Abstract
BMP15 (Bone morphogenetic protein 15) is an oocyte-secreted growth factor required for ovarian follicle development and ovulation in mammals, but its effects on reproduction in chickens are unclear. In this study, the association between BMP15 polymorphisms and reproduction traits were analyzed, and its expression characteristics in different tissues were explored in LaiWu Black chickens. Three single nucleotide polymorphisms (SNPs) were identified in four hundred LaiWu Black chickens. One SNP (NC_006091.3:g.1773T>C) located in exon 2 which was significantly associated with egg weight at first egg (EWFE) (P = 0.0389), was novel. Diplotypes based on the three SNPs were found to be significantly associated with egg weight at age of 43W (EW43) (P = 0.0058). The chickens with H3H3 diplotype had their first egg 0.57 days later than chickens with H5H5 diplotype and 1.21 days-3.96 days earlier than the other five diplotype chickens. The egg production at age of 43W (E43), egg production at age of 46W (E46) and egg production at age of 48W (E48) for chickens with H3H3 diplotype were the highest among all the chickens, and the E48 of chickens with H3H3 diplotype had 11.83 eggs higher than chickens with H1H5 diplotype. RT-qPCR results showed that the expression level of BMP15 gene in ovarian follicle was in the order of 4 mm>6 mm -8 mm> 15 mm -19 mm> 23 mm -29 mm > 33 mm -34 mm in diameter. The mRNA level in follicles of 4 mm and 6–8 mm in diameter were significantly higher than that in the other follicles (P<0.01). In the same week, the highest mRNA level was found in the ovary, and it was significantly different from that found in the liver and oviduct (P<0.01). Our results indicate that BMP15 plays a vital role in the development of ovary and follicles, especially in the development of primary follicles. H3H3 may be an potential advantageous molecular marker for improving reproduction traits in chickens.
Introduction
Bone morphogenetic protein 15 (BMP15) is a member of bone morphogenetic proteins (BMPs), which are multi-functional growth factors that belong to the transforming growth factor beta (TGF-beta) superfamily [1,2]. BMPs play critical roles in heart, neural, cartilage development, bone formation, and ovarian follicle development [3,4,5].
There is extensive evidence that BMP15 regulates granulosa cells proliferation and differentiation, ovarian folliculogenesis and appears to be crucial for female reproduction in mammals [6–9]. Mouse BMP15 is specifically expressed in the oocyte, beginning at the one-layer primary follicle stage and continuing through ovulation [10]. Knockout mouse technology also confirms the function of BMP15. BMP15 knockout female mice (Bmp15(-/-)) are sub fertile with decreased ovulation and fertilization rates [11]. Immature transgenic mice that overexpress BMP-15 exhibited accelerated follicle growth with decreased primary follicles and an increase in secondary follicles. Adult mice had normal litter sizes but an increased number of atretic antral follicles [12]. BMP15 is also found to be a pivotal factor inducing cumulus cell expansion [13]. BMP15 stimulates the expression of EGF-like growth factors in mouse cumulus cells as well as series of molecules downstream of EGF-like growth factor signaling, which are necessary for normal cumulus expansion [14]. BMP15 expression pattern is associated specifically with the period of cumulus cell expansion during in vitro maturation of buffalo [15]. The most important function of BMP15 for ovarian follicular development in sheep [16], zebrafish [17], cattle [18], human [19] and pig [20,21] have also been confirmed. Untill now, eight genetic mutations with major effects on ovulation rate and litter size have been identified in BMP15. They are FecXI in Inverdale sheep, FecXB in Belclare sheep, FecXG in Cambridge sheep, FecXH in Hanna sheep, FecXL in Lacaune sheep, FecXR in Rasa Aragonesa sheep and FecXGr FecX° in the Grivette and Olkuska sheep. However, these mutations are not applicable to all breeds of sheep [22–27].
In chicken, BMP15 is found to be preferentially expressed in the ovary, low in the brain, but not found in other tissues. The BMP15 expression was maintained during hierarchical follicular maturation in the germinal disc region and then progressively decreased after ovulation [28]. Despite identifying BMP15 biological functions in mammals and other animals, its biological functions in chicken are still unclear. Also, nothing is known about its genetic mutations in chicken. Hence, this study was designed to analyze the expression characteristics of BMP15 gene in different tissues, discover the potential molecular genetic markers that are related to reproduction traits in LaiWu Black chicken, and explore the biological effect of BMP15 on chicken reproduction traits.
Materials and Methods
Ethics Statement
All experiments were approved by the Animal Care Committee of the Academy of Agricultural Sciences, Shandong Province, Ji’nan, China. The care and use of experimental animals were carried out in accordance with the Directory Proposals on the Ethical Treatment of the Experimental Animals, established by the Ministry of Science and Technology (Beijing, China).
Animals and Reproduction traits
Four hundred LaiWu Black chickens were used for the detection of SNPs in BMP15 and the analysis of the relationship between the SNPs and the reproduction traits. All the birds were selected randomly from the conservation population maintained by Local Breed Genetic Resources Bank of Shandong Province. All birds were hatched on the same day, reared in a stair-step cage under the same nutritional and environmental conditions, and transferred later to the single cage at the age of 100 d. Six reproduction traits were measured according to The Poultry Production Performance Terms and Measurement Statistics Method (NY/T823-2004). These traits include age at first egg (AFE), egg weight at first egg (EWFE), egg weight at age of 43W (EW43), egg production at age of 43W (E43), egg production at age of 46W (E46) and egg production at age of 48W (E48). Venous blood samples were collected from all four hundred LaiWu Black chickens by venipuncture. The genomic DNA was isolated with the TIANamp Blood DNA Kit (DP318, Tiangen, Beijing, China) according to the manufacturer’s instructions, and then stored at -20°C for genotype.
Five LaiWu Black chickens were randomly selected for the tissue sampling of the liver (L), oviduct (S) and ovary (O) at the age of 19W, 23W and 40W, respectively. Also, all levels of follicles were obtained at the age of 40W. The follicles were classified according to Onagbesan (2009) [29] as F1 (33mm-34mm diameter), F2/F3 (23mm-29mm diameter), F4/F5 (9mm-15mm diameter), yellow follicles (6mm-8mm diameter) and white follicles (4mm diameter). Total RNA was isolated from the tissues using Ultrapure RNA Kit (CW0581, CWbio.Co.Ltd, Beijing, China) according to the manufacturer’s instructions, and then stored at -80°C for tissue expression analysis.
Detection of SNPs, Genotype, and Construction of haplotypes and diplotypes
Six DNA pools based on 180 samples were constructed to detect the genetic mutations of BMP15. Each pool was made up of 30 DNA samples in equal concentration (5 μL of genomic DNA (50ng/μL) for each sample). According to the CDS sequence of BMP15 (GenBank Accession no. NC_006091.3, GI: 358485508), two sets of PCR primers (BMP15P1 and BMP15P2, Table 1) were designed with the Primer Premier5.0 to amplify the DNA pool for the identification of SNPs in BMP15. The PCR was performed in 15 μL volume system containing 1 μL of genomic DNA (50ng/μL), 0.45 μL of each primer (10pmol/μL), 5.6 μL ddH2O, and 7.5 μL of 2×MasterMix (Tiangen, Beijing, China). The PCR cycle conditions used include one denaturation step at 94°C for 5 min; 35 cycles at 94°C for 30 s, 58.4°C or 59.4°C (depends on the primer pair used) for 30 s, and 72°C for 60s; and a final elongation at 72°C for 10 min. The PCR products were purified with AxyPrepTM DNA Gel Extraction Kit (Axygen, Union City, CA, USA) and sequenced by Jinan Li Ge Technology Co., Ltd (Jinan, China). All sequences were analyzed with DNAMAN7.0 and Chromas 2.31 software.
Table 1. Primer parameters for SNPs identification and genotyping of BMP15.
| Primer | Sequences (5′–3′) | Length of products/bp | Annealing temp (°C) | Usage |
|---|---|---|---|---|
| BMP15P1 | F: GGGACCTCTTTCTGCTTTACCR: CATCACCCATTGCCACCA | 406 | 58.4 | SNPs identification |
| BMP15P2 | F: GGACAACAAGGGCAAGGGR: ATCGCATCGAGTGGAGACAA | 1096 | 59.4 | SNPs identification |
| NC_006091.3:g.474A>G | F: GGGACCTCTTTCTGCTTTACCR: CATCACCCATTGCCACCA | 406 | 58.4 | Genotyping with HhaI |
| NC_006091.3:g.594C>T | F: GGGTTTTAGCCCTGATCTTGCACTCR: ATCACCCATTGCCACCACCTTACCT | 517 | 58 | Genotyping with Alw21I |
| NC_006091.3:g.1773T>C | F: TGAGCACCTTCTCCGTGTCAR: ATCCAATGGTCCCAACCC | 462 | 59.6 | Genotyping with MvaI |
Then, three genetic mutations (NC_006091.3:g.474A>G, NC_006091.3:g.594C>T and NC_006091.3:g.1773T>C) were found and genotyped by PCR-RFLP with primers (Table 1). The primer of g.594C>T was referenced in Li et al. (2012) [30]. The PCR were performed as described above. PCR-RFLP reactions were performed in a 15 μL volume system containing 0.5 μL of restriction enzymes (10U/μL) (HhaI, Alw21I or MvaI, New England Biolabs, Inc., USA), 1 μL of 10× NEBuffer, 8 μL of PCR productions and 5.5 μL ddH2O. The PCR products were digested at 37°C or 65°C for 4 h, and then the digested products were separated on a 2.5% agarose gel for 30 min at 120V. After the genotyping, two samples for each genotype of each SNP were sequenced by Jinan Li Ge Technology Co., Ltd (Jinan, China) to confirm the variation. Haplotypes and diplotypes were constructed based on the SNPs identified in all 400 birds using the PHASE 2.0 software.
Real-time quantitative PCR
The total RNA isolated from tissues was assessed using the A260/A280nm ratio with the expected values falling between 1.8 and 2.0 (Eppendorf, Hamburg, Germany). 1μg of total RNA was used for cDNA synthesis with HiFi-MMLV cDNA Kit (CW0744, CWbio.Co.Ltd, Beijing, China) according to the manufacturer’s instructions.
The chicken β-actin gene was used as a control for RT-qPCR. According to the mRNA sequences of BMP15 (GenBank Accession no. NM_001006589.2, GI: 55742820) and β-actin (GenBank Accession no. NM_205518.1, GI: 45382926), the primers for real-time PCR (Table 2) were designed with the Primer Premier5.0. The real-time quantitative PCR reactions were conducted with Roche LC-480II (Roche, California, USA) in a 20μL-volume system containing 10 μL UltraSYBR Mixture (with Rox) (CW0956, CWbio.Co.Ltd, Beijing, China), 0.4 μL of each primer (10pmol/μL), 2 μL cDNA, and 7.2 μL ddH2O. The PCR cycle condition is as follows: one denaturation step at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The specificity of the PCR productions was checked by a final melting curve analysis. The cycle threshold value of control gene was used to normalize the target gene signals in each sample. Standard curves were generated for each gene by serial dilution to quantify the amplified products. Each sample was amplified in triplicate, and the mean value of each triplicate was used for further analysis. The 2-△△CT method was used to calculate the relative expression of transcripts between the target gene and the control gene.
Table 2. Primer parameters for RT-qPCR.
| Gene | Primer sequence(5’-3’) | Annealing temp (°C) | Product size(bp) |
|---|---|---|---|
| BMP15 | F: TTGATGCTTGGTGGGTGGTTR: CACCATAGACTGCCTCGTTC | 60 | 177 |
| β-actin | F: CCATCTATGAAGGCTACGCR: CTCGGCTGTGGTGGTGAA | 60 | 124 |
Statistical analysis
The Hardy-Weinberg equilibrium was evaluated by an χ2 test. The association between SNPs or diplotypes with reproduction traits were analyzed by the GLM procedures of SAS8.12 (SAS Inst. Inc., Cary NC, USA), and the estimated genotypes values were compared by Duncan’s Multiple Range Test (SAS8.12).The statistical model was shown as following:
Where Y = the phenotypic value of traits, μ = the population mean, G = fixed effects of genotype or diplotype, and e = random residual error. Multiple comparisons were performed with the least squares means. One-way ANOVA was used to examine the mRNA expression differences among different tissues in chicken. All the values were considered significant at P < 0.05 and are presented as least square means ± standard error means.
Results
Genotype, allele frequencies, and association analysis
The three genetic mutations (NC_006091.3:g.474A>G, NC_006091.3:g.594C>T and NC_006091.3:g.1773T>C) were genotyped by PCR-RFLP, and a total of eight genotypes were detected. By sequencing, the nucleotide variation at each locus was further confirmed. Relative to GenBank Accession No. NC_006091.3, an A→G mutation at position 474 nucleotide (NC_006091.3:g.474A>G) and a C→T mutation at position 594 nucleotide (NC_006091.3:g.594C>T) were located on exon 1, and a T→C mutation at position 1773 nucleotide (NC_006091.3:g.1773T>C) was located on exon 2. All the three SNPs did not cause amino acid change.
Genotypes and alleles frequency analysis showed that (Table 3) in NC_006091.3:g.474A>G, A was the advantageous allele. In NC_006091.3:g.594C>T, the frequency of CT was higher than that of the CC and TT genotypes, and allele C was dominant. In NC_006091.3:g.1773T>C, C was the dominant allele since the CC genotype (0.957) occurred much more frequently than the other genotypes. The genotype distributions for all the three SNPs fit the Hardy-Weinberg equilibrium with the P-value higher than 0.05.
Table 3. Genotypes and alleles frequencies at site NC_006091.3: g.474A>G, NC_006091.3:g.594C>T and NC_006091.3:g.1773T>C of BMP15 gene.
| SNPs | Location | Genotypes frequency | Allele frequency | P-value a | |||
|---|---|---|---|---|---|---|---|
| NC_006091.3:g.474A>G | Exon1 | AA | AG | GG | A | G | |
| 0.742 | 0.235 | 0.023 | 0.860 | 0.140 | 0.6299 | ||
| NC_006091.3:g.594C>T | Exon1 | CC | CT | TT | C | T | |
| 0.265 | 0.505 | 0.230 | 0.518 | 0.482 | 0.8221 | ||
| NC_006091.3:g.1773T>C | Exon2 | TT | TC | CC | T | C | |
| 0.000 | 0.043 | 0.957 | 0.021 | 0.979 | 0.6641 | ||
a P-value is the probability of the χ2-text for the Hardy-Weinberg equilibrium
The association between genotypes and six reproduction traits were estimated with a total of 398 chickens because traits recording of two chickens went missing. The results are summarized in Table 4. For NC_006091.3:g.474A>G, the association was not significant with EW43 although a trend was observed (P = 0.0519). For NC_006091.3:g.594C>T, the genotypes had no significant assocaitons with AFE (P = 0.0534), EW43 (P = 0.0529) and E48 (P = 0.0683). For NC_006091.3:g.1773T>C, there was a significant association with EWFE (P = 0.0389). The AFE, EW43, E46 and E48 of chickens with N1N2 genotype were superior to chickens with N2N2 genotype, but no significant difference was observed between them (P>0.05).
Table 4. Least-squares means and standard errors for reproduction traits of different genotypes in SNPs.
| Traits | P -Value | NC_006091.3:g.474A>G | ||
|---|---|---|---|---|
| H1H1 (295) | H1H2 (94) | H2H2(9) | ||
| EWFE(g) | 0.9359 | 30.47±0.25 | 30.29±0.44 | 30.22±1.45 |
| AFE | 0.6978 | 139.55±0.52 | 140.02±0.93 | 137.44±3.01 |
| EW43(g) | 0.0519 | 47.62±0.21a | 47.47±0.38a | 44.34±1.32b |
| E43 | 0.8166 | 117.85±1.25 | 116.78±2.20 | 114.00±7.44 |
| E46 | 0.7993 | 130.82±1.38 | 129.83±2.43 | 125.87±8.21 |
| E48 | 0.7956 | 139.19±1.47 | 137.81±2.59 | 134.50±8.74 |
| NC_006091.3:g.594C>T | ||||
| M1M1(105) | M1M2(201) | M2M2 (92) | ||
| EWFE (g) | 0.9881 | 30.37±0.42 | 30.45±0.30 | 30.42±0.45 |
| AFE | 0.0534 | 140.99±0.87a | 139.69±0.63a | 137.88±0.93b |
| EW43(g) | 0.0529 | 47.76±0.36a | 47.76±0.25a | 46.66±0.39b |
| E43 | 0.1772 | 114.60±2.06 | 117.87±1.50 | 120.25±2.28 |
| E46 | 0.1352 | 127.21±2.27b | 130.70±1.66ab | 134.00±2.52a |
| E48 | 0.0683 | 134.80±2.42b | 138.95±1.76ab | 143.19±2.68a |
| NC_006091.3:g.1773T>C | ||||
| N1N2(17) | N2N2(381) | |||
| EWFE (g) | 0.0389* | 28.29±1.05b | 30.51±0.22a | |
| AFE | 0.4846 | 138.11±2.19 | 139.68±0.46 | |
| EW43(g) | 0.2865 | 46.63±0.85 | 47.56±0.19 | |
| E43 | 0.8110 | 118.70±5.10 | 117.45±1.10 | |
| E46 | 0.7426 | 132.29±5.62 | 130.40±1.21 | |
| E48 | 0.5950 | 141.88±5.99 | 138.61±1.29 | |
The least square means within a row lacking a common lowercase superscript differ significantly (P<0.05).
The numbers in the brackets are the chicken individuals of respective genotypes.
Construction of haplotypes and association analysis
The parameters of haplotypes and diplotypes based on the three SNPs are shown in Table 5. A total of six haplotypes were obtained, and A-T-C, A-C-C, and G-C-C were the main haplotypes, accounting for 97.625% of the observations. Eleven diplotypes were obtained based on the six haplotypes. To ensure that the analysis was accurate, four haplotypes with a frequency lower than 2% were not used in the further association analysis.
Table 5. Haplotypes and diplotypes inferred based on the 3single nucleotide polymorphisms.
| Haplotype | NC_006091.3:g.474A>G | NC_006091.3:g.594C>T | NC_006091.3:g.1773T>C | Frequency (%) | Diplotype | Frequency(%) | Diplotype | Frequency(%) |
|---|---|---|---|---|---|---|---|---|
| H1 | A | C | C | 36.125 | H1H1 | 12.750 | H3H5 | 12.500 |
| H2 | A | C | T | 2.000 | H1H2 | 1.000 | H3H6 | 0.250 |
| H3 | A | T | C | 47.750 | H1H3 | 36.000 | H4H6 | 0.250 |
| H4 | A | T | T | 0.125 | H1H5 | 9.750 | H5H5 | 2.250 |
| H5 | G | C | C | 13.750 | H2H3 | 2.250 | ||
| H6 | G | T | C | 0.250 | H2H5 | 0.750 | ||
| H3H3 | 22.250 |
The least squares mean multiple comparisons of diplotypes are shown in Table 6. Diplotypes was found to be highly significantly associated with EW43 (P = 0.0058). With the increase in chickens’ age, the association between the diplotypes and egg production increased. The AFE of chickens with H3H3 diplotype was 138.01 days old, which was 0.57 days later than chickens with H5H5 diplotype but 1.21 days-3.96 days earlier than the other five diplotype chickens. Meanwhile, the E43, E46 and E48 of chickens with H3H3 diplotype were the highest among all the chickens, and the E48 of chickens with H3H3 diplotype had 11.83 eggs higher than that of chickens with H1H5 diplotype. The H1H5 chickens had the highest AFE and lowest E43, E46, E48 among the other chickens.
Table 6. Association of diplotypes of chicken BMP15 gene with the reproductive traits.
| Traits | EWFE (g) | AFE | EW43 (g) | E43 | E46 | E48 |
|---|---|---|---|---|---|---|
| P value | 0.8798 | 0.2606 | 0.0058** | 0.4918 | 0.3473 | 0.1953 |
| H1H1(50) | 30.98±0.61 1 | 141.26±1.27 | 48.57±0.52 1 | 114.98±2.98 | 127.90±3.28 | 135.50±3.48 |
| H1H3(143) | 30.50±0.36 | 139.82±0.75 | 47.97±0.30 | 117.13±1.80 | 129.56±1.98 | 137.62±2.10 |
| H1H5(39) | 30.07±0.70 | 141.97±1.44 1 | 47.59±0.59 | 112.71±3.42 2 | 124.86±3.76 2 | 131.78±4.00 2 |
| H2H3(9) | 28.77±1.45 2 | 139.22±3.00 | 46.04±1.15 | 118.00±7.02 | 131.77±7.73 | 141.22±8.21 |
| H3H3(89) | 30.43±0.46 | 138.01±0.95 | 46.62±0.40 | 120.74±2.34 1 | 134.48±2.57 1 | 143.61±2.73 1 |
| H3H5(50) | 30.58±0.61 | 139.40±1.27 | 47.33±0.51 | 119.85±3.04 | 133.77±3.34 | 142.43±3.55 |
| H5H5(9) | 30.22±1.45 | 137.44±3.00 2 | 44.34±1.31 2 | 114.00±7.45 | 125.87±8.20 | 134.50±8.71 |
1 Bold values represent the advantageous diplotypes
2 Italic values represent the negative diplotypes.
** P ≤ 0.01
Tissue expression analysis of BMP15
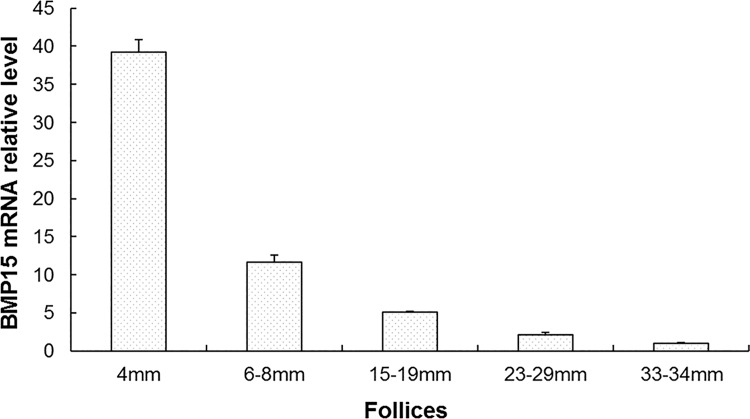
The mRNA relative expression of BMP15 in different levels of follicles is shown in Fig 1. The expression level of BMP15 in ovarian follicle was in the order of 4 mm>6 mm -8 mm> 15 mm -19 mm> 23 mm -29 mm > 33 mm -34 mm in diameter. The mRNA level of BMP15 in follicles with a diameter of 4 mm and 6–8 mm were significantly higher than that in the other follicles (P<0.01).
Fig 1. Relative expression level of BMP15 mRNA in follicles of 40w Laiwu Black chicken.
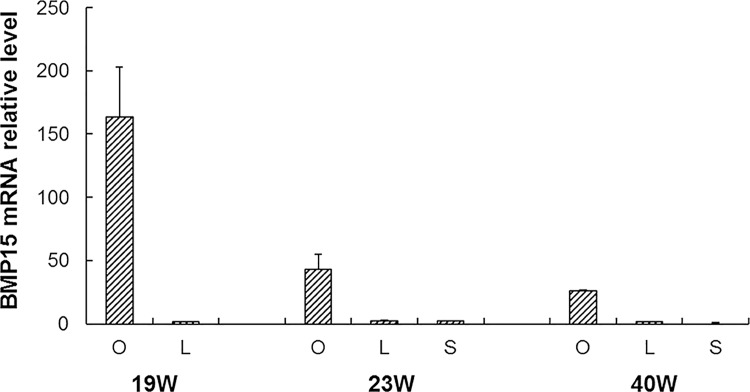
The differential expression pattern of BMP15 in different tissues in the same week is presented in Fig 2. In the same week, the trends of the mRNA level in various tissues were ovary > oviduct > liver. The highest mRNA level was found in the ovary, and it was significantly different from that found in the liver and oviduct (P<0.01).
Fig 2. The expression of a chicken BMP15 gene in liver, ovary and oviduct at the age of the same week.
Discussion
The oocyte factor, bone morphogenetic protein-15 (BMP-15) has proven to be critical for normal fertility in female mammals [31,32]. Wu (2009) detected a T→A mutation of exon 2; total number born and number born alive for BB pigs were significantly lower than AA and AB pigs for both Xiaomeishan pig and Large White pig (P<0.05) [33]. Li L et al. (2009) found two SNPs in rabbit BMP15, and the mean litter size of BB genotypes was higher than that of AA and AN genotype in angora rabbit flock (P<0.05) [34]. Shabir M, et al. (2013) found that SNP "C" of mutations in exon-2 of the designated genotype AC was observed to produce a significant effect on litter size with average litter size going up by 0.63, as compared with the nearest genotype AB with the litter size of 1.29±0.05 [35]. In sheep, eight genetic mutations with major effects on ovulation rate and litter size have been identified in BMP15 [22–27]. In Shaobo hens, Huang HY, et al. (2015) detected three SNPs in each of BMP15 (A111G, C231T and C34T) and GDF9 (G593A, T824C and C896T), and found that the C34T had an effect on total egg production at 300 d of age (EN) and age at first laying (AFE), G593A affected EN and both C231T and C896T influenced AFE [36]. But our results showed that only NC_006091.3:g.594C>T had a trend of association with AFE (P = 0.0534), EW43 (P = 0.0529) and E48 (P = 0.0683), but the association was not significant (P>0.05). The difference between results may be caused by the different experimtal chicken populations we used, and the different SNPs we detected. The C34T they detected leads to the substitution of Leu by Phe, which was predicted to affect protein function. But In this study, all the three SNPs (NC_006091.3:g.474A>G and NC_006091.3:g.594C>T and NC_006091.3:g.1773T>C) were synonymous mutations. NC_006091.3:g.1773T>C was significantly associated with EWFE (P = 0.0389). Synonymous mutations do not change the sequence or structure of the protein, once was thought to be functionally neutral, but evidence now indicates it is shaped by evolutionary selection and affects other aspects of protein biogenesis beyond specifying the amino acid sequence of the protein [37]. Studies analyzing the consequences of synonymous codon changes in different organisms have revealed that they impact nucleic acid stability, protein levels, structure and function without altering amino acid sequence [38]. Furthermore, gene expression is correlated with synonymous codon usage bias [39,40]. Thus for the SNPs, the condon choice may affect the BMP15 gene expression or the coordinated expression of functionally related genes, further affect the follicle development, ovulation and the reproduction traits in chicken. However, much remains unknown about the molecular mechanisms connecting synonymous codon usage to efficient protein biogenesis and proper cell physiology [37]. So, the mechanism of the three SNPs of BMP15 to reproduction traits in chicken should be further studied.
Diplotypes was found to be significantly associated with EW43 (P = 0.0058), and the association between them increased with the increase in chickens’ age (in days). The AFE of H3H3 chickens was 138.01 days old, which was 0.57 days later than H5H5 chickens but 1.21 days-3.96 days earlier than the other five diplotype chickens. Meanwhile, the E43, E46 and E48 of H3H3 chickens were the highest among all the chickens, and the E48 of H3H3 was 11.83 eggs higher than that of the H1H5 chickens. So, the H3H3 was regarded as the advantageous diplotype for reproduction trait. The H1H5 chickens had the highest AFE and lowest E43, E46, E48 among the other chickens. As a result, it was regarded as the detrimental diplotype for chicken reproduction traits, suggesting that it should be deleted during the cultivation of new varieties. These findings showed that BMP15 has a significant effect on the reproduction traits in chicken, similar to its function in the other animals.
The function of BMP15 in chicken reproduction was further confirmed by analyzing the mRNA expression pattern. Bone morphogenetic protein 15 is a major and well-known oocyte-secreted growth factor, required for follicular development and ovulation [41,42]. Eckery et al. (2002) found that the BMP15 protein was observed in oocytes from the primordial stage of follicular formation and the results suggest a possible role for these proteins in the maintenance of primordial follicles, as well as playing a key role during follicular development in the brushtail possum [43]. Using real-time PCR, Paradis et al. (2009) revealed that BMP15 mRNA was most abundant in the oocyte; its expression remained relatively constant during follicular development in pig [44]. Sun (2010) found that BMP15 was presented in oocytes and granulosa cells of all follicles in mice and porcine [45]. Our results showed that BMP15 mRNA was expressed in all ovarian follicles of Laiwu Black chickens, and strongly expressed in follicles with 4 mm and 6–8 mm diameter. Moreover, in contrast to oviduct and liver, BMP15 was preferentially expressed in the ovary during the same week, which was consistent with the findings of He X L, et al. (2009) [46], Wang Y, et al. (2012) [47] and Sudiman J et al. (2014) [48]. Collectively, the findings suggest that BMP15 might be involved in ovarian follicular growth and development, and plays a major role in primordial follicular recruitment in chicken.
In Conclusion, the expression level of BMP15 was different in different tissues; the expression level in the ovary was higher than that in the other tissues. Also, the highest mRNA level was found in 4 mm follicles, indicating that BMP15 played a vital function in the development of ovary and follicles, especially in the development of primary follicles. H3H3 could be used as a potential advantageous molecular marker for reproduction traits in chickens. This research provides exciting new opportunities for understanding the role of the oocyte-secreted factor BMP-15 on ovarian follicular growth and development in chicken.
Acknowledgments
The authors gratefully acknowledge D. L. Cao and H. L. Han for their help in managing the birds and collecting the data.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was financially supported by the Youth Fund Project of Shandong Academy of Agricultural Sciences (2014QNM14, 2015YQN53), the Major Natural Science Foundation of Shandong Province, China (ZR2014CZ003), the Joint Funds of the Natural Science Foundation of Shandong Province, China (ZR2014YL024), and Major Agricultural Stock Breeding Project of Shandong Province (New Excellent Poultry Breeding). The funders had no role in study design, data collection and analysis, preparation or the publication of the manuscript.
References
- 1. Auclair S, Rossetti R, Meslin C, Monestier O, Di Pasquale E, Pascal G, et al. Positive selection in bone morphogenetic protein 15 targets a natural mutation associated with primary ovarian insufficiency in human. PLoS One. 2013;8: e78199 10.1371/journal.pone.0078199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mullen MP, Hanrahan JP, Howard DJ, Powell R. Investigation of prolific sheep from UK and Ireland for evidence on origin of the mutations in BMP15 (FecX(G), FecX(B)) and GDF9 (FecG(H)) in Belclare and Cambridge sheep. PLoS One. 2013;8: e53172 10.1371/journal.pone.0053172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22: 233–241. 10.1080/08977190412331279890 [DOI] [PubMed] [Google Scholar]
- 4. Knight PG, Glister C. TGF-beta superfamily members and ovarian follicle development. Reproduction. 2006;132: 191–206. 10.1530/rep.1.01074 [DOI] [PubMed] [Google Scholar]
- 5. Persani L, Rossetti R, Di Pasquale E, Cacciatore C, Fabre S. The fundamental role morphogenetic protein 15 in ovarian function and its involvement in female fertilitydisorders. Hum Reprod Update. 2014;20: 869–883. 10.1093/humupd/dmu036 [DOI] [PubMed] [Google Scholar]
- 6. Otsuka F, Yao Z, Lee T, Yamamoto S, Erickson GF, Shimasaki S. Bone morphogenetic protein-15. Identification of target cells and biological functions. J Biol Chem. 2000;275: 39523–39528. 10.1074/jbc.M007428200 [DOI] [PubMed] [Google Scholar]
- 7. Otsuka F, Yamamoto S, Erickson GF, Shimasaki S. Bone morphogenetic protein-15 inhibits follicle-stimulating hormone (FSH) action by suppressing FSH receptor expression. J Biol Chem. 2001;276: 11387–11392. 10.1074/jbc.M010043200 [DOI] [PubMed] [Google Scholar]
- 8. Otsuka F, Shimasaki S. A negative feedback system between oocyte bone morphogenetic protein 15 and granulosa cell kit ligand: its role in regulating granulosa cell mitosis. Proc Natl Acad Sci U S A. 2002;99: 8060–8065. 10.1073/pnas.122066899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laitinen M, Vuojolainen K, Jaatinen R, Ketola I, Aaltonen J, Lehtonen E, et al. A novel growth differentiation factor-9 (GDF-9) related factor is co-expressed with GDF-9 in mouse oocytes during folliculogenesis. Mech Dev. 1998;78: 135–140. [DOI] [PubMed] [Google Scholar]
- 10. Dube JL, Wang P, Elvin J, Lyons KM, Celeste AJ, Matzuk MM. The bone morphogenetic protein 15 gene is X-linked and expressed in oocytes. Mol Endocrinol. 1998;12: 1809–1817. 10.1210/mend.12.12.0206 [DOI] [PubMed] [Google Scholar]
- 11. Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15: 854–866. 10.1210/mend.15.6.0662 [DOI] [PubMed] [Google Scholar]
- 12. McMahon HE, Hashimoto O, Mellon PL, Shimasaki S. Oocyte-specific overexpression of mouse bone morphogenetic protein-15 leads to accelerated folliculogenesis and an early onset of acyclicity in transgenic mice. Endocrinology. 2008;149: 2807–2815. 10.1210/en.2007-1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Su YQ, Wu X, O'Brien MJ, Pendola FL, Denegre JN, Matzuk MM, et al. Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte-cumulus cell complex in mice: genetic evidence for an oocyte-granulosa cell regulatory loop. Dev Biol. 2004;276: 64–73. 10.1016/j.ydbio.2004.08.020 [DOI] [PubMed] [Google Scholar]
- 14. Yoshino O, McMahon HE, Sharma S, Shimasaki S. A unique preovulatory expression pattern plays a key role in the physiological functions of BMP-15 in the mouse. Proc Natl Acad Sci U S A. 2006;103: 10678–10683. 10.1073/pnas.0600507103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kathirvel M, Soundian E, Kumanan V. Differential expression dynamics of Growth differentiation factor9 (GDF9) and Bone morphogenetic factor15 (BMP15) mRNA transcripts during in vitro maturation of buffalo (Bubalus bubalis) cumulus-oocyte complexes. Springerplus. 2013;2: 206 10.1186/2193-1801-2-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Juengel JL, Hudson NL, Heath DA, Smith P, Reader KL, Lawrence SB, et al. Growth differentiation factor 9 and bone morphogenetic protein 15 are essential for ovarian follicular development in sheep. Biol Reprod. 2002;67: 1777–1789. [DOI] [PubMed] [Google Scholar]
- 17. Tan Q, Zagrodny A, Bernaudo S, Peng C. Regulation of membrane progestin receptors in the zebrafish ovary by gonadotropin, activin, TGF-beta and BMP-15. Mol Cell Endocrinol. 2009;312: 72–79. 10.1016/j.mce.2009.03.011 [DOI] [PubMed] [Google Scholar]
- 18. Hosoe M, Kaneyama K, Ushizawa K, Hayashi KG, Takahashi T. Quantitative analysis of bone morphogenetic protein 15 (BMP15) and growth differentiation factor 9 (GDF9) gene expression in calf and adult bovine ovaries. Reprod Biol Endocrinol. 2011;9: 33 10.1186/1477-7827-9-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peng J, Li Q, Wigglesworth K, Rangarajan A, Kattamuri C, Peterson RT, et al. Growth differentiation factor 9:bone morphogenetic protein 15 heterodimers are potent regulators of ovarian functions. Proc Natl Acad Sci U S A. 2013;110: E776–785. 10.1073/pnas.1218020110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhai B, Liu H, Li X, Dai L, Gao Y, Li C, et al. BMP15 prevents cumulus cell apoptosis through CCL2 and FBN1 in porcine ovaries. Cell Physiol Biochem. 2013;32: 264–278. 10.1159/000354435 [DOI] [PubMed] [Google Scholar]
- 21. Lin ZL, Li YH, Xu YN, Wang QL, Namgoong S, Cui XS, et al. Effects of growth differentiation factor 9 and bone morphogenetic protein 15 on the in vitro maturation of porcine oocytes. Reprod Domest Anim. 2014;49: 219–227. 10.1111/rda.12254 [DOI] [PubMed] [Google Scholar]
- 22. Galloway SM, McNatty KP, Cambridge LM, Laitinen MP, Juengel JL, Jokiranta TS, et al. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat Genet. 2000;25: 279–283. 10.1038/77033 [DOI] [PubMed] [Google Scholar]
- 23. Hanrahan JP, Gregan SM, Mulsant P, Mullen M, Davis GH, Powell R, et al. Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries). Biol Reprod. 2004;70: 900–909. 10.1095/biolreprod.103.023093 [DOI] [PubMed] [Google Scholar]
- 24. Bodin L, Di Pasquale E, Fabre S, Bontoux M, Monget P, Persani L, et al. A novel mutation in the bone morphogenetic protein 15 gene causing defective protein secretion is associated with both increased ovulation rate and sterility in Lacaune sheep. Endocrinology. 2007;148: 393–400. 10.1210/en.2006-0764 [DOI] [PubMed] [Google Scholar]
- 25. Chu M, Sun J, Chen H, Fang L. Detection of the FecX~ L mutation of BMP15 gene in sheep. Chinese Agricultural Science Bulletin. 2007;23: 85–88. [Google Scholar]
- 26. Martinez-Royo A, Jurado JJ, Smulders JP, Martí JI, Alabart JL, Roche A, et al. A deletion in the bone morphogenetic protein 15 gene causes sterility and increased prolificacy in Rasa Aragonesa sheep. Anim Genet. 2008;39: 294–297. 10.1111/j.1365-2052.2008.01707.x [DOI] [PubMed] [Google Scholar]
- 27. Demars J, Fabre S, Sarry J, Rossetti R, Gilbert H, Persani L, et al. Genome-wide association studies identify two novel BMP15 mutations responsible for an atypical hyperprolificacy phenotype in sheep. PLoS Genet 2013;9: e1003482 10.1371/journal.pgen.1003482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elis S, Dupont J, Couty I, Persani L, Govoroun M, Blesbois E, et al. Expression and biological effects of bone morphogenetic protein-15 in the hen ovary. J Endocrinol. 2007;194: 485–497. 10.1677/joe-07-0143 [DOI] [PubMed] [Google Scholar]
- 29. Onagbesan O, Bruggeman V, Decuypere E. Intra-ovarian growth factors regulating ovarian function in avian species: a review. Anim Reprod Sci. 2009;111: 121–140. 10.1016/j.anireprosci.2008.09.017 [DOI] [PubMed] [Google Scholar]
- 30. Li CM, Li SF, Zhao ZH, Huang HY, Xue LG. Detection of SNPs of Bone Morphogenetic Protein 15 Gene Exon1 and Its Association with Egg Production Traits in Female Line of Shaobo Chicken. Chin J Anim Vet Sci. 2012;11: 024. [Google Scholar]
- 31. Erickson GF, Shimasaki S. The role of the oocyte in folliculogenesis. Trends in Endocrinology & Metabolism. 2000;11: 193–198. [DOI] [PubMed] [Google Scholar]
- 32. Fortune JE. The early stages of follicular development: activation of primordial follicles and growth of preantral follicles. Anim Reprod Sci. 2003;78: 135–163. [DOI] [PubMed] [Google Scholar]
- 33. Wu JS, Zhu ML, Chen C, Chen M, Li GH. Effect of gene Bcu I polymorphism of bone morphogenetic protein-15 on the litter size of pigs. Anim Hus&vet Med. 2009;141. [Google Scholar]
- 34. Lin L, Tao C, Peng W, Jia-tong D. Study on Relationship between Single Nucleotide Polymorphism of BMP15 Gene and Litter Size of Rabbits. Journal of Anhui Normal University (Natural Science). 2009;6: 012. [Google Scholar]
- 35. Shabir M, Ganai TA, Misra SS, Shah R, Ahmad T. Polymorphism study of growth differentiation factor 9B (GDF9B) gene and its association with reproductive traits in sheep. Gene. 2013;515: 432–438. 10.1016/j.gene.2012.12.018 [DOI] [PubMed] [Google Scholar]
- 36. Huang HY, Liang Z, Li SF, Li CM, Zhao ZH, Wang QB. Polymorphism identification in BMP15 and GDF9 genes and their association with egg production in chickens. Br Poult Sci. 2015;56(3): 277–283. 10.1080/00071668.2015.1019829 [DOI] [PubMed] [Google Scholar]
- 37. Chaney JL, Clark PL. Roles for Synonymous Codon Usage in Protein Biogenesis. Annu Rev Biophys. 2015;44: 143–66. 10.1146/annurev-biophys-060414-034333 [DOI] [PubMed] [Google Scholar]
- 38. Bali V, Bebok Z. Decoding mechanisms by which silent codon changes influence protein biogenesis and function. Int J Biochem Cell Biol. 2015. July; 64:58–74. 10.1016/j.biocel.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williford A, Demuth JP. Gene expression levels are correlated with synonymous codon usage, amino acid composition, and genearchitecture in the red flour beetle, Tribolium castaneum. Mol Biol Evol. 2012;29(12): 3755–66. 10.1093/molbev/mss184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De La Torre AR, Lin YC, Van de Peer Y, Ingvarsson PK. Genome-wide analysis reveals diverged patterns of codon bias, gene expression, and rates of sequence evolution in picea gene families. Genome Biol Evol. 2015;7(4): 1002–15. 10.1093/gbe/evv044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laissue P, Christin-Maitre S, Touraine P, Kuttenn F, Ritvos O, Aittomaki K, et al. Mutations and sequence variants in GDF9 and BMP15 in patients with premature ovarian failure. Eur J Endocrinol. 2006;154: 739–744. 10.1530/eje.1.02135 [DOI] [PubMed] [Google Scholar]
- 42. Di Pasquale E, Rossetti R, Marozzi A, Bodega B, Borgato S, Cavallo L, et al. Identification of new variants of human BMP15 gene in a large cohort of women with premature ovarian failure. J Clin Endocrinol Metab. 2006;91: 1976–1979. 10.1210/jc.2005-2650 [DOI] [PubMed] [Google Scholar]
- 43. Eckery DC, Whale LJ, Lawrence SB, Wylde KA, McNatty KP, Juengel JL. Expression of mRNA encoding growth differentiation factor 9 and bone morphogenetic protein 15 during follicular formation and growth in a marsupial, the brushtail possum (Trichosurus vulpecula). Mol Cell Endocrinol. 2002;192: 115–126. [DOI] [PubMed] [Google Scholar]
- 44. Paradis F, Novak S, Murdoch GK, Dyck MK, Dixon WT, Foxcroft GR. Temporal regulation of BMP2, BMP6, BMP15, GDF9, BMPR1A, BMPR1B, BMPR2 and TGFBR1 mRNA expression in the oocyte, granulosa and theca cells of developing preovulatory follicles in the pig. Reproduction. 2009;138: 115–129. 10.1530/rep-08-0538 [DOI] [PubMed] [Google Scholar]
- 45. Sun RZ, Lei L, Cheng L, Jin ZF, Zu SJ, Shan ZY, et al. Expression of GDF-9, BMP-15 and their receptors in mammalian ovary follicles. J Mol Histol. 2010;41: 325–332. 10.1007/s10735-010-9294-2 [DOI] [PubMed] [Google Scholar]
- 46. He XL, Gao LZ, Wang F, Liu YB, Tian CY. Analysis on the expression of mRNA coded by bone morphogenetic protein 15 gene in different tissues of cattle. Animal husbandry and feed science. 2009;2009: 129–131. [Google Scholar]
- 47. Wang Y, Chen AQ, Yang ZG, Liu ZW, Guo ZH. Cloning and tissue expression of GDF9 and BMP15 genes in Carassius auratus cuvieri. Journal of Shanghai ocean university. 2012;21: 693–700. [Google Scholar]
- 48. Sudiman J, Sutton-McDowall ML, Ritter LJ, White MA, Mottershead DG, Thompson JG, et al. Bone morphogenetic protein 15 in the pro-mature complex form enhances bovine oocyte developmental competence. PLoS One. 2014;9: e103563 10.1371/journal.pone.0103563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.