KanMX:UAS10:leftborder) and pBFF I-SceI or LBA2556 p14-2GKX and pBFF VirD2-I-SceI. Different yeast strains with and without I SceI recognition sites between the uracil autotrophy marker and the UAS promoter sequences making up the target locus (Figure 2A, 2C) were compared, here indicated as I-Sce RS+ and I-SceI RS−. A horizontal line is used to mark the presence of galactose (inducing nucleosome removal UAS regions at the target locus) or glucose (inducing nucleosome formation). Every bar represents that average of four independent experiments (n = 4). The p-values were calculated performing a two-tailed heteroscedastic Student's T-test. Error bars indicate the SEM. The asterisk indicates statistical significance with α = 0.05. The transformation frequencies were calculated by dividing the total number of G418 resistant colonies by an estimate of the total number of yeast cells.
Figure 5. Assay on uracil auxotrophy of G418 resistant yeast transformants.
As a consequence of the integration of the T-DNA (UAS10:kanMX:UAS7) mediated by HR, the uracil autotrophy marker of the target locus can be replaced by the KanMX marker gene leading to Ura auxotrophy (here indicated as URA3−). The off-target integration of the T-DNA leaves the URA3 gene intact (here indicated as URA3+). Each bar represents the average of fifty G418 resistant colonies assayed for growth on SD-ura.
Interestingly, this positive effect of translocated NLS-I-SceI-VirFCT on T-DNA integration was only found when galactose had been present during the cocultivation period but not when glucose was added to the induction medium instead. This observation suggests that the I-SceI nuclease gains much better access to its recognition sites in the genome if nucleosomes are absent due to cultivation on galactose medium. The formation of DSBs in turn enhances T-DNA integration at the target locus. Reversely, addition of glucose apparently leads to reduced accessibility of the target locus for the I-SceI fusion protein.
To verify that the majority of the integration events indeed took place at the target locus, G418 resistant colonies were assessed for loss of uracil autotrophy. As shown for colonies resulting from three different cultivations (Figure 5), the majority of these cells had lost the URA3 marker, indicative of HR-mediated gene replacement. The few colonies that retained uracil autotrophy possibly represent single cross-overs at the target locus or at native UAS7 or UAS10 promoter sequences that are part of the cognate GAL genes.
Discussion
Using a yeast model system, we investigated possible effects of different chromatin states on gene targeting by means of Agrobacterium mediated transformation (AMT). We also assessed the contribution of co-delivered fusion proteins containing I-SceI nuclease domains to facilitate targeted integration of T-DNA during the AMT. Based on our findings, we conclude that enhancing or decreasing nucleosome occupancy of a target locus has no significant impact on the integration efficiency in the absence of a site specific nuclease. For T-DNA integration, the presence of nucleosomes might not be limiting because T-DNA tends to integrate at sites where the genomic DNA is broken. In these situations the DNA repair mechanisms are likely to remove the nucleosomes during the process of DNA repair. However, T-DNA integration at intended target sites was clearly enhanced by simultaneous transfer of I-SceI nuclease to galactose-grown yeast cells. This effect might very well be attributed to galactose-induced removal of nucleosomes from the nuclease target sites, thus making them more accessible for the nuclease to induce the double stranded breaks (DSBs) that are required for enhancement of the T-DNA integration efficiency. However, it cannot be excluded that other changes occurring during growth on galactose containing medium might play a role as well. In any case, the obtained results clearly demonstrated that the combined translocation of nuclease proteins and T-strands during AMT can facilitate HR mediated integration of T-DNA.
After evaluating the performance of the experimental tools that were generated, experiments were performed to determine if the nucleosome occupancy of a target locus would have an impact on the efficiency of T-DNA integration via the HR pathway. As described above, we used promoter sequences from genes that are part of the yeast galactose pathway as the regions homologous to the incoming T-strand at the target locus. The nucleosome occupancy of the UAS type promoter sequences used has been well characterized. For UAS10 it has been shown that galactose-induced association of the transcriptional activator Gal4, rapidly recruits the nucleosome remodeling complex SWI/SNF which then removes nucleosomes from this promoter26. In another study, a Gal4 binding site was transferred to a histidine promoter located on a yeast episome40. On galactose-induced expression of Gal4, nucleosomes adjacent to the Gal4 binding site were removed. These findings demonstrate that Gal4-mediated nucleosome removal was independent of the sequences flanking the Gal4 binding site. Concerning the studies mentioned above, it should be safe to assume that the unaltered UAS promoters used still exhibit the reported galactose- and glucose dependent regulation of nucleosome occupancy.
Our experiments showed that HR-mediated integration of T-DNA at the target locus was independent of the presence or absence of nucleosomes or other effects that a change of carbon source might have. This is an intriguing observation since for yeast cells transformed with linear dsDNA using a LiAc/ssDNA/PEG transformation protocol, the presence of nucleosomes at the genomic locus carrying sequence homology to the incoming DNA fragment is known to negatively affect the transformation efficiency41. Our findings however are in line with earlier research showing that integrated T-DNAs are not enriched at genomic sites that have a methylation status indicative for transcriptional activity10.
The results regarding the effects of nuclease proteins translocated concomitantly with T-strands into yeast cell showed that of the proteins tested, translocation of NLS-I-SceI-VirFCT clearly enhanced T-DNA integration, but only when I-SceI recognition sites were present at the target locus (Figure 4). The majority of the integration events were correctly targeted and the presence of I-SceI recognition sites at the target locus even led to a slight increase in correctly targeted integration events (Figure 5). These data provide evidence that it is indeed feasible to translocate nucleases simultaneously with T-strands via the T4SS of Agrobacterium to introduce DSBs in the host genome during in order to enhance gene targeting. The improvements regarding gene targeting perhaps remained relatively modest because a yeast model was used that already has a very effective HR pathway. However, in systems where the HR mechanism is much less proficient, delivery of targetable nuclease proteins might very well serve to improve gene targeting. Procedures used thus far to improve T-DNA targeting, have always relied on the introduction of gene constructs encoding nucleases of interest, but our methodology of “protein therapy” in combination with a gene targeting construct has the advantage that no extra nucleic acids need to be introduced besides the gene targeting construct.
Strikingly, as can be observed in Figure 4, the enhanced transformation efficiency that was obtained by the Agrobacterium mediated translocation of NLS-I-SceI-VirFCT was abolished if glucose was present in the medium instead of galactose. As is illustrated in Figure 3, the presence of galactose or glucose had no effect on the transformation efficiency when the T-DNA was homologous to the PDC6 locus. Since the PDC6 locus is not part of the galactose pathway and the nucleosome occupancy should therefore not be affected by the carbon source, this experiment excluded a general effect of the utilized carbon source on the T-DNA integration frequency. Therefore, although additional changes might have occurred, repression of AMT frequency in the presence of glucose is likely to be indicative of glucose induced nucleosome formation at the UAS target sites, thereby reducing the availability of the I-SceI binding sites for digestion by NLS-I-SceI-VirFCT. The consequent reduction in DSB formation at the target locus would then account for the reduced transformation efficiency.
While an in vivo assay demonstrated that NLS-VirD2-I-SceI-VirFCT still possessed nuclease activity, attempts to use this chimerical protein for T-strand delivery as well as DSB induction resulted in very low transformation efficiencies, yielding too few yeast transformants to give a reliable estimate of the T-DNA targeting efficiency (Figure 4). The observed low transformation efficiency could be due to reduced T4SS mediated translocation of NLS-VirD2-I-SceI-VirFCT compared to WT VirD2 or to reduced relaxase activity of VirD2 when fused to other protein moieties. Both issues would lead to less T-strands being transferred, resulting into lower transformation efficiencies. However, the chimerical VirD2 proteins can still support enzyme activity for DNA metabolism as shown for the ability of NLS-VirD2-I-Cre-VirFCT to support in vivo Cre-mediated recombination on a yeast target locus. In parallel experiments performed in plant cells, it was shown that also NLS-VirD2-I-SceI-VirFCT can be transferred during AMT, while retaining nuclease activity23. Therefore, unless bottlenecks can be adequately addressed, the collected data indicate that the most effective approach to transfer nuclease proteins to induce DSBs concomitant with AMT is the separate translocation of the nuclease proteins rather than constructing more complex VirD2 fusion proteins. When only assessing the transfer of a very similar NLS-Cre-VirFCT protein to yeast cells containing a loxP flanked locus, it was previously found that up to 1% of yeast target cells exhibited the recombined genotype after AMT22.
In summary, the data obtained using the yeast model system for AMT indicated that Agrobacterium mediated translocation of the homing endonuclease I-SceI has a positive effect on the integration of T-DNA via the homologous repair (HR) pathway. We found no evidence that the nucleosome occupancy status of the future integration site had an impact on the transformation efficiency. However, we did observe that the presence of glucose, that is known to lead to an elevation of the nucleosome occupancy of the UAS sequences present at the target locus, removed the positive effect of Agrobacterium mediated transfer of specific nucleases on T-DNA integration at the target locus. For an optimal contribution of DSB induction by specific nucleases to gene targeting, it is apparently pivotal to take the nucleosome occupancy of the intended target site into account.
Acknowledgments
This work was supported by the Top grant Chemical Sciences programme of the Netherlands Organization for Scientific Research (700.56.303) and by the Top Technology Institute for Green Genetics (4CC057RP).
Footnotes
Author Contributions M.R. performed all experiments and wrote the main manuscript. B.Z. and M.R. conceived the experiments. B.Z. and P.H. reviewed the manuscript.
References
- Yadav N. S., Vanderleyden J., Bennett D. R., Barnes W. M. & Chilton M. D. Short direct repeats flank the T-DNA on a nopaline Ti plasmid. Proc Natl Acad Sci U S A 79, 6322–6326 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrenberger F., Crameri A., Hohn B. & Koukolikova-Nicola Z. Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc Natl Acad Sci U S A 86, 9154–9158 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper F., Koncz C., Schell J. & Steinbiss H. H. Agrobacterium T-strand production in vitro: sequence-specific cleavage and 5' protection of single-stranded DNA templates by purified VirD2 protein. Proc Natl Acad Sci U S A 91, 694–698 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. E. et al. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J Biol Chem 263, 5804–5814 (1988). [PubMed] [Google Scholar]
- Tinland B. The integration of T-DNA into plant genomes. Trends Plant Sci. 1, 178–184 (1996). [Google Scholar]
- Chilton M. D. & Que Q. Targeted integration of T-DNA into the tobacco genome at double-stranded breaks: new insights on the mechanism of T-DNA integration. Plant Physiol 133, 956–965, 10.1104/pp.103.026104 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T., Frankman L. R., Vaidya M. & Citovsky V. Site-specific integration of Agrobacterium tumefaciens T-DNA via double-stranded intermediates. Plant Physiol 133, 1011–1023, 10.1104/pp.103.032128 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon S. & Puchta H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. The EMBO journal 17, 6086–6095, 10.1093/emboj/17.20.6086 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z. & Tzfira T. In vivo formation of double-stranded T-DNA molecules by T-strand priming. Nat Commun 4, 2253, 10.1038/ncomms3253 (2013). [DOI] [PubMed] [Google Scholar]
- Kim S. I., Veena & Gelvin S. B. Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J 51, 779–791 (2007). [DOI] [PubMed] [Google Scholar]
- Offringa R. et al. Extrachromosomal homologous recombination and gene targeting in plant cells after Agrobacterium mediated transformation. The EMBO journal 9, 3077–3084 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H. Gene replacement by homologous recombination in plants. Plant Mol Biol 48, 173–182 (2002). [PubMed] [Google Scholar]
- Puchta H., Dujon B. & Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res 21, 5034–5040 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto J., Molina R. & Montoya G. Molecular scissors for in situ cellular repair. Crit Rev Biochem Mol Biol 47, 207–221, 10.3109/10409238.2011.652358 (2012). [DOI] [PubMed] [Google Scholar]
- Ozawa K. A high-efficiency Agrobacterium-mediated transformation system of rice (Oryza sativa L.). Methods Mol Biol 847, 51–57, 10.1007/978-1-61779-558-9_5 (2012). [DOI] [PubMed] [Google Scholar]
- van Attikum H., Bundock P. & Hooykaas P. J. Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. The EMBO journal 20, 6550–6558, 10.1093/emboj/20.22.6550 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A. et al. Suppression of Ku70/80 or Lig4 leads to decreased stable transformation and enhanced homologous recombination in rice. New Phytol 196, 1048–1059, 10.1111/j.1469-8137.2012.04350.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y. et al. Increasing frequencies of site-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNA repair pathways. Genome Res 23, 547–554, gr.145557.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. Q. et al. Regulation of an endogenous locus using a panel of designed zinc finger proteins targeted to accessible chromatin regions. Activation of vascular endothelial growth factor A. J Biol Chem 276, 11323–11334, 10.1074/jbc.M011172200 (2001). [DOI] [PubMed] [Google Scholar]
- Zhang L. et al. Synthetic zinc finger transcription factor action at an endogenous chromosomal site. Activation of the human erythropoietin gene. J Biol Chem 275, 33850–33860, 10.1074/jbc.M005341200 (2000). [DOI] [PubMed] [Google Scholar]
- Vergunst A. C. et al. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci U S A 102, 832–837 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrammeijer B., den Dulk-Ras A., Vergunst A. C., Jurado Jacome E. & Hooykaas P. J. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res 31, 860–868 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kregten M., Hooykaas P. J. & van der Zaal B. J. Agrobacterium-mediated delivery of a meganuclease into target plant cells. PhD thesis Leiden University (2011). [Google Scholar]
- van Kregten M., Lindhout B. I., Hooykaas P. J. & van der Zaal B. J. Agrobacterium-mediated T-DNA transfer and integration by minimal VirD2 consisting of the relaxase domain and a type IV secretion system translocation signal. Mol Plant Microbe Interact 22, 1356–1365, 10.1094/MPMI-22-11-1356 (2009). [DOI] [PubMed] [Google Scholar]
- van Attikum H. & Hooykaas P. J. Genetic requirements for the targeted integration of Agrobacterium T-DNA in Saccharomyces cerevisiae. Nucleic Acids Res 31, 826–832 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant G. O. et al. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS Biol 6, 2928–2939, 08-PLBI-RA-3549 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S. & Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuteboom L. W., Lindhout B. I., Saman I. L., Hooykaas P. J. & van der Zaal B. J. Effects of different zinc finger transcription factors on genomic targets. Biochem Biophys Res Commun 339, 263–270, 10.1016/j.bbrc.2005.11.011 (2006). [DOI] [PubMed] [Google Scholar]
- Meijer A. H., Ouwerkerk P. B. & Hoge J. H. Vectors for transcription factor cloning and target site identification by means of genetic selection in yeast. Yeast 14, 1407–1415, (1998). [DOI] [PubMed] [Google Scholar]
- Hohmann S. Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J Bacteriol 173, 7963–7969 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S. PDC6, a weakly expressed pyruvate decarboxylase gene from yeast, is activated when fused spontaneously under the control of the PDC1 promoter. Curr Genet 20, 373–378 (1991). [DOI] [PubMed] [Google Scholar]
- Schiestl R. H. & Petes T. D. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 88, 7585–7589 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., Willems A. R. & Woods R. A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360, 10.1002/yea.320110408 (1995). [DOI] [PubMed] [Google Scholar]
- Offringa R. Homologous recombination between T-DNAs after co-introduction into tobacco protoplasts via Agrobacterium. Thesis Chapter 2, 29–47 (1992). [Google Scholar]
- Beijersbergen A., Dulk-Ras A. D., Schilperoort R. A. & Hooykaas P. J. Conjugative Transfer by the Virulence System of Agrobacterium tumefaciens. Science 256, 1324–1327, 256/5061/1324 (1992). [DOI] [PubMed] [Google Scholar]
- Jurado-Jacome E. Translocation of the Agrobacterium tumefaciens VirD2 and VirE3 proteins into eukaryotic host cells. Thesis Chapter 2, 39–60 (2003). [Google Scholar]
- Vergunst A. C. et al. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290, 979–982 (2000). [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller R. & Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122, 0378111995000377 (1995). [DOI] [PubMed] [Google Scholar]
- Bundock P., den Dulk-Ras A., Beijersbergen A. & Hooykaas P. J. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. The EMBO journal 14, 3206–3214 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L. & Morse R. H. Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol Cell Biol 19, 5279–5288 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslankoohi E. et al. Nucleosomes affect local transformation efficiency. Nucleic Acids Res 40, 9506–9512, 10.1093/nar/gks777 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]