SUMMARY
The intestinal mucus layer provides a barrier limiting bacterial contact with the underlying epithelium. Mucus structure is shaped by intestinal location and the microbiota. To understand how commensals modulate gut mucus, we examined mucus properties under germ-free (GF) conditions and during microbial colonization. Although the colon mucus structure of GF mice was similar to conventionally raised (Convr) mice, the GF inner mucus layer was penetrable to bacteria-sized beads. During colonization, in which GF mice were gavaged with Convr microbiota, the small intestine mucus required five weeks to be normally detached and colonic inner mucus six weeks to become impenetrable. The composition of the small intestinal microbiota during colonization was similar to Convr donors until three weeks when Bacteroides increased, Firmicutes decreased, and segmented filamentous bacteria became undetectable. These findings highlight the dynamics of mucus layer development and indicate that studies of mature microbe-mucus interactions should be conducted weeks after colonization.
Keywords: MUC2, Mucin, Mucus, Bacteria, Colonization, Commensals, Colon, Small Intestine, Glycosylation, Glycosyltransferase, Proteomics, Mass Spectrometry
INTRODUCTION
The intestine is typically colonized with 1013–1014 bacteria residing in the lumen and in the mucus with only limited contact with the epithelium (Backhed et al., 2005; Johansson et al., 2013). The MUC2 mucin is the main component of the mucus in mice and humans and responsible for this separation (Johansson et al., 2008). However, the mucus is differently structured in the small and large intestine. In the small intestine, the mucus allows limited diffusion of bacteria and anti-bacterial peptides and keeps the epithelium clean by moving detached mucus with trapped bacteria forward for expulsion in the feces (Ermund et al., 2013; Vaishnava et al., 2011). In the large intestine on the other hand, the inner attached mucus layer acts as a physical barrier that does not allow bacteria to pass due to their size (Johansson et al., 2008). Some bacterial contact with the epithelial cells is tolerated, but massive exposure triggers inflammation as found in inflammatory bowel diseases (IBD) (Johansson et al., 2014).
Both host and bacteria have adapted to each other and their coevolution has created an intricate symbiotic system that has triggered significant interest during the last 10 years. The main reason has been the capability to identify and characterize the intestinal microbiota by powerful DNA-sequencing methods revealing that a fraction of the bacteria were known using only culture methods (Kuczynski et al., 2012). This development has been further fueled by the understanding that the intestinal microbiota has implications for the development of some of todays’ major problems, such as obesity, diabetes and allergies (Ley et al., 2005). The interdependence of host and bacteria is also obvious from the emerging understanding of mucus and its properties. Germ free (GF) mucus was observed to be different both in the small and large intestine and recently the colon mucus was found to be dependent on the bacterial composition (Johansson et al., 2008; Schütte et al., 2014; Johansson et al., 2014; Jakobsson et al., 2015). It is today evident that there is a lot of interaction and communication between host and bacteria although they only have limited contact.
Studies of GF rodents have become a necessary tool for the analysis of bacteria-host interactions (Falk et al., 1998; Backhed et al., 2005). Adult GF mice can be colonized with limited or full mouse caecal or human fecal flora and studied over a short period usually not exceeding two weeks (Lecuit et al., 2007). As we knew that bacteria affect the intestinal mucus system, we undertook a systematic study colonizing GF mice with caecal flora and followed these animals over eight weeks. The results showed that it takes about 5 weeks until the small intestinal mucus becomes detached and about 6 weeks for the colon inner mucus layer to become fully impenetrable as in conventionally raised (Convr) mice. The microbiota showed dramatic transient alterations during this time and the composition did not reach the conventionalized ones until after 8 weeks. These observations have strong implications on how to perform studies including colonization of GF mice to study host-microbe interactions.
RESULTS
Germ free mice have a mucus system that differs from conventionally raised mice
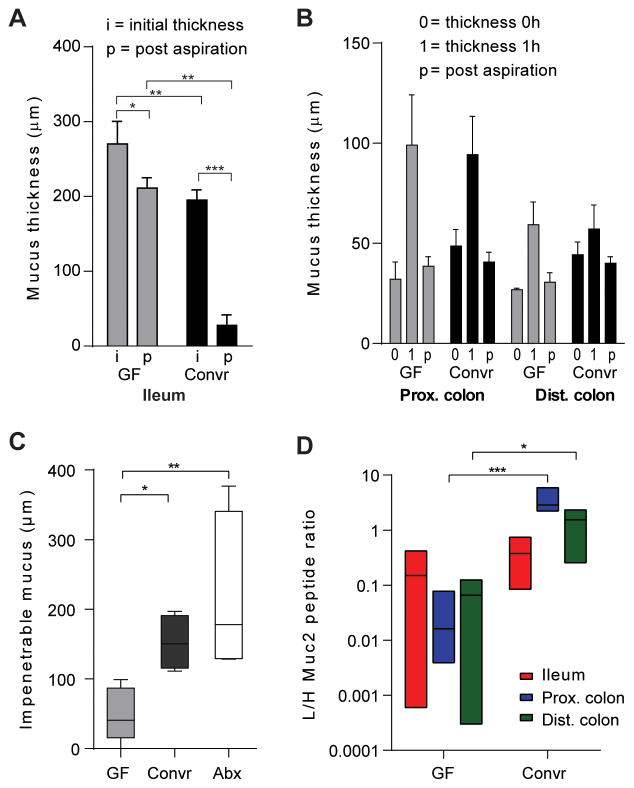
Normal wild-type (WT) mice have mucus that provides protection of the epithelium by creating a diffusion barrier with antimicrobial peptides in the small intestine and a physical barrier restricting access of the microbiota in colon (Johansson et al., 2008; Ermund et al., 2013). The mucus system in ileum and colon of GF mice was compared to Convr animals. The thickness of the stimulated mucus on small intestinal explants showed a slight increase in GF mice (Fig. 1A, i, initial thickness). However, a more pronounced difference was that the GF ileum mucus could not be aspirated off as in Convr mice (Fig. 1A, p, post-aspiration). The mucus thickness after aspiration was only decreased 23% in GF mice as compared to Convr mice, where most of the mucus could be removed (85%). In colon, no significant differences in thickness, growth, and attachment to the epithelium were observed (Fig. 1B). Instead, major differences were observed in the mucus properties. The mucus secreted on top of the colon explants was overlaid with bacterium-size fluorescent beads, these were allowed to sediment into the mucus, and the penetration of these into the mucus was estimated from confocal Z-stacks. Beads penetrated deep into the mucus of GF mice while the mucus in Convr mice was impenetrable to all beads (Fig. 1C). Depleting most of the intestinal bacteria in Convr mice by treatment with antibiotics for 3 weeks did not make the mucus penetrable (Fig. 1C). The relative amounts of Muc2 in the mucus were estimated by quantitative proteomics, and found to be significantly lower in colon of GF compared to Convr mice (Fig. 1D). The mucus in both small and large intestine thus depends on bacteria to become mature and normal.
Figure 1. Mucus in germ free and conventionalized mice.
(A) Mucus thickness in distal small intestine of germ free (GF) or conventionally raised (Convr) mice was measured as initial thickness (i) or post-aspiration (p). The mucus thickness was significantly different before and post-aspiration. The mucus remaining after aspiration was significantly different between GF and Convr mice revealing attached mucus in GF mice. (B) Mucus thickness measured in distal colon during 1h. The results are reported as the thickness of the initially attached mucus (0), mucus thickness after 1h (1) and the attached mucus after 1h post aspiration (p). No differences between GF animals and Convr controls were observed. (C) Mucus penetrability to beads the size of bacteria (0.5–2 μm) was measured by adding beads to newly secreted mucus and allowing sedimentation for 40 min followed by collecting confocal Z-stack images. The impenetrable mucus fraction was analyzed in distal colon and revealed significant differences between GF and Convr mice. Treatment with antibiotics for 3 weeks did not impair the mucus penetrability (Abx). (D) Muc2 protein amounts were analyzed by mass spectrometry with heavy labelled peptides and significantly higher amounts of Muc2 were observed in colon of Convr mice compare to GF (*= P<0.05, **= P<0.01, ***= P<0.001, ****= P<0.0001).
Mucus layer thicknesses and penetrability during colonization
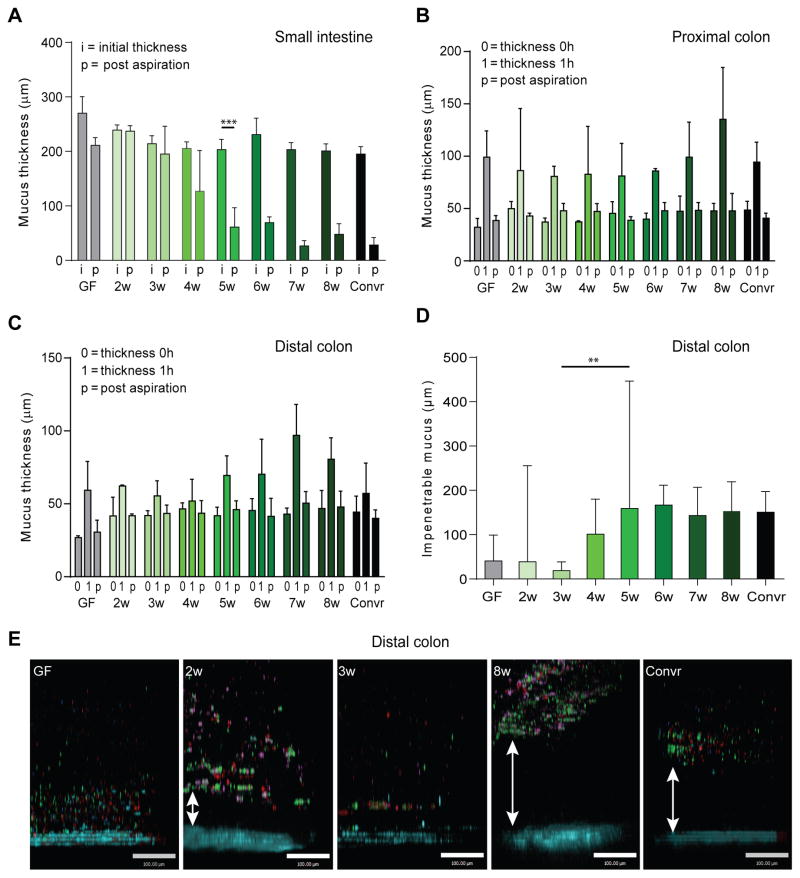
The different mucus properties in GF and Convr mice suggest a mucus maturation process dependent on bacteria. To further address this, colonization of GF mice with caecal microbiota from mice with well-developed impenetrable mucus was studied weekly from two to eight weeks after colonization. A minimum of three mice were used for each experiment and week. The experiments were repeated three times and showed consistent and reproducible results. The initial mucus (i) thickness as well as the mucus remaining after aspiration (p) was measured on explants from distal small intestine (Fig. 2A). The attached mucus in colonized GF mice was preserved during the first three weeks of colonization and did not resume a Convr-like phenotype until 4–5 weeks after colonization when the mucus became detached and possible to aspirate. From five weeks of conventionalization the mucus appeared as in Convr mice.
Figure 2. Mucus during colonization with a complex microbiota.
(A) Mucus thickness in distal small intestine was measured as in Fig. 1A during 2 to 8 weeks of colonization. The mucus was removable after 4 weeks of colonization. (B–C) The mucus thickness was measured over 1 h in proximal (B) and distal (C) colon as in Fig. 1B during 2–8 weeks of colonization. (D) Mucus penetrability was analyzed in distal colon during colonization as in Fig. 1C. An impenetrable mucus developed after 6 weeks. (E) Representative pictures of confocal Z-stacks from the penetrability experiments with beads (0.5 μm red, 1 μm purple, 2 μm green) close to the epithelium (blue) in GF, 2 and 3-week colonized mice. Mucus separated the beads from the epithelium in Convr mice and after 8 weeks of conventionalization. The scale bar represents 100 μm. **, P=0.016, ***, P=0.002.
The proximal and distal colon mucus were studied and three parameters were quantified: the thickness of the attached mucus at time zero (0); the thickness of the mucus at one hour due to growth over this time (1); and the thickness of the attached mucus at 1 hour remaining after taking away the loose and non-attached outer mucus (p) (Fig. 2B and C). There were no major differences in the initial and final inner mucus layer thicknesses (0, p), nor in the growth over this hour (1). The penetrability of beads the size of bacteria (0.5–2 μm) was analyzed on colon explants from mice 2–8 weeks after colonization and compared to GF and Convr mice (Fig. 2D and E). The distance between the top of the impenetrable mucus and the epithelium (mucus thickness) was measured (Fig. 2D). Representative combined z-stacks are shown in Fig. 2E. At two weeks, the mucus was penetrable as in GF mice with a large individual variation. Different from the GF animals, the beads showed increased aggregation and clumping suggesting altered mucus properties at this time. At three weeks, the mucus was highly penetrable and had beads in close contact with the epithelium in all samples. After this time point, the inter-individual variability increased and a shift towards more impenetrable mucus emerged during weeks 4–5. From week 6 and onwards the mucus penetrability was essentially normal and similar to Convr mice. Together the results show that normalization of the mucus layer after colonization of GF mice takes about 5 weeks in the small intestine and about 6 in colon.
Bacteria localization in mucus during colonization
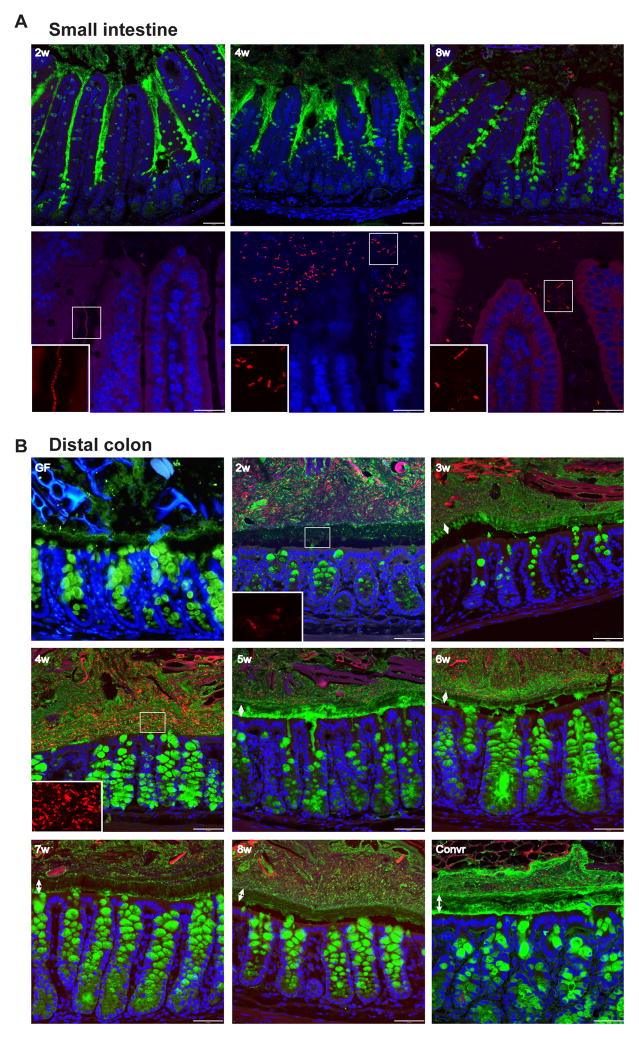
To illustrate the mucus maturation over an 8 week period, Carnoy-fixed tissue sections that preserve the mucus were stained for Muc2 (green) and bacteria (by in situ hybridization with a probe detecting most bacteria; red), and cells were visualized by their nuclei (blue) (Fig. 3). During the first weeks of small intestinal colonization, more secreted mucus was observed compared to the later time points and to the Convr mice (Fig. 3A. 2–4w vs 8w-Convr). At the same time the number of filled goblet cells was lower, something that could suggest a larger mucus secretion at weeks 2–4. The number of bacteria in the lumen was lowest at week 2 and 8. At week 4, an increased number of bacteria was observed (Fig. 3A: 4w). In all samples, bacteria were found between the villi, but never in the small intestinal crypts.
Figure 3. Immunohistology of small intestine and colon during conventionalization.
Immunostaining of sections from distal small intestine (A) and distal colon (B) of Muc2 (green) with FISH detecting bacteria (red) and Hoechst counterstain of DNA (blue). Double arrows indicate the inner mucus layer. Inserts show the bacteria magnified in the boxed areas. Typically, the middle part of the inner mucus layer is not stained as well as Muc2 at other locations. Scale bars in A, upper panel and B, 50 μm; A, lower panel, 25 μm.
In the colon, an inner mucus layer was observed in all animals, both GF and Convr (Fig. 3B). However, the structure and thickness varied at different time points. The inner mucus layer appeared thinnest in the GF mice and was best developed 7–8 weeks of colonization. At 2 weeks, the mucus did not restrict the bacteria from being in close contact with the epithelial cells (Fig. 3A, 2w), while an inner mucus layer with only few bacteria was developed after 3 weeks (Fig. 3B, 3w). At this time point, the goblet cells showed decreased intracellular staining of stored mucus in the theca, probably reflecting enhanced secretion. At week 4, the highest amounts of bacteria in the mucus were observed and had contact with the epithelium (Fig. 3B, 4w). Mucus secretion in the crypt was observed at week 4–6. From week 6, the mucus looked as in Convr mice (Fig. 3B, 6w–8w). Together the tissue staining showed a picture compatible with what was observed in the explant system at similar time points.
Proteomic analyses of Muc2 and other proteins in the mucus during colonization
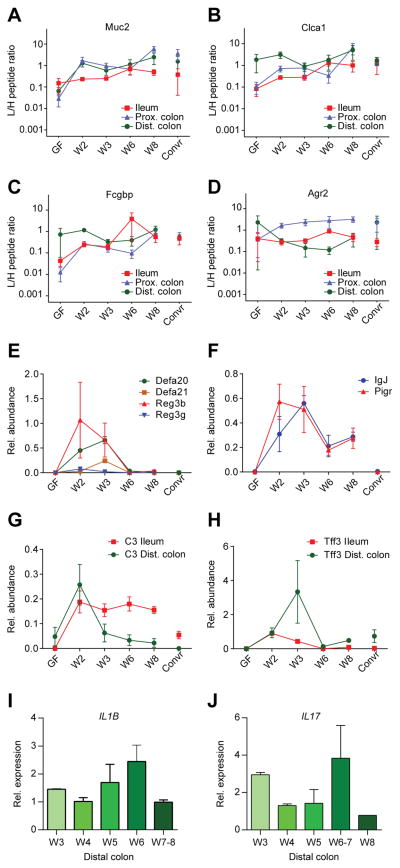
To address mucus protein alterations during colonization, the ileal and colon mucus from the explants used for mucus measurements were isolated, digested by trypsin, and analyzed by mass spectrometry (Rodriguez-Pineiro et al., 2013). Labeled peptides from abundant core mucus proteins were added and the relative amounts of the proteins Muc2, Clca1, Fcgbp and Agr2 were compared (Fig. 4A–D, Table S2). The first three proteins, but not Agr2, showed a tendency for increased amount from the levels in GF during the first two weeks, after which they remained relatively constant over the next 6 weeks (Fig. 4A–D). For Muc2 the increase was most pronounced in the colon, whereas for Clca1 and Fcgbp ileum and proximal colon showed larger increments. The Muc2 mRNA expression levels in weeks 3–9 were stable over time (Fig. S1, Table S1).
Figure 4. Alterations in mucus proteins after colonization of germ free mice.
Amounts of the mucus protein Muc2 (A), Clca1 (B), Fcgbp (C), and Agr2 (D) relative to supplemented labelled internal standard peptides in the ileum, proximal colon and distal colon as determined by proteomics of aspirated mucus from explants. (E–F) Alterations in relative amounts of antibacterial peptides/proteins (E), immunoglobulin J chain (IgJ) and the polymeric immunoglobulin receptor (Pigr) (F) in ileum as determined by proteomics of aspirated mucus from explants. (G–H) Alterations of the protein amounts of complement factor 3 (C3) and trefoil factor 3 (Tff3) in ileum and distal colon as determined by proteomics of aspirated mucus from explants. (I–J) mRNA levels of IL1β (I) and IL17 (J) as determined by Q-PCR in whole distal colon tissue. See also Fig. S1, S5, and Table S2.
The exposure of bacteria to the intestinal cells can be expected to increase proteins involved in host responses. This was shown to be the case when the ileal mucus proteome was analyzed for secreted antimicrobial proteins. The Reg3β and Reg3γ proteins increased fast from GF levels to a peak at week 2, whereas the alpha-defensins Defa20 and Defa21 peaked slightly later (Fig. 4E). Interestingly, the increased amounts of these proteins then quickly returned to the same low levels as seen in the GF mice as well as in the Convr mice. The levels of the immunoglobulin J chain (IgJ) and the polymeric immunoglobulin receptor (Pigr) also peaked in ileum at weeks 2 and 3, respectively. After this they decreased to lower levels at week 6–8, but still not to the same low level as for the Convr mice (Fig. 4F). The ileal amounts of the complement factor C3 peaked at week 2 and remained at an elevated level during the colonization period. In contrast, the distal colon mucus levels of C3 showed a peak in week 2 before returning to low levels at week 3 and onwards (Fig. 4G). Finally, the wound healing component trefoil factor 3 (Tff3) showed an ileal peak at week 2 after which it returned to levels similar to the Convr mice, whereas in the distal colon it peaked at week 3 and then returned to levels similar to the Convr mice at weeks 6–8 (Fig. 4H). Together this shows that different host response proteins all show increased levels following colonization, however with variable peak times in the small and large intestine.
To further understand the host response to bacterial colonization, the mRNA expression levels of the proinflammatory IL1β; IL10, IL13, IL17, and IFNγ cytokines were analyzed in colon. IL1β showed a slight increase at weeks 5–6, whereas IL17 had a peak in weeks 6–7 (Fig. 4I–J, Table S1). Thus, cytokines peaked later during colonization as compared to other host response proteins.
Alterations in the mucin oligosaccharides during colonization
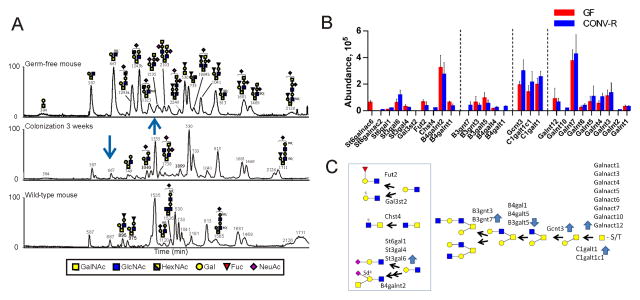
Most glycans of the colon mucus emanate from the Muc2 mucin that was semipurified from midcolon, the O-glycans released, and analyzed by capillary-LC mass spectrometry. The obtained base peak chromatograms of GF, 3 weeks-colonized mice and Convr mice are shown in Fig. 5A and Table S3. The glycans were different in the GF mice, but already at three weeks the glycans were similar to previous studies of proximal and distal colon mouse Muc2 that are characterized by their molecular masses m/z 1535, 530, 733, 813, 1681, 1469, and 1711 (Fig. 5A) (Thomsson et al., 2002; Holmen-Larsson et al., 2013). The most obvious GF mucus difference was the abundant core 2 trisaccharide with m/z 667 and the two monosialylated core 2 isomers with m/z 1243a and 1243b in that all decreased up to week 3 (Fig. 5A). Analyses of week 4, 6 and 8 showed similar Muc2 glycosylation patterns as week 3, while at week 2 the Muc2 glycosylation pattern was intermediate between GF and 3 weeks (not shown).
Figure 5. Glycosylation alterations during colonization.
(A) Base peak chromatograms of the capillaryLC-MS/MS analyses of the mouse midcolon Muc2 O-glycans of a GF mouse (top), GF conventionalized for 3 weeks (middle), and Convr WT mouse (bottom). The most abundant glycans in the LC profiles are annotated in the figure. Arrows point to major alterations. (B) Relative abundance of glycosyltransferases in the epithelial cells of GF and Convr mice as determined by proteomic analyses. (C) Proposed biosynthetic pathways and corresponding glycosyltransferases for the observed Muc2 glycans. Blue arrows show increased or decreased levels of the transferases upon colonization. See also Table S3–S4.
Next we determined the relative levels of the glycosyltransferases in the epithelial cells of GF and Convr mice by proteomics (Fig. 5B, Table S4). Nine different peptidyl-GalNAc transferases initiating the O-glycosylation on the peptide core were observed, with the Galnt7 as the most abundant. All transferases required for generating the observed colon Muc2 glycans were identified. A majority of the midcolon glycans were based on the core 2 substructure (Galβ1-3(GlcNAcβ1-6)GalNAcol) formed by the beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase 3 (Gcnt3), an enzyme increased by conventionalization (Fig. 5B and C). The m/z 1535 glycan with a terminal Sda/Cad epitope was also increased upon conventionalization as was the ST3gal6 transferase and became the most abundant glycan in Convr mice. The altered levels of the individual glycosyltransferase were in accordance with the altered glycans, although many details of the interdependence and specificities of the transferases have not been studied. Thus colonization probably causes alterations in both the glycan structures and in the specific serines and threonines glycosylated.
Intestinal microbiota development during colonization
FISH-staining of bacteria during colonization suggested microbial alterations of especially the mucus-associated bacteria residing at the outer border or in the attached inner colon mucus. To analyze the bacterial composition, luminal material (called lumen), and the remaining tissue containing the attached mucus (called mucus) were sequenced for bacterial 16S rDNA. The way the experiments were performed, the bacteria in the mucus (mucosa) could theoretically also reside within the tissue, but this was never observed by staining with DAPI or FISH.
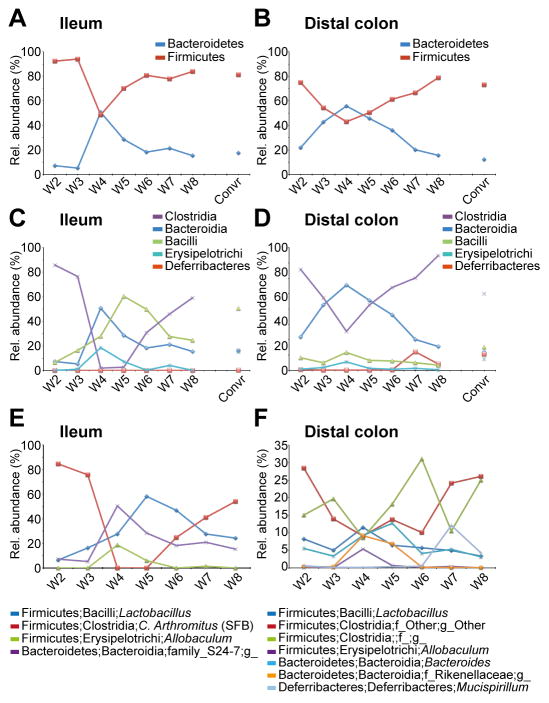
In the small intestine, Firmicutes dominated over the Bacteroidetes at week 2 and in later weeks, as well as in Convr (Fig. 6A). The only exception was week 4, where a main reconfiguration took place and the relative abundance of the Bacteroidetes increased and reached the same level as the Firmicutes that declined (Figs. 6A, S2C, and Table S5). This Firmicutes/Bacteroidetes shift observed at 4–5 weeks was found in both lumen and mucus samples, but it was most pronounced in the mucus (Figs. 6A, S2A, S3A, and Table S5). At the class level in the mucus, the increased Bacteroidetes corresponds to an increase in the Bacteroidia class and the reduction in Firmicutes is explained by a dramatic decrease in the relative amount of the Clostridia class (Figs. 6C, S2D, and Table S5). The Erysipelotrichi class increased at weeks 4–5 in both ileum lumen and mucus and returned to week 2-levels at weeks 6–8 (Figs. 6C, S2BD). The segmented filamentous bacteria (SFB) genus Candidatus Arthromitus was as expected by its epithelial cell attachment largely found in the ileum mucus fraction (Fig. 6E, S4D, and Table S5). Interestingly, this genus almost disappeared at weeks 4–5, but as for other bacteria returned to levels found in the first weeks of colonization (Fig. 6E, S4D, Table S5). SFB in the lumen also followed the same pattern with a decrease at week 4 (Fig. S4D). Concomitant with the decrease in SFB at weeks 4–5, there was an increase in several genera, such as Lactobacillus, Allobaculum, and an unknown genus of the class Bacteroidia (family S24-7) (Figs. 6E and S4A, E, F, Table S5). Lactobacillus was a dominant genus in ileum lumen and followed the mucus levels, as was also the case for the two other bacteria except Lactobacillus at week 4 (Fig S4A).
Figure 6. Microbiota alterations in ileum and distal colon mucus during colonization.
(A–B) The relative abundance (%) of the Firmicutes and Bacteroidetes phyla in ileum mucus (A) and distal colon mucus (B) at weeks 2–8 after colonization. (C–D) The relative abundance (%) of classes found in ileum mucus (C) and distal colon mucus (D) at weeks 2–8 after colonization. (E–F) The relative abundance (%) of selected, most abundant, genera in ileum mucus (E) and distal colon mucus (F) at weeks 2–8 after colonization. n=2–3 mice/time point. For details regarding bacteria and mice, see also Figs. S2–S4, S6, Table S5.
The return to the Convr microbiota composition coincided in time with the transformation of the small intestinal mucus from an attached to released phenotype. We have previously observed that the Muc2 mucin is released by the protease meprin β and that this enterocyte anchored meprin β was not released into the mucus in GF animals (Schütte et al., 2014). To analyze the period studied here, we searched the mucus proteome for meprin β. As before, no meprin β was found in the GF mucus, but surprisingly meprin β peptides were found already at week 2, before the Muc2 mucin was released (Fig. S5). This may suggest that the released meprin β remains inactive, as it has to undergo proteolytic cleavage to become activated.
In distal colon there was a shift with less Firmicutes and more Bacteroidetes at week 4 in the mucus and week 5 in the lumen (Fig. 6B, S2GI, S3C, and Table S5). This is due to a relative increase in the Bacteroidia class and a relative decrease in the Clostridia class (Fig. 6D, S2HJ, and Table S5). In the mucus, a few dominant genera within the Firmicutes phylum were decreased at week 4, e.g. two undefined genera within the Clostridia class (Fig. 6F and Fig. S4B–C). The genera Lactobacillus, Bacteroides, an unknown genus within the family Rikenellaceae, and Allobaculum were all increased at week 4 in distal colon mucus (Fig. 6F and S4AEGH). The phylum and class Deferribacteres, with Mucispirillum (known to be mucus associated) as the major genus, were found in increased relative levels in the distal colon mucus at weeks 7–8, suggesting a special niche in the well-developed mucus (Fig. 6F, S2I-J, and Table S5). This phylum, class, and genus were also found in ileal mucus, the lumen in distal colon, as well as in caecum, however in very low levels (Table S5). This genus was only found in ileal mucus at weeks 7 and 8, suggesting that a special niche for Mucispirillum was developed at this time point. In caecum the microbiota composition was stable throughout the colonization experiment, however, the shifts seen in ileum and distal colon can also be observed less pronounced in caecum (Figs. S2EF, S3B, and Table S5). The microbiota of the Convr mice was almost identical to that of week 8, although a careful comparison suggests that the microbiota was still not completely developed. The microbiota diversity was more stable in distal colon and caecum than in the ileum (Fig. S6).
DISCUSSION
Humans are considered to be born bacterially sterile, however immediately following birth there is an extensive colonization of all body sites (Walker, 2013). There is an intimate contact between the bacteria that reside within the human body and internal surfaces, for example the mucosal surface within the GI tract. In human infants, a high variability is observed within the gut microbiota during the first year of life that starts to stabilize at two years of age (Jakobsson et al., 2014; Backhed et al., 2015). Little is, however, known about the interaction of the bacteria on the development of the mucus layer in the GI tract. By colonizing GF mice we aimed at understanding the interplay between gut microbiota and the mucus of ileum and distal colon.
Similar to the gut microbiota, the mucus system also varies along the length of the intestine. In the small intestine a single mucus layer covers the epithelial cells whereas the large intestine is covered by two layers (Ermund et al., 2013; Johansson et al., 2013). The mucus of the GF small intestine is anchored to the goblet cells and require the meprin β enzyme to be released (Schütte et al., 2014). Further studies here show that the conversion to this normal colonized phenotype takes 4–5 weeks. The large intestine has a two-layered mucus system also in GF animals (Johansson et al., 2008), but as shown here the inner mucus layer was fully penetrable to beads the size of bacteria. The outer non-attached mucus layer is continuously formed from the inner mucus layer also in the GF mice. Two weeks after colonization the inner mucus layer thickness had increased to normal values, penetrable to bacterial-sized beads. The thickness of the impenetrable part of the inner mucus started to increase at week 4, was highly variable at week 5, and reached normal levels at week 6. The mucus properties at weeks 2–3 were different, although difficult to visualize and measure. At this time the mucus was stickier and adhered to the glass pipet at the same time as the fluorescent beads had a strong tendency to aggregate. This type of mucus has not been observed in any other experiments. The transition from GF to the Convr mucus phenotype is thus not linear and involves processes probably controlled by the host’s epithelial cells.
Regional differences in O-glycan patterns have been observed in Convr mice. For example, the proximal and distal colonic Muc2 mucin carries different glycan repertoires as compared to the small intestine (Holmen-Larsson et al., 2013). The present study was performed in the midcolon, which was shown to have an intermediate pattern compared to the distal and proximal colon (Holmen-Larsson et al., 2013). Although human intestine shows a different glycan repertoire compared to the mouse, they also have a mucin O-glycan gradient from ileum to the distal colon (Robbe et al., 2003). Studies have further shown that there is a region-specific bacterial colonization in the different parts of the gastrointestinal tract, suggesting that the mucin O-glycans can have a role in the selection of bacterial microbiota (Sekirov et al., 2010). Six different sialyltransferases were detected, although the products of only a few of these could be suggested. Two sulfotransferases were observed together with expected sulfated GlcNAc and Gal residues. Most glycans in midcolon contain the core 2 substructure, something that is in accordance with the levels of C1galt1, C1galtc1 and Gcnt3, all increasing upon colonization. Altered levels of the required glycosyltransferases were observed during colonization, although these appear rather modest in comparison with the observed alterations in the levels of O-glycans. These observations suggest a complex interrelation of glycan genes and their secondary products.
Already 3 weeks after the introduction of intestinal bacterial microbiota into GF mice, the glycosylation pattern appeared to be normalized and the levels and activities of glycosyl- and sialyltransferases seemed to reach normal values. This suggests that the mouse glycosylation machinery is a rather robust system that responds fast to the bacteria and their secreted products. Mucin O-glycosylation has also been previously shown to be a dynamic system and glycan patterns are commonly altered as secondary effects to infection and inflammation. The Fut2 enzyme is known to be upregulated in the mouse small intestine upon infection and in cystic fibrosis (Bry et al., 1996; Holmen et al., 2002; Thomsson et al., 2002). Patients with ulcerative colitis have been shown to have a transient altered glycosylation of MUC2 that was reversed to normal when the patient went into remission (Holmén Larsson et al., 2011). Together it can be suggested that the glycosylation of mucins and other glycoconjugates is modulated by infection and inflammation, something that is also observed here.
The human and mouse gut is heavily colonized by different bacteria, however only two major phyla dominate: the Firmicutes and the Bacteroidetes. This was seen also in the present study where these two phyla constituted the major gut microbiota at all locations and during the entire length of the colonization. As seen in human infants (Jakobsson et al., 2014; Backhed et al., 2015), the Bacteroidetes compose a large part of the gut microbiota at early time points followed by a decline after four weeks. In the present mouse study, a transient shift between Firmicutes and Bacteroidetes with a peak of Bacteroidetes at week 4 was observed first in the small intestinal mucus. Samples from lumen and caecum showed a similar pattern. Members of the Clostridia class that had been very dominant for the first 2–3 weeks were reduced during the expansion of the Bacteroidetes. Once the Bacteroidetes had declined, the members of the Clostridia class expanded back to normal levels. This is similar to what was observed in human infants (Jakobsson et al., 2014). Ileum was quickly and heavily colonized by segmented filamentous bacteria (SFB) and interestingly this bacterial group disappeared at weeks 4–5 after colonization. SFB expanded back again at week 6 as the relative levels of Bacteroidetes declined.
At eight weeks following colonization the microbiota composition was similar to that of the Convr mice, although it is evident that the microbiota at week 8 still undergoes some compositional modifications. The bacterial diversity was higher in colon than in ileum as previously observed, but there was a higher individual variation in the ileum (Booijink et al., 2010).
After 4–6 weeks the inner colon mucus started to show the normal phenotype and went from a penetrable to impenetrable mucus layer, something that influenced the bacterial niches. This was evident for Deferribacteres, with its major species Mucispirillum present in low relative amounts during the first six weeks and suddenly expanding in weeks 7–8. This was most evident in the distal colon where the inner mucus layer is best developed. This suggests a special mucus niche for this bacterium, something that has also been observed before (Robertson et al., 2005; Vereecke et al., 2014; Belzer et al., 2014).
The microbial alterations were larger in the ileal mucus. Several of the bacterial changes were first observed in the ileal mucus and then slightly delayed also in ileal lumen and distal colon mucus. These observations suggest that the small intestine and its mucus properties might be important for bacterial selection. The bacterial composition at week 2 primarily reflects the donor mice microbiota. Earlier studies have shown a rapid microbial alteration associated with immune responses at week 4 and onwards like Reg 3 β/γ in our study (El Aidy et al., 2012; El Aidy et al., 2013). Their study also suggests that the bacterial alterations cause a metabolic reorientation. The protein levels of the defensins, IgJ as a marker for IgA and the transporter for immunoglobulins over the epithelium peak at week 2–3 as a response to the bacteria. During weeks 2 and 3 the mucus remained attached to the epithelium. This probably facilitated the expansion of the Bacteroidetes during weeks 3 and 4 and outcompeted the SFB that require close accession and attachment to the epithelial cells (Ivanov and Littman, 2010). This situation dramatically changed when the small intestinal mucus was altered and became detached from the epithelial cells during weeks 4 and 5. This changed the situation and the Bacteroidetes started to return to lower more normal levels at the same time as the Firmicutes increased (including the SFB). The bacterial composition returned to a more ‘normal’ status after this. It is very likely that the alteration in mucus properties might drive this bacterial shift. We can conclude that the mechanism of releasing the Muc2 mucin and thus the mucus is very important for controlling the small intestinal microbiota and thus also the colonic one.
The mucus of the small intestine requires an active and released meprin β protease (Schütte et al., 2014). This enzyme is cleaving at least two sites in the N-terminal part of the MUC2 mucin and by this releasing the MUC2 mucin from its anchor (Schütte et al., 2014). Meprin β is made by small intestinal enterocytes where it is inserted in the apical brush border membrane (Lottaz et al., 1999). GF mice express meprin β, but it remained anchored to the enterocyte membrane and thus not released into the mucus and not able to detach the mucin (Schütte et al., 2014). However, peptides diagnostic for meprin β were found in relatively high amounts in the mucus at weeks 2–3. These two meprin β peptides declined at the same time as the MUC2 mucin was detached from its epithelial attachment. Meprin β is made as an inactive proenzyme and requires proteolytic cleavage for activation (Sterchi et al., 2008). This cleavage is not fully understood when it comes to localization and enzyme, but occurs around the two meprin β peptides that declined at week 4. It can be proposed that meprin β is released from its anchor during the initial colonization, but that its activation is delayed until weeks 4–5. The processes of meprin β activation and processing of the MUC2 mucin are thus not fully understood and require further demanding studies due to the complex and insoluble nature of the Muc2 mucin and the presence of both endogenous and bacterial proteases during these time points.
Little is known about the molecular mechanisms that control the colon mucus properties. As Muc2 forms the skeleton of the mucus it is understandable that the lower amounts of Muc2 present in the GF mucus could make this more penetrable to bacteria. However, the mucus proteome and glycome is normalized around week 3 after which the mucus is still penetrable to bacteria. There are thus additional unknown mechanisms involved, but it is known that the immune system, exemplified by IL10, is important as the IL10−/− mice have a normal mucus thickness that is fully penetrable to bacteria (Johansson et al., 2014). Also other cytokines could be involved as the colonic IL1β and IL17 levels peaked at late colonization stages with impenetrable mucus. Obviously this mucus maturation process is driven by bacteria, but the mechanisms involved are still far from understood. Lipopolysaccharides (LPS) and peptidoglycans (PGN) are likely to be involved as they can stimulate mucus secretion in GF mice (Petersson et al., 2011). However, this is not that simple as shown in a recent study where the inner mucus layer was penetrable in mice with one bacterial composition, but not another (Jakobsson et al., 2015). This suggests that certain bacteria or groups of bacteria are better in triggering a non-penetrable inner mucus whereas others might have the opposite effect. This complex host-bacteria interaction is not only based on LPS and PGN and might for example involve one or several of the numerous small molecules secreted by intestinal bacteria (Donia and Fischbach, 2015). The host response to bacteria seems also to contain some long-lasting mechanisms, as ablation of most bacteria from Convr mice by four antibiotics did not render the inner mucus penetrable after three weeks. This again suggest a complex, well programmed, and intricate system of intestinal interaction between host and microbe.
The present study shows that conventionalization of GF mice is a slow and complex process. The mucus system of the small and large intestinal tract takes 5 and 6 weeks, respectively, to come close to a normalized mucus phenotype. To reach a bacterial composition similar to the Convr mice, additional weeks are needed and around 8 weeks of colonization are required. This time span is not normally used for conventionalization studies of GF animals as the standard time typically used is rather two weeks. Two weeks is the time when the small intestinal mucus peaks in defensin amounts and remains anchored, and when the large intestinal mucus is partly penetrable and stickier. The present results suggest that conventionalization experiments of adult GF animals addressing bacterial-host interaction should be performed for longer times. How human microbiota might influences the mouse mucus system remains to be addressed.
EXPERIMENTAL PROCEDURES
Animals
GF female mice (8–16 weeks) were colonized with caecal bacteria from C57BL/6 mice with normal mucus phenotype. Each experiment and week was performed on a minimum of 3 mice and repeated 3 times. Convr C57BL/6 mice were given ampicillin (1 g/l, Doktacillin, AstraZeneca), metronidazole (1 g/l, Sigma) vancomycin (0.5 g/l, Hospira) and neomycin trisulfate (1 g/l, Sigma) in the drinking water for 3 weeks (Fu et al., 2011).
Explant and mucus thickness and penetrability measurements
Gastrointestinal tissue was mounted in a horizontal perfusion chamber (Gustafsson et al., 2012b). The thickness of the intestinal mucus was measured (Gustafsson et al., 2012a; Gustafsson et al., 2012b). Mucus penetrability was measured (Gustafsson et al., 2012b; Johansson et al., 2014).
Tissue fixation and immunostaining
Intestine with fecal material were fixed in water free-Carnoy (methanol) and stained with hematoxylin and eosin, Alcian blue/periodic acid–Schiff (PAS) or hybridized with 10 ng/μL of a general bacterial 16S rRNA gene probe (EUB 338) and immunostained for Muc2 using the MUC2C3 antiserum, and DNA by Hoechst 34580 (Life technologies) (Johansson et al., 2008).
Proteomic analyses
Mucus samples removed after thickness measurements were solubilized in a guanidinium hydrochloride-based buffer, following the filter-aided sample preparation (FASP) and mass spectrometry with the addition of isotopicall internal standard peptides for Muc2, Clca1, Fcgbp and Agr2 (Rodriguez-Pineiro et al., 2013). Intestinal tissues were incubated in PBS containing 3 mM EDTA and 1 mM DTT at 37°C for 60 min and epithelial cells dissociated by vigorous shaking in PBS and pelleted at 1000g, 5 min.
Mucin oligosaccharide analysis
Mucus from midcolon of GF, Convr, and conventionalized mice (2, 3, 4, 6 and 8 weeks after gavage) was scraped and the insoluble Muc2 mucin semipurified by repeated 6M guanidinium hydrochloride (GuHCl) extraction, mucins separated by Ag-PAGE composite gel electrophoresis and the Muc2 mucin O-glycans analyzed by mass spectrometry after reductive β-elimination (Holmen-Larsson et al., 2013).
Quantitative real-time PCR
Distal colon (10–30 mg) with fecal material removed were snap-frozen and analyzed by Q-PCR for Muc2, IL1β, IL10, IL13, IL17, and IFNgamma.
Microbiota analysis
DNA was extracted from 80 mg of caecal sample and 10–130 mg of lumen or mucosal tissue from each mice (Jakobsson et al., 2015). Bacterial 16S rRNA gene sequences were amplified using the primers 27F (5′ AGAGTTTGATCCTGGCTCAG 3′) with Titanium Adaptor B and 338R (5′ TGCTGCCTCCCGTAGGAGT 3′) with Titanium Adaptor A and a sample-specific barcode sequence (Fierer et al., 2008) consisting of twelve nucleotides targeting the V1–V2 hypervariable region of the 16S rRNA gene. Samples were sequenced using Roche 454 FLX and Titanium chemistry (Roche) at SciLife (Solna, Sweden). Finally, data analysis was done using the QIIME software (1.7.0) package (Caporaso et al., 2010).
Statistical analysis
For all mucus measurements data comparing two independent groups was analyzed using a two-tailed Mann-Whitney U test. Multiple independent group comparisons were performed using Kruskal-Wallis ANOVA with Dunn’s correction for multiple comparisons. GraphPad Prism 6 was used for plotting the results. A P-value <0.05 (*) was regarded as statistically significant (**= P<0.01, ***= P<0.001, ****= P<0.0001). Only significant differences are marked in graphs. The peptide ratios from the proteomics data were analyzed for significant differences by ANOVA with 250 randomizations and FDR correction at 1%. Multiscatter between all samples and correlation calculations using Pearson’s correlation coefficient were performed. For the microbiota analysis, statistical significance testing for over- and under-representation of the bacterial lineages was made at phylum, class and genus (3% dissimilarity) levels. Significant differences were analyzed using the Wilcoxon rank-sum test, and P-values were converted to false discovery rate values (q-values) to correct for multiple testing in the R software (http://www.r-project.org/).
Supplementary Material
Acknowledgments
This work was supported by the Swedish Research Council, The Swedish Cancer Foundation, The Knut and Alice Wallenberg Foundation, IngaBritt and Arne Lundberg Foundation, Sahlgren’s University Hospital (LUA-ALF), Wilhelm and Martina Lundgren’s Foundation, Clas Groschinsky Foundation, T. Söderberg Foundation, Sahlgrenska Academy, National Institute of Allergy and Infectious Diseases (U01AI095473), and Swedish Foundation for Strategic Research - The Mucus-Bacteria-Colitis Center (MBC) of the Innate Immunity Program. We acknowledge CCI and Karin Ahlman for technical help.
Abbreviations
- GF
germ free
- Convr
conventionally raised
- WT
wild type
- GI
gastro-intestinal
Footnotes
Supplemental Information including Complete Methods and Supplemental Tables S1–S5 and Figures S1–6 can be found online.
AUTHOR CONTRIBUTION
MEVJ and GCH conceived the original idea; MEVJ, HEJ, JHL, AS, AE, AR-P, LA, FS, FB, and GCH designed the experiments; MEVJ, HEJ, JHL, AS, AE, AR-P, LA, CW, FS performed the experiments; MEVJ, HEJ, JHL, AS, AE, AR-P, LA, CW, FS, FB, and GCH analyzed the data; MEVJ, HEJ, JHL, AR-P, LA, FB and GCH wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-Bacterial Mutualism in the Human Intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Belzer C, Gerber GK, Roeselers G, Delaney M, DuBois A, Liu Q, Belavusava V, Yeliseyev V, Houseman A, Onderdonk A, et al. Dynamics of the Microbiota in Response to Host Infection. PLoS ONE. 2014;9:e95534. doi: 10.1371/journal.pone.0095534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booijink CC, El-Aidy S, Rajilic-Stojanovic M, Heilig HG, Troost FJ, Smidt H, Kleerebezem M, de Vos WM, Zoetendal EG. High temporal and inter-individual variation detected in the human ileal microbiota. Environ Microbiol. 2010;12:3213–3227. doi: 10.1111/j.1462-2920.2010.02294.x. [DOI] [PubMed] [Google Scholar]
- Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Fischbach MA. Small molecules from the human microbiota. Science. 2015:349. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Aidy S, Merrifield CA, Derrien M, van Baarlen P, Hooiveld G, Levenez F, Doré J, Dekker J, Holmes E, Claus SP, et al. The gut microbiota elicits a profound metabolic reorientation in the mouse jejunal mucosa during conventionalisation. Gut. 2013;62:1306–1314. doi: 10.1136/gutjnl-2011-301955. [DOI] [PubMed] [Google Scholar]
- El Aidy S, van Baarlen P, Derrien M, Lindenbergh-Kortleve DJ, Hooiveld G, Levenez F, Dore J, Dekker J, Samsom JN, Nieuwenhuis EES, et al. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 2012;5:567–579. doi: 10.1038/mi.2012.32. [DOI] [PubMed] [Google Scholar]
- Ermund A, Schutte A, Johansson MEV, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastroint Liver Physiol. 2013;305:G341–G347. doi: 10.1152/ajpgi.00046.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62:1157–1170. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Wei B, Wen T, Johansson MEV, Xiaowei L, Bradford E, Thomsson KA, McGee S, Mansour L, Tong M, et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis. J Clin Invest. 2011;121:1657–1666. doi: 10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson JK, Ermund A, Ambort D, Johansson MEV, Nilsson HE, Thorell K, Hebert H, Sjovall H, Hansson GC. Bicarbonate and functional CFTR channel is required for proper mucin secretion and link Cystic Fibrosis with its mucus phenotype. J Exp Med. 2012a;209:1263–1272. doi: 10.1084/jem.20120562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson JK, Ermund A, Johansson MEV, Schutte A, Hansson GC, Sjovall H. An ex vivo method for studying mucus formation, properties and thickness in human colonic biopsies and mouse small and large intestinal explants. Am J Physiol Gastrointest Liver Physiol. 2012b;302:G430–G438. doi: 10.1152/ajpgi.00405.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmén Larsson JM, Karlsson H, Gråberg Crespo J, Johansson MEV, Eklund L, Sjövall H, Hansson GC. An Altered O-glycosylation Profile of the MUC2 Mucin Occurs in Active Ulcerative Colitis and is Associated with Increased Inflammation. Infl Bowel Dis. 2011;17:2299–2307. doi: 10.1002/ibd.21625. [DOI] [PubMed] [Google Scholar]
- Holmen JM, Olson FJ, Karlsson H, Hansson GC. Two glycosylation alterations of mouse intestinal mucins due to infection caused by the parasite Nippostrongylus braseliensis. Glycoconj J. 2002;19:67–75. doi: 10.1023/a:1022589015687. [DOI] [PubMed] [Google Scholar]
- Holmen-Larsson JM, Thomsson KA, Rodriguez-Pineiro AM, Karlsson H, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. III Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am J Physiol Gastroint Liver Physiol. 2013;305:G357–G363. doi: 10.1152/ajpgi.00048.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Littman DR. Segmented filamentous bacteria take the stage. Mucosal Immunol. 2010;3:209–212. doi: 10.1038/mi.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Bjorksten B, Engstrand L, Andersson AF. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- Jakobsson HE, Rodriguez-Pineiro AM, Schütte A, Ermund A, Boysen P, Sommer F, Bäckhed F, Hansson GC, Johansson MEV. The gut microbiota composition impairs the colon inner mucus layer barrier. EMBO Reports. 2015;16:164–177. doi: 10.15252/embr.201439263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjövall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and in patients with ulcerative colitis. Gut. 2014;213:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Phillipson M, Petersson J, Holm L, Velcich A, Hansson GC. The inner of the two Muc2 mucin dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Sjovall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nature Rev Gastroenterol Hepatol. 2013;10:352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet. 2012;13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit M, Sonnenburg JL, Cossart P, Gordon JI. Functional Genomic Studies of the Intestinal Response to a Foodborne Enteropathogen in a Humanized Gnotobiotic Mouse Model. J Biol Chem. 2007;282:15065–15072. doi: 10.1074/jbc.M610926200. [DOI] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottaz D, Hahn D, Muller S, Muller C, Sterchi EE. Secretion of human meprin from intestinal epithelial cells depends on differential expression of the alpha and beta subunits. Eur J Biochem. 1999;259:496–504. doi: 10.1046/j.1432-1327.1999.00071.x. [DOI] [PubMed] [Google Scholar]
- Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg J, Roos S, Holm L, Phillipson M. Importance and Regulation of the Colonic Mucus Barrier in a Mouse Model of Colitis. Am J Physiology. 2011;300:G327–G333. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbe C, Capon C, Maes E, Rousset M, Zweibaum A, Zanetta JP, Michalski JC. Evidence of Regio-specific Glycosylation in Human Intestinal Mucins: Presnece of an acidic gradient along the gastrointestinal tract. J Biol Chem. 2003;278:46337–46348. doi: 10.1074/jbc.M302529200. [DOI] [PubMed] [Google Scholar]
- Robertson BR, O’Rourke JL, Neilan BA, Vandamme P, On SLW, Fox JG, Lee A. Mucispirillum schaedleri gen. nov., sp nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. International Journal of Systematic and Evolutionary Microbiology. 2005;55:1199–1204. doi: 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Pineiro AM, Bergstrom JH, Ermund A, Gustafsson JK, Schutte A, Johansson MEV, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. II Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am J Physiol Gastroint Liver Physiol. 2013;305:G348–G356. doi: 10.1152/ajpgi.00047.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütte A, Ermund A, Becker-Pauly C, Johansson MEV, Rodriguez-Pineiro AM, Bäckhed F, Müller S, Lottaz D, Bond JS, Hansson GC. Microbial Induced Meprin beta Cleavage in MUC2 Mucin and Functional CFTR Channel are Required to Release Anchored Small Intestinal Mucus. Proc Natl Acad Sci USA. 2014;111:12396–12401. doi: 10.1073/pnas.1407597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Russel SL, Antunes LCM, Finlay B. Gut Microbiota in Health and Disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Sterchi EE, Stocker W, Bond JS. Meprins, membrane-bound and secreted astacin metalloproteinases. Mol Aspects Med. 2008;29:309–328. doi: 10.1016/j.mam.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsson KA, Hinjosa-Kurtzberg M, Axelsson KA, Domino SE, Lowe JB, Gendler SJ, Hansson GC. Intestinal Mucins from Cystic Fibrosis Mice show Increased Fucosylation due to an Induced Fuca1-2 Glycosyltransferase. Biochem J. 2002;367:609–616. doi: 10.1042/BJ20020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The Antibacterial Lectin RegIIIg Promotes the Spatial Segregation of Microbiota and Host in the Intestine. Science. 2011;33:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereecke L, Vieira-Silva S, Billiet T, van Es JH, Mc Guire C, Slowicka K, Sze M, van den Born M, De Hertogh G, Clevers H, et al. A20 controls intestinal homeostasis through cell-specific activities. Nat Commun. 2014;5 doi: 10.1038/ncomms6103. [DOI] [PubMed] [Google Scholar]
- Walker WA. Initial Intestinal Colonization in the Human Infant and Immune Homeostasis. Annals of Nutrition and Metabolism. 2013;63(suppl 2):8–15. doi: 10.1159/000354907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.