Abstract
Aim:
Toxic metals including lead and mercury are associated with adverse pregnancy outcomes. This study aimed to assess the association between miRNA expression in the cervix during pregnancy with lead and mercury levels.
Materials & methods:
We obtained cervical swabs from pregnant women (n = 60) and quantified cervical miRNA expression. Women's blood lead, bone lead and toenail mercury levels were analyzed. We performed linear regression to examine the association between metal levels and expression of 74 miRNAs adjusting for covariates.
Results:
Seventeen miRNAs were negatively associated with toenail mercury levels, and tibial bone lead levels were associated with decreased expression of miR-575 and miR-4286.
Conclusion:
The findings highlight miRNAs in the human cervix as novel responders to maternal chemical exposure during pregnancy.
Keywords: : blood, bone, cervix, delivery, labor, lead, mercury, miRNA, patella, tibia
Background
Environmental contaminants are associated with numerous adverse pregnancy outcomes including infertility, pregnancy loss and preterm delivery [1]. Specifically, maternal exposure to lead and mercury may contribute to increased risk of spontaneous abortion [2–4] and preterm delivery [5,6]. Exposure to lead and mercury is widespread in US and Mexican populations [7,8]. In Mexico, lead exposure occurs mainly through use of glazed pottery, lead paint and air pollution [8,9]. Maternal mercury exposure commonly occurs through maternal diet (fish consumption), coal combustion and personal care product use in Mexico [10–12].
Environmental exposures likely contribute to tissue-specific pathophysiology in the placenta and uterus, including the cervix. Studies have demonstrated that reproductive tissues are susceptible to environmental contaminants [13,14]. Epigenetic changes could contribute to the onset of labor via regulation of uterine contraction and quiescence [15,16]. Cervical dilation is a necessary step for labor and parturition to occur, yet few studies have examined the impact of environmental exposures on epigenetic changes in the cervix during pregnancy.
Many environmental contaminants including toxic metals have been associated with epigenetic modifications including DNA methylation, histone modification and miRNA expression [17,18]. MiRNAs are small noncoding RNAs that posttranscriptionally control gene expression. In previous reports, miR-16, miR-21 and miR-146a were downregulated in placentas exposed to cigarette smoke compared with unexposed controls [19,20]. The same investigators have linked these changes to reduced fetal growth [21]. Moreover, a number of miRNAs have been identified as associated with ovarian and cervical cancers (as reviewed in [22]) demonstrating that these biomarkers are biologically active in the reproductive system. Previously, we have shown that metal exposure affects the expression of specific miRNAs in blood from occupationally exposed individuals [23] and that miRNAs in the cervix during pregnancy are associated with subsequent gestational age at delivery [24], but metal-altered miRNA expression has not yet been investigated with respect to tissues involved in parturition and labor.
In this study, we assessed the association between maternal lead and mercury exposures and miRNA expression in the cervix during the second trimester of pregnancy in a subset of 60 women enrolled in a prospective cohort. We used swabs to collect cervical cells during a Papanicolaou (Pap) smear and analyzed the expression profiles of 800 miRNA using the NanoString nCounter assay. This study aimed to identify miRNAs that were significantly associated with blood lead, patellar or tibial bone lead, or toenail mercury. Lead was selected as a prioritized metal of interest based on previously observed associations between prenatal lead exposure and preterm birth [6,25–26]. Mercury was also prioritized since limited literature suggests a relationship may exist with increased preterm birth risk [5]. The findings herein have the potential to enhance the current understanding of the complex molecular systems that govern environmentally associated alterations in the cervix during pregnancy.
Materials & methods
Study design
This study was conducted on a subcohort of 60 Mexican women aged 18–40 years participating in the PROGRESS birth cohort in Mexico City, and who consented to a cervical swab during pregnancy thereby participating in the PROGRESS Cervix Study. Full details of enrollment for the parent cohort and subcohort are published elsewhere [24,27]. Briefly, women in their second trimester were recruited between 2007 and 2011 through the Mexican social security system (Instituto Mexicano del Seguro Social). The parent cohort consists of 1054 mothers and 948 mother–infant pairs. Because the Cervix Study was conceived after the majority of recruitment had occurred only the last 100 women enrolled in the parent cohort were approached. Eighty enrollees provided written informed consent for an obstetrician to obtain a cervical swab mid-pregnancy (16–19-week gestation) for miRNA analysis. Women were offered a free Pap smear in return for their participation, which is the standard of care during pregnancy in the USA, but is not routinely offered in Mexico to pregnant women. The IRBs of the participating institutions approved this study. For funding reasons, 60 (80%) of the 80 samples collected were randomly selected for miRNA profiling analysis. All 60 women had blood lead exposure data and were included in this analysis. Forty (67%), 44 (73%) and 45 (75%) women had toenail mercury, patella and tibia lead data and were included in each subanalysis.
Participant data collection
Demographics and birth outcomes were collected as part of the parent study including maternal age, parity, self-reported prepregnancy BMI and years of education. Staff conducted in-person interviews, which included a question about household smoke exposure. Household smoke exposure was dichotomized as yes/no based on the mother's report that at least one household member smoked. All participants reported that they did not smoke during pregnancy. Parity was dichotomized as multiparous, if a woman had a prior live-born infant versus nulliparous if she had not. A histopathologic assessment for evidence of inflammation on the Pap smear served as a proxy for shifts in the cell-type mixture. The cervical swab sample collection contains mostly cervical epithelial cells (ectocervical and endocervical cells), and also leukocytes, which can be categorically quantified by a trained histopathologist. Although there were too few cells to perform detailed cell sorting, the clinical pathologist provided a qualitative assessment of the inflammatory cells noted on the Pap smear thereby allowing a subanalysis to adjust for cell type. We dichotomized Pap smear inflammation as yes/no based on the histopathologist's blinded interpretation of the smear. Twenty-four women (44%) had evidence of inflammation on the Pap smear. We performed a sensitivity analysis and confirmed that regression results were unchanged when adjusting for Pap inflammation (data not shown). This is an important consideration in future studies given that cell type and/or inflammation could impact miRNA expression profiles.
Lead & mercury exposure assessment
Venous whole blood samples and toenail samples were prepared and analyzed at the Trace Metals Laboratory of Harvard TH Chan School of Public Health in Boston as we have previously reported [28,29]. Lead concentrations were measured with a dynamic reaction cell-inductively coupled plasma mass spectrometer (Elan 6100; PerkinElmer, CT, USA). Toenail mercury analysis was performed by using direct mercury analysis methods for atomic absorption (DMA-80, Milestone Inc., CT, USA). Maternal tibia (cortical) and patella (trabecular) bone lead levels were measured within 1 month of delivery using a spot-source 109Cd K-shell x-ray fluorescence (K-XRF) instrument (ABIOMED, MA, USA) [30]. Bone lead measurements were calculated as the average of two measures (one from each leg), weighted by the inverse proportion of measurement error. This process can generate negative values, which were included in the analyses, as we have previously reported in other studies [31].
Cervical miRNA collection & extraction
Cervical cells were collected in a method similar to a standard Pap smear protocol, where a cotton swab was used to collect cells from the endocervix of the external os. The sample was immediately placed in RNALater (Qiagen, CA, USA) and the specimen was frozen at -80°C until subsequent analysis. Total RNA was extracted using the Exiqon miRCURY kit (Exiqon, MA, USA) according to the manufacturer's protocol. A cleanup step was then performed by using an Amicon Ultra 0.5 ml cleanup kit (EMD Millipore, MA, USA). MiRNAs were quantified by using a NanoPhotometer P-300 (Implent GmbH, CA, USA).
NanoString nCounter assay for miRNA expression
MiRNA expression was assessed using the NanoString nCounter system (NanoString Technologies, WA, USA). This method enables multiplexed direct digital counting of miRNA molecules [32]. This method measured a total of 800 probes that were available for analysis at the time of this study, and included both endogenous human-associated miRNAs as well as viral miRNAs that are expressed in human cells [33–35]. We performed a feasibility pilot with ten initial samples, including two technical replicates, which we previously reported showed strong correlation (r = 0.98) [24].
The raw count data from the 60 samples were normalized by using the NanoStringNorm R package [36]. Data were background-corrected by subtracting the mean of the six negative controls included on the platform, and normalized using the geometric mean of the ten probes with the lowest coefficients of variation – which were used to calculate a scaling factor as suggested by the package guidelines. A priori we required that probes be detectable in at least 60% of the samples. This resulted in 74 probes that were included in the analyses. Individual probes with expression levels below the LOD were assigned a nominal value of 1. Note that the proportion of samples below detect for each miRNA is reported in Supplementary Table 2. The distributions of miRNA expression and model residuals showed that our selection of a linear model was appropriate for these data.
Statistical analysis
To examine the association between miRNA expression levels and biomonitored lead or mercury, we used linear regression models. We used separate models to estimate the mean doubling of expression (log2 unit increase) of each miRNA associated with a unit (bone measures) or tenfold unit (toenail mercury and blood lead) increase metal exposure. To satisfy linear model assumptions, blood lead and toenail mercury levels were log transformed whereas untransformed patellar and tibial bone lead measures were normally distributed and thus we kept them untransformed. We chose covariates a priori and included maternal age, education, parity and smoke exposure inside the home. Parity and smoke exposure were dichotomized as described above. Maternal education was categorized as having less than high school, high school or greater than high school education. Both adjusted and unadjusted regression models were performed to examine the association between each metal exposure and the 74 probes’ log2-transformed miRNA levels. The reported beta coefficients represent the fold change (doubling or halvings) in expression per unit change in exposure. To transform from fold-change into raw expression change, the beta coefficients can be back transformed by using the antilog of 2. For example, the equation (2beta - 1) × 100 yields the percent change in raw miRNA expression. Because expression data are conventionally interpreted in log2, we present the beta coefficients that are easily interpreted in doublings or halvings. p-values and Storey's false discovery rate (FDR) q-values were calculated to estimate significance [37]; p < 0.05 was considered statistically significant. MiRNAs with an FDR q-value <0.1 that met these requirements in the adjusted model were retained for downstream target prediction and pathways enrichment analyses. A sensitivity analysis was performed that also adjusted for evidence of inflammation on the Pap smear. An additional sensitivity analysis with only the two bone lead measures also adjusted for prepregnancy BMI categorized as normal (BMI: 18.5–25 kg/m2), overweight (BMI: 25–30 kg/m2) and obese (≥30 kg/m2).
We performed two sensitivity analyses to assess the influence of probes with expression levels below the LOD. First, we treated probes with expression levels below the LOD as missing and excluded these probes from the analysis. Second, we assigned probes below the LOD to the minimum expression measured for a specific miRNA.
Prediction of miRNA targets
To predict downstream mRNA targets, the set of differentially expressed miRNAs which passed p < 0.05 and FDR q-value < 0.1 in the adjusted linear regression model were uploaded into the Ingenuity Pathway Analysis (IPA) tool (Ingenuity® Systems, CA, USA). Putative miRNA–mRNA relationships were identified using the IPA miRNA Target Filter, based on a knowledge-base of predicted and experimentally observed relationships. Where possible, we stringently selected for only the experimentally observed miRNA–mRNA relationships, and the resulting target gene list was analyzed for functional network and pathway analysis. If no experimentally observed relationships were identified, we analyzed the highly predicted mRNA targets.
Functional network & pathway enrichment analysis of miRNA transcripts
Functional analysis was carried out to identify molecular networks and biological functions significantly associated with the mRNA target gene set. Analysis of molecular network mapping, physiological system function enrichment and reproductive system disease and function enrichment was performed by using the IPA tool. Within the reproductive system disease and function enrichment analysis, the results were further filtered by excluding explicitly male reproductive categories (e.g., prostate and sperm) or breast-related results. The IPA proprietary database curates gene–phenotype associations, molecular interactions, regulatory events and chemical knowledge to provide a global molecular network. Related networks were algorithmically constructed based on connectivity. Statistical significance of each biological function was calculated by using Fisher's exact text with an alpha set at 0.05.
Results
Characteristics of study participants
Demographics of the 60 Mexican women participating in the PROGRESS Cervix Study are presented in Table 1. Generally, this was a lean cohort of women, the majority of whom reported no smoke exposure in the home, had at least 12 years of education and were multiparous. Second trimester whole blood lead levels were detectable in every sample (n = 60). Six women (10%) had blood lead levels greater than 5 μg/dl, the US Centers for Disease Control and Prevention reference level for pregnant women [38]. Postpartum bone lead levels were measured in 44 and 45 women, respectively, and toenail mercury levels were measured in a subset of 40 women. Spearman correlation of the exposures showed that patella and blood lead levels were positively correlated (r = 0.49; p = 0.0008); none of the other pair-wise correlations of exposure measures were significantly related (Supplementary Table 1). Descriptive statistics for each of the 74 miRNAs measured in cervical cells are provided in Supplementary Table 2.
Table 1. . Maternal demographics for 60 pregnant women participating in the PROGRESS cervix study.
| Participant characteristics | n (%) |
|---|---|
| Education: | |
| – <12 years | 19 (32) |
| – 12 years | 24 (40) |
| – >12 years |
17 (28) |
| Smoke in home: | |
| – No | 42 (70) |
| – Yes |
18 (30) |
| Parity: | |
| – Multiparous | 34 (57) |
| – Nulliparous |
26 (43) |
| BMI: | |
| – Normal | 30 (50) |
| – Overweight | 19 (32) |
| – Obese |
11 (18) |
| Maternal age (years), mean ± SD (range) |
27.9 ± 5.7 (18–40) |
| Blood lead† (μg/dl), mean ± SD (range) |
2.85 ± 1.63 (0.87–9.38) |
| Patella lead† (n = 44), mean ± SD (range) |
4.16‡ ± 6.99 (-6.85–20.90) |
| Tibia lead† (n = 45), mean ± SD (range) |
1.45‡ ± 8.39 (-32.60–19.45) |
| Toenail† mercury (n = 40); μg/g, mean ± SD (range) | 0.17 ± 0.09 (0.03–0.47) |
†The number of samples measured for each exposure marker varied as follows: blood lead (n = 60), patella lead (n = 44), tibia lead (n = 45) and toenail mercury (n = 40).
‡The reported mean and standard deviation for patellar and tibial lead include measurements that had negative-weighted average values (n = 15). The mean and standard deviation excluding negative values were 7.28 ± 6.20 and 5.66 ± 4.18 for patellar and tibial lead.
SD: Standard deviation.
Mid-pregnancy cervical miRNA expression was associated with mercury exposure
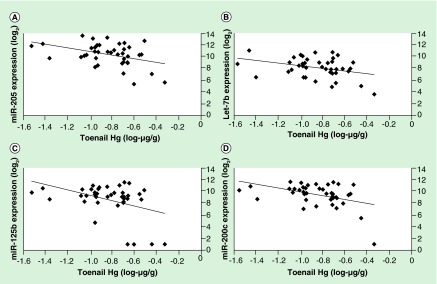
Using adjusted linear regression models, we identified 17 miRNAs that were associated with toenail mercury levels, which included let-7a and let-7b, and miRs-205, 125b, 200c, 342, 203, 24, 22, 23b, 375, 23a, 210, 200b, 99a, 21 and 193b (p < 0.05, and FDR q-value < 0.07) (Table 2). All of the miRNAs had lower expression among the higher mercury-exposed women. The beta coefficients that indicate the respective fold-change in miRNA expression per tenfold increment of mercury exposure and their interpreted values are presented in Table 2. For example, a beta coefficient of -4.0 for miR-205 represents a fourfold decrease (i.e., four halvings) in expression per tenfold increase in toenail mercury level. For completeness, the beta coefficients can be back transformed by using the antilog of 2. For example, 2-4.03 yields a value of 0.062, which is interpreted as a 94% decrease in raw miRNA expression. Because expression data are conventionally interpreted in log2, we present the beta coefficients that are interpreted as doublings or halvings. When we compare the 25–75th percentiles (interquartile range) of toenail mercury for the significant miRNAs, the differences in miRNA expression correspond to a one- to twofold change (data not shown). Representative plots of the four most significant mercury-associated miRNAs, miR-205, miR-125b, let-7b and miR-200c are shown in Figure 1.
Table 2. . Differentially expressed miRNA associated with blood lead, bone lead or toenail mercury (p < 0.05)†.
| Biomarker | miRNA | Adjusted beta (95% CI) | Interpreted beta as % expression change (95% CI) | p-value | q-value | Predicted targets (n) |
|---|---|---|---|---|---|---|
| Blood lead (n = 60) | hsa-miR-297 | 0.88 (0.21–1.55) | 84.0 (15.7–192.8) | 0.01 | 0.45 | 38 |
| |
hsa-miR-188 |
0.57 (0.11–1.03) |
48.5 (7.9–104.2) |
0.01 |
0.45 |
40 |
| Patella lead (n = 44) | hsa-miR-320e | -0.07 (-0.11 to -0.02) | -4.7 (-7.3 to -1.4) | 0.003 | 0.17 | 64 |
| hsa-miR-22–3p | -0.07 (-0.13 to -0.01) | -4.7 (-8.6 to -0.7) | 0.020 | 0.23 | 597 | |
| hsa-miR-93–5p | -0.10 (-0.19 to -0.01) | -6.7 (-12.3 to -0.7) | 0.025 | 0.23 | 1235 | |
| hsa-miR-769–5p | -0.08 (-0.15 to -0.01) | -5.4 (-9.9 to -0.7) | 0.030 | 0.23 | 82 | |
| hsa-miR-297 | 0.03 (0.00–0.06) | 2.1 (0.0–4.2) | 0.031 | 0.23 | 38 | |
| hsa-miR-425–5p | -0.10 (-0.19–0.00) | -6.7 (-12.3–0.0) | 0.046 | 0.23 | 238 | |
| |
hsa-miR-361–3p |
0.04 (0.00–0.08) |
2.8 (0.0–5.7) |
0.047 |
0.23 |
539 |
| Tibia lead (n = 45) | hsa-miR-575 | -0.06 (-0.1 to -0.02) | -4.1 (-6.7 to -1.4) | 0.003 | 0.08 | 82 |
| hsa-miR-4286 | -0.13 (-0.21 to -0.05) | -8.6 (-13.5 to -3.4) | 0.003 | 0.08 | 227 | |
| hsa-miR-15a-5p | 0.10 (0.02–0.19) | 7.2 (1.4–14.1) | 0.018 | 0.36 | 1452 | |
| hsa-miR-142–3p | 0.08 (0.01–0.16) | 5.7 (0.7–11.7) | 0.029 | 0.40 | 368 | |
| hsa-miR-193b-3p | -0.11 (-0.20 to -0.01) | -7.3 (-12.9 to -0.7) | 0.033 | 0.40 | 340 | |
| |
hsa-miR-494 |
-0.06 (-0.12–0.00) |
-4.1 (-8.0–0.0) |
0.044 |
0.46 |
581 |
| Toenail mercury (n = 40) | hsa-miR-205–5p | -4.03 (-6.48 to -1.58) | -93.9 (-98.9 to -66.6) | 0.001 | 0.02 | 426 |
| hsa-miR-125b-5p | -5.86 (-9.59 to -2.14) | -98.3 (-99.9 to -77.3) | 0.002 | 0.02 | 985 | |
| hsa-let-7b-5p | -2.90 (-4.85 to -0.94) | -86.6 (-96.5 to -47.9) | 0.004 | 0.02 | 1180‡ | |
| hsa-miR-200c-3p | -3.52 (-5.97 to -1.08) | -91.3 (-98.4 to -52.7) | 0.005 | 0.02 | 1074‡ | |
| hsa-miR-342–3p | -3.10 (-5.39 to -0.81) | -88.3 (-97.6 to -43.0) | 0.008 | 0.03 | 337 | |
| hsa-miR-203 | -3.39 (-5.95 to -0.83) | -90.5 (-98.4 to -43.7) | 0.009 | 0.03 | 870 | |
| hsa-let-7a-5p | -4.21 (-7.42 to -0.99) | -94.6 (-99.4 to -49.7) | 0.010 | 0.03 | 1180‡ | |
| hsa-miR-24–3p | -3.79 (-6.99 to -0.60) | -92.8 (-99.2 to -34.0) | 0.020 | 0.05 | 741 | |
| hsa-miR-22–3p | -2.81 (-5.21 to -0.41) | -85.7 (-97.3 to -24.7) | 0.022 | 0.05 | 597 | |
| hsa-miR-23b-3p | -4.14 (-7.76 to -0.52) | -94.3 (-99.5 to -30.3) | 0.025 | 0.05 | 1172‡ | |
| hsa-miR-375 | -2.29 (-4.32 to -0.25) | -79.6 (-95.0 to -15.9) | 0.027 | 0.05 | 233 | |
| hsa-miR-23a-3p | -3.50 (-6.64 to -0.37) | -91.2 (-99.0 to -22.6) | 0.028 | 0.05 | 1172‡ | |
| hsa-miR-210 | -3.77 (-7.20 to -0.34) | -92.7 (-99.3 to -21.0) | 0.031 | 0.05 | 78 | |
| hsa-miR-200b-3p | -3.33 (-6.40 to -0.26) | -90.1 (-98.8 to -16.5) | 0.034 | 0.05 | 1074‡ | |
| hsa-miR-99a-5p | -3.55 (-6.89 to -0.20) | -91.5 (-99.2 to -12.9) | 0.038 | 0.05 | 73 | |
| hsa-miR-21–5p | -5.17 (-10.20 to -0.14) | -97.2 (-99.9 to -9.2) | 0.044 | 0.05 | 340 | |
| hsa-miR-193b-3p | -3.77 (-7.53 to -0.01) | -92.7 (-99.5 to -0.7) | 0.050 | 0.06 | 340 |
Each multivariable regression model was adjusted for maternal age, education, smoke exposure in the home and parity.
†The adjusted beta values represent the fold-change in miRNA per tenfold increase in toenail mercury and blood lead, or per one unit increase for bone lead measures.
‡Let-7b and let-7a, miR-23a and miR-23b, and miR-200b and miR-200c share sequence homology and have identical target mRNAs.
Figure 1. . Representative plots of the top four mercury-associated miRNAs.
Plots shown include: (A) miR-205; (B) let-7b; (C) miR-125b and (D) miR-200c. Unadjusted regression lines are shown and Pearson correlation estimates are: (A) -0.41; (B) -0.35; (C) -0.40 and (D) -0.43.
We performed two sensitivity analyses that treated probes with expression levels below the LOD as missing and excluded these probes from the analysis or assigned probes below the LOD to the minimum expression measured for a specific miRNA. The results were similar for the second sensitivity analysis; however, some miRNAs lost statistical significance when probes below detect were excluded from analysis probes (e.g., miR-125b and let-7a) (Supplementary Table 3).
Mid-pregnancy cervical miRNA expression was associated with lead biomarkers
We identified distinct sets of miRNAs that were associated with maternal blood or bone lead (Table 2). Using adjusted linear regression models, we identified two miRNAs, miR-297 and miR-188, that had increased expression associated with blood lead levels (p < 0.05). We identified seven miRNAs that were associated with patellar bone lead levels including miR-320e, miR-22, miR-93, miR-769, miR-297, miR-425 and miR-361. All but miR-297 had decreased expression with increasing patellar lead levels. Notably, miR-297 was common to both the blood lead and patellar bone subsets and had increased expression with increasing blood lead and patellar bone lead. To show that the difference in identified miRNAs associated with blood lead from bone lead measures was not due to shifts in the sample population, we performed a subanalysis of blood lead associations restricted to the 44 individuals with both bone lead measures and the results were similar (Supplementary Table 4). None of the blood or patella lead-associated miRNAs reached statistical significance after FDR correction. In addition, we identified six miRNAs associated with tibial bone lead that included miR-575, miR-4286, miR-15a, miR-142, miR-193b and miR-494 (p < 0.05). MiR-575, miR-4286, miR-193b and miR-630 showed decreased expression with increasing tibial lead levels, and miR-15a and miR-142 had increased expression. Both miR-575 and miR-4286 had significantly decreased expression with increasing tibial lead levels (p < 0.05 and FDR q-value < 0.1) and thus we retained them for downstream functional analysis. The beta coefficients in Table 2 indicate the respective fold-change in miRNA expression per one unit increment of bone lead exposure. For example, a beta coefficient of -0.06 for miR-575 represents approximately a 0.06-fold decrease or 4.1% decrease in raw expression per unit increase in tibia lead level (Table 2). A beta coefficient of 0.10 for miR-15a represents a 0.1-fold increase or a 7.2% increase in raw expression. When we compare the 25th to 75th percentiles (interquartile range) of bone lead for the significant miRNAs, the differences in miRNA expression correspond to modest approximately 0.5-fold changes (data not shown).
Because the relationship between miRNA expression and bone lead measures may additionally be confounded by BMI, we performed a sensitivity analysis adjusted for prepregnancy BMI in addition to the primary set of covariates (Supplementary Table 5); the top identified miRNAs were similar.
Metal-associated miRNAs have known & predicted mRNA targets
For each of the metal-associated miRNAs, we report the number of experimentally observed or highly predicted mRNA targets regulated by the metal-associated miRNAs (Table 2). The blood lead, patella lead, tibia lead and toenail mercury-associated miRNAs had a total of 78, 2793, 3050 and 8446 downstream gene targets. Note that among the 8446 downstream gene targets of the 17 mercury-associated miRNAs, three pairs of miRNAs share sequence homology and have identical target mRNAs including let-7b and let-7a, miR-23a and miR-23b and miR-200b and miR-200c. Together these six miRNAs potentially coregulate over 3000 genes.
Functional network & pathway analysis of miRNA target genes
The 17 mercury-associated and two tibia lead-associated miRNAs that passed FDR correction (q-value < 0.1) in the linear model were selected for subsequent functional pathway analysis. We identified over 8000 mRNAs that were experimentally observed or highly predicted targets of the 17 miRNAs (Table 2). When the data were stringently filtered for experimentally observed targets only, we found that 15 of the miRNAs are known to regulate a total of 362 genes consisting of 330 unique mRNA targets. The 330 mercury-associated mRNA targets mapped to several molecular networks (Supplementary Table 6). The top two networks were enriched for organismal, cellular and cardiovascular system development (p = 1 × 10-55), as well as cell cycle, cancer and gene expression (p = 1 × 10-38). Physiological system development and function analysis showed enrichment for organismal survival (p < 5.0 × 10-18), cardiovascular system development and function (p < 4.3 × 10-18), organismal development (4.9 × 10-13), tissue morphology (p < 6.2 × 10-14) and embryonic development (p < 4.9 × 10-13). When the list of associated diseases and functions was filtered for reproductive system disease or function, a group of 60 genes was enriched for reproductive system development (p < 5.76 × 10-23) and another group of 40 genes was enriched for abnormal morphology of the reproductive system (p < 5.05 × 10-15). Specifically, a majority (56%, n = 26) of the gene subset enriched for abnormal morphology is regulated by three miRNAs: let-7, miR-125b and miR-24. Also of note, 18 miRNA-regulated molecules are involved in aryl hydrocarbon receptor (AHR) signaling (p = 6 × 10-12) (Supplementary Table 7). The AHR signaling pathway has known links to environmentally mediated epigenetic modification [39] and plays a critical role in the female reproductive system [40]. Previously, we have shown that cervical miRNAs associated with subsequent gestational age were also enriched for the AHR pathway signaling [24].
None of the mRNA targets of the tibia lead-associated miRNAs, miR-575 and miR-4286 had experimentally observed relationships with target genes. Therefore, we analyzed a total of 309 highly predicted targets for the two miRNAs (Table 2). The tibia lead-associated predicted gene targets showed enrichment for several networks including carbohydrate metabolism, small-molecule biochemistry and DNA replication (p = 1 × 10-43), as well as behavior, lipid metabolism and posttranslational modification (p = 1 × 10-27) (Supplementary Table 8). Physiological system development and function analysis showed enrichment for organ and tissue development (p < 0.02), hematological system development and function (p < 0.02), hematopoiesis (p < 0.02) and cardiovascular system development and function (p < 0.01). When we filtered the list of associated disease and functions for reproductive system disease or function, a group of 14 mRNA targets was enriched for preeclampsia (p < 8.80 × 10-3) [41] and two genes were associated with formation of placenta in mouse models [42,43]. Ten of the preeclampsia-associated genes are the predicted targets of miR-4286, and four are the predicted targets of miR-575. Interestingly, miR-4286 is predicted to target three genes involved in AHR signaling including AHR repressor, prostaglandin E synthase 3, and tumor protein P73.
Discussion
Maternal exposure to lead or mercury has been associated with numerous adverse birth outcomes [1]; however, the molecular mechanisms by which lead or mercury influence the female reproductive system during pregnancy are unknown. Changes in cervical miRNA expression are a potential mechanism that could alter gene expression leading to aberrant changes in cervix tissue function and subsequently impact parturition. In this cohort study of pregnant women, we identified distinct miRNAs measured in cervical samples during pregnancy that are associated with maternal metal exposure. Our study identified 17 miRNAs that were significantly associated with maternal mercury exposure in the second trimester. Separate and largely distinct subsets of two, seven and six miRNAs were also identified in association with maternal second trimester blood lead, and postpartum lead measures in patellar and tibial bone, respectively. We computationally predicted the downstream mRNA targets for the most significantly metal-associated miRNAs, and subsequent functional enrichment analyses revealed that mercury-associated miRNA gene targets are involved in reproductive system development and morphology, lead-associated gene targets are enriched for preeclampsia, and molecular targets of both the mercury- and lead-associated miRNAs are involved in the AHR signaling pathway.
No previous studies have evaluated the effects of metals on cervical miRNA expression; however, some similar patterns have been identified between other tissue-specific or circulating miRNAs and metals. For example, we report that miR-21 had decreased expression in association with maternal mercury levels. While miR-21 has not been previously studied with respect to mercury, miR-21 was downregulated in placentas exposed to cigarette smoke, which contains lead and cadmium, compared with controls [19]. Additionally, urinary miR-21 had a negative relationship with blood lead levels in adolescents [44]. In the present study, cervical miR-21 had marginally significantly decreased expression with respect to patella lead and was not associated with tibia lead or blood lead levels. Previously, we have shown that miR-146a expression in blood was significantly decreased in association with lead- and cadmium-rich particulate matter in adult males [45]; however, miR-146a was not detected in cervical samples in the present study. A study of fibroblasts exposed to a mixture of arsenic, cadmium and lead computationally predicted, then confirmed upregulation of six candidate miRNAs: miRs-154, 222, 379, 10, 375, 204 and 133 [46]. We report that miR-375 was significantly negatively associated with toenail mercury levels, and found no association with blood or bone lead. The only previous study of mercury-altered miRNAs exposed neuronal/glial cells derived from NT2 carcinoma pluripotent stem cells to methyl mercury chloride and showed upregulated levels of miR-302b, 367, 372, 196b and 141 [47], none of which overlap with miRNAs reported here.
Our study identified 17 miRNAs that have significantly decreased expression with maternal mercury exposure. Notably, miR-205, miR-125b and let-7 are miRNAs known as oncomirs, which are involved in a number of human cancers and directly regulate oncogenes including phosphatase and tensin homolog, tumor protein P53 and RAS, respectively [48]. Supported by our functional enrichment analysis, the concerted action of these oncomirs and the other identified miRNAs have known impacts on a number of cell cycle and proliferation pathways that could affect parturition.
It is not surprising then that a number of the mercury-associated miRNAs and their mRNA targets have also been identified with respect to reproductive system diseases. Our functional analysis showed distinct subsets of mRNA targets that are involved in reproductive system development and morphology. Specifically, the gene subset enriched for abnormal morphology had mRNA targets predominantly regulated by let-7, miR-125b and miR-24. For example, miR-205, miR-24 and miR-21 are highly expressed in cervical cancers compared with normal controls [49]; and miR-205 is thought to play a role in cervical cancer or serve as a diagnostic marker in plasma [22,50]. Downregulation of miR-203 and upregulation of let-7 and miR-21 are associated with early stage and invasive cervical carcinoma [51]. Furthermore, we noted a negative association between miRs-200c and 200b with increasing maternal mercury levels. The miR-200 family has been shown to increase in myometrium expression levels as pregnancies approach term, and in turn negatively regulate several contraction-associated genes including zinc finger E-box binding homeobox-1 and -2, and connexin 43 [15]. We note that although miRNAs typically act to negatively regulate gene expression, evidence also demonstrates that some miRNAs act by increasing target gene expression [15]. Given the paucity of data on normal cervix expression outside of cancer-based studies with environmental contaminants, additional studies on the biological role of the identified miRNAs and their role in normal pregnancies and parturition are needed.
Analysis of the blood and bone lead exposures identified largely distinct subsets of lead-associated miRNAs with the exception of miR-297, which had increased expression with increasing blood lead and patellar bone lead. Additional studies are needed to ascertain whether miR-297 is a robust biomarker for acute and/or chronic lead exposure. Tibia lead was more strongly associated with changes in cervical miRNA expression than patellar lead. Among the tibia lead-associated targets, miR-575 and miR-4286 had significantly decreased expression. It is notable that 14 of the downstream targets we identified were differentially expressed in a study of genome-wide expression using decidua basilis (placenta) tissue from preeclamptic versus normal pregnancies [41]. Moreover, miR-4286 is predicted to regulate three genes involved in AHR signaling, which is a vital pathway in the female reproductive system that is influenced by environmental contaminants such as cigarette smoke and dioxins [40]. Taken together, these findings are suggestive of a possible environmentally mediated mechanism of miRNA gene regulation during pregnancy. Additional mechanistic studies using tandem mRNA and miRNA transcriptional profiling are needed.
The tibia consists mainly of cortical bone, which has a lower turnover rate and longer half-life with respect to lead compared with the patella, which consists mostly of trabecular bone [30]. Therefore, patella lead is likely to track closely with bone remodeling, while tibia lead represents cumulative exposure. Our data support these conclusions given the positive correlation between blood lead and patella lead as bone lead stores are a major source of maternal blood lead. Mobilization of lead stores via bone turnover is considered the major source of circulating lead in absence of ongoing external exposure sources [30]. Thus, during pregnancy, patellar lead might exert the potential greatest impact on miRNA expression, if the mechanism required only mobilization as a result of bone demineralization. In contrast, our data support a stronger association between tibia lead (chronic lead exposure) and miRNA expression, suggesting that cumulative lead exposure among women may explain our findings more than concurrent pregnancy exposures.
To our knowledge, this is the first study to examine the association between cervical miRNA expression and environmental exposures and has many strengths. We sampled the cervix during the second trimester, a relevant time point for miRNAs potentially involved in pregnancy outcomes or reproductive system pathophysiology. miRNA expression is tissue-specific, and investigating a key tissue for delivery initiation is critical to understand mechanisms that may regulate signaling cascades, as we have previously reported in this cohort and the association with subsequent gestational age [24]. Our primary outcome of interest in the previous study was gestational age, because pathophysiologically we hypothesized that metal-induced alteration in the cervix would not be associated with fetal growth, unlike those in the placenta, which biologically might mediate a metal-fetal growth relationship. We prospectively enrolled women during pregnancy who were participating in an ongoing population-based cohort study with carefully collected covariate data. Given the cohort design of our study, we were able to adjust for potential confounding variables. We assessed total mercury in toenails, which is a valid biomarker for chronic mercury exposure that correlates with methylmercury levels from hair [52,53]. Additionally, in this study, we examined human cervical miRNA responses to acute (i.e., blood levels) and chronic (i.e., tibia bone levels) environmentally relevant levels of lead. Studies that compare the acute versus chronic miRNA response to exposure, as we observed here between the lead biomarkers are useful in understanding the impact and contributions of each type of population-level exposure. The levels of metals assessed in this study are comparable to those reported in previous cohorts. For example, the levels observed were slightly higher than the geometric mean of 0.68 μg/dl among 253 pregnant women in the USA participating in the Fourth Report on Human Exposure to Environmental Chemicals (NHANES IV) [54]. The levels of toenail mercury reported here were comparable to levels observed in two previous studies among adult populations, which reported average mercury toenail levels of 0.212 μg/g (geometric mean) [55] and 0.25 μg/g [56]. Only six women in this study had blood lead levels greater than 5 μg/dl; the Centers for Disease Control and Prevention reference level for pregnant women [38], there is no current guideline for toenail mercury levels during pregnancy.
Our study also has several limitations. The PROGRESS cohort is a racially/ethnically homogeneous population, which may limit external generalizability, although results may likely be better generalized to Mexican–Americans. We sampled the cervix just once mid-pregnancy and thus cannot identify from this study the impact of toxic metals on miRNA expression throughout pregnancy. Differences in the miRNAs identified in this study compared with previous studies may originate from differences in the tissue type, exposure levels and source populations, as well as the type of quantification platform used, and time of sample collection (during pregnancy vs not). Our source population was largely a healthy group of pregnant women in Mexico City. Our selection of miRNAs was informed by the NanoString platform. The NanoString platform analyzed 800 human and viral miRNA at the time of this study. Many of these targets (n = 726) were detectable in fewer than 60% of cervical samples, and thus were not included in our analyses. Our study, therefore, was unable to detect subtle differences in less highly expressed miRNAs. This is a known characteristic of the NanoString platform that has less sensitive probe detection than other platforms [57]. Therefore, we chose to retain miRNAs that were below the limit of detection in less than 40% of our participants and we performed sensitivity analyses to examine how our approach affected the magnitude and direction of observed associations. However, a strength of the NanoString platform is that it provides quantitative assessment of miRNAs from samples with low overall RNA yields such as the samples obtained with a single cervical swab. We did not have enough biologic material to analyze our samples in duplicate using another platform. While rigorous computational methods were employed to identify potential miRNA-mediated mRNA expression, this study did not directly measure the downstream changes on RNA expression. The future characterization of miRNA control of gene expression and protein translation in cervical tissue and in vivo models will provide information on how cervical cells respond to lead or mercury that could contribute to adverse pathophysiology. Additional studies from larger populations and mechanistic studies are needed to replicate the findings here.
Conclusion
Our findings support a miRNA-mediated response to environmental contaminants in the cervix, in an obtainable tissue type during human pregnancy. We observed mercury- and lead-associated miRNAs in the cervix during pregnancy. To our knowledge, this is one of the first studies to link molecular changes in cervical tissue during pregnancy to maternal toxic metal exposure.
Future perspective
Understanding the pathophysiological role of mercury- and lead-altered miRNAs in the cervix and other relevant tissues is a priority for future studies. Replication in human cohorts followed by in vitro/vivo mechanistic studies should be prioritized to further validate these findings. The contributions of environmentally altered molecular programming in the cervix to adverse birth outcomes such as spontaneous abortion and preterm delivery, as well as the role in transgenerational reproductive system effects remain to be studied.
Executive summary.
Seventeen mercury-associated miRNAs were identified from the cervix during pregnancy (p < 0.05; false discovery rate q-value < 0.1).
Two tibia lead-associated miRNAs were identified (p < 0.05; false discovery rate q-value < 0.1).
The mercury-associated miRNA gene targets were involved in reproductive system development and morphology.
The tibia lead-associated gene targets were enriched for preeclampsia.
Molecular targets of both the mercury- and lead-associated miRNAs are involved in the aryl hydrocarbon receptor signaling pathway.
Our findings indicate a miRNA-mediated response to environmental contaminants in the cervix, in an obtainable tissue type during human pregnancy.
The contributions of environmentally altered molecular programming in the cervix to adverse birth outcomes such as spontaneous abortion and preterm delivery are a priority for future studies.
Supplementary Material
Acknowledgements
The authors thank Erroll Reuckert at NanoString Technologies for his assistance with the miRNA expression profiling and analysis. The authors also thank the ABC Medical Center in Mexico City for providing facilities during data collection.
Footnotes
Financial & competing interests disclosure
This work was supported in part by Pilot Project funding from the HSPH-NIEHS Center for Environmental Health (ES000002) and NIH/NIEHS: K23ES022242, K99ES023450, P42ES016454, P30ES23515, R01ES013744, R01ES020268, R01ES021357, the Klarman Scholars Program at Beth Israel Deaconess Medical Center, the Harvard Catalyst D-MaPS Program and the National Institute of Public Health/Ministry of Health of Mexico. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The IRBs of the participating institutions approved this study: Brigham and Women's Hospital # 2006-P-001416 and P001792, Icahn School of Medicine at Mount Sinai human subjects management #12-00751 and Instituto Nacional de Salud Publica project #560. Written informed consent was obtained from women participating in the PROGRESS study.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Silbergeld EK, Patrick TE. Environmental exposures, toxicologic mechanisms, and adverse pregnancy outcomes. Am. J. Obstet. Gynecol. 2005;192(5 Suppl.):S11–S21. doi: 10.1016/j.ajog.2004.06.117. [DOI] [PubMed] [Google Scholar]
- 2.Savitz DA, Sonnenfeld NL, Olshan AF. Review of epidemiologic studies of paternal occupational exposure and spontaneous abortion. Am. J. Ind. Med. 1994;25(3):361–383. doi: 10.1002/ajim.4700250306. [DOI] [PubMed] [Google Scholar]
- 3.Aschengrau A, Zierler S, Cohen A. Quality of community drinking water and the occurrence of late adverse pregnancy outcomes. Arch. Environ. Health. 1993;48(2):105–113. doi: 10.1080/00039896.1993.9938403. [DOI] [PubMed] [Google Scholar]
- 4.Anttila A, Sallmen M. Effects of parental occupational exposure to lead and other metals on spontaneous abortion. J. Occup. Environ. Med. 1995;37(8):915–921. doi: 10.1097/00043764-199508000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Xue F, Holzman C, Rahbar MH, Trosko K, Fischer L. Maternal fish consumption, mercury levels, and risk of preterm delivery. Environ. Health. Perspect. 2007;115(1):42–47. doi: 10.1289/ehp.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perkins M, Wright RO, Amarasiriwardena CJ, Jayawardene I, Rifas-Shiman SL, Oken E. Very low maternal lead level in pregnancy and birth outcomes in an Eastern Massachusetts population. Ann. Epidemiol. 2014;24(12):915–919. doi: 10.1016/j.annepidem.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Fourth National Report on human exposure to environmental chemicals, updated tables. 2013. www.cdc.gov/exposurereport/pdf/FourthReport_UpdatedTables_Sep2013.pdf
- 8.Romieu I, Palazuelos E, Hernandez Avila M, et al. Sources of lead exposure in Mexico City. Environ. Health. Perspect. 1994;102(4):384–389. doi: 10.1289/ehp.94102384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caravanos J, Dowling R, Tellez-Rojo MM, et al. Blood lead levels in Mexico and pediatric burden of disease implications. Ann. Glob. Health. 2014;80(4):269–277. doi: 10.1016/j.aogh.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Weldon MM, Smolinski MS, Maroufi A, et al. Mercury poisoning associated with a Mexican beauty cream. West. J. Med. 2000;173(1):15–18. doi: 10.1136/ewjm.173.1.15. discussion 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agency for Toxic Substances and Disease Registry (ATSDR) Toxicological profile for Mercury. 1999. http://www.atsdr.cdc.gov/toxprofiles/tp.asp?id=115&tid=24
- 12.Oken E, Guthrie LB, Bloomingdale A, et al. Assessment of dietary fish consumption in pregnancy: comparing one-, four- and thirty-six-item questionnaires. Public. Health. Nutr. 2014;17(9):1949–1959. doi: 10.1017/S1368980013001985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leino O, Kiviranta H, Karjalainen AK, et al. Pollutant concentrations in placenta. Food. Chem. Toxicol. 2013;54:59–69. doi: 10.1016/j.fct.2011.10.058. [DOI] [PubMed] [Google Scholar]
- 14.Boverhof DR, Kwekel JC, Humes DG, Burgoon LD, Zacharewski TR. Dioxin induces an estrogen-like, estrogen receptor-dependent gene expression response in the murine uterus. Mol. Pharmacol. 2006;69(5):1599–1606. doi: 10.1124/mol.105.019638. [DOI] [PubMed] [Google Scholar]
- 15.Renthal NE, Chen CC, Williams KC, Gerard RD, Prange-Kiel J, Mendelson CR. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc. Natl Acad. Sci. USA. 2010;107(48):20828–20833. doi: 10.1073/pnas.1008301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams KC, Renthal NE, Condon JC, Gerard RD, Mendelson CR. MicroRNA-200a serves a key role in the decline of progesterone receptor function leading to term and preterm labor. Proc. Natl Acad. Sci. USA. 2012;109(19):7529–7534. doi: 10.1073/pnas.1200650109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ray PD, Yosim A, Fry RC. Incorporating epigenetic data into the risk assessment process for the toxic metals arsenic, cadmium, chromium, lead, and mercury: strategies and challenges. Front. Genet. 2014;5:201. doi: 10.3389/fgene.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsit CJ. Influence of environmental exposure on human epigenetic regulation. J. Exp. Biol. 2015;218(Pt 1):71–79. doi: 10.1242/jeb.106971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maccani MA, Avissar-Whiting M, Banister CE, Mcgonnigal B, Padbury JF, Marsit CJ. Maternal cigarette smoking during pregnancy is associated with downregulation of miR-16, miR-21, and miR-146a in the placenta. Epigenetics. 2010;5(7):583–589. doi: 10.4161/epi.5.7.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study highlights the downregulation of placental miRNA expression in response to maternal cigarette exposure.
- 20.Maccani MA, Knopik VS. Cigarette smoke exposure-associated alterations to non-coding RNA. Front. Genet. 2012;3:53. doi: 10.3389/fgene.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maccani MA, Marsit CJ. Exposure and fetal growth-associated miRNA alterations in the human placenta. Clin. Epigenetics. 2011;2(2):401–404. doi: 10.1007/s13148-011-0046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The authors of this review propose a novel hypothesis that placental miRNAs may serve as mediators between environmental exposures and fetal growth.
- 22.Logan M, Hawkins SM. Role of microRNAs in cancers of the female reproductive tract: insights from recent clinical and experimental discovery studies. Clin. Sci. (Lond.) 2015;128(3):153–180. doi: 10.1042/CS20140087. [DOI] [PubMed] [Google Scholar]
- 23.Motta V, Angelici L, Nordio F, et al. Integrative analysis of miRNA and inflammatory gene expression after acute particulate matter exposure. Toxicol. Sci. 2013;132(2):307–316. doi: 10.1093/toxsci/kft013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders AP, Burris HH, Just AC, et al. microRNA expression in the cervix during pregnancy is associated with length of gestation. Epigenetics. 2015;10(3):221–228. doi: 10.1080/15592294.2015.1006498. [DOI] [PMC free article] [PubMed] [Google Scholar]; • We observed associations between miRs-21, 30e, 142, 148b, 29b and 223 obtained from cervical samples in pregnancy and subsequent length of gestation in the PROGRESS Cervix Study.
- 25.Andrews KW, Savitz DA, Hertz-Picciotto I. Prenatal lead exposure in relation to gestational age and birth weight: a review of epidemiologic studies. Am. J. Ind. Med. 1994;26(1):13–32. doi: 10.1002/ajim.4700260103. [DOI] [PubMed] [Google Scholar]
- 26.Jelliffe-Pawlowski LL, Miles SQ, Courtney JG, Materna B, Charlton V. Effect of magnitude and timing of maternal pregnancy blood lead (Pb) levels on birth outcomes. J. Perinatol. 2006;26(3):154–162. doi: 10.1038/sj.jp.7211453. [DOI] [PubMed] [Google Scholar]
- 27.Burris HH, Braun JM, Byun HM, et al. Association between birth weight and DNA methylation of IGF2, glucocorticoid receptor and repetitive elements Line-1 and Alu. Epigenomics. 2013;5(3):271–281. doi: 10.2217/epi.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mordukhovich I, Wright RO, Hu H, et al. Associations of toenail arsenic, cadmium, mercury, manganese, and lead with blood pressure in the normative aging study. Environ. Health. Perspect. 2012;120(1):98–104. doi: 10.1289/ehp.1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crinnion WJ. The CDC Fourth National Report on human exposure to environmental chemicals: what it tells us about our toxic burden and how it assist environmental medicine physicians. Altern. Med. Rev. 2010;15(2):101–109. [PubMed] [Google Scholar]
- 30.Hu H, Rabinowitz M, Smith D. Bone lead as a biological marker in epidemiologic studies of chronic toxicity: conceptual paradigms. Environ. Health. Perspect. 1998;106(1):1–8. doi: 10.1289/ehp.981061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tellez-Rojo MM, Hernandez-Avila M, Lamadrid-Figueroa H, et al. Impact of bone lead and bone resorption on plasma and whole blood lead levels during pregnancy. Am. J. Epidemiol. 2004;160(7):668–678. doi: 10.1093/aje/kwh271. [DOI] [PubMed] [Google Scholar]
- 32.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008;26(3):317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 33.Stern-Ginossar N, Elefant N, Zimmermann A, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317(5836):376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cullen BR. Five questions about viruses and microRNAs. PLoS Pathog. 2010;6(2):e1000787. doi: 10.1371/journal.ppat.1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411(2):325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waggott D, Chu K, Yin S, Wouters BG, Liu FF, Boutros PC. Nanostringnorm: an extensible R package for the pre-processing of nanostring mRNA and miRNA data. Bioinformatics. 2012;28(11):1546–1548. doi: 10.1093/bioinformatics/bts188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Centers for Disease Control and Disease. Low level lead exposure harms children: a renewed call for primary prevention. 2012. www.cdc.gov/nceh/lead/acclpp/final_document_030712.pdf
- 39.Joubert BR, Haberg SE, Nilsen RM, et al. 450k epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health. Perspect. 2012;120(10):1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernandez-Ochoa I, Karman BN, Flaws JA. The role of the aryl hydrocarbon receptor in the female reproductive system. Biochem. Pharmacol. 2009;77(4):547–559. doi: 10.1016/j.bcp.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loset M, Mundal SB, Johnson MP, et al. A transcriptional profile of the decidua in preeclampsia. Am. J. Obstet. Gynecol. 2011;204(1):84., e1–e27. doi: 10.1016/j.ajog.2010.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morasso MI, Grinberg A, Robinson G, Sargent TD, Mahon KA. Placental failure in mice lacking the homeobox gene Dlx3. Proc. Natl Acad. Sci. USA. 1999;96(1):162–167. doi: 10.1073/pnas.96.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Nes J, De Graaff W, Lebrin F, Gerhard M, Beck F, Deschamps J. The Cdx4 mutation affects axial development and reveals an essential role of Cdx genes in the ontogenesis of the placental labyrinth in mice. Development. 2006;133(3):419–428. doi: 10.1242/dev.02216. [DOI] [PubMed] [Google Scholar]
- 44.Kong AP, Xiao K, Choi KC, et al. Associations between microRNA (miR-21, 126, 155 and 221), albuminuria and heavy metals in Hong Kong Chinese adolescents. Clin. Chim. Acta. 2012;413(13–14):1053–1057. doi: 10.1016/j.cca.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 45.Bollati V, Marinelli B, Apostoli P, et al. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ. Health. Perspect. 2010;118(6):763–768. doi: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A prior study of metal-altered miRNAs in an adult population found that miR-222 and miR-146a were differentially expressed with respect to lead exposure.
- 46.Martinez-Pacheco M, Hidalgo-Miranda A, Romero-Cordoba S, Valverde M, Rojas E. mRNA and miRNA expression patterns associated to pathways linked to metal mixture health effects. Gene. 2014;533(2):508–514. doi: 10.1016/j.gene.2013.09.049. [DOI] [PubMed] [Google Scholar]
- 47.Pallocca G, Fabbri M, Sacco MG, et al. miRNA expression profiling in a human stem cell-based model as a tool for developmental neurotoxicity testing. Cell. Biol. Toxicol. 2013;29(4):239–257. doi: 10.1007/s10565-013-9250-5. [DOI] [PubMed] [Google Scholar]
- 48.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer. Res. 2007;67(16):7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Tang S, Le SY, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE. 2008;3(7):e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng H, Zhang L, Zhao Y, et al. Plasma miRNAs as diagnostic and prognostic biomarkers for ovarian cancer. PLoS ONE. 2013;8(11):e77853. doi: 10.1371/journal.pone.0077853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JW, Choi CH, Choi JJ, et al. Altered microRNAs expression in cervical carcinomas. Clin. Cancer. Res. 2008;14(9):2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 52.Bjorkman L, Lundekvam BF, Laegreid T, et al. Mercury in human brain, blood, muscle and toenails in relation to exposure: an autopsy study. Environ. Health. 2007;6:30. doi: 10.1186/1476-069X-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alfthan GV. Toenail mercury concentration as a biomarker of methylmercury exposure. Biomarkers. 1997;2(4):233–238. doi: 10.1080/135475097231607. [DOI] [PubMed] [Google Scholar]
- 54.Woodruff TJ, Zota AR, Schwartz JM. Environmental chemicals in pregnant women in the United States: NHANES 2003–2004. Environ. Health. Perspect. 2011;119(6):878–885. doi: 10.1289/ehp.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xun P, Liu K, Morris JS, Jordan JM, He K. Distributions and determinants of mercury concentrations in toenails among American young adults: the CARDIA Trace Element Study. Environ. Sci. Pollut. Res. Int. 2013;20(3):1423–1430. doi: 10.1007/s11356-012-1126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gibb H, Haver C, Kozlov K, et al. Biomarkers of mercury exposure in two Eastern Ukraine cities. J. Occup. Environ. Hyg. 2011;8(4):187–193. doi: 10.1080/15459624.2011.556984. [DOI] [PubMed] [Google Scholar]
- 57.Knutsen E, Fiskaa T, Ursvik A, et al. Performance comparison of digital microRNA profiling technologies applied on human breast cancer cell lines. PLoS ONE. 2013;8(10):e75813. doi: 10.1371/journal.pone.0075813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.