Abstract
High throughput sequencing is poised to change all aspects of the way antibodies and other binders are discovered and engineered. Millions of available sequence reads provide an unprecedented sampling depth able to guide the design and construction of effective, high quality naïve libraries containing tens of billions of unique molecules. Furthermore, during selections, high throughput sequencing enables quantitative tracing of enriched clones and position-specific guidance to amino acid variation under positive selection during antibody engineering. Successful application of the technologies relies on specific PCR reagent design, correct sequencing platform selection, and effective use of computational tools and statistical measures to remove error, identify antibodies, estimate diversity, and extract signatures of selection from the clone down to individual structural positions. Here we review these considerations and discuss some of the remaining challenges to the widespread adoption of the technology.
Introduction
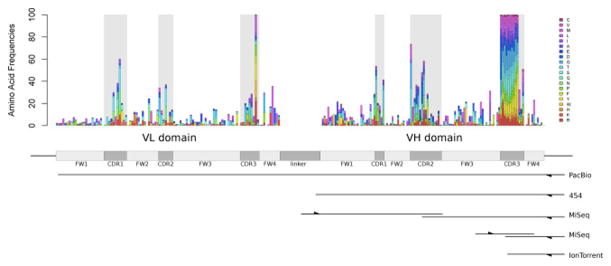
Next generation sequencing (NGS) has transformed genomics. Its impact in antibody library selection projects has been slower, but is likely to be equally disruptive. In many ways, the display technologies and deep sequencing are approaching a perfect match as sequencing technologies improve. For library analysis, total numbers of bases sequenced is less important than the number of reads and their length. Present sequencing technology is able to generate up to 40 million reads from a single MiSeq run (figure 1). A naïve antibody (or other binding scaffold) library could potentially have a diversity at least 25 fold greater (≥109), the true diversity of which can be estimated using the methods described below. However, once these libraries are subject to selection by phage or yeast display, diversity is reduced to ~106 after a single round, allowing comprehensive analysis of the complete diversity of dozens of different selections in a single MiSeq run. After two or more rounds of selection, diversity is reduced still further, and the percentage of positive clones increases significantly; making analysis of ≥100 selections in a single run relatively straightforward. Read lengths vary, depending upon the technology (figure 1). Although 454 and PacBio provide the longest reads, the higher read number and low cost have made paired end MiSeq (2x300bp) or Ion Torrent (400bp) sequencing the most commonly used for library analysis. While MiSeq will completely cover variable domains, encompassed by ≤600 bp (e.g. single Ig-like domain – VH domain of a scFv, camelid VHH’s or fibronectin domains, smaller DARPINs, affibodies), it is presently insufficient to completely cover both the VH and VL chains found in an scFv in a single read. We expect this problem to be overcome as read lengths increase with further technology development.
Figure 1.
NGS sequencing on scFv genes. Variability plots for representative VL and VH genes are shown, with the CDRs shaded in grey. Length coverage for the most popular NGS platforms and scFv-based libraries targeted regions are shown. For each platform, single or double directional arrows indicate single or paired-end sequencing, respectively.
The convergence of these technologies is important in structural biology for the increased use of antibody fragments [1] and other binders [2–4], as crystallization chaperones. While such chaperones were originally derived from immunized animals, recombinant display techniques using immunized or naïve binder sources as starting materials has broadened the nature of molecules used to include synthetic recombinant Fabs [5,6], designed ankyrin repeat proteins (DARPINs) [7–9], fibronectin domains [10] and nanobodies [11]. Any method that simplifies the generation of suitable crystallization chaperones is to be welcomed, and it is anticipated that the combination of NGS with display technologies will facilitate the development of effective chaperones, particularly if selection strategies can be specifically designed to select such molecules directly.
Here we review the technology and the informatic analyses required before describing the insights that can be gained from the use of next generation sequencing in library selection projects.
The technologies
The ability to assess the entire diversity of an antigen-specific sub-library allows the identification of all unique species in a sub-library, independently of their relative enrichment during the selection process. In fact, the wide span of relative abundances within a selected population is a known bias in the random screening process [12,13]. NGS technologies can successfully interrogate, at the deepest levels, theoretically every individual molecule, hence their increasing use in the screening of selected sub-libraries.
Several NGS platforms, each with specific advantages and, usually, preferred applications, are available. As a general consideration, read length and depth of sequencing are inversely proportional: technologies that provide the longest read lengths have the lowest throughputs, and vice versa for platforms that favor depth over length. The choice of sequencing usually lies in the nature of the selected library to be investigated: single scaffold synthetic libraries are generally easy to analyze because the sequencing can be focused on that limited region of the scaffold molecule that encodes the diversity. NGS platforms generating short reads are preferred in this case. Antibody-derived molecules, such as scFvs (single chain Fragment variable), represent a more complex scenario, where diversity is spread along a ~800bp-long gene. In this specific case, full-length gene sequencing would be ideal. Sanger sequencing provides sufficient read length, to cover the entire gene, but the low-throughput and high expense only allows a very limited snapshot of the true diversity of the selection process. At present among the different NGS platforms, PacBio is the only one able to provide sufficiently long read lengths to cover the entire scFv, but to the detriment of throughput [14] and read quality, where the ability to discriminate minimal sequence differences with certainty[15] has had limited adoption.
For selection projects, where depth of sequencing is preferred, platforms that provide shorter reads become the obvious alternative. However, this imposes a choice on the region of the gene that is to be analyzed. Roche’s 454 and Illumina’s MiSeq paired-end sequencing allow the coverage of the entire VL or VH domain [14,16]. The cheapest and fastest sequencing runs are provided by IonTorrent, and MiSeq single or paired-end reads; here, the main drawback is represented by the read length, which in the case of the single reads is sufficient to only partially cover one of the domains. In this case, the general consensus is to analyze the heavy chain VH domain, as it contains the complementarity determining region 3 (CDR3) as the primary signature of clonality, as well as amino acid variation in H1 and H2, framework mutations, biochemical liabilities in the variable domain and the identity of the V-gene and J-gene scaffold elements. As shown in Figure 1, CDR3s have the highest variability of all CDRs in both variable light (VL) and heavy (VH) domains, with HCDR3 being considered the principal determinant of specificity in antigen binding and, consequently, a surrogate for scFv identity in a naive library[17,18] due to its diversity in length and aa composition[19,20]. The light chains are often sequenced as well, but given their relatively low diversity, are usually insufficiently diverse to reliably and uniquely indicate clonality. Efforts to utilize paired-end reads in the VH and the VL could provide a means of tracking VL diversity for each HCDR3. Table 1 summarizes the features of some of the most popular NGS platforms in selection projects.
Table 1.
Read lengths, number of reads, cost (per read), time taken and error rate for each technology.
Comparison of the different next generation sequencing platforms available. Some platforms can be used in different ways, and the read lengths and number of reads, costs and error rates are indicated appropriately. In the case of PacBio, the error rate of a single long read (8500 bp) is very high at 11–15%. However, if a shorter DNA fragment is read multiple times (e.g. 850bp ten times), the error rate improves to ≤99.999%, by the creation of a consensus sequence.
| Platform | Type of sequencing | Max read length (bp) | Throughput | Cost (lowest) | Accuracy | Time | Type of error |
|---|---|---|---|---|---|---|---|
| MiSeq (Illumina) v2/v3 | 2 x 300 | 600 | 25x106/lane | $1750/lane | >70% reads at 99.9% | 55h | Substitution |
| 2 x 150 | 300 | 16x106/lane | $1100/lane | >80% reads at 99.9% | 24h | ||
|
| |||||||
| IonTorrent (LifeTech)-316 | 1 x 400 | 400 | 2x106/chip | $900/chip | > 99% | 5 h | InDel |
| 1 x 200 | 200 | 3 h | |||||
|
| |||||||
| PacBio-RSII | 1 x 8500 | 8500 | 47,000 | $1050 | 11–15% | ≤4 h | InDel |
| e.g. 10 passes of 850 bp | 99.999% depends on no. passes | ||||||
|
| |||||||
| 454 (Roche)-GS-FLWX+ | 1 x 700 | 700 | 50,000 in 1/8 plate | $2400/1/8 plate | 99.997% | 23 h | InDel |
| 1 x 450 | 450 | $1900/1/8 plate | 99.995% | 10 h | |||
The genetic material used to perform the NGS analysis on scFv-based libraries is usually a plasmid preparation of the selected sublibrary, from which the relevant immunoglobulin coding regions can be extrapolated by PCR amplification; the relevant coding regions, can consist of an entire scFv, an entire domain or just a portion of it, according to the chosen platform.
The amplification step requires the design of primers complementary to the target regions of interest; when the entire scFv gene or domain are sequenced (by PacBio, 454 or MiSeq paired ends), the use of external primers (mapping on the plasmid or linker flanking regions) is to be preferred, as these anneal to constant sequence elements. This provides the least biased means of amplification and makes the entire variable domain accessible. Shorter amplicons have also been designed around specific regions of interest by using multiple primers mapping upstream of the desired region (i.e. HCDR3), but these primers need to be carefully designed, in order to avoid amplification bias: due to the diversity of the antibody variable region frameworks, the primers are usually designed for families of antibody genes (consensus sequences) able to detect the highest number of gene segments[21]. As physical amplicon tolerances and read lengths increase, invariant vector primers have emerged as a standard.
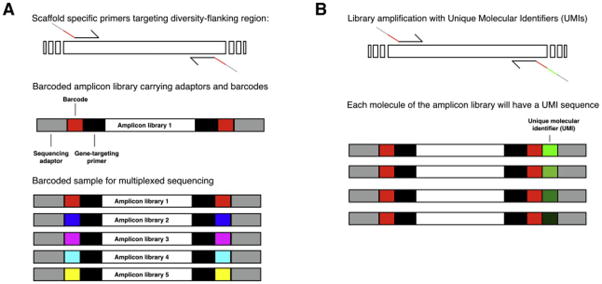
As a general consideration, the primers are designed to allow the amplification of the target sequence and to carry adapters. These are platform-specific sequences that allow: i) annealing of sequencing primers; ii) anchoring of the amplicon to beads or other solid substrate during sequencing; iii) amplification of the single DNA molecule on the solid substrate. The primers can be further modified to allow multiplexing: the ability to sequence multiple selection outputs in a single run (Figure 2). Different selections are distinguished from one another with short sequences, unique for each selection inserted into the primers. This allows for significant reductions in costs as (except for naïve library analysis) platform throughputs vastly exceed diversity in most selection outputs, providing sufficient depth to allow comprehensive analysis.
Figure 2.
A) Schematic representation of NGS barcoded primers. Primers on conserved regions flanking the diversity carry barcode sequencing and NGS-specific adapters. The PCR product contains a unique identifier (barcode) for a specific library. Multiple amplicon libraries can be pooled in a unique sample and sequenced together. Each single sequenced DNA fragment is associated to a specific library based on its barcode. B) An amplicon library can also be generated with the inclusion of a Unique Molecular Identifier (UMI) sequence, which are added at an initial step (e.g., first-strand cDNA synthesis) resulting in each molecule of a library being tagged with a UMI. Similar to A), amplicons can be sequenced in a multiplex fashion but following NGS, sequences with identical UMIs are grouped together for consensus building-based error correction.
Multiplexing is achieved by adding unique DNA sequences (usually 6–8 bp) at the 5′ end of the gene-specific region on the primer (Figure 2). The sequencing of the barcode, along with the gene sequence, allows for the association of a read to a specific sub-library within the sequenced sample. Over 100 samples can easily be barcoded using such schemes, and by extending the barcode length multiplexing can be extended still further, and arbitrary numbers of samples could theoretically be generated, with primer costs becoming the primary limitation. For high number of multiplexed samples, the most efficient method is combinatorial barcoding: different barcodes are added to each end of the sub-library-specific amplicon, thus allowing for hundreds of different sub-libraries to be pooled and sequenced in a single run. The NGS platforms most suitable for multiplexing are IonTorrent and Illumina, due to their higher sequencing depths: a 10-fold coverage of the estimated sub-library diversity is a desirable feature that allows the minimization of the effects of PCR amplification and sequencing errors. Alternatively, when evaluating selections, a simple rule of thumb can be applied: every sample should be performed in replicate to a depth of 100k reads. The 100k read depth will allow any sequence occurring at a frequency of 1e-4 (one in 10k reads) to be observed 10 times in each replicate on average, irrespective of how diverse the background library may be. As a consequence, treating 1e-4 as a threshold of meaningful enrichment, this simple rule allows all samples from all panning rounds to be processed identically, sequenced to equal depth, and analyzed in a comparable manner.
For antibody research a key aspect is to obtain the entire sequence of the highly diverse antibody variable regions, which allows precise definition of the antibody [22]. At present, none of the existing NGS platforms can provide sufficient accuracy and read length to characterize full-length scFv genes in a large sub-library. To overcome these limitations, some methods have been proposed and successfully applied to the characterization of naïve and immune repertoires: in one instance [14,23], two independent MiSeq paired-end sequencings have been used to sequence the entire VL and VH domains, while a third sequencing, coupled with appropriate bioinformatics tools (discussed in the next paragraph), aims to bridge the VL and VH pair. Alternatively, the sequencing of a full-length scFv could be achieved in a single run by using the same “bridging” approach, with 2 primers sequencing sequentially from the 2 ends, and a third primer (or set of primers) bridging the gap starting from the framework region 3 of the VL domain. The method is yet to be tested for feasibility.
Roche’s 454 is able to generate long reads, currently around 700 bp, making it well suited for V region analysis, with the limitations of a limited number of reads (Table 1) and significantly higher cost per run. MiSeq is cheaper, has much higher throughput and a faster turnaround compared to 454. However, read lengths are shorter, making it more appropriate for analyses of single variable domains. The reads obtainable by Ion Torrent range from 35 to 400 bp, enough to cover the CDR3 region as well as a single V domain (which will provide sequences of the other CDRs and help identify the antibody family). The lower quality of the sequences and current read length are the major drawbacks, while the main advantages are speed and low price per run.
Pacific Bio is a single DNA molecule sequencing platform that gives very long reads, but with error rates >10%. While this is not a problem for genome assembly, where it is now usually combined with other platforms [24–26], it has not been used successfully for antibody analysis in a single pass mode. More recently, accuracy has been significantly increased by circularizing DNA and sequencing it multiple times [15]. However, throughput remains relatively low.
In a recent paper [21] we compare the use of 454, MiSeq and Ion Torrent to sequence the same antibody library samples, and find that each method has its advantages, as outlined above.
Informatic analysis of naïve libraries and panning selections
The high-throughput sequence analysis of both naïve libraries and antibody library selections follows a well-established series of common steps. First, any paired-end reads are assembled. Next, all reads are screened to distinguish reads bearing antibody-like content from off-target content [20]. Antibody analysis typically begins with identification of the V, D and J region segments found in each antibody using a known germline reference set (all methods). This reference set can be from any source, but in practice is most often obtained from IMGT [27]. Identification of reference segments enables somatic hypermutation analysis and statistical analysis of selective forces favoring individual segments [28]. Segment analysis is insufficient for selection interpretation, as antigen-driven selection acts not on the genetic elements directly, but rather on the translated CDRs. Consequently, it is critical to identify the correct frame of translation and obtain the translated CDR sequences.
The analysis of selection outputs is more straightforward than naïve library analysis. Once individual reads have been analyzed, the relative abundance of all clones in a selection is of great interest. To ensure accuracy, multiple sources of error, including PCR error, read error, bioinformatic classification ambiguity and variable read lengths, need to be accounted for. Once error processing is complete, clonal clustering can be used to gather de-facto identical clones, trace affinity maturation lineages of related clones, and even identify convergent paratopes emerging after a selection. The analysis of the clonal enrichment and clonal diversity of this final data can be used to trace individual clones across different panning rounds or selection pressures, estimate the total diversity of responding clones after a selection, and even estimate the total diversity of the selection output and the functional fitness of every amino acid at every homologous position [18,29].
In the identification of the best lead antibodies to pursue, the translation of frameworks and CDRs additionally enables annotation of biochemical and immunological liabilities – these include non-synonymous framework mutations, N-linked glycosylation sites, deamination sites, acid hydrolysis sites, free cysteines, known aggregation and destabilizing variation, domain truncations and other gross-defects caused by library assembly.
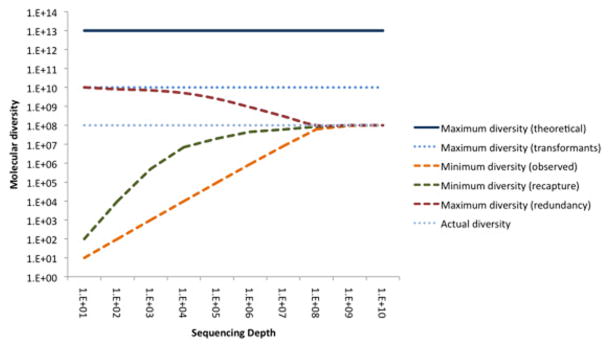
The analysis of naïve unselected libraries, and assessments of total diversity, is far more challenging, since total potential diversity (i.e. all VH+VL combinations) almost always exceeds deep sequencing capacity (<108) (figure 3). Accumulation analysis (i.e. counting number of unique clones observed) provides a lower bound estimate of how many sequences exist in a library, and is necessarily incomplete when the entire scFv or Fab fragment cannot be observed in a single sequence. While accumulation analysis of diversity on individual CDRs is effective for H1, H2, L1, L2 and L3, the H3 diversity alone can easily be greater than that of sequencing depth, and in our experience is probably increased ten fold when diversity in the remaining CDRs and frameworks are accounted for [20]. Furthermore, extrapolation of total library diversity from individual CDR observations requires either strong assumptions of positional independence, or sophisticated mathematical models of positional relationships. More effective measures for library diversity estimation can be borrowed from field ecology – the Fisher’s capture recapture [30] and the Chao statistic [31] can both be used to estimate the number of unseen species on the basis of the number and diversity of observed species, although both will likely return lower bound estimates. To complement lower bound estimates, a higher-bound estimate can be obtained by saturation analysis: subtracting the fraction of the repertoire taken up by observed high frequency clones. However, these species richness estimators are hindered by the presence of errors that inflate the number of rare species in the dataset (see next section). Used together, the methods provide a low-bound and high-bound of diversity, allowing for a sensitive detection of library defects that reduce effective library size below 109. (Figure 3).
Figure 3.
Estimating upper and lower diversity bounds as a function of sequencing depth. Maximum theoretical diversity is the total number of unique molecules that could exist in a library of this design if the number of transformants were infinite. Maximum transformant diversity is the maximum library size if every molecule in the library was non-redundant. Minimum observed diversity is the accumulated diversity observed from sequencing: the number of different clones actually seen. Minimum diversity estimated by capture-recapture methods more rapidly approaches the true diversity of the library, by anticipating library diversity from subsample overlap. Maximum diversity can be calculated by extracting known library redundancy from the transformation size. Actual diversity is the number of unique clones in the library. All measures convergence on true diversity with increasing sampling depth, although libraries with “long tails” of rare clones will converge slowly.
Overcoming challenges of antibody repertoire Informatic analysis
Annotating antibody sequences
The diversity of repertoires poses a number of unique bioinformatics challenges, compared with most other high-throughput sequence analysis applications (genomic sequencing, transcriptomics, chip-SEQ, microbiome analysis, virome analysis, etc). These involve mapping tens of millions of reads to a relatively finite reference set of segments that is on the order of thousands to hundred of thousands of segments (genes, exons, cDNAs, bacterial and viral genomes). In contrast, an antibody library can easily contain a billion antibodies, drawn from a VDJ rearrangement capacity exceeding 100 trillion possible combinations in most known organisms [32], and a nearly infinite molecular diversity (1050) when considering somatic hypermutation, or synthetic libraries created by highly diverse oligonucleotides. As a consequence, some mapping shortcuts cannot be performed, and each read must be analyzed individually at early stages of repertoire analysis. This additional computational burden is addressed either through distributed computing, typically on commercial cloud computing environments, or research into novel parsimonious algorithms (regex-based methods [21], the VDJ challenge [33]).
Segment identification is confounded by somatic hypermutation, codon-reoptimized frameworks, read error, incomplete reads, and incomplete allele coverage. Incorrect segment assignment is best avoided, as it can lead to artificial separation of variants of the same clonal family in downstream analysis. For natural encodings using segments from a well-characterized reference species, the majority of reads can be reliably identified by best-blast based methods [34]. Read error will have little effect on segment classification, as the majority of segments are easily distinguishable even given the burden of read error typical of high throughput sequencing technologies. To improve accurate identification of segments in clones with high degrees of somatic hypermutation or incomplete reads, a probabilistic classifier can be used to assign confidence to the top hit, and reduce the resolution of assignment when necessary [20,28]. When working with codon reoptimized frameworks, a novel reference segment database or segment assignment at the amino acid level can be attempted. The D-segment, given its short length and vulnerability to trim back, can often not be reliably classified in even the best circumstances.
Multiple analysis toolkits exist for the analysis of antibody library selections. The LANL Antibody Mining toolbox [21] operated through nucleotide-level pattern recognition of HCDR3 boundary elements. It is limited to frequency analysis of CDR3 sequences from naturally encoded human libraries, but is the fastest of all of the above algorithms, able to parse millions of reads in less than a minute, and thus well-suited for analyzing enrichment of CDR-H3 clones from a naïve library within minutes from a single computer. Most of the other toolkits provide analysis of segment identities and VDJ junctional boundaries and alignments, but at an additional computational cost that requires distributed computing capabilities to process millions of sequences efficiently. iHMMunalign uses a nucleotide-level Bayesian Hidden Markov Model to assign probabilities to segment identities and VDJ junctional boundaries [35]. The NIH/NIAID/CIT Joinsolver operates through a combination of nucleotide-level CDR3 boundary motif recognition and parsed segment alignments [36]. IMGT’s V-Quest benefits from wide breadth, performing analysis on BCRs, TCRs and multiple species including human, mouse, rat rabbit and pig, as well as a powerful reference database, classifying input sequences to their definitive reference IMGT set [37]. IMGT has also expanded their analysis support by offering High V-Quest, a web-based NGS compatible version currently able to handle up to 500k batches of sequencings, providing full annotation of VDJ segments, CDR regions, and somatic hypermutations [38]. The NIH Igblast performs blast-blast segment classification and boundary recognition [34]. The VDJFasta algorithm is the most generalizable tool, adapted for continued utility when analyzing very mutated antibodies, engineered antibodies and novel species [20,28,29]. VDJFasta utilizes amino acid profile Hidden Markov Models to identify CDRs and performs alignments by amino acid homology, then assigns segments by a probabilistic classifier. It is able to operate on any species without requiring a segment reference, as well as codon reoptimized frameworks and other heavily engineered monoclonal libraries. It can be run either in a very fast CDR3 discovery mode, or in a more complete analytical mode that recovers alignments, segments, CDR boundaries and biochemical liabilities. It provides affinity maturation tree construction as an embedded feature of the toolkit, unique among the other tools. ImmunediveRsity, is another stand-alone pipeline for the analysis of antibody repertoire data, providing quality filtering, noise correction and repertoire reconstruction based on VDJ assignment, clonal origin and unique VH identification. Finally, the very recently published open-access software MiXCR uses an advanced alignment algorithm that enables rapid annotation of germline segments, CDRs, SHMs, and error correction (see next section for more), processing 107 sequence reads in minutes [39].
In addition to academically available command line resources, a set of industrial platforms, including Adaptive Analytics and the Distributed Bio AbGenesis platform, have also emerged as solutions for non-technical users. Open source community portals, such as receptormarker.com, have emerged as free academic user interfaces for specific applications [40].
The amino acid diversity of immunoglobulins presents a challenge for accurate CDR identification: even 10% of the Kabat database is estimated to be mis-numbered by their own classification system [41]. Motif-based CDR boundary recognition methods can often be used to recognize CDRs, but they will fail in more heavily mutated antibodies, engineered antibodies, and antibodies from novel species. Profile Hidden Markov Model (HMM) based Bayesian methods have emerged as powerful tools for CDR recognition, given their ability to recognize homology signatures of the frameworks to aid in contextualizing CDR diversity [20]. Such tools can operate at the nucleotide [35] or amino acid [20] level. However, they tend to be slower than motif based CDR recognition, and require longer CDR flanking sequences to function, when considering naturally encoded human antibody libraries [21]. HMMs provide a substantial advantage when analyzing codon optimized libraries, novel species, or highly affinity matured antibodies such as broadly neutralizing HIV repertoires, as the amino acid homology signatures are more robust to mutation and do not require nucleotide motif re-definition with each new species.
Error correction
Another challenge in repertoire analysis, particularly the assessment of naïve library diversity, is the presence of errors, as all sequencing technologies are susceptible to read error [42]. Compounding errors introduced by the sequencing platform is the fact that antibody library preparation requires PCR amplification, where DNA polymerase can also produce additional errors. Furthermore, unlike genome or transcriptome sequencing where errors can often be simply corrected by read-based consensus building or alignment with a reference genome/transcriptome, antibodies undergo somatic hypermutation making it nearly impossible to distinguish between technical errors versus true biological mutations without an informatics-based error correction method.
Fortunately, the analysis of antibody repertoires, and in particular selections, is somewhat unique in that many types of read error have minimal impact on quantifiable features. Read error has little effect on segment identification, as it introduces proportionally less variation than somatic hypermutation into the underlying sequences. In analysis of positional amino acid frequencies in a total library, the read error rate will typically introduce less than 1% noise to observed frequencies. In selections, the reads of greatest interest will have the greatest depth of coverage, having expanded in the pool and thus receiving greater proportional read depth.
The greatest challenges lie in analysis of non-expanded clones or accurate total repertoire diversity. Errors in the CDR3 region could alter clonal diversity measurements, as 100% CDR3 identity is a common definition for clonality. The fact that CDR3 regions themselves are considered hotspots for somatic hypermutation adds further complexity to this problem. Recently the impact of errors was comprehensively evaluated where high-throughput sequencing (Illumina HiSeq) was performed on a “model repertoire” consisting of seven monoclonal cell lines expressing antibodies or TCRs [43]. Following, sequencing and annotation of CDR3 regions, there was a large number of false positive clones detected, which would have resulted in a drastic overestimation of clonal diversity. Indeed our results corroborate this. Following duplicate (using two different barcodes) Ion Torrent sequencing of the HCDR3 of a single VH region >99.5% of the ~80,000 sequences in each were correct. The remaining 0.5% comprised 166 unique HCDR3 false positives. ~40% were found in both barcodes, and the remaining sequences were unique to each barcode. Although the majority of these were 1, 2 or 3 amino acid mutations away from the original HCDR3, there were also a number of unrelated HCDR3s, thought to be the result of contamination (unpublished data Bradbury group).
Despite the presence of errors, simple methods of informatics processing and filtering can be utilized to achieve partial correction. One example is CDR-based clustering at the amino acid level (used in the example above), a method that minimizes the footprint of read error to non-synonymous mutations in the paratope and accounts for the majority of read error, which will be single nucleotide mutations away from a true clone [20]. Another is frequency-based consensus building. At the depth of sequencing now available, higher frequency clones will often result in hundreds (or thousands) of reads, while the majority of their read error variants will typically appear as singletons that are often only a single nucleotide away from the correct higher-frequency read. Thus these singletons can be dropped from analysis or corrected by consensus alignment to the higher frequency clone [16]. Another method to alleviate overestimation of clonal diversity is to apply clonotyping, which is the grouping of similar CDR3s (e.g., 80%, 90% identity) [44], although, measuring intraclonal diversity or the number of somatic variants would still not be possible. Tree-based single-linkage clustering methods are useful in libraries derived from natural repertoires where in vivo affinity maturation will generate complex SHM trees as well as read errors that may not resolve accurately by other clustering approaches [28]. In addition, another approach to partially overcome errors would be to perform replicate sequencing, in such a case only clones present in both replicates would be considered reliable [45–47]. However, this would not correct for reproducible sequencing or PCR hotspot errors; for example in our aforementioned sequencing experiment that resulted in 166 false positive HCDR3s, common false HCDR3s were found in both datasets. Others have also observed this phenomenon of reproducible systematic errors in NGS [43,48]. So while these methods are easy to implement and do improve repertoire accuracy, they still fail to provide fully accurate measurements of somatic hypermutation or clonal diversity, thus making more advanced methods necessary
In order to correct for errors in NGS, several variations of an advanced method have been developed which rely on library preparation with unique molecular identifiers (UMIs, which are also known as unique identifiers, barcodes, molecular identifiers groups, primer IDs); UMIs are a stretch of degenerate nucleotides (e.g., NNNNNNNN) that are typically added to mRNA or cDNA molecules via reverse transcription or ligation [49,50]. Thus when PCR or sequencing introduces errors, these can be corrected by grouping sequences that share a common UMI and correcting variant sequences to the group’s consensus sequence [51] (Figure 2B). The consensus is typically the correct sequence since all sequences with a common UMI are assumed to be derived from the same original template molecule. Recently UMI addition has also been applied for antibody repertoire sequencing. In one example, UMIs were incorporated into forward and reverse primers and added during first and second strand cDNA synthesis, which was combined with replicate sequencing to improve the accuracy of human B cell repertoires obtained from vaccinated individuals [52]. While UMI-based consensus building enables correction of sequencing errors, it does not address all PCR errors. For example, a polymerase-introduced error in an early PCR cycle that ends up becoming the majority positional nucleotide for that UMI group would result in a false consensus built sequence variant. While this might be considered a rare event, several reports have identified that this is more common than once believed [48,53]. To date the only method that has been developed to correct for PCR errors is based on UMI-labeling of cDNA followed by a read-gain/loss secondary correction (filtering) step [43]. Here, the original sequences or clone read counts are compared to those after consensus building. Since these early PCR errors tend to be systematic and reproducible, they will often appear in multiple UMI groups in later rounds of amplification. This phenomena leads to an overall greater number of erroneous variants being corrected, resulting in a net loss of erroneous sequence variants. This method when applied to a control repertoire was able to achieve nearly absolute error correction (removal of all false positive CDR3 variants) [43]. All UMI-based error correction methods require oversampling of UMIs (each UMI ≥ 3 reads). However, this has been challenging to accomplish, as it either requires a very high read depth or precise sample preparation and quantification methods to achieve adequate, but not excessive oversampling. This sample preparation precision has yet to be fully standardized for antibody sequencing. Finally, it has also emerged that errors introduced to the UMIs themselves are substantially present and will thus need to be addressed in the future [53,54].
Applications
Naïve library size estimation
The size of naïve antibody libraries has been generally assessed by counting the number of bacterial colonies on a dilution plate after transformation, and multiplying accordingly, making the assumption that each bacterial colony represents a unique antibody. While this assumption may be reasonable in synthetic libraries, in which potential diversity usually exceeds actual diversity by orders of magnitude, this may be less true for libraries prepared from natural sources in which clonal dominance may occur if the number of donors is limited. The ability to sequence millions of antibodies in a naïve library allows a far more accurate assessment of diversity. The heavy chain variable region, and in particular, the heavy chain CDR3, are considered to be the most important determinants of recognition [17,18,55], exemplified by experiments in which HCDR3 sequences from an anti-lysozyme VHH antibody (VHH) [56] were transplanted into neocarzinostatin [57] and sfGFP [58], conferring lysozyme binding activity. HCDR3’s have even been harvested as diversity elements [59,60] and binders have been selected from libraries in which they provided the only diversity [18,58]. As a result, deep sequencing initially concentrated on HCDR3, expanding to VH as read lengths increased. One somewhat surprising result from naïve natural library sequencing [20,21], is that lower bound estimates of heavy chain diversity, 3x106 unique clones (40 donors, ref [21]) and as little as 2x105 (~654 donors, ref [20]) paratopes differing by at least 2 amino acids to any other, assessed using different methods, are significantly less than the almost limitless potential diversity of human VH rearrangement [32], and far closer to the VH diversity found in any single person (106–7) [32]. Given that most VH sequences are unique to an individual [14,28], unlike VL sequences, which are more commonly public [14], one would expect the number of unique VH sequences in a natural library to increase with the number of donors. That this does not appear to be the case is likely to be a consequence of library construction methods. In both the described cases of naïve natural library sequencing [20,21], V region amplifications were carried out on pools of B cell cDNA from different donors using pools of specific primers [61]. The extent of multiplex primer bias in repertoire sequencing has recently been carefully evaluated using 56 synthetic templates of all human TCR V-alpha genes, which revealed substantial bias, as in some cases entire V genes were not amplified at all [62]. Therefore multiplex PCR with pools of primers may be expected to bias amplification towards templates with more favorable primer-specific regions, as well as VH genes that are more abundant. One way to overcome bias would be to use both optimized primer concentrations and informatics correction, however this requires rather in-depth and sophisticated characterization studies [62]. Another possibility would be to use individual primer pairs on each individual, or small pools of individuals, rather than pooled, B cell cDNA. Although one very large library has been created using this approach [63], it has not been analyzed by deep sequencing.
For synthetic libraries, theoretical diversity in the heavy chain (but not usually the light chain) can vastly exceed the diversity achievable by bacterial transformation, depending on the design. This is confirmed by NGS: in an appropriately designed library, the vast majority of clones occurs only once, even when applying strict paratope distance measures to ensure that read error isn’t artificially inflating the result [12,29,64,65].
For both natural and synthetic libraries, if VL and VH chains are assorted independently, library diversity increases enormously. This makes most sequences unique, and validates the colony counting approach to estimate library size. However, if construction methods (e.g. assembly PCR) are used where opportunities for clonal dominance exist, diversity may be overestimated by colony counting, and can only be assessed by NGS.
Quality control and library design
In addition to assessing diversity, NGS has been proposed as a straightforward method to quality control libraries after they have been created [64]. Sequencing provides accurate information on the percentages of clones that are non-functional due to stop codons or frame shifts; assesses how close actual diversity of synthetic libraries is to designed diversity; assesses the randomness of VH/VL linkage; and can assess library redundancy due to contaminants or clonal dominance.
Even when clones appear correct on the basis of sequence, they may be nonfunctional. For synthetic libraries, diversity may be functionally reduced by sequences that prevent correct folding, or are polyreactive, due to inappropriate amino acid choices in complementarity determining regions (CDRs) [66]. In the case of natural libraries, reduced functionality may be caused by excessively mutated V regions which display poorly [67], or by the restricted recognition properties of libraries with reduced VH diversity, given that VH is the major determinant of antibody recognition. The deep sequencing and analysis of well (and poorly) folding, or non-aggregating [68], antibody variable regions will result in the gradual accumulation of data on amino acid preferences at different positions (e.g. see refs [69,70]), which in turn will feed back into library design and more sophisticated functional quality control analyses that go beyond the mere identification of open reading frames.
For both library classes incompatible VH/VL pairs are also likely to reduce functional diversity. While there is likely to be significant individual variation, this may be mitigated by choosing known functional VH/VL pairs[65]. In addition to analyzing final library diversity, NGS can be used to monitor fidelity of components during construction [29]. It is expected that its continued use after each step of library construction will allow the direct analysis of the roles of different construction strategies in the generation, or loss, of diversity in the future, allowing more efficient library construction. The unexpected relatively low final VH diversity described in the two libraries above, could be better understood if NGS was applied to intermediate steps in the construction process, and indicate the insight NGS can bring to library creation methods.
Selections
Target specific
One of the paradoxes in the early days of selection from antibody libraries was the inconsistency between the number of identified unique positive clones selected from large libraries, and the number of clones expected from theoretical calculations [71], or the scale up of the selection result from small libraries [72,73]. While the number of unique positive clones will depend upon the complexity of the antigen, the threshold affinity and the number of clones screened, practical experiments [72,73] indicate that it should be possible to select 1–5 positive clones from libraries with a diversity of 107, suggesting that libraries 100 fold greater in size (~109) should yield 100–500 unique binders. In general, this has not been the case, unless extreme efforts have been taken to carry out selection under many different conditions [74]. Deep sequencing of selection outputs reveals that this is a combination of libraries not being as diverse as anticipated, and also that the recovery of unique clones poses a sampling problem: when only 96–384 clones are tested in different selection experiments, sometimes not even the ten most abundant clones can be identified [12,75]. Furthermore, when 96/384 clones are randomly picked, most are duplicates of abundant clones, while others represent single copies of far less abundant clones [76]. In our experience, such rare clones may individually comprise less than 0.001% of the selection output, and yet still be positive for the target, indicating that the only way to identify the full spectrum of binding antibodies after selection is to sequence and rank the complete output. This of course makes it even more paramount to apply error correction methods, as otherwise true rare clones would not be able to be distinguished from errors. However, sequencing needs to be sufficiently deep that such clones are seen multiple times as read error correction cannot be carried out on single sequences. Sequence identification, however, is not the same as clone isolation. Once identified, clones can be isolated with inverse PCR [77], using the HCDR3 as a barcode for outward facing primers [78,79]. In order to reduce the numbers of primers required, arbitrary screening can be used initially. This usually provides many of the commonest clones, as well as a random selection of rarer ones, which, after individual sequencing, can be mapped back to the ranked list of antibodies. Inverse PCR can then be used to isolate missed clones.
NGS has also shown that the pattern of diversity found in selection outputs against different targets can be very variable. In some cases selections are dominated by single HCDR3s (or VHs), while in other cases, diversity is far broader. However, even when responses are relatively monoclonal, less abundant clones isolated by inverse PCR, are positive. Given their low abundance, NGS is the only way that these rare clones can be identified, as they cannot be found by standard screening methods. In these cases NGS is able to rescue selections that would otherwise have been considered failures due to their apparent limited diversities.
Identification of clones with desirable properties
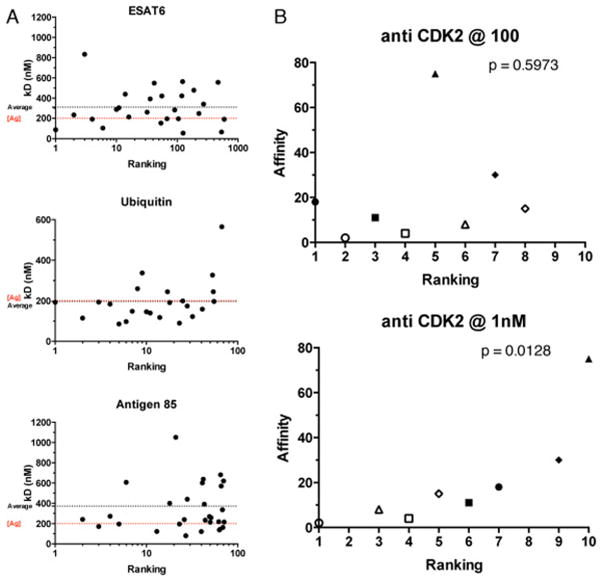
Initial experiments in which antibodies were ranked for abundance after phage/yeast display selection and deep sequencing surprisingly revealed no correlation between affinity and abundance for all targets we have tested (fig 4a). In these experiments target concentration was kept relatively high (~200nM), in order to preserve binding diversity, but as can be seen, the antibodies with the highest affinities (lowest Kd) are usually the less abundant ones. We believe this is because all antibodies with Kds lower than the target concentration used for sorting will bind approximately similar amounts of antigen, providing them with no selective advantage over antibodies with better affinities. As target concentration is reduced, only those yeast displaying antibodies with lower Kds are able to capture target. Further analysis revealed that at the lowest target concentration at which positive yeast can be identified by flow cytometry, there is a far better correlation between abundance rank and affinity (fig 4b). Consequently, we believe NGS can be used to identify antibodies with the best affinities in a binding population, by sorting with diminishing target concentrations, and sequencing the output of the lowest target concentration that yields a positive population. Under these conditions, the most abundant antibodies will tend to be those with the lowest Kds. It is likely that similar approaches can be taken to similarly optimize individual steps in the selection process, including washing, temperature, incubation times and elution methods for phage display as well.
Figure 4.
Relative abundance and affinity. Panel A: the experimentally measured kDs (nM) of selected clones are plotted in relation to their ranking position in the sequenced selection output (i.e. the clone in ranking position 1 has the highest relative abundance in the selection output). Average affinity of the clones (black) and antigen concentration used in the selection process (red) are shown as dotted lines. Data collected for three different antigen selections (ESAT6, Antigen85, and Ubiquitin) are reported. Panel B: ranking and affinity plots are shown for anti CDK2 selection at different antigen concentrations. The affinities of identical clones (identified by the same symbol in the 2 plots) found in sequenced populations selected at different antigen concentrations are shown in relation to their ranking position. P-values for significant correlation are reported.
In addition to sequencing the outputs of target-specific selections, it will also be possible to apply deep sequencing to the analysis of common desirable antibody traits, such as thermostability [80], binding to protein A [81] or high display/expression levels [82], and other developability traits. We anticipate this can be carried out on individual target specific selections, or by sequencing complete libraries subjected to particular selection gates. In the latter case it may be possible to identify common sequence features correlated with desired properties, which could then be used to build and improve subsequent libraries, as described above. Such approaches could be powerfully combined with structural modeling and prediction using in silico prediction tools (e.g., Rosetta, MOE, Discovery studio) [83–86].
Identification of common clones
When display methods are used to generate antibodies, the selection targets are more complex than assumed. Antigens are usually biotinylated [87], which introduces additional complications: the presence of the biotin, the chemical moiety linking the biotin to the protein and the streptavidin (which itself may or may not be modified). Further, many targets are expressed recombinantly, and include common domains, such as peptide tags recognized by antibodies, fusion protein or His tags (see [88] for a review). All these additional common components can themselves, become targets for selection, potentially leading to antibodies that do not recognize the specific target but the common feature. Although appropriate controls and negative selections usually allow the elimination of such apparently cross-reactive antibodies, NGS can also be used to identify them after selection. In a recent paper [89], polyclonal antibodies selected from a large naïve library [90] created by recombination [91] using phage/yeast display [76,92] against a series of in vitro biotinylated proteins were found to be strongly cross-reactive with other targets. Careful analysis of the cross-reactivity revealed the polyclonal antibody pool recognized proteins biotinylated using a particular kit (Lightening Link), but not if biotinylated with other kits or in vivo. Deep sequencing of the antibody populations showing this cross-reactivity identified one common antibody in all the selections, which when tested, was found to recognize the Lightening Link biotinylation site [89]. A similar approach could be adapted to the identification of antibodies recognizing epitopes in common between different targets and enable informatics based library removal: e.g. human and murine versions of the same protein, or related therapeutic targets.
Conclusion
Next generation sequencing has been introduced to the study of molecular diversity libraries only relatively recently. However, its power, quantitative nature, and analytical depth and breadth is likely to make it an essential investigative tool in the generation and use of libraries based on antibodies and other scaffolds. As more advanced sequencing and informatics tools become available, we anticipate that it will only become more valuable to integrate NGS with antibody engineering.
Table 2.
Repertoire analysis and tree clustering with VDJFasta
Repertoire analysis and error/tree clustering with VDJFasta. A) MiSeq paired end reads are converted to joined fasta. B) Reads are split into subsets of 10,000 reads per file for parallel processing, and submitted to an openLAVA queueing service. C) Analyzed output files are joined and paratope clustered to gather read errors and construction affinity maturation trees.
| Converting paired end Illumina to joined fasta |
fastq2fasta.pl –-file=seq_R1.fastq > seq_R1.fasta
fastq2fasta.pl –-file=seq_R2.fastq > seq_R2.fasta
fasta-hiseq-join.pl --forward=seq_R1.fasta \
--reverse=seq_R2.fasta \
--outputfile=seq_join.fa
|
| Splitting of reads into 10k subunits for distributed analysis |
fasta-split.pl -–file=seq_join.fa for file in *split.fa do bsub “fasta-vdj-pipeline.pl –file=$file” done |
| Joining and SHM tree construction |
fasta-join.pl –file=seq_join fasta-cluster.pl –file=seq_join.VDJ.H3.L3.CH1.dnaH3.fa |
Highlights.
Explanation of next generation sequencing technologies
Use of next generation sequencing in selection from display libraries
Discussions of appropriate informatic analyses, errors and error correction
Future directions sequencing technologies will impact display technologies
Acknowledgments
This work was supported by: NIH U54 Grant “Technology Development for New Affinity Reagents Against the Human Proteome (U54) RFA-RM-10–018,” grant number 1-U54-DK093500-01 (to AB); the Swiss National Science Foundation SystemsX.ch – AntibodyX RTD project grant (to STR).
Footnotes
Conflict of interest
Jacob Glanville is the CSO and co-founder of Distributed Bio, a company that provides the AbGenesis antibody repertoire analysis package as a commercial service.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- *1.Ostermeier C, Iwata S, Ludwig B, Michel H. Fv fragment-mediated crystallization of the membrane protein bacterial cytochrome c oxidase. Nat Struct Biol. 1995;2:842–846. doi: 10.1038/nsb1095-842. One of the first examples of the use of recombinant antibodies as crystallization chaperones. [DOI] [PubMed] [Google Scholar]
- 2.Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam AY, Pardon E, Korotkov KV, Hol WG, Steyaert J. Nanobody-aided structure determination of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus. J Struct Biol. 2009;166:8–15. doi: 10.1016/j.jsb.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korotkov KV, Pardon E, Steyaert J, Hol WG. Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure. 2009;17:255–265. doi: 10.1016/j.str.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uysal S, Vasquez V, Tereshko V, Esaki K, Fellouse FA, Sidhu SS, Koide S, Perozo E, Kossiakoff A. Crystal structure of full-length KcsA in its closed conformation. Proc Natl Acad Sci U S A. 2009;106:6644–6649. doi: 10.1073/pnas.0810663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *6.Ye JD, Tereshko V, Frederiksen JK, Koide A, Fellouse FA, Sidhu SS, Koide S, Kossiakoff AA, Piccirilli JA. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc Natl Acad Sci U S A. 2008;105:82–87. doi: 10.1073/pnas.0709082105. The application of an antibody as a crystallization chaperone to study RNA structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schweizer A, Roschitzki-Voser H, Amstutz P, Briand C, Gulotti-Georgieva M, Prenosil E, Binz HK, Capitani G, Baici A, Pluckthun A, et al. Inhibition of caspase-2 by a designed ankyrin repeat protein: specificity, structure, and inhibition mechanism. Structure. 2007;15:625–636. doi: 10.1016/j.str.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Huber T, Steiner D, Rothlisberger D, Pluckthun A. In vitro selection and characterization of DARPins and Fab fragments for the co-crystallization of membrane proteins: The Na(+)-citrate symporter CitS as an example. Journal of structural biology. 2007;159:206–221. doi: 10.1016/j.jsb.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Kohl A, Amstutz P, Parizek P, Binz HK, Briand C, Capitani G, Forrer P, Pluckthun A, Grutter MG. Allosteric inhibition of aminoglycoside phosphotransferase by a designed ankyrin repeat protein. Structure (Camb) 2005;13:1131–1141. doi: 10.1016/j.str.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. High-affinity single-domain binding proteins with a binary-code interface. Proc Natl Acad Sci U S A. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Low C, Yau YH, Pardon E, Jegerschold C, Wahlin L, Quistgaard EM, Moberg P, Geifman-Shochat S, Steyaert J, Nordlund P. Nanobody mediated crystallization of an archeal mechanosensitive channel. PLoS One. 2013;8:e77984. doi: 10.1371/journal.pone.0077984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravn U, Gueneau F, Baerlocher L, Osteras M, Desmurs M, Malinge P, Magistrelli G, Farinelli L, Kosco-Vilbois MH, Fischer N. By-passing in vitro screening--next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res. 2010;38:e193. doi: 10.1093/nar/gkq789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Niro R, Ziller F, Florian F, Crovella S, Stebel M, Bestagno M, Burrone O, Bradbury AR, Secco P, Marzari R, et al. Construction of miniantibodies for the in vivo study of human autoimmune diseases in animal models. BMC Biotechnol. 2007;7:46–55. doi: 10.1186/1472-6750-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **14.DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. 2014 doi: 10.1038/nm.3743. First high throughput sequence analysis of paired heavy and light chain human antibodies using a microfluidic approach. [DOI] [PubMed] [Google Scholar]
- 15.Travers KJ, Chin CS, Rank DR, Eid JS, Turner SW. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res. 2010;38:e159. doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **16.Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH, Chrysostomou C, Hunicke-Smith SP, Iverson BL, Tucker PW, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol. 2010;28:965–969. doi: 10.1038/nbt.1673. One of the initial papers that demonstrated the ability to use NGS of antibody repertoires for monoclonal antibody discovery. [DOI] [PubMed] [Google Scholar]
- *17.Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. Shows the importance of the heavy chain CDR3 for antibody specificity in anin vivo model. [DOI] [PubMed] [Google Scholar]
- 18.Mahon CM, Lambert MA, Glanville J, Wade JM, Fennell BJ, Krebs MR, Armellino D, Yang S, Liu X, O’Sullivan CM, et al. Comprehensive interrogation of a minimalist synthetic CDR-H3 library and its ability to generate antibodies with therapeutic potential. J Mol Biol. 2013;425:1712–1730. doi: 10.1016/j.jmb.2013.02.015. [DOI] [PubMed] [Google Scholar]
- *19.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HW, Jr, Kirkham PM. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. One of the best early analyses of heavy chain CDR3 in mouse and man. [DOI] [PubMed] [Google Scholar]
- *20.Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, Ni I, Mei L, Sundar PD, Day GM, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A. 2009;106:20216–20221. doi: 10.1073/pnas.0909775106. One of the first applications of deep sequencing to the study of natural antibody repertoires, including a discussion of the issues in estimating diversity from relatively limited read numbers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Angelo S, Glanville J, Ferrara F, Naranjo L, Gleasner CD, Shen X, Bradbury AR, Kiss C. The antibody mining toolbox: an open source tool for the rapid analysis of antibody repertoires. MAbs. 2014;6:160–172. doi: 10.4161/mabs.27105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **22.Bradbury A, Pluckthun A. Reproducibility: Standardize antibodies used in research. Nature. 2015;518:27–29. doi: 10.1038/518027a. A commentary on the problems with research antibodies and a proposed solution. [DOI] [PubMed] [Google Scholar]
- 23.Dekosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM, Varadarajan N, Giesecke C, Dorner T, Andrews SF, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamada M, Hase S, Sato K, Toyoda A, Fujiyama A, Sakakibara Y. Whole genome complete resequencing of Bacillus subtilis natto by combining long reads with high-quality short reads. PLoS One. 2014;9:e109999. doi: 10.1371/journal.pone.0109999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One. 2012;7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribeiro FJ, Przybylski D, Yin S, Sharpe T, Gnerre S, Abouelleil A, Berlin AM, Montmayeur A, Shea TP, Walker BJ, et al. Finished bacterial genomes from shotgun sequence data. Genome Res. 2012;22:2270–2277. doi: 10.1101/gr.141515.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehrenmann F, Lefranc MP. IMGT/DomainGapAlign: the IMGT(R) tool for the analysis of IG, TR, MH, IgSF, and MhSF domain amino acid polymorphism. Methods Mol Biol. 2012;882:605–633. doi: 10.1007/978-1-61779-842-9_33. [DOI] [PubMed] [Google Scholar]
- 28.Glanville J, Kuo TC, von Budingen HC, Guey L, Berka J, Sundar PD, Huerta G, Mehta GR, Oksenberg JR, Hauser SL, et al. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Natl Acad Sci U S A. 2011;108:20066–20071. doi: 10.1073/pnas.1107498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *29.Zhai W, Glanville J, Fuhrmann M, Mei L, Ni I, Sundar PD, Van Blarcom T, Abdiche Y, Lindquist K, Strohner R, et al. Synthetic antibodies designed on natural sequence landscapes. Journal of molecular biology. 2011;412:55–71. doi: 10.1016/j.jmb.2011.07.018. Application of deep sequencing to the study and creation of a synthetic antibody library. [DOI] [PubMed] [Google Scholar]
- *30.Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. One of the first applications of deep sequencing to the study of natural antibody repertoires. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi Q, Liu Y, Cheng Y, Glanville J, Zhang D, Lee JY, Olshen RA, Weyand CM, Boyd SD, Goronzy JJ. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci U S A. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnaout R, Lee W, Cahill P, Honan T, Sparrow T, Weiand M, Nusbaum C, Rajewsky K, Koralov SB. High-resolution description of antibody heavy-chain repertoires in humans. PLoS One. 2011;6:e22365. doi: 10.1371/journal.pone.0022365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakhani KR, Boudreau KJ, Loh PR, Backstrom L, Baldwin C, Lonstein E, Lydon M, MacCormack A, Arnaout RA, Guinan EC. Prize-based contests can provide solutions to computational biology problems. Nat Biotechnol. 2013;31:108–111. doi: 10.1038/nbt.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaeta BA, Malming HR, Jackson KJ, Bain ME, Wilson P, Collins AM. iHMMune-align: hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics. 2007;23:1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- 36.Souto-Carneiro MM, Longo NS, Russ DE, Sun HW, Lipsky PE. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J Immunol. 2004;172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- **37.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–508. doi: 10.1093/nar/gkn316. Reference for the widely used IMGT database of immunological sequences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alamyar E, Giudicelli V, Li S, Duroux P, Lefranc MP. IMGT/HighVQUEST: the IMGTs web portal for immunoglobulin (IG) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Res. 2012;8:26. [Google Scholar]
- 39.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, Chudakov DM. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 40.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abhinandan KR, Martin AC. Analysis and improvements to Kabat and structurally correct numbering of antibody variable domains. Mol Immunol. 2008;45:3832–3839. doi: 10.1016/j.molimm.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 42.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. Performance comparison of benchtop high-throughput sequencing platforms. Nature biotechnology. 2012 doi: 10.1038/nbt.2198.. [DOI] [PubMed] [Google Scholar]
- **43.Shugay M, Britanova OV, Merzlyak EM, Turchaninova MA, Mamedov IZ, Tuganbaev TR, Bolotin DA, Staroverov DB, Putintseva EV, Plevova K, et al. Towards error-free profiling of immune repertoires. Nat Methods. 2014;11:653–655. doi: 10.1038/nmeth.2960. Demonstrated with a control repertoire that NGS resulted in large number of false positive clonal (HCDR3) variants, then provided an experimental-bioinformatic UMI-based approach to achieve error correction. [DOI] [PubMed] [Google Scholar]
- 44.Wine Y, Boutz DR, Lavinder JJ, Miklos AE, Hughes RA, Hoi KH, Jung ST, Horton AP, Murrin EM, Ellington AD, et al. Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proc Natl Acad Sci U S A. 2013;110:2993–2998. doi: 10.1073/pnas.1213737110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greiff V, Menzel U, Haessler U, Cook SC, Friedensohn S, Khan TA, Pogson M, Hellmann I, Reddy ST. Quantitative assessment of the robustness of next-generation sequencing of antibody variable gene repertoires from immunized mice. BMC Immunol. 2014;15:40. doi: 10.1186/s12865-014-0040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Menzel U, Greiff V, Khan TA, Haessler U, Hellmann I, Friedensohn S, Cook SC, Pogson M, Reddy ST. Comprehensive evaluation and optimization of amplicon library preparation methods for high-throughput antibody sequencing. PLoS One. 2014;9:e96727. doi: 10.1371/journal.pone.0096727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robasky K, Lewis NE, Church GM. The role of replicates for error mitigation in next-generation sequencing. Nat Rev Genet. 2014;15:56–62. doi: 10.1038/nrg3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brodin J, Mild M, Hedskog C, Sherwood E, Leitner T, Andersson B, Albert J. PCR-induced transitions are the major source of error in cleaned ultra-deep pyrosequencing data. PLoS One. 2013;8:e70388. doi: 10.1371/journal.pone.0070388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc Natl Acad Sci U S A. 2011;108:20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. Practical innovations for high-throughput amplicon sequencing. Nat Methods. 2013;10:999–1002. doi: 10.1038/nmeth.2634. [DOI] [PubMed] [Google Scholar]
- 51.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *52.Vollmers C, Sit RV, Weinstein JA, Dekker CL, Quake SR. Genetic measurement of memory B-cell recall using antibody repertoire sequencing. Proc Natl Acad Sci U S A. 2013;110:13463–13468. doi: 10.1073/pnas.1312146110. Combined UMI-tagging and replicate sequencing to achieve error correction for human antibody libraries. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brodin J, Hedskog C, Heddini A, Benard E, Neher RA, Mild M, Albert J. Challenges with using primer IDs to improve accuracy of next generation sequencing. PLoS One. 2015;10:e0119123. doi: 10.1371/journal.pone.0119123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deakin CT, Deakin JJ, Ginn SL, Young P, Humphreys D, Suter CM, Alexander IE, Hallwirth CV. Impact of next-generation sequencing error on analysis of barcoded plasmid libraries of known complexity and sequence. Nucleic Acids Res. 2014;42:e129. doi: 10.1093/nar/gku607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kabat EA, Wu TT. Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J Immunol. 1991;147:1709–1719. [PubMed] [Google Scholar]
- 56.Desmyter A, Transue TR, Ghahroudi MA, Thi MH, Poortmans F, Hamers R, Muyldermans S, Wyns L. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol. 1996;3:803–811. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 57.Nicaise M, Valerio-Lepiniec M, Minard P, Desmadril M. Affinity transfer by CDR grafting on a nonimmunoglobulin scaffold. Protein Sci. 2004;13:1882–1891. doi: 10.1110/ps.03540504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai M, Temirov J, Pesavento E, Kiss C, Velappan N, Pavlik P, Werner JH, Bradbury AR. Using T7 phage display to select GFP-based binders. Protein Eng Des Sel. 2008;21:413–424. doi: 10.1093/protein/gzn016. [DOI] [PubMed] [Google Scholar]
- 59.Kiss C, Fisher H, Pesavento E, Dai M, Valero R, Ovecka M, Nolan R, Phipps ML, Velappan N, Chasteen L, et al. Antibody binding loop insertions as diversity elements. Nucleic Acids Res. 2006;34:e132. doi: 10.1093/nar/gkl681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Venet S, Kosco-Vilbois M, Fischer N. Comparing CDRH3 diversity captured from secondary lymphoid organs for the generation of recombinant human antibodies. MAbs. 2013;5:690–698. doi: 10.4161/mabs.25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *61.Sblattero D, Bradbury A. A definitive set of oligonucleotide primers for amplifying human V regions. Immunotechnology. 1998;3:271–278. doi: 10.1016/s1380-2933(97)10004-5. One of the most widely used primer sets for amplication of human variable regions. [DOI] [PubMed] [Google Scholar]
- 62.Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, Steen MS, LaMadrid-Herrmannsfeldt MA, Williamson DW, Livingston RJ, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- *63.Schofield DJ, Pope AR, Clementel V, Buckell J, Chapple S, Clarke KF, Conquer JS, Crofts AM, Crowther SR, Dyson MR, et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 2007;8:R254. doi: 10.1186/gb-2007-8-11-r254. Demonstrates the utility of in vitro antibody selection at high throughput. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ravn U, Didelot G, Venet S, Ng KT, Gueneau F, Rousseau F, Calloud S, Kosco-Vilbois M, Fischer N. Deep sequencing of phage display libraries to support antibody discovery. Methods. 2013;60:99–110. doi: 10.1016/j.ymeth.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 65.Tiller T, Schuster I, Deppe D, Siegers K, Strohner R, Herrmann T, Berenguer M, Poujol D, Stehle J, Stark Y, et al. A fully synthetic human Fab antibody library based on fixed VH/VL framework pairings with favorable biophysical properties. MAbs. 2013;5:445–470. doi: 10.4161/mabs.24218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Birtalan S, Zhang Y, Fellouse FA, Shao L, Schaefer G, Sidhu SS. The Intrinsic Contributions of Tyrosine, Serine, Glycine and Arginine to the Affinity and Specificity of Antibodies. J Mol Biol. 2008;377:1518–1528. doi: 10.1016/j.jmb.2008.01.093. [DOI] [PubMed] [Google Scholar]
- 67.Saggy I, Wine Y, Shefet-Carasso L, Nahary L, Georgiou G, Benhar I. Antibody isolation from immunized animals: comparison of phage display and antibody discovery via V gene repertoire mining. Protein engineering, design & selection : PEDS. 2012;25:539–549. doi: 10.1093/protein/gzs060. [DOI] [PubMed] [Google Scholar]
- 68.Dudgeon K, Rouet R, Kokmeijer I, Schofield P, Stolp J, Langley D, Stock D, Christ D. General strategy for the generation of human antibody variable domains with increased aggregation resistance. Proc Natl Acad Sci U S A. 2012;109:10879–10884. doi: 10.1073/pnas.1202866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jung S, Spinelli S, Schimmele B, Honegger A, Pugliese L, Cambillau C, Pluckthun A. The importance of framework residues H6, H7 and H10 in antibody heavy chains: experimental evidence for a new structural subclassification of antibody V(H) domains. J Mol Biol. 2001;309:701–716. doi: 10.1006/jmbi.2001.4665. [DOI] [PubMed] [Google Scholar]
- 70.Wang N, Smith WF, Miller BR, Aivazian D, Lugovskoy AA, Reff ME, Glaser SM, Croner LJ, Demarest SJ. Conserved amino acid networks involved in antibody variable domain interactions. Proteins. 2009;76:99–114. doi: 10.1002/prot.22319. [DOI] [PubMed] [Google Scholar]
- **71.Perelson AS, Oster GF. Theoretical studies of clonal selection: minimal antibody repertoire size and reliability of self-non-self discrimination. J Theor Biol. 1979;81:645–670. doi: 10.1016/0022-5193(79)90275-3. Highly cited paper on the theoretical background to selection from antibody repertoires. [DOI] [PubMed] [Google Scholar]
- **72.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. The first paper describing selection from naïve human phage antibody libraries. [DOI] [PubMed] [Google Scholar]
- 73.Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, Kontermann RE, Jones PT, Low NM, Allison TJ, et al. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13:3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edwards BM, Barash SC, Main SH, Choi GH, Minter R, Ullrich S, Williams E, Du Fou L, Wilton J, Albert VR, et al. The remarkable flexibility of the human antibody repertoire; isolation of over one thousand different antibodies to a single protein, BLyS. J Mol Biol. 2003;334:103–118. doi: 10.1016/j.jmb.2003.09.054. [DOI] [PubMed] [Google Scholar]
- 75.Di Niro R, Ferrara F, Not T, Bradbury AR, Chirdo F, Marzari R, Sblattero D. Characterizing monoclonal antibody epitopes by filtered gene fragment phage display. Biochem J. 2005;388:889–894. doi: 10.1042/BJ20041983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferrara F, D’Angelo S, Gaiotto T, Naranjo L, Tian H, Graslund S, Dobrovetsky E, Hraber P, Lund-Johansen F, Saragozza S, et al. Recombinant renewable polyclonal antibodies. MAbs. 2015;7:32–41. doi: 10.4161/19420862.2015.989047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoskins RA, Stapleton M, George RA, Yu C, Wan KH, Carlson JW, Celniker SE. Rapid and efficient cDNA library screening by self-ligation of inverse PCR products (SLIP) Nucleic Acids Res. 2005;33:e185. doi: 10.1093/nar/gni184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.D’Angelo S, Kumar S, Naranjo L, Ferrara F, Kiss C, Bradbury AR. From deep sequencing to actual clones. Protein Eng Des Sel. 2014;27:301–307. doi: 10.1093/protein/gzu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spiliotopoulos A, Owen JP, Maddison BC, Dreveny I, Rees HC, Gough KC. Sensitive recovery of recombinant antibody clones after their in silico identification within NGS datasets. J Immunol Methods. 2015 doi: 10.1016/j.jim.2015.03.005.. [DOI] [PubMed] [Google Scholar]
- 80.Orr BA, Carr LM, Wittrup KD, Roy EJ, Kranz DM. Rapid method for measuring ScFv thermal stability by yeast surface display. Biotechnol Prog. 2003;19:631–638. doi: 10.1021/bp0200797. [DOI] [PubMed] [Google Scholar]
- 81.Hillson JL, Karr NS, Oppliger IR, Mannik M, Sasso EH. The structural basis of germline-encoded VH3 immunoglobulin binding to staphylococcal protein A. J Exp Med. 1993;178:331–336. doi: 10.1084/jem.178.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shusta EV, Kieke MC, Parke E, Kranz DM, Wittrup KD. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J Mol Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- 83.Sircar A, Kim ET, Gray JJ. RosettaAntibody: antibody variable region homology modeling server. Nucleic acids research. 2009;37:W474–479. doi: 10.1093/nar/gkp387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weitzner BD, Kuroda D, Marze N, Xu J, Gray JJ. Blind prediction performance of RosettaAntibody 3. 0: grafting, relaxation, kinematic loop modeling, and full CDR optimization. Proteins. 2014;82:1611–1623. doi: 10.1002/prot.24534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li L, Kumar S, Buck PM, Burns C, Lavoie J, Singh SK, Warne NW, Nichols P, Luksha N, Boardman D. Concentration dependent viscosity of monoclonal antibody solutions: explaining experimental behavior in terms of molecular properties. Pharm Res. 2014;31:3161–3178. doi: 10.1007/s11095-014-1409-0. [DOI] [PubMed] [Google Scholar]
- 86.Sydow JF, Lipsmeier F, Larraillet V, Hilger M, Mautz B, Molhoj M, Kuentzer J, Klostermann S, Schoch J, Voelger HR, et al. Structure-based prediction of asparagine and aspartate degradation sites in antibody variable regions. PLoS One. 2014;9:e100736. doi: 10.1371/journal.pone.0100736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hawkins RE, Russell SJ, Winter G. Selection of phage antibodies by binding affinity. Mimicking affinity maturation. Journal of molecular biology. 1992;226:889–896. doi: 10.1016/0022-2836(92)90639-2. [DOI] [PubMed] [Google Scholar]
- 88.Malhotra A. Tagging for protein expression. Methods Enzymol. 2009;463:239–258. doi: 10.1016/S0076-6879(09)63016-0. [DOI] [PubMed] [Google Scholar]
- 89.Ferrara F, Naranjo LA, D’Angelo S, Kiss C, Bradbury AR. Specific binder for Lightning-Link(R) biotinylated proteins from an antibody phage library. J Immunol Methods. 2013;395:83–87. doi: 10.1016/j.jim.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *90.Sblattero D, Bradbury A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat Biotechnol. 2000;18:75–80. doi: 10.1038/71958. Method to make extremely large antibody libraries using site specific recombination. [DOI] [PubMed] [Google Scholar]
- 91.Sblattero D, Lou J, Marzari R, Bradbury A. In vivo recombination as a tool to generate molecular diversity in phage antibody libraries. Reviews in Mol Biotech. 2001;74:303–315. doi: 10.1016/s1389-0352(01)00022-8. [DOI] [PubMed] [Google Scholar]
- 92.Ferrara F, Naranjo LA, Kumar S, Gaiotto T, Mukundan H, Swanson B, Bradbury AR. Using phage and yeast display to select hundreds of monoclonal antibodies: application to antigen 85, a tuberculosis biomarker. PLoS One. 2012;7:e49535. doi: 10.1371/journal.pone.0049535. [DOI] [PMC free article] [PubMed] [Google Scholar]