Abstract
The effective delivery of DNA locally could increase the applicability of gene therapy in tissue regeneration and therapeutic angiogenesis. One promising approach is through use of porous hydrogel scaffolds that incorporate and deliver DNA in the form of nanoparticles to the affected sites. While we have previously reported on Caged nanoparticle Encapsulation (CnE) to load DNA polyplexes within hydrogels at high concentrations without aggregation, frequent issues with limited polyplex release following CnE have been encountered. In this study, we report two alternative approaches to polyplex presentation for decreasing aggregation in porous hydrogels. The first approach reduces polyplex aggregation by utilizing polyethylene glycol modification of the gene carrier polymer polyethyleneimine (sPEG-PEI) to mitigate charge-charge interactions between polyplexes and the scaffold during gelation. The second approach electrostatically presents polyplexes on the surfaces of scaffold pores as opposed to an encapsulated presentation. The sPEG-PEI polymer formed a smaller, less toxic, and more stable polyplex that exhibited less aggregation within HA gels when compared to the traditionally used linear PEI (LPEI) polymer. Surface-coated polyplexes also resulted in a more homogenous distribution of polyplexes in hydrogels. Furthermore, sPEG-PEI polyplexes retained transfection abilities comparable to LPEI in 3D surface-coated transfections. These results demonstrate a significant improvement in scaffold-mediated gene delivery and show promise in applications to multi-gene delivery systems.
Keywords: Hydrogel, gene delivery, non-viral, PEGylation, porous, aggregation
1 Introduction
Scaffolds for tissue repair aim to promote the repair of diseased or injured tissues through the use of a biocompatible material that support cellular infiltration and contain bioactive signals that guide invading cells through tissue formation1,2. Because cells can sense and respond to mechanical properties of their environment, a general approach to designing a scaffold for tissue engineering is to choose a material that mimics the extracellular matrix and provides structural support that can resist tensile and compressive stresses3. In addition to the structural characteristics of the scaffold, the effective delivery of bioactive signals is necessary to induce regeneration. Delivery of growth factors from scaffolds is a commonly utilized approach; however, encapsulation or incorporation of growth factors into scaffolds can often result in protein denaturation or degradation and the delivery method must be optimized for each protein4. An alternative approach in guiding cellular response is through DNA delivery. Naked DNA delivery has been shown to achieve transgene expression and guide regeneration in vivo5,6, but often results in poor delivery efficiency because the plasmid’s negative charge and large hydrodynamic radius triggers plasmid degradation, ineffective internalization and trafficking by the cell and induces immune responses due to the bacterial elements of the plasmids7. This motivated the use of encapsulated DNA inside nanoparticles as a means to improve DNA stability, delivery, intracellular trafficking and reduce immune interactions with the host. Cationic polymers are one type of DNA encapsulation approach that is commonly used. Through charge interactions of the positively charged polymer (amines) to the negatively charged DNA (phosphates) an aggregate forms (polyplex) that can protect the DNA from degradation and improve delivery. Poly(ethylene imine) (PEI) is one such cationic polymer that has been shown to successfully transfect cells both in vitro and in vivo and serves as the gold standard for non-viral gene delivery8.
We have been interested in incorporating PEI/DNA polyplexes inside hyaluronic acid (HA) hydrogel scaffold to guide soft tissue repair. HA is an anionic, non-sulfated polysaccharide that is widely distributed throughout the ECM of connective tissues9. HA is an exciting scaffold candidate for tissue engineering therapies due to its high biocompatibility and low immunogenicity7,10,11. HA interacts with CD44, RHAMM, and ICAM-1 surface receptors that contribute to cell proliferation and migration, which are processes that are necessary for tissue regeneration to occur12,13. Moreover, HA chains have been known to promote matrix metalloproteinase (MMP) expression14–16. MMPs are typically present in high concentration as diseased sites and are upregulated in tissue repair and microenvironment remodeling and can therefore serve as a trigger for bioactive signal delivery17,18.
Semi-synthetic HA hydrogels, which can be degraded my matrix metalloproteinases (MMPs) and hyaluronidases, have been previously developed for culturing stem cells in 3D19–21. Although we have previously demonstrated the ability to deliver DNA-PEI polyplexes from HA gels, direct polyplex encapsulation resulted in aggregation when concentrations exceeding 0.3 µg/µL22. To mitigate this issue, Lei et. al. developed a caged nanoparticle encapsulation (CnE) technique that utilized neutral saccharides (sucrose) and polysaccharides (agarose) to protect the polyplexes from inactivation and aggregation during hydrogel formation, respectively22,23. This technique was coupled with the introduction of micron-sized pores within gels in an attempt to promote increased cell migration and infiltration to the scaffold. Although gene delivery was achieved both in vitro and in vivo, transgene expression levels remained low24–26. We hypothesize that CnE and acetone processing during micron-size pore hydrogel formation, which causes an increase in gel stiffness, reduces pore size, resulting in a slower rate of gel degradation and polyplex release.
In order to maintain high DNA loading concentrations in HA gels without using CnE, it is necessary to overcome aggregation issues that occur during gelation and at high DNA polyplex concentrations. Herein, we present two approaches to prevent polyplex aggregation within porous HA hydrogels. Our first approach utilizes polyethylene glycol modification of PEI to mitigate charge-charge interactions between polyplexes and the scaffold (sPEG-PEI) (Figure 1A). The second introduces DNA polyplexes during hydrogel formation via encapsulation within the hydrogel or after hydrogel formation by presentation of polyplexes on the pore surface.
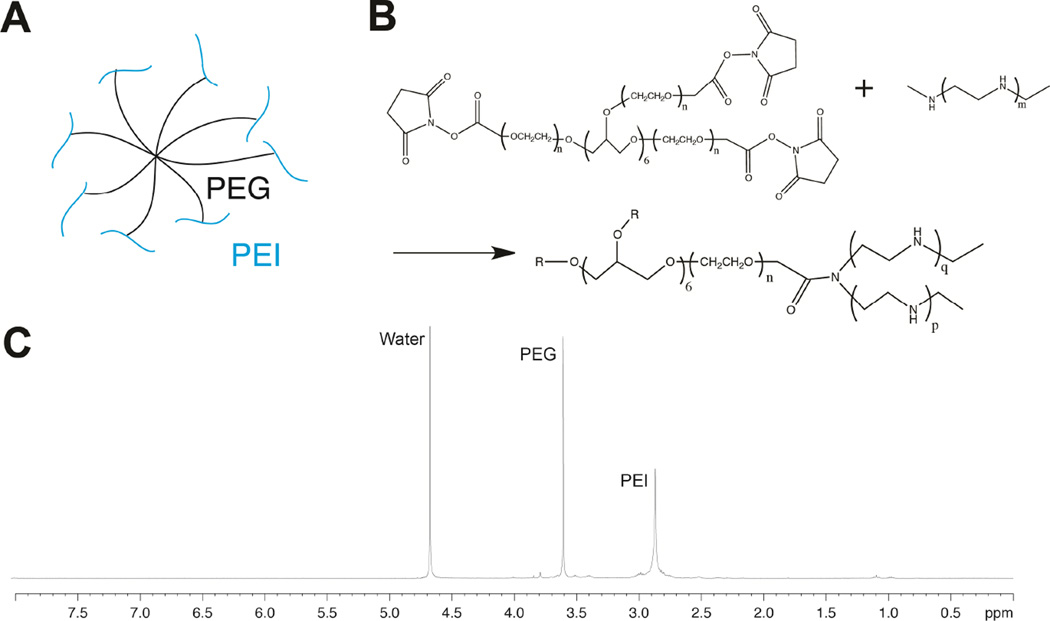
Fig. 1.
Schematic of PEI conjugated to 8-arm PEG (sPEG-PEI) (A). Reaction scheme of sPEG-PEI synthesis (B). 1H-NMR spectra of sPEG-PEI (C).
2 Materials and methods
2.1 Materials
Peptides Ac-GCRDGPQGIWGQDRCG-NH2 (HS-MMP-SH) and Ac-GCGYGRGDSPG-NH2 (RGD) were purchased from Genscript (Piscataway, NJ). Sodium hyaluronan (HA) was a gift from Genzyme Corporation (60 kDa, Cambridge, MA). High molecular weight linear poly(ethylene imine) (LPEI, 25kDa) and low molecular weight linear poly(ethylene imine) (LMW-PEI, 2.5kDa) were purchased from Polysciences (Warrington, PA). 8-arm poly(ethylene gycol) succinimidyl carboxyl methyl ester (PEG-SCM, 10kDa) was purchased from Creative PEGWorks (Winston Salem, NC). Vectors for the mammalian expression of Gaussia luciferase (pGluc) and secreted embryonic alkaline phosphatase (pSEAP) were purchased from New England Biolabs (Ipswich, MA) and BD Biosciences (San Jose, CA), respectively. Both vectors were expanded using a Giga Prep kit from Qiagen (Valencia, CA) per manufacturer’s protocol. All other chemicals were purchased from Fisher Scientific (Pittsburgh, PA) unless otherwise noted.
2.2 Methods
Synthesis of sPEG-PEI
LMW-PEI was conjugated to the 8-arm PEG-SCM to using amine to carboxylic acid chemistry. LMW-PEI (190.8mg, 0.07632 mmol) was dissolved in MES buffer (100uM, pH 5.5), and once fully dissolved the pH was increased to 7.4. After the PEI had fully dissolved, 50 mg of PEG-SCM (0.00477mmol) was dissolved in separate MES buffer pH 7.4. The PEG-SCM solution was added dropwise to the PEI solution while maintaining a constant pH, and the mixture was allowed to react overnight. The product was purified by dialysis (10,000 MWCO) against water for five days with two water changes per day to remove any unreacted PEI, and the product was analyzed using 1H-NMR (D2O). 1HNMR (D2O) spectroscopy indicated attachment of PEI to every arm of the 8-arm PEG-SCM by assessing the ratio of the PEI peak (δ = 2.85) to the PEG-SCM peak (δ = 3.6).
Polyplex formation and characterization
To form polyplexes, 3 µg of plasmid DNA was diluted in 150 µl of nuclease free water and the desired amount of either LPEI or sPEG-PEI, depending on the required N/P ratio (ratio of the number of nitrogen groups on the polymer to the number of phosphate groups on the DNA backbone), was diluted into a separate 150 µl of nuclease free water. For polyplexes formed at N/P 7, 4 µg sPEG-PEI was used and 2.73 µg LPEI was used. For polyplexes formed at N/P 12, 6.87 µg sPEG-PEI was used and 4.69 µg LPEI was used. The PEI solution (either LPEI or sPEG-PEI) was drop wise added to the DNA while vortexing, and each sample was incubated at room temperature for 15 min. 150mM NaCl or PBS was then added to each polyplex solution and the size and ζ-potential of the polyplexes were determined by photon correlation spectroscopy (Malvern Zetasizer, Malvern Instruments Ltd., U.K.). The measurements were performed at 25°C.
Agarose gel retardation assay
An agarose hydrogel retardation assay was performed in order to assess the N/P ratio at which the polyplexes are fully condensed. Polymer/pDNA complexes were prepared at N/P ratios 1–7 in nuclease free water per the aforementioned protocol, with naked DNA containing no polymer as the control. The polyplexes were electrophoresed through a 1% (w/v) agarose hydrogel containing a 1/10,000 dilution of SYBR® Safe DNA Stain (Life Technologies, Grand Island, NY) in 1X Tris-acetate-EDTA (TAE) buffer at 80 V for 30 min. The hydrogel was then analyzed on a Hydrogel Doc EZ Imager (Bio Rad, Hercules, CA) to observe the fluorescence of each polyplex relative to naked DNA.
Cell culture
HEK293T cells were a kind gift from Lonnie Shea of the University of Michigan, and HEK293T-MMP2 cells were a kind gift from Jeffrey Smith from the Burnham Institute for Medical Research. Mouse-bone-marrow-derived mesenchymal stem cells (D1, CRL12424) were purchased from ATCC (Manassas, VA). HEK293T, HEK293T-MMP2, and D1 cells and cultured in Dulbecco’s modified Eagle medium (DMEM) (Invitrogen, Grand Islands, NY) supplemented with 10% bovine growth serum (BGS, Hyclone, Logan, UT) and 1% penicillin/streptomycin (Invitrogen) at 37°C and 5% CO2. The cells were passaged using trypsin following standard cell culture protocols every 2–3 days.
Cytotoxicity of sPEG-PEI and LPEI polyplexes
An MTT assay (CellTiter 96R AQueous One Solution Cell Proliferation Assay, Promega, Madison, WI) was used to quantify the metabolic activity of cells exposed to sPEG-PEI or LPEI polyplexes in order to correlate metabolic activity to cell viability. 12,000 D1 cells were cultured in a 96-well plate for 16 hours, then transfected with 20 µL of either sPEG-PEI or LPEI polyplexes at DNA concentrations ranging from 0–0.2 µg/µL and N/P ratios 7 and 12. After a 2 day incubation, the media in each well was aspirated and cells were then incubated with 20 µL of MTT reagent and 100 µL of DMEM for 2 hours at 37°C. Following the 2 hour incubation, 25 µL of 10% sodium dodecyl sulfate was added to each well to stop the reaction, the solutions were transferred to a new plate, and the absorbance was measured at 490 nm using a standard plate reader.
In vitro 2-D bolus transfection
To assess the tranfection efficiency of sPEG-PEI compared to LPEI, pGluc or pSEAP polyplexes of N/P ratios 7 and 12 were created using both polymers. D1 cells were seeded on a 48-well plate at a density of 40,000 cells/well in 500 µL of media, and allowed to incubate for 16 hours. Following this incubation, the media from each well was removed and replaced with fresh media. 50 µL of polyplex solution (0.02 µg DNA/µL in 150 mM NaCl) was added to each well. The cells were allowed to incubate for 2 days, after which the media was collected and frozen at −20C until assayed. To quantify secreted Gaussia luciferase levels in the media, the samples were thawed on ice and assayed using a BioLux Gaussia Luciferase Assay Kit (New England Biolabs, Ipswich, MA) as per the manufacturer’s protocol. Briefly, 20 µL of each sample were mixed with 50 µL of substrate solution, pipetted for 2 to 3 seconds to mix, and read for luminescence with a 5 s integration time using a Modulus Fluorometer (Turner BioSystems, South San Francisco, CA). To quantify secreted embryonic alkaline phosphatase levels in the media, the samples were thawed on ice and assayed using a pSEAP Assay Kit (Life Technologies, Grand Island, NY). Briefly, 100 µL of sample was incubated with 200 µL of dilution buffer and incubated at 65°C for 30 min. 100 µL of the diluted sample was then mixed with 100 µL of assay buffer and incubated for 20 min. 100 µL of reaction buffer was then added to each sample, incubated for 20 min, and samples were read for luminescence with a 1 s integration time.
Internalization of DNA over different periods of polyplex exposure
D1 mouse mesenchymal stem cells were seeded in a tissue culture-treated 48-well plate at a seeding density of 20,000 cells per well 16 h prior to transfection. 1.25 µg plasmid DNA was radiolabelled with 32P-dCTP (250µCi, PerkinElmer, Waltham, MA) using a Nick translation kit (Roche, Indianapolis, IN) according to the manufacturer’s protocol. The reaction mixture was purified using the DNA Clean and Concentrator kit from Zymo Research (Irvine, CA) and mixed with 498.75 µg non-radiolabelled DNA to make a 0.25% “hot” plasmid DNA solution. Polyplexes were then formed with the radiolabelled DNA and either sPEG-PEI or L-PEI at N/P ratios of 7 and 12. 1 µg of DNA’s worth of polyplexes was added to each well and incubated at 37C for 2, 4, 8 h. At each of the timepoints, the cells were washed with PBS + CaCl2, CellScrub buffer (Genlantis, San Diego, CA) to remove any un-internalized polyplexes, and then PBS twice before trypsinizing. The cells were then harvested and added to 2 mL Bio-Safe II scintillation cocktail (Research Products International Corp., Mt. Prospect, IL) and measured using a scintillation counter at the UCLA chemistry facility. The readout was analyzed using a standard curve.
Internalization of DNA and transgene expression under inhibition of endocytosis pathways
D1 cells were seeded in a tissue culture-treated 48-well plate at a seeding density of 20,000 cells per well 16 h prior to treatment. Clathrin-mediated endocytosis was inhibited with 10 µg/mL chlorpromazine (Fisher Scientific), caveolae-mediated endocytosis with 200 µM genistein (Sigma Aldrich, St. Louis, MO), and macropinocytosis with 100 µM amiloride hydrochloride (Sigma Aldrich, St. Louis, MO), for 1 h at 37C. Polyplexes were formed with either radiolabelled DNA as described above to track cell internalization or pGLuc as described above to assess transgene expression and added to the wells in the presence of inhibitors for a 4-h incubation. The cells exposed to radiolabelled DNA were then washed with PBS + CaCl2, CellScrub buffer, and then PBS twice before trypsinizing, harvesting, and reading with a scintillation counter as described above. For the pGLuc transfections, the media was replaced with fresh media after 4 h. Gaussia luciferase expression was assayed as described above.
Hyaluronic acid modification
Sodium hyaluronan was modified to contain acrylate function groups via a two-step reaction as previously described25. HA (2 g, 60 kDa) was reacted with 36.77 g (211.07 mmol) adipic acid dihydrazide (ADH) at pH 4.75 with 4 g 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) overnight. The product was purified through dialysis (8000 MWCO) against a NaCl gradient for 1 day. Further dialysis was done in DI water for 4 days. The purified product HA-ADH was then lyophilized and analyzed with 1H-NMR. All of the HA-ADH was then reacted with 4.46 g N-acryloxysuccinimide (NHS-Ac) in 10mM HEPES, 150mM NaCl, 10mM EDTA at pH 7.4 overnight at room temperature before purification via dialysis against a NaCl gradient for 1 day and in DI water for 4 days. The purified product HA-Ac was then lyophilized and analyzed with 1H-NMR (D2O). 1H-NMR indicated a 54.67% modification of the carboxyl groups on the HA backbone to ADH groups by taking the ratio of peaks δ = 1.6 and 23, which correspond to the eight H of the methylene groups on the ADH to the singlet peak of the acetyl methyl protins in HA (δ = 1.88). After the second step in which HA-ADH was reacted with N-acryloxysuccinimide (NHS-Ac), 1H-NMR (D2O) spectroscopy confirmed 11.42% acrylate modification (HA-Ac) by taking the ratio of the multiplet peak at δ = 6.2 corresponding to the acrylate H to the singlet peak of the acetyl methyl protons in HA (δ = 1.88).
Polyplex lyophilization by caged nanoparticle encapsulation (CnE)
For CnE, plasmid DNA (8.3 µg) and either sPEG-PEG or LPEI (13.4 µg or 9.1 µg, respectively) were mixed in 3.5 mL water in the presence of 3.5mg (0.01 mmol) of sucrose (Ultrapure, MP Biomedicals, Santa Ana, CA) and incubated at room temperature for 15 min. Low-melting point agarose (0.1 mg, Ultrapure Agarose, T – 34.5–37.5°C, Invitrogen, Grand Islands, NY) in 150 mL water was added before lyophilization. Each aliquot was intended for a 10 µL hydrogel.
Porous hydrogel design template using PMMA microspheres
Chemically sintered microsphere templates were prepared as previously described26. Briefly, polymethyl methacrylate (PMMA) microspheres (53–63 µm, Cospheric, Santa Barbara, CA) were suspended in sintering solution (70% ethanol, 1% acetone) at a concentration of 0.4444mg µL−1, and 75 µL of this bead solution was then added to each well of 6-mm diameter flexiPERM molds (Sigma-Aldrich, St. Louis, MO) adhered to glass slides treated with Sigmacote (Sigma-Aldrich, St. Louis, MO). The molds were sintered at 37°C for 2 hours before use.
Porous and nonporous hydrogel formation
Porous and nonporous HA hydrogels were prepared as previously described25. Hydrogels were formed by Michael-addition of acrylate functionalized HA (HA-Ac) with bis-cysteine containing MMP peptide crosslinkers at pH 7.6–7.8. Prior to the reaction, a hydrogel precursor solution was made by mixing HA-Ac with a lyophilized aliquot of the cell adhesion peptide RGD for 30 min at 37°C. After incubation, HA-RGD was mixed with the remaining HA-Ac and 0.3M triethanolamine (TEOA, pH 8.8), for a final hydrogel concentration of 3.5% weight/volume% HA and 100uM RGD. Finally, lyophilized aliquots of the crosslinker (0.8mg HS-MMP-SH) were diluted in 16 µL of TEOA buffer, pH 8.2, immediately before addition to the hydrogel precursor solution. This hydrogel precursor solution was then mixed with either lyophilized (CnE) or fresh (direct encaspulation) DNA/polymer polyplexes for hydrogels containing polyplexes. For direct encapsulation, DNA and sPEG-PEI or LPEI were mixed according to the aforementioned protocol. For nonporous hydrogels, 20uL of the hydrogel solution was added between 2 slides with a 1mm spacer to separate the two slides. Hydrogels were incubated at 37°C for 30 min, then hydrated in phosphate-buffered saline (PBS) and left in PBS until used. For the porous hydrogels, 20 µL of the hydrogel solution was then added directly on top of a PMMA microsphere template and perfused into the template by centrifugation at 500 g for 15min at 4°C. The template was then incubated at 37°C for an additional 20 min to induce complete crosslinking. Once complete, the hydrogels were removed from the flexiPERM molds and placed directly into 100% acetone for 48 h to dissolve the PMMA microsphere template. The acetone solution was replaced 3 times a day for the 48 h wash. The hydrogels were then serially hydrated in PBS, and stored in PBS until used.
Characterization of HA hydrogel mechanical properties
The storage and loss modulus of nonporous and microporous hydrogels were measured with a plate-to-plate rheometer (Physica MCR, Anton Paar, Ashland, VA) using a 8 mm plate under a constant strain of 0.1% and frequency ranging from 0.1 to 10 rad/s. Nonporous and porous hydrogels were synthesized according the aforementioned protocol and cut to 8 mm using an 8 mm biopsy punch. To prevent the hydrogel from drying, a humidity hood was utilized and the stage was set to 37°C.
Surface coating of porous hydrogels and polyplex visualization
Polyplexes were visualized within nonporous and porous gels to determine if sPEG-PEI polyplexes aggregate less than LPEI polyplexes. Prior to the formation of polyplexes, plasmid DNA was tagged with a 500 nm tag using a PromoFluor Nick Translation kit (Promokine, Heidelberg, Germany) per the manufacturer’s protocol. The DNA was purified using the DNA concentrator kit from Zymo Research (Irvine, CA) following the manufacturer’s instructions. sPEG-PEI or LPEI polyplexes were formed per the aforementioned protocol, and incorporated into CnE, direct encapsulation, or surface coated hydrogels. Porous hydrogels were surface coated with DNA polyplexes by incubating hydrated porous hydrogels in a 0.2 µg/µL DNA polyplex solution for two hours with agitation applied every 20 minutes. Gels were then washed in PBS three times before imaging. Hydrogels were imaged over a constant z-plane using an epifluorescence microscope (Oberkochen, Germany).
DNA release from surface coated hydrogels
In order to determine the overall the extent of release of surface coated polyplexes, gels were formed and surface coated using the aforementioned protocols with 0.25% radiolabeled DNA polyplexes. Gels were then placed in 200 µL of either 1 U/mL collagenase type 1 solution (Col I) (Worthington Biochemical, Lakewood, NJ) or PBS (control). The solutions were collected and replenished daily or every three days, and DNA concentrations were measured using a scintillation counter at the UCLA chemistry facility. The readouts were analyzed using a standard curve.
3D transfection from surface coated porous HA hydrogels
To assess sPEG-PEI vs. LPEI transfection efficiency in 3-D, 3.5% HA porous hydrogels were synthesized via the aforementioned protocol. After fully hydrating the hydrogels in PBS, each hydrogel was cut to a diameter of 4 mm using a 4-mm biopsy punch and placed in individual 1.5 mL tubes. 50 µL of a 0.2ug/ul pGluc/polymer polyplex solution was added to each tube and incubated for 2 hr at room temperature, flicking every 20 min. Hydrogels were then washed three times with PBS to remove any unbound polyplexes. Hydrogels were then seeded with D1 cells by incubating gels in 250 µL/gel of media with suspended cells at 1000000 cells/mL for 3 hr with flicking every 20 min, and washed to remove any unbound cells. At 2, 4 and 7 days, conditioned medium was collected from all samples and Gaussia luciferase expression was determined for both polymers via the aforementioned protocol. To determine how many cells were seeded in the gels, porous hydrogels without polyplexes were seeded with D1 cells via the aforementioned method and allowed to incubate in media for 12 h to allow for cell attachment. The CyQUANT assay (Life Technologies, Grand Island, NY) was used to quantify the number of cells seeded in the gel according to the manufacturer’s protocol.
Two-gene delivery system from porous hydrogels
In order to demonstrate a dual-gene delivery system, pGluc polyplexes were synthesized per the aforementioned protocol and incorporated into the hydrogel precursor solution at a concentration of 0.2 µg/µL. Porous gels were then formed with this hydrogel precursor solution per the aforementioned protocol. After the PMMA beads had dissolved and the hydrogels had been swelled, pSEAP polyplexes were synthesized per the aforementioned protocol. Each gel was surface coated with 50 µL of 0.2 µg/µL polyplex solution per the aforementioned protocol. Finally, 40,000 HEK293T-MMP2 cells in 250 µL media were surface coated on the porous gels per the aforementioned protocol. Media was collected at days 2, 4, 7, 10, and 14 and assayed for pGluc and pSEAP expression as per the aforementioned protocols.
Statistical analysis
Statistical analyses were performed using Prism (GraphPad, San Diego, CA). Data were analyzed using a one-way analysis of variance (ANOVA) test followed by a Tukey post-hoc test. The results are presented as mean ± SD. Polyplex size data was analyzed using a t-test, and the results are presented as mean ± SD. Single, double, and triple asterisks represent p < 0.05, p < 0.01, p < 0.001, respectively. A p value of < 0.05 was considered statistically significant.
3 Results and Discussion
sPEG-PEI synthesis
sPEG-PEI was synthesized using the reaction between the activated carboxylic acid in 8-arm PEG-Succinimidyl carboxyl methyl ester (PEG-SCM) and the amines in PEI. Due to insoluble nature of PEI at basic pH, LMW-PEI was first dissolved in acidic conditions and once fully dissolved the pH of this solution was slowly increased to 7.4 in order to prevent precipitation. The extent of conjugation was calculated through NMR by comparing the integration of the observed peaks of LMW-PEI and PEG-SCM at δ=2.85 and δ=3.6, respectively, which indicated that 94.9% of the PEG arms were conjugated with PEI (Figure 1C).
Polyplex Characterization
Polyplexes of either sPEG-PEI or LPEI with plasmid DNA were formed at an N/P ratio of 7 and 12 to study the influence of polymer composition on particle size and stability. After the addition of salt to physiological salt conditions, sPEG-PEI polyplexes doubled in size but showed no significant increase in size thereafter until day 5 (Figure 2A). Conversely, LPEI polyplexes experienced a significant seven-fold increase after the addition of salt, and showed a 130-fold increase from the initial size just after day 1 (Figure 2B). To assess polyplex zeta potential, sPEG-PEI and LPEI polyplexes were synthesized in either 150 mmol NaCl or PBS solutions. The average charge of sPEG-PEI polyplexes N/P 7 in 150 mmol NaCl and PBS was 10.47 mV and 2.16 mV, respectively, and for polyplexes N/P 12 in 150 mmol NaCl and PBS was 22.17 mV and 5.68 mV, respectively (Figure 2D). The average charge of LPEI polyplexes N/P 7 in 150 mmol NaCl and PBS was 25.00 mV and 11.03 mV, respectively, and for polyplexes N/P 12 in 150 mmol NaCl and PBS was 30.43 mV and 12.83 mV, respectively (Figure 2D).
Fig. 2.
Size and stability of sPEG-PEI polyplexes (A) and LPEI polyplexes (B) over 5 days. Agarose gel retardation assay to assess the condensation abilities of sPEG-PEI and LPEI polymers with pDNA (C). Average zeta potential of sPEG-PEI and LPEI polyplexes formed at N/P 7 and 12 in 150 mmol NaCl and PBS (D).
The efficiency of the polymer/DNA interactions was evaluated by determining the amount of conjugate required to retard the migration of DNA through an agarose gel over a range of N/P ratios 0–7. As shown in Figure 2C, complete retardation of sPEG-PEI polyplexes was observed at an N/P ratio of 6 while complete retardation of LPEI polyplexes was observed at an N/P ratio of 4. In addition, the complete shielding of DNA by sPEG-PEI occurs at a higher N/P ratio than by LPEI.
These results indicate sPEG-PEI forms a smaller, more stable polyplex when complexed to pDNA compared to LPEI (Figure 2A and B). The charges of the LPEI polyplexes were higher than their relative sPEG-PEI counterparts, which can be attributed to the higher number of nitrogen groups on the LPEI backbone compared to the sPEG-PEI backbone (25.25 nmol N/1 µg LPEI, 15.63 nmol N/1 µg sPEG-PEI) (Figure 2D). The less charged nature of sPEG-PEI polyplexes may be the reason for smaller particle size upon addition of salt when compared to LPEI polyplexes, which aggregate with a much higher tendency in a highly charged environment. This is an important property difference that we hypothesize may be the underlying reason for differences seen in characterizing polyplex loading in hydrogels as well as in transgene expression profile differences upon transfection. Furthermore, the ability of LPEI to more tightly complex DNA may be due to the higher number of nitrogen groups on the LPEI backbone that can interact with the phosphate groups on the DNA backbone. This is supported by the complete shielding of DNA by LPEI occurring at a lower N/P ratio than by sPEG-PEI. The presence of PEG groups on sPEG-PEI may also interfere with complexation due to steric hindrance.
Cell-polyplex interactions
An MTT assay was performed to compare the toxicity of sPEG-PEI to LPEI polyplexes (Figure 3A). Since the concentrations used in this study were high for in vitro culture systems, some toxicity was expected. Cells exposed to sPEG-PEI polyplexes of N/P 7 show no decrease in metabolic activity up to 200 ng/µL, while cells exposed to LPEI at N/P 7 had a significant decrease starting at 100ng/mL (p < 0.001). Similarly, cell exposed to sPEG-PEI polyplexes at N/P 12 show no decrease in metabolic activity up to 100 ng/µL, while cells exposed to LPEI N/P 12 had a significant decrease starting at 50ng/mL (p < 0.001). In addition, cell viability was overall higher with exposure to sPEG-PEI polyplexes than with LPEI polyplexes with equal dosing of DNA, indicating that the administration of sPEG-PEI as a complexing agent may be less cytotoxic than LPEI. Electrostatic interactions between sPEG-PEI and DNA are relatively weak compared to between LPEI and DNA, which may result in more free sPEG-PEI being present in the cellular microenvironment due to more random decomplexation, posing concerns of cytotoxicity. However, sPEG-PEI complexes were actually less cytotoxic than LPEI complexes.
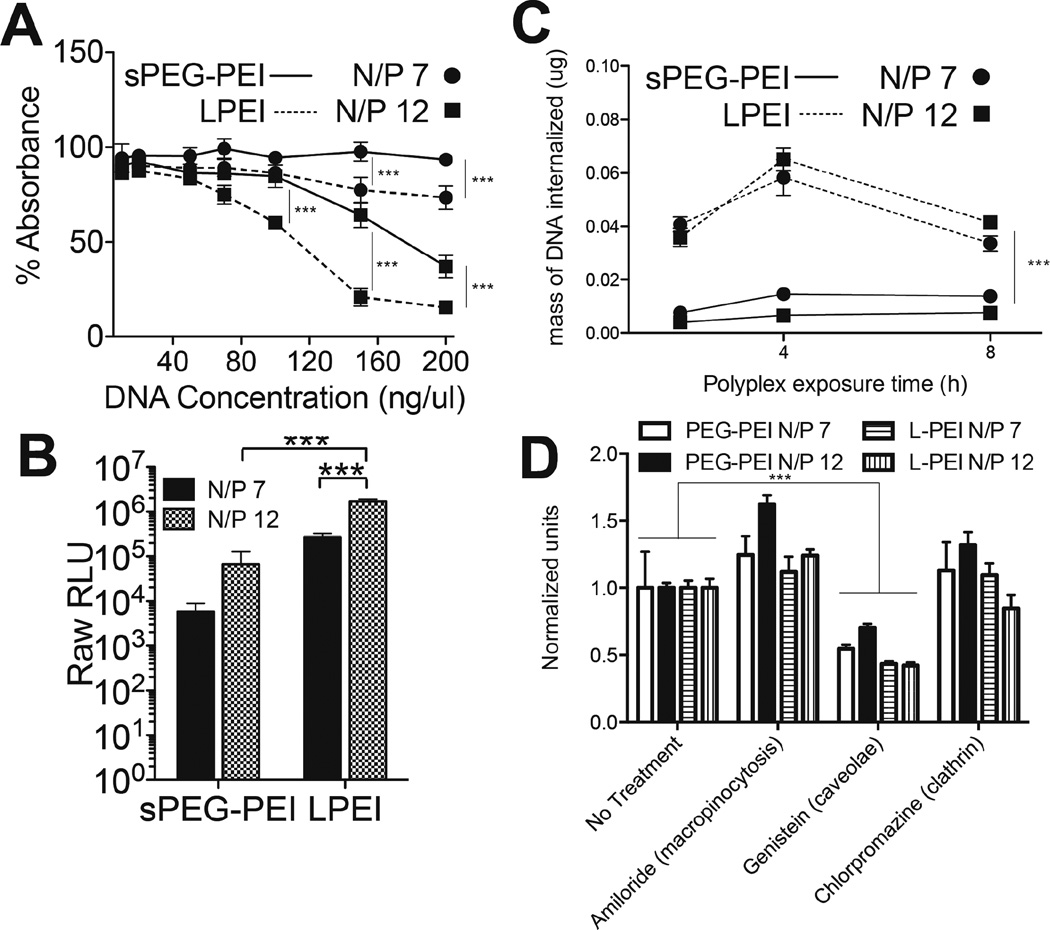
Fig. 3.
Cell-polyplex interactions. MTT assay assessing cytotoxicity of polyplexes formed with sPEG-PEI and L-PEI (A). Relative GLuc expression after 48 h of a 2-D bolus transfection with polyplexes formed with sPEG-PEI and L-PEI (B). Internalized radiolabelled DNA over different polyplex exposure times (C). Internalized radiolabelled DNA after 4 h exposure to polyplexes with the inhibition of different endocytosis pathways (D).
A bolus transfection using the Gaussia luciferase vector was performed to compare the transfection abilities of sPEG-PEI and LPEI polyplexes to D1 cells in 2D. Transgene expression at both N/P 7 and N/P 12 for sPEG-PEI were significantly lower than those for LPEI at the same N/P ratios (p < 0.001) (Figure 3B). In addition, a control bolus transfection experiment was performed to evaluate the transfection efficiency of a simple mixture of the components of sPEG-PEI (non-functionalized 8-arm PEG and 2.5kDa PEI) compared to the reacted product sPEG-PEI (Supplementary Figure 1). While not significant, there was an increase in transgene expression with sPEG-PEI compared to the simple mixture, to using 2.5kDa PEI only, and to the use of no gene carrier polymer (naked DNA).
To understand whether the difference in transgene expression between using sPEG-PEI and LPEI polyplexes was due to a difference in the extent of internalization of those polyplexes, D1 cells were plated and transfected over different exposure times using the four different polyplex compositions with radiolabelled DNA to determine how much DNA is being internalized at each time point (Figure 3C). It was observed that the mass of DNA internalized when transfecting with LPEI polyplexes was significantly higher over all exposure times than when transfecting with sPEG-PEI. A surprising observation in Figure 3C was the drop in internalized DNA at the 8-h time point compared to the 4-h time point when using LPEI as the cationic polymer for complexing. This decrease may suggest substantial amounts of DNA polyplexes that are unable to escape the endosome into the cytoplasm. Instead, this subset of total internalized polyplexes during the first 4 hours was eventually exocytosed.
In addition, to examine whether there is a difference in the dependence of the uptake of the different polyplexes on different internalization pathways, macropinocytosis, caveolae-mediated endocytosis, or clathrin-mediated endocytosis was pharmacologically inhibited (Figure 3D). Dhaliwal et al reported no effects on cell viability after four hours of exposure to each of the inhibitors used in this study, permitting the use of these inhibitors to study internalization after four hours of exposure27. When caveolae-mediated endocytosis was inhibited with genistein, the amount of DNA internalized was significantly lower than a no-treatment transfection for both sPEG-PEI and LPEI polyplexes. There was no decrease in amount of internalized DNA observed when macropinocytosis was inhibited using amiloride or when clathrin-mediated endocytosis was inhibited with chlorpromazine with either sPEG-PEI or LPEI-mediated transfection. Within treatments, there was no difference in the relative amounts of DNA internalized among the different polyplex compositions.
These results show that while the use of LPEI as the complexing polymer led to more DNA internalized, there was no difference in the dependence on specific endocytotic pathways for internalization between the two polymers. This suggests that the differences in both the chemical structure of the gene carrier polymers and the physical properties of the DNA polyplexes resulting from those polymers do not affect the dominant internalization pathway involved in polyplex uptake. Instead, this difference in extent of uptake may be due to the difference in surface charge densities of the two types of polyplexes, with the more highly charged LPEI polyplexes electrostatically binding more strongly to the cell membrane to result in higher uptake rates.
Visualization of polyplex aggregation
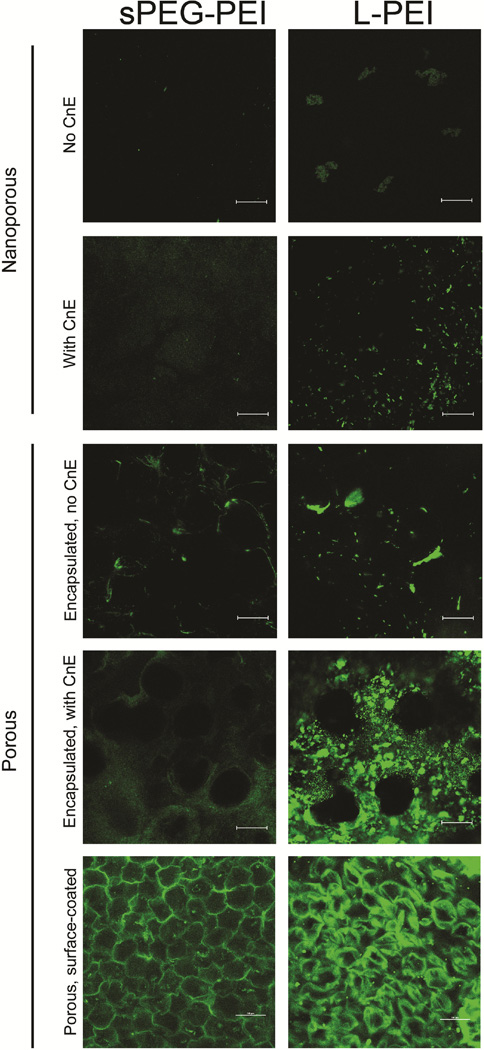
sPEG-PEI and LPEI polyplexes were incorporated into nonporous and porous hyaluronic acid hydrogels either by surface-association with the pore surfaces or by encapsulation within the gel phase. The storage (G’) and loss (G”) moduli of the hydrogels were analyzed using a plate-to-plate rheometer. Results showed that the G’ and G” did not cross at any measured frequency (0.1 – 10 Hz) and were frequency-independent for both porous and nonporous hydrogels, both of which are consistent with typical hydrogel characteristics. We reported G’ of 523 ± 32 Pa and 222 ± 23 Pa for our porous and nonporous hydrogels, respectively. This difference in mechanical stiffness was statistically significant between the two gel types.
To assess aggregation, porous and nonporous hydrogels containing either sPEG-PEI or LPEI polyplexes were stained and imaged (Figure 4). Complexing DNA with sPEG-PEI resulted in noticeably less aggregation than with LPEI after encapsulating these DNA polyplexes in both nonporous and porous hydrogels. This observation is consistent with and without the use of the caged nanoparticle encapsulation (CnE) technique. This difference in aggregation behavior may be attributed to the decrease in surface charge density when using sPEG-PEI polyplexes, which may result in a lower tendency for the occurrence of particle-particle charge-based interactions. By using sPEG-PEI polyplexes, it is possible to load DNA into hydrogels via encapsulation at higher DNA loading concentrations when compared to LPEI polyplexes. By surface coating porous hydrogels with either sPEG-PEI or LPEI polyplexes, a more homogenous surface distribution of DNA polyplexes was observed throughout the hydrogel when compared to the aggregation seen in porous hydrogels with encapsulated LPEI polyplexes.
Fig. 4.
Visualization of polyplex aggregation in polyplex-loaded HA hydrogels. Fluorescently labelled DNA polyplexes were incorporated into porous and nonporous hydrogels and imaged.
PEG-PEI and LPEI polyplex studies with surface-coated porous hydrogels
To more fully characterize the use of surface coating porous hydrogels as a method for DNA loading, the amount of DNA that was loaded into the porous gels using sPEG-PEI or LPEI as the gene carrier was first quantified. With a 0.2 µg/µL DNA polyplex solution, 3.45 ± 0.19 µg and 4.26 ± 1.20 µg DNA were loaded using sPEG-PEI N/P 7 and 12, respectively, and 3.91 ± 1.62 µg and 4.56 ± 0.70 µg DNA were loaded using LPEI N/P 7 and 12, respectively. Due to the lower charge density of sPEG-PEI/DNA polyplexes, we expected less DNA to be loaded with sPEG-PEI than with LPEI. However, there was no significant difference between the amount of DNA loaded with sPEG-PEI and LPEI when synthesized at identical N/P ratios.
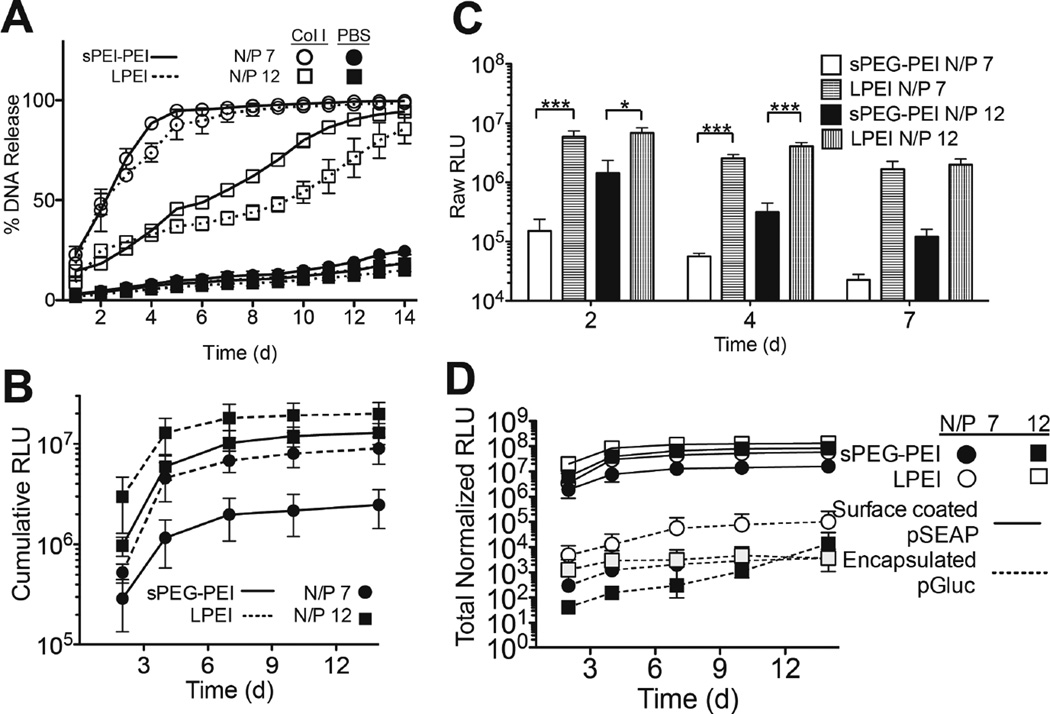
The release of surface-bound polyplexes due to enzymatic degradation of the porous hydrogel was then assessed by submerging radiolabelled polyplex-bound porous gels in solutions of collagenase I or PBS to study desorption (Figure 5A). A sustained release of polymer polyplexes was observed in the presence of Col I over a 14-day period, with less than 25% released cumulatively in PBS (Figure 5A). sPEG-PEI polyplexes of N/P’s 7 and 12 released at a quicker rate than their LPEI polyplex counterpart, which we attribute to weaker electrostatic interactions with the hydrogel due to the more neutral charge of sPEG-PEI polyplexes. For gels suspended in Col I solution, sPEG-PEI and LPEI polyplexes N/P 7 exhibited 90% release by days 5 and 7, respectively. Furthermore, sPEG-PEI polyplexes N/P 12 exhibited 90% release by day 12, while LPEI polyplexes N/P 12 did not achieve 90% release by the end of the study at 14 days.
Fig. 5.
Surface-coated porous hydrogel studies and in vitro two-gene delivery system. Cumulative DNA release from HA porous hydrogels with surface-coated polyplexes with Col I treatment and in PBS (A). Cumulative (B) and per-day (C) 3-D transfection profile of cell-seeded surface-coated hydrogels over 14 d. 2-gene delivery transfection profile in porous hydrogels with surface-coated polyplexes containing pSEAP and encapsulated polyplexes containing pGLuc (D).
To compare polymer transfection abilities in 3D, porous HA gels were first surface coated with polyplexes followed by coating of 40,000 D1 cells as determined by a CyQUANT assay. Results indicate sustained transfection over the period of 14 days for sPEG-PEI and LPEI polyplexes at N/P 7 and 12, with the highest individual levels of transgene expression occurring at day 2 for all conditions (Figure 5C). We observe that LPEI polyplexes N/P 12 led to the most transgene expression at each time point and in terms of cumulative expression, with a total cumulative expression of 1.995 × 107 RLU (Figure 5B and C). Comparably, sPEG-PEI polyplexes N/P 12 exhibited a total cumulative expression of 1.289 × 107 RLU (Figure 5B and C), which is not statistically significant in comparison to LPEI polyplexes N/P 12. This same conclusion is met when comparing the kinetic and cumulative expression of LPEI and sPEG-PEI polyplexes N/P 7, which resulted in cumulative expressions of 2.474 × 106 and 9.010 × 106 RLU, respectively.
In this study, the differences in cell-polyplex interactions were explored in both two- and three-dimensional cell culture. With results indicating that DNA polyplexes complexed with sPEG-PEI at both N/P ratios of 7 and 12 were less toxic to cells at higher levels of DNA exposure, sPEG-PEI polyplexes can serve as a less toxic alternative to LPEI polyplexes in systems where polyplex toxicity presents issues. In two-dimensional cell culture, it was also seen that while sPEG-PEI polyplexes were less toxic, their usage also resulted in lower levels of transgene expression when compared to LPEI polyplexes at both N/P ratios of 7 and 12 (Figure 3B). A similar difference was also seen in three-dimensional culture in surface-coated porous HA hydrogels (Figure 5C). These findings suggest that in order to obtain comparable levels of transgene expression when using sPEG-PEI polyplexes as LPEI polyplexes, it will be necessary to increase DNA polyplex loading concentrations, but to a much smaller extent with surface-coated porous hydrogels as the culturing platform. These results, when coupled with the lower toxicity observed with sPEG-PEI polyplexes, suggest that sPEG-PEI can serve as an alternative transfection polymer to LPEI that is less toxic and still retains high levels of expression. In 3D hydrogel-mediated transfections, this conclusion is further supported by presenting the DNA polyplexes by surface coating rather than by encapsulation within the hydrogel.
The delivery of multiple genes from a single system can have applications in tissue engineering and regenerative medicine28–30. To demonstrate the use of a two-gene delivery system with different expression profiles as a function of presentation method, pGluc plasmid was encapsulated within the nanoporous region of porous gels, and pSEAP was surface coated onto the pores of the porous gel. HEK293T-MMP2 cells that over express MMP2 were surface coated onto the pores of the hydrogel at a concentration of 40,000 cells/hydrogel, and were used instead of D1 cells in order to ensure hydrogel degradation and subsequent polyplex release. Because pGluc and pSEAP are different vectors that result in different transfection levels at a constant DNA concentration, a 2D transfection was performed to normalize the expression levels of the two vectors in this two-gene delivery assay (data not shown). We observed sustained transfection over 14 days of both vectors from all four polyplex conditions (Figure 5D). Expression of the gene that was encapsulated within the porous gel scaffold was consistently lower than expression of the surface-coated gene, a finding that is in agreement with previous findings31. This is likely due to more direct exposure of surface-coated DNA to infiltrating cells, while exposure of encapsulated DNA to cells is more dependent on the progress of cell-mediated gel degradation. Analogous to the 3D surface-coated transfection results, there was no significant difference in total expression of surface-coated pSEAP at 14 days between sPEG-PEI and LPEI polyplexes N/P 7 or sPEGPEI and LPEI polyplexes N/P 12. By either presenting a gene in a different loading method or by complexing with a different polymer, it is possible to tune the transgene expression profiles of each gene of interest to be delivered in addition to being able to decrease the aggregation encountered when loading LPEI polyplexes via encapsulation.
4 Conclusion
In this study, using PEGylated PEI and surface-coating DNA along pores of porous hydrogels were explored as alternatives to LPEI-polyplex loading in hydrogels via caged nanoparticle encapsulation as means to decrease polyplex aggregation when loading DNA at higher concentrations. Using PEGylated PEI resulted in lower toxicity to cells upon exposure and less aggregation when encapsulating in HA hydrogels, but also led to decreased transgene expression levels, which was attributed to lower amounts of DNA being internalized by cells when complexed with sPEG-PEI. By surface coating hydrogels with both sPEG-PEI and LPEI polyplexes, it is also possible to decrease aggregation, thereby maintaining spatial homogeneity throughout the surfaces of the porous hydrogel. The options explored in this study can be used to further inform multi-gene delivery systems and tune individual transgene expression profiles for a wide range of therapeutic applications.
Supplementary Material
A 2-D bolus transfection was performed on 40,000 D1 cells per well, with GLuc expression assayed at 48 h after initial exposure to conditions. DNA was complexed with LPEI, sPEG-PEI, a simple mixture of nonfunctionalized 8-arm PEG and 2.5kDa PEI, 2.5 kDa PEI, and no carrier.
Acknowledgements
The authors would like to thank the Segura lab members for their help and support, with special thanks to Dr. Donald Griffin for his help with the sPEG-PEI chemistry. We would also like to thank funding from the National Institutes of Health (R01HL110592).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lavik E, Langer R. Tissue engineering: current state and perspectives. Applied microbiology and biotechnology. 2004;65(1):1–8. doi: 10.1007/s00253-004-1580-z. [DOI] [PubMed] [Google Scholar]
- 2.Nerem RM, Sambanis a. Tissue engineering: from biology to biological substitutes. Tissue engineering. 1995;1(1):3–13. doi: 10.1089/ten.1995.1.3. [DOI] [PubMed] [Google Scholar]
- 3.Lutolf MP, Hubbell Ja. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nature biotechnology. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 4.Cam C, Segura T. Matrix-based gene delivery for tissue repair. Current opinion in biotechnology. 2013:1–9. doi: 10.1016/j.copbio.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonadio J, Smiley E, Patil P, Goldstein S. Localized, direct plasmid gene delivery in vivo: prolonged therapy results in reproducible tissue regeneration. Nature medicine. 1999;5(7):753–759. doi: 10.1038/10473. [DOI] [PubMed] [Google Scholar]
- 6.Jang JH, Rives CB, Shea LD. Plasmid delivery in vivo from porous tissue-engineering scaffolds: Transgene expression and cellular transfection. Molecular Therapy. 2005;12(3):475–483. doi: 10.1016/j.ymthe.2005.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wieland Ja, Houchin-Ray TL, Shea LD. Non-viral vector delivery from PEG-hyaluronic acid hydrogels. Journal of Controlled Release. 2007;120(3):233–241. doi: 10.1016/j.jconrel.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. Journal of controlled release. 1999;60:149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 9.Lam J, Truong NF, Segura T. Design of cell-matrix interactions in hyaluronic acid hydrogel scaffolds. Acta biomaterialia. 2013 doi: 10.1016/j.actbio.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peattie Ra, Nayate aP, Firpo Ma, Shelby J, Fisher RJ, Prestwich GD. Stimulation of in vivo angiogenesis by cytokine-loaded hyaluronic acid hydrogel implants. Biomaterials. 2004;25(14):2789–2798. doi: 10.1016/j.biomaterials.2003.09.054. [DOI] [PubMed] [Google Scholar]
- 11.Seidlits SK, Khaing ZZ, Petersen RR, Nickels JD, Vanscoy JE, Shear JB, Schmidt CE. The effects of hyaluronic acid hydrogels with tunable mechanical properties on neural progenitor cell differentiation. Biomaterials. 2010;31(14):3930–3940. doi: 10.1016/j.biomaterials.2010.01.125. [DOI] [PubMed] [Google Scholar]
- 12.Ahrens T, Assmann V, Fieber C, Termeer CC, Herrlich P, Hofmann M, Simon JC. CD44 is the principal mediator of hyaluronic-acid-induced melanoma cell proliferation. Journal of Investigative Dermatology. 2001;116(1):93–101. doi: 10.1046/j.1523-1747.2001.00236.x. [DOI] [PubMed] [Google Scholar]
- 13.Sherman L, Sleeman J, Herrlich P, Ponta H. Hyaluronate receptors: key players in growth, differentiation, migration and tumor progression. Current opinion in cell biology. 1994;6(5):726–733. doi: 10.1016/0955-0674(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 14.Rooney P, Wang M, Kumar P, Kumar S. Angiogenic oligosaccharides of hyaluronan enhance the production of collagens by endothelial cells. Journal of cell science. 1993;105(Pt 1):213–218. doi: 10.1242/jcs.105.1.213. [DOI] [PubMed] [Google Scholar]
- 15.Gao F, Liu Y, He Y, Yang C, Wang Y, Shi X, Wei G. Hyaluronan oligosaccharides promote excisional wound healing through enhanced angiogenesis. Matrix biology. 2010;29(2):107–116. doi: 10.1016/j.matbio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Pardue EL, Ibrahim S, Ramamurthi A. Role of hyaluronan in angiogenesis and its utility to angiogenic tissue engineering. Organogenesis. 2008;4(4):203–214. doi: 10.4161/org.4.4.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue. Nat Rev Mol Cell Biol. 2007;8(3):221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parks WC. Matrix metalloproteinases in repair. Wound Repair and Regeneration. 1999;7(6):423–432. doi: 10.1046/j.1524-475x.1999.00423.x. [DOI] [PubMed] [Google Scholar]
- 19.Lei Y, Segura T. DNA delivery from matrix metalloproteinase degradable poly(ethylene glycol) hydrogels to mouse cloned mesenchymal stem cells. Biomaterials. 2009;30(2):254–265. doi: 10.1016/j.biomaterials.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JKJ, Kim ISKIS, Hwang SJHSJ, Kim HCKHC, Park YPY, Sun KSK. Bone regeneration using MMP sensitive-hyaluronic acid based hydrogels; 2009 IEEE 35th Annual Northeast Bioengineering Conference; 2009. [Google Scholar]
- 21.Kim J, Park Y, Tae G, Lee KB, Hwang SJ, Kim IS, Noh I, Sun K. Synthesis and characterization of matrix metalloprotease sensitive-low molecular weight hyaluronic acid based hydrogels. Journal of Materials Science: Materials in Medicine. 2008;19(11):3311–3318. doi: 10.1007/s10856-008-3469-3. [DOI] [PubMed] [Google Scholar]
- 22.Lei Y, Huang S, Sharif-Kashani P, Chen Y, Kavehpour P, Segura T. Incorporation of active DNA/cationic polymer polyplexes into hydrogel scaffolds. Biomaterials. 2010;31(34):9106–9116. doi: 10.1016/j.biomaterials.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei Y, Rahim M, Ng Q, Segura T. Hyaluronic acid and fibrin hydrogels with concentrated DNA/PEI polyplexes for local gene delivery. Journal of controlled release. 2011;153(3):255–261. doi: 10.1016/j.jconrel.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokatlian T, Cam C, Siegman SN, Lei Y, Segura T. Design and characterization of microporous hyaluronic acid hydrogels for in vitro gene transfer to mMSCs. Acta biomaterialia. 2012;8(11):3921–3931. doi: 10.1016/j.actbio.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tokatlian T, Cam C, Segura T. Non-viral DNA delivery from porous hyaluronic acid hydrogels in mice. Biomaterials. 2014;35(2):825–835. doi: 10.1016/j.biomaterials.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tokatlian T, Cam C, Segura T. Porous Hyaluronic Acid Hydrogels for Localized Nonviral DNA Delivery in a Diabetic Wound Healing Model. Advanced Healthcare Materials. 2015 doi: 10.1002/adhm.201400783. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhaliwal A, Maldonado M, Han Z, Segura T. Differential uptake of DNA-poly(ethylenimine) polyplexes in cells cultured on collagen and fibronectin surfaces. Acta biomaterialia. 2010;6(9):3436–3447. doi: 10.1016/j.actbio.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen FM, Zhang M, Wu ZF. Toward delivery of multiple growth factors in tissue engineering. Biomaterials. 2010;31(24):6279–6308. doi: 10.1016/j.biomaterials.2010.04.053. [DOI] [PubMed] [Google Scholar]
- 29.Zisch AH, Lutolf MP, Hubbell JA. Biopolymeric delivery matrices for angiogenic growth factors. Cardiovascular Pathology. 2003;12(6):295–310. doi: 10.1016/s1054-8807(03)00089-9. [DOI] [PubMed] [Google Scholar]
- 30.Chen RR, Silva EA, Yuen WW, Mooney DJ. Spatio-temporal VEGF and PDGF delivery patterns blood vessel formation and maturation. Pharmaceutical Research. 2007;24(2):258–264. doi: 10.1007/s11095-006-9173-4. [DOI] [PubMed] [Google Scholar]
- 31.Saul JM, Linnes MP, Ratner BD, Giachelli CM, Pun SH. Delivery of non-viral gene carriers from sphere-templated fibrin scaffolds for sustained transgene expression. Biomaterials. 2007;28(31):4705–4716. doi: 10.1016/j.biomaterials.2007.07.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A 2-D bolus transfection was performed on 40,000 D1 cells per well, with GLuc expression assayed at 48 h after initial exposure to conditions. DNA was complexed with LPEI, sPEG-PEI, a simple mixture of nonfunctionalized 8-arm PEG and 2.5kDa PEI, 2.5 kDa PEI, and no carrier.