Synopsis
In spite of a rapidly expanding understanding of head and neck tumor biology as well as optimization of radiation, chemotherapy, and surgical treatment modalities, head and neck squamous cell carcinoma (HNSCC) remains a major cause of cancer related morbidity and mortality. Although our biologic understanding of these tumors had largely been limited to pathways driving proliferation, survival, and differentiation, the identification of HPV as a major driver of HNSCC, specifically oropharyngeal SCC, as well as recent genomic sequencing analyses of HNSCC has dramatically influenced our understanding of the underlying biology behind carcinogenesis, and in part, our approach to therapy. In particular, we are at a major molecular and clinical crossroads with an explosion of promising diagnostic and therapeutic agents that hold great promise. Here, we summarize our current understanding of HNSCC biology, including a review of recent sequencing analyses, and identify promising areas for potential diagnostic and therapeutic agents.
Keywords: head and neck cancer, squamous cell carcinoma, molecular biology, targeted therapy, synthetic lethality, genomic sequencing, intratumor heterogeneity
Introduction
Despite advances in our understanding of tumor biology including its evolutionary refinements as well as radiation, chemotherapy, and surgical treatments, head and neck squamous cell carcinoma (HNSCC) remains the sixth leading cause of cancer related morbidity and mortality, with 600,000 new cases diagnosed each year (1, 2). These tumors arise from mucosal epithelium in the oral cavity, oropharynx, larynx, and hypopharynx, which together represent 75% of diagnosed cancers (3).
HNSCC tumors can be broadly divided into those that are HPV− and associated with alcohol and tobacco consumption (4) and those that are HPV+ and due to HPV infection primarily with serotype 16 (5,6). While HPV− cancers arise via field cancerization and clonal progression in the setting of repetitive carcinogen application, HPV+ tumors harbor few mutations and are driven by a fundamentally distinct pathophysiologic mechanisms that rely on E6 and E7 viral proteins to inactivate or bypass cellular tumor suppressive responses (7). Although recent vaccines against HPV (Gardasil, Cervarix) will influence the prevalence of HPV+ HNSCC in the decades to follow, for now, the incidence of HPV+ HNSCC continues to rise. Current estimates suggest that 45-90% of OPSCC are HPV+ with 90% associated with HPV serotype 16 (8, 9). The division of HNSCC into two fundamentally distinct tumor cohorts with widely disparate survival rates based on HPV status represents one of the most significant developments of the past decade in head and neck cancer research and treatment.
Treatment for HNSCC is most often chosen based on the primary tumor subsite, TNM staging, and predicted functional outcomes following different treatment modalities. In general, early stage (I or II) HNSCC is treated with local therapy, taking advantage of the ability of surgical removal or radiation to offer a curative modality. Advanced disease (stage III or IV) requires multimodality treatment with surgery, radiation, and/or chemotherapy (10). Although the influence of treatment-related medical complications on mortality has declined (11) and some improvements in head and neck survival have been documented, these are largely related to the increasing incidence of HPV+ cancers rather than substantive gain in the clinical management of HNSCC. Treatment failure in HNSCC relates to resistance of tumor cells to primary or adjuvant chemoradiation therapy as well as residual undetectable microscopic disease that remains after surgical resection.
Recent whole exome sequencing of HNSCC offers several lessons into how these tumors will need to be treated to improve upon traditional therapeutic modalities. First, the sequencing of such a large number of tumors from numerous institutions demonstrates the successful endeavor of a multi-institutional collaborative effort to molecularly characterize the biology of head and neck tumors. Second, these analyses have validated that p53 inactivating mutations remain the predominant genetic defect identified, substantiating prior studies and emphasizing the observation that the majority of tumors harbor loss of function mutations. Third, sequencing data separates HPV+ and HPV− tumors into distinct groups with completely different mutational profiles. Fourth, we have learned that HNSCC will be challenging to treat – there is no singular target for these tumors. Intratumor heterogeneity will also remain a challenge as we attempt to advance our therapeutic approaches.
In this review, we will briefly discuss the molecular pathways driving HNSCC as identified using traditional genetics and biochemistry, but focus primarily on the new and interesting scientific advances in the field. In particular, we will emphasize insights from recent whole exome sequencing analyses of HNSCC, discuss lesson learned from analyses of intratumor heterogeneity, and explore the implications of recent studies on future therapeutic approaches.
Tumor suppressors frequently drive HNSCC but are difficult to target
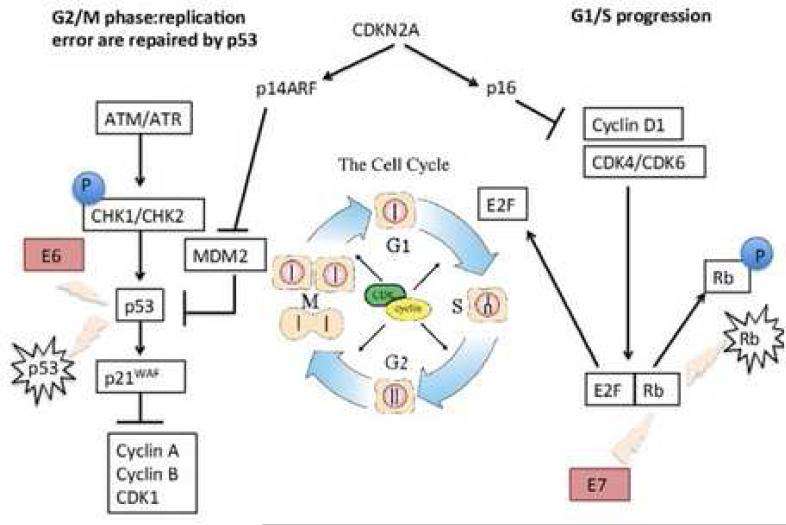
p53 is a ubiquitous tumor suppressor which is critically altered in a number of human cancers (12), with up to two-thirds of HNSCC harboring mutations in exons 5-8 (13, 14). Mutations in p53 dysregulate the cell cycle and monitoring of genomic integrity, thereby leading to aberrant proliferation, disrupted apoptosis, and defective DNA repair, while the HPV viral oncogene E6 targets p53 for degradation (Figure 1). Clinically, alterations in p53 function are associated with resistance to radiation and cisplatin-based chemotherapeutics (15), emphasizing the importance of this master regulator in HNSCC pathogenesis.
Figure 1.
Cell cycle signaling is interrupted in HPV+ HNSCC through disruption of multiple cell cycle checkpoints.
From Machiels JP, Lambrecht M, Hanin FX et al. Advances in the management of squamous cell carcinoma of the head and neck. F1000 Prime Rep. 2014; 6(44): eCollection; with permission.
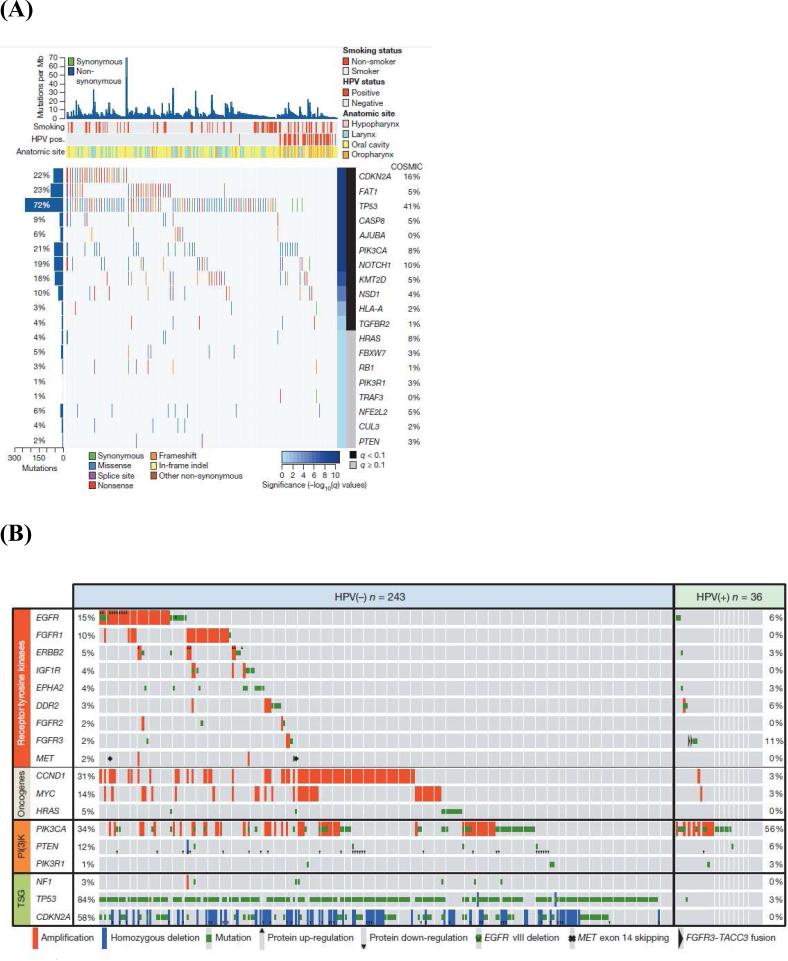
Recent whole exome sequencing analyses have validated these observations in cell lines and in vitro models, confirming that p53 mutations are common in HNSCC with loss of function mutations predominating. Stransky and collegues analyzed 74 tumor-normal pairs with their analysis suggesting 63% contained mutations or deletions in p53 (16). Analyses from the Cancer Genome Atlas of 279 HNSCCs identified mutations in p53 in 84% of HPV− tumors, with only 3% (one of 36) of HPV+ tumors containing a p53 mutation (Figure 2) (17). Similarly, inactivating mutations in the cell cycle regulator CDKN2A were found in 58% of HPV− tumors (17). Thus, a major conclusion of these whole exome sequencing analyses has been validating the near universal loss-of-function of p53 and CDKN2A inactivation in smoking/alcohol-related HNSCC. The challenge with p53and CDKN2A loss-of-function mutations is reactivation and/or replacing these critical cell cycle regulators. Adenoviral gene therapy, chemical activators of mutated genes, and antagonists of endogenous p53 inhibitors are all possibilities, but pre-clinical and clinical trials hold variable promise (18) and these strategies suffer the inherent limitations of targeting tumor suppressor genes including efficient delivery, tumor cell target specificity, and public resistance to gene therapy.
Figure 2.
Whole exome sequencing of HNSCC reveals novel insights into tumor pathogenesis and mutational profile. (A) Significantly mutated genes in HNSCC. (B) Candidate therapeutic targets and driver oncogenic events. (C) Deregulation of signaling pathways and transcription factors.
From Cancer Genome Atlas. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015; 517: 576-582; with permission.
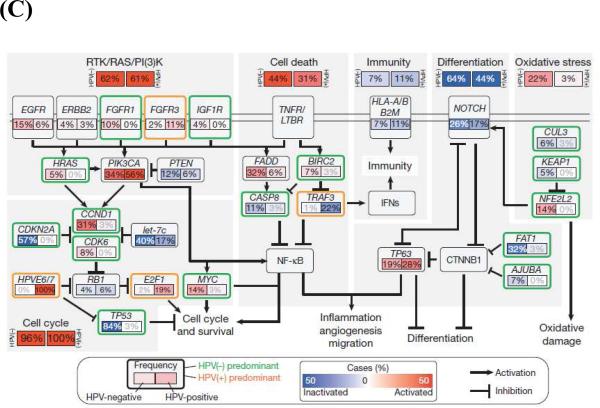
Alteration of differentiation pathways through the loss of TGFβR/SMAD signaling may also promote the transformation of aerodigestive mucosa to invasive SCC by critically altering tumor suppressor pathways (Figure 2 and 3). Loss of function mutations in TGFβR2 as well as in SMAD2 and SMAD4 have been identified (19, 20). Interestingly, data from cutaneous SCC suggests that TGFβ may play a dual role in oncogenesis, initially acting as a tumor suppressor to prevent the transformation to invasive SCC, but subsequently promoting the epithelial-mesenchymal transition and supporting metastasis (21). Animal data from mice confirm this complex signaling dichotomy: Conditional deletion of SMAD4 triggers genomic instability through activation of TGFβ1 and other SMADs (22), while deletion of TGFβR2 acts cooperatively with KRAS to promote metastases (23). In whole exome sequencing, comparison of mutations by subset analyses of anatomical site revealed unique mutations in TGFβR2 in oral cavity tumors, consistent with previous described functions in animal models (17). Given that TGFβ inhibitors are readily available and already being used in clinical trials for non-small cell lung cancer, colorectal cancer, and prostate cancer (24), inhibition of these differentiation pathways in HNSCC may be an accessible and exciting avenue for novel therapeutics.
Figure 3.
Key cellular and molecular pathways implicated in HNSCC tumorigenesis.
From Rothenberg SM and Ellisen LW. The molecular pathogenesis of head and neck squamous cell carcinoma. Jnl of Clin Invest 2012; 122(6): 1951-1957; with permission.
Sequencing data has also provided a rich array of data implicating loss of function mutations in additional pathways of differentiation in HNSCC, defining major new potential therapeutic targets. NOTCH1 loss-of-function mutations, for example, were noted in 11-19% of tumors, with another 11-14% containing NOTCH2 or NOTCH3 mutations (Figure 2A) (16, 17). Interestingly, these same tumors had mutations in gene sets associated with differentiation such as IRF6 and TP63, implying that these genes may act together with NOTCH1 to ultimately trigger the development of immature, dedifferentiated, highly proliferative basaloid cells. Additional mutations were identified in less well-characterized genes such as SYNE1 and 2, which control nuclear polarity, and RIMS2 and PCLO, which regulate calcium sensing during terminal squamous cell differentiation. Thus, dysregulation of programs involving cellular differentiation appears to be a critical component of HNSCC tumor biology. In addition, inhibition of NOTCH activation has been associated with an increased risk of cutaneous SCC, with a recent phase III trial with the gamma-secretase inhibitor semagacestat halted due to an increased rate of cutaneous SCC in the treatment arm compared to placebo (25). Together, these data suggest that differentiation pathways may be a major regulator of HNSCC tumorigenesis, raising the possibility that dysregulation of differentiation identified in other SCC of the lung, esophagus, and cervix may be relevant to head and neck biology.
Other tumor suppressor pathways have also been identified using unbiased approaches: FAT1, which has well described roles in aberrant Wnt signaling, was mutated in 12-23% of tumors (Figure 2A) (16, 17). Prior studies have shown that FAT1 encodes a cadherin-related protein which suppresses the nuclear localization of β-catenin and thereby inhibits proliferation (26). In addition, FAT1 appears to regulate cell migration and invasiveness (Figure 3) (27), suggesting that there may be multiple effects of FAT1 mutations on HNSCC tumorigenesis. Additional mutations and deletions were identified in apopotosis-related genes (CASP8, DDX3X), histone methyltransferases (PRDM9, EZH2, NSD1) as well as Ajuba, a centrosomal protein that regulates cell division and vertebrate ciliogenesis in an EGFR-RAS-MAPK-dependent manner (Figure 2A and C) (16, 17); however, further work is required to biologically characterize the mechanism and impact of these mutations on tumorigenesis. Nevertheless, identification of these mutations emphasizes the major role tumor suppressor pathways play in HNSCC pathogenesis.
Targeting tumor suppressor pathways is significantly more challenging than inhibiting oncogenic signaling, as it requires reactivation of tumor suppressor genes or their downstream effectors rather than simple chemical or biological inhibition. In this setting, the concept of synthetic lethality may prove useful for making therapeutic advances. Loss of a tumor suppressor gene may evoke unique susceptibilities to inhibition of a second gene or pathway that is normally not observed. Synthetic lethality leverages this principle that inhibition of two genes is lethal, while in contrast, inhibition of either gene alone is not.
There is now great interest in identifying the synthetic lethal partners of inactive genes such as tumor suppressors to help identify novel therapeutic targets (28). A recent data-driven computational approach for genome wide identification of synthetic lethal interactions has been developed (28), with the ability to identify synthetic lethal partners of both oncogenes and tumor suppressors. This approach has been utilized to develop a network analysis of synthetic lethal interactions which predicts which genes are essential and likely to be efficient pharmaceutical targets. Although this analysis has not been completed in HNSCC, it will be critical to move forward swiftly with such an approach, taking advantage of genome wide short hairpin RNA and small interfering RNA-mediated drug sensitization and small molecular inhibitor screens (29). By characterizing the synthetic lethal genes across a broad array of head and neck tumors, one could identify the gene targets worthy of aggressive drug targeting. Such an approach leverages the bioinformatics power of network analyses, rather than trying to simply inhibit or activate a single gene target, an effect which may ultimately be escaped through evolution of the cancer cells themselves.
In more practical terms, McLornan and colleagues have broadly characterized the pathways that may be non-overlapping and unique to cancer cells; thus, targeting these aspects of cell biology may allow for selective targeting of cancer cells through synthetic lethality (Table 1) (29). For example, although DNA damage response pathways generally provide a “unified guard” against genomic instability, many malignancies have defects in aspects of DNA repair mechanisms. For example, mutations in the DNA repair gene MSH2 or MLH1 become synthetically lethal when combined with inhibitors of DNA polymerase due to the accumulation of double strand breaks (30). Indeed, methotrexate has been shown to induce DNA damage in MSH2-mutant cells compared to wildtype, which has been the basis for methotrexate trials in MSH2-deficient colorectal cancer (31).
Table 1.
Mechanisms for achieving synthetic lethality in cancer cells.
Adapted from McLornan DP1, List A, Mufti GJ. Applying synthetic lethality for the selective targeting of cancer. N Engl J Med. 2014; 371(18): 1725-35; with permission.
| Mechanisms for achieving synthetic lethality in cancer cells |
|---|
| Targeting oncogenic drivers |
| Exploiting DNA-repair or cell-cycle defects |
| Using new drug combinations derived from screen |
| Using altered drug timing and sequencing |
| Exploring the tumor-cell environment |
| Targeting the stroma |
| Exploiting the altered metabolome |
| Targeting the altered proteome |
| Exploiting nononcogene “addiction” |
Targeting cells with p53 loss of function, which is mutated in 72% of HNSCC (17), may be one especially important context where synthetic lethality approaches may prove valuable. In the context of p53 loss-of-function, cancer cells lose the normal mechanisms of p53-dependent G1-S cell cycle arrest and become dependent on G2-M checkpoint arrest for DNA repair and survival. Thus, targeting G2-M checkpoint proteins can induce mitotic catastrophe and synthetic lethality in p53 loss-of-function cells. For example, inhibition of stress-activated p38 mitogen-activated protein kinase MAPKAP kinase 2 (MK2), ATM, and SGK2 or PAK3 may sensitize tumor cells to chemotherapy (MK2, ATM) or induce autophagy (SGK2) or apopotosis (PAK3) (32-34). Recent computational analyses have identified multiple candidate kinase genes which serve as synthetic lethal partners of p53 mutants including polo-like kinase 1, cyclin-dependent kinase 1, and aurora kinase A (35). Within head and neck oncology, this approach has been put into practice with an RNAi kinome viability screen in p53 mutant HNSCC cells to identify oncogenes that may be targeted in this mutational context (36). Using this method to screen primary human HNSCC tumors as well as tissue from murine models, several “critical survival kinases” were identified, including the kinase WEE1. Small molecule inhibition of WEE1 using the compound MK-1775 revealed durable effects on HNSCC viability and apoptosis, while also potentiating the efficacy of cisplatin in a mouse xenograft model. This inhibitor is now part of a phase I clinical trial to determine whether it may be useful in combination with neoadjuvant weekly docetaxel and cisplatin prior to surgery in p53 mutant HNSCC. Exploring these approaches further is likely to yield additional novel therapeutics for loss of function mutations that have remained difficult to target.
Oncogene mutations are uncommon in HNSCC with limited potential for targeted therapy in specific contexts
Unlike other malignancies such as breast cancer or chronic myelogenous leukemia (CML), which stand as examples of cancers driven by oncogenes (Epidermal Growth Factor Receptor (EGFR) and BCR-Abl, respectively) which can be inhibited with profound effects on clinical outcomes, HNSCC does not appear to demonstrate significant oncogene addiction. In vitro studies have identified a role for EGFR signaling in HNSCC, but sequencing analyses suggest only 6% of HPV− and 15% of HPV+ contain mutations or amplification of EGFR (Figure 2B) (17). EGFR is a transmembrane tyrosine kinase receptor in the HER/erbB family of proteins that triggers Ras and PI3K signaling (Figure 2C). In HNSCC, candidate sequencing studies have shown that EGFR is overexpressed most commonly through gene amplification and increased copy number (37), rather than activating mutations or truncation mutants such as EGFRvIII.
Based on the limited dependence of HNSCC on EGFR signaling, it is not surprising that inhibitors of EGFR have had variable success. EGFR overexpression appears predictive of poor clinical prognosis and resistance to radiation (38-40), with data suggesting improved overall survival when cetuximab, a monocolonal antibody against EGFR, is combined with radiation or chemotherapy (41, 42). However, response to cetuximab does not correlate with the degree of overexpression and as a monotherapy, the benefits of cetuximab are limited to a 6-13% response rate (3, 41). Similarly, the Radiation Therapy Oncology Group recently completed a Phase III trial exploring the effects of cetuximab in patients with Stage III or IV HNSCC who were undergoing concurrent accelerated fractionated radiotherapy and cisplatin treatment (43). This group found no differences in patient outcomes (mortality, progression free survival, overall survival, locoregional failure, or distant metastasis) with the addition of cetuximab. These findings suggest that other mechanisms may be activated upon EGFR inhibition or redundant activators of cell survival may limit treatment efficacy, consistent with whole exome studies suggesting oncogenes have low mutant allele frequencies and rarely drive HNSCC (Figure 2C) (16, 17). Thus, while there has been substantial interest in kinase inhibitors of EGFR in treating HNSCC, these agents have limited clinical impact in a significant portion of HNSCC tumors.
One exception for targeted therapy may be activating Ras or PI3K mutations which occur at higher frequency in HPV+ cancers, offering a specific context in which targeted therapy may facilitate de-intensification of chemoradiation. PI3K signaling is frequently altered in HNSCC through several mechanisms including: loss of function mutations in PTEN which negatively regulate PI3K (40% of HNSCC) and activating mutations in PI3KCA (6-11% of HNSCC) (44-47). Recent data suggests that the PTEN gene may exhibit a gene dosage effect (48), with loss of a single allele promoting tumor growth. Interestingly, in the case PI3KCA, mutations may be associated with HPV+ OPSCC (49), raising the possibility that PI3K acts synergistically with HPV E6 and E7 proteins in this HNSCC subset.
Ras signaling may work in collaboration with PI3K activation or independently to promote HNSCC (Figure 2C). While KRAS is frequently mutated in other cancers, HNSCC is associated primarily with HRAS mutations especially in patients with extensive tobacco exposure (50, 51). Like PI3K, HRAS mutations are also associated with HPV+ tumors (52). However, Ras family members have proven recalcitrant to inhibition with therapeutic strategies primarily aimed at targeting downstream effectors.
Recent sequencing analyses have validated these in vitro observations identifying amplifications or mutations (specifically the exon 9 helical domain) of PI3K in 56% of HPV+ HNSCC and 34% of HPV− tumors (Figure 2B) (17). Activating mutations in HRAS were also described in 5-8% of HNSCC tumors (Figure 2A) (16, 17). There are numerous ongoing trials evaluating small molecule inhibitors of PI3K (53). Such molecular inhibitors may be one exception in HNSCC where oncogene targeting may prove valuable.
HPV infection and integration alters tumor biology and triggers carcinogenesis by divergent biologic mechanisms than smoking and alcohol-related HNSCC
Over a decade of research has made it clear that HPV+ and HPV− HNSCC are distinct entities, with unique etiology, patient demographics, pathophysiology, and clinical outcomes (7). We now know that HPV− cancers are those that are driven by traditional risk factors such as smoking and alcohol, with carcinogenesis dependent on the acquisition of multiple epigenetic and genetic alterations yielding a premalignant progenitor which then undergoes additional alterations to become an invasive malignancy. This concept, known as field cancerization, posits that exposure of aerodigestive mucosa to alcohol and tobacco develops genetically distinct fields where additional mutations may cause transformation. Consistent with this, Slaughter and Southwick made the early observation that 11.2% of HNSCC primaries present with a second primary (54). Similarly, Sidransky and colleagues examined 87 HNSCC including analysis of pre-invasive lesions using microsatellite analyses for loss of heterozygosity (LOH) at 10 distinct loci. These analyses revealed progressive chromosomal loss when comparing benign hyperplasia to dysplasia to carcinoma in situ to invasive cancer, suggesting a common clonal progenitor and clonal expansion (55).
In contrast, HPV+ tumors are driven by HPV infection, usually by serotype16, with the integration of viral DNA into the host genome and the activation of specific and consistent molecular regulators including p16 (INK4A) and viral proteins E6 and E7 (Figure 1). Cell lines transfected with p16 and the alternate transcript p14arf displayed markedly inhibited growth (56), arresting in G1consistent with a role for p16 in blocking the G1-S transition. Indeed, transfection of p16-INK4A adenovirus demonstrated a 96% reduction in proliferation of HNSCC cell lines and in vivo studies in nude mice showed a significant decline in xenograft tumor growth (57). More recent studies have linked p16 positive IHC (best defined as ≥ 70% cytoplasmic and nuclear staining) with HPV+ tumors (58) and suggested that p16 overexpression be utilized as an independent factor to risk stratify OPSCC (59). Interestingly, epigenetic regulation of p16 through hypermethylation may also play an important role in predicting clinical prognosis and outcomes (60, 61).
The biologic mechanisms explaining differences in clinical outcomes with HPV status are likely multifactorial. The absence of field cancerization certainly reduces the incidence of locoregional recurrence and second primaries, while the persistence of functional p53 may explain the improved response to chemotherapy and radiation (62, 63). There is also growing evidence that tumor-immune interactions may explain the improved response of HPV+ tumors: HPV positivity is associated with a more substantial lymphocyte response (64-66) and animal models suggest that immunocompetence is essential for complete tumor eradication (67).
More recent genome wide studies of HPV+ and HPV− tumors reveal a clear divergence of these tumors at the genomic level. Compared to HPV− tumors, HPV+ HNSCC has lower rates of mutations and less frequent copy number alterations, indicating that there is less genomic instability in this cohort (16, 17). In a recent study, Akagi and colleagues have subsequently demonstrated that HPV integrants flank extensive regions of the host genome resulting in amplifications and rearrangement (68). In addition, looping of HPV integrant-mediated replication leads to viral-host concatemers, thereby triggering oncogenesis. However, by DNA analysis there is no consistency in the site of HPV integration: Interrogation of RNA transcripts demonstrated transcription across the viral-human integration locus with no recurrent genes identified, suggesting there are diverse mechanisms related to HPV integration, adding another potential source of intra- and inter-tumor heterogeneity (17).
At the level of individual genes, recent whole exome sequencing studies suggest that HPV+ tumors have infrequent mutations in p53 and CDKN2A in stark contrast to HPV− tumors where these genes are commonly altered (16, 17). Interestingly, HPV+ are distinguished by recurrent deletions and truncation mutations in TNF receptor-associated factor 3 (TRAF3) (17), which has been implicated in innate and acquired viral response to EBV, HPV, and HIV. Loss of TRAF3 promotes aberrant NFκB signaling with diverse downstream effects on cytokine signaling and cell death (69). Thus, there is now unequivocal evidence that mechanistically separates HPV+ and HPV− HNSCC, clarifying the biological basis for the distinct clinical behavior of these distinct head and neck tumors.
Intratumor heterogeneity poses unique therapeutic challenges and opportunities in HNSCC
Although many tumors are fairly homogeneous, HNSCC is characterized by tremendous intratumor diversity and heterogeneity. Early studies characterizing intratumor heterogeneity utilized microsatellite marker testing of distinct areas of tumor to demonstrate intratumor heterogeneity at the molecular level (70). These findings were validated with dual-fluorescence in situ hybridization studies (FISH) which demonstrated changes in DNA ploidy and intra-sample heterogeneity in 68 out of 89 tumors (71). Interestingly, heterogeneity was more substantial in primary tumors compared to metastatic samples. These early efforts established the presence of intratumor heterogeneity at the genetic level.
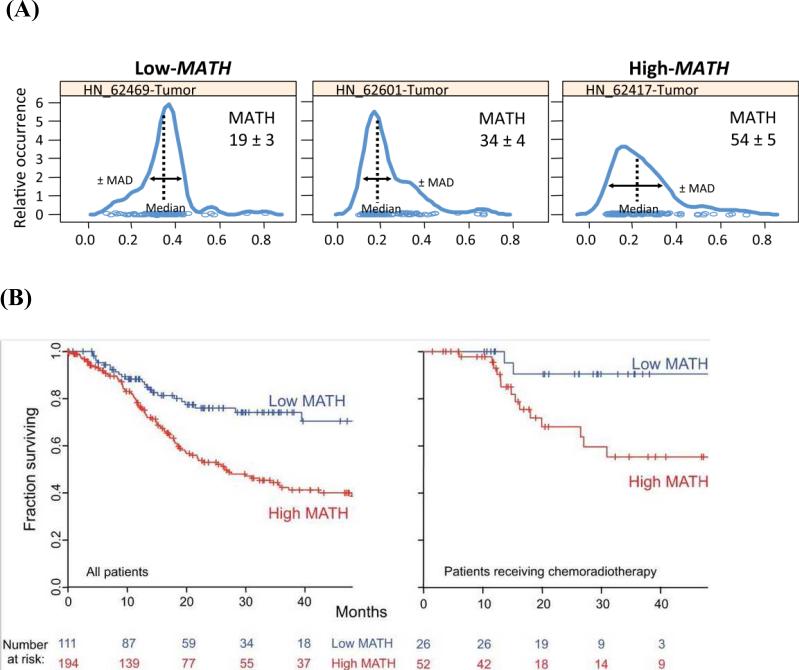
To determine the impact of intratumor genetic heterogeneity on clinical outcomes, more recent studies have introduced novel measures of genetic heterogeneity and correlated these findings with primary patient data. For example, we have defined a mutant allele tumor heterogeneity (MATH) score which is defined as the ratio of the width to the center of the distribution of mutant-allele fractions at tumor-specific mutated loci (Figure 4A) (72). MATH scores were calculated for 74 HNSCC with publicly available next-generation sequencing data, revealing higher scores in three well-established patient cohorts with poor outcomes, namely tumors with inactivating mutations in TP53 (compared to wildtype or non-disruptive mutations), HPV− tumors (compared to HPV+ tumors), and HPV− tumors from smokers with higher pack-years of smoking. Additional analyses demonstrated that higher MATH scores corresponded with shorter overall survival as well as adverse treatment outcomes in clinically high-risk patients (Figure 4B) (73). Together, these findings serve as the first clinical correlation of genetic intratumor heterogeneity to poor patient outcomes, providing an ideal biomarker that could be used to quantify intratumor genetic heterogeneity.
Figure 4.
(A) Representative mutant allele distributions from three HNSCC. A heterogenous tumor with a high MATH score will have a broader distribution, shorter peak, and lower median mutant allele fraction compared to a more homogeneous, low MATH score tumor.
From Rocco JW. Mutant Allele Tumor Heterogeneity (MATH) and Head and Neck Squamous Cell Carcinoma. Head Neck Pathol. 2015; 9(1): 1-5; with permission.
(B) Relationship of intra-tumor heterogeneity as captured by MATH score with overall survival in HNSCC.
From Mroz EA, Tward AM, Hammon RJ et al. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015; 12(2): eCollection; with permission.
More recently, we have applied this analysis of intratumor heterogeneity to the Cancer Genome Atlas database of 305 patients with HNSCC (74). Tumor MATH scores were calculated based on whole-exome sequencing data, revealing a substantiating association between high MATH scores and decreased overall survival (hazard ratio of 2.2 for high vs. low heterogeneity). This difference was independent of other clinical or biologic differences such as patient age, HPV status, tumor grade, TP53 mutations, and nodal disease. Based on analyses using MATH, a substantial improvement in overall prognostication compared to traditional staging analyses was demonstrated using multivariate analyses, establishing MATH as a useful predictor of tumor behavior and patient outcomes. Collectively, these studies emphasize the importance of intratumor heterogeneity as a major influence on tumor progression, treatment resistance, and metastatic potential, with implications for patient care and prognosis.
Unfortunately, our prior studies do not pinpoint a biological explanation as to why intratumor heterogeneity and higher MATH scores correlate with poorer clinical outcomes. Detailing the genetic and biochemical basis of intratumor heterogeneity in HNSCC remains one of the major challenges and opportunities within head and neck oncology. New sequencing technology may enable high-fidelity studies of HNSCC and allow the identification of distinct cellular subpopulation and cancer cell subcohorts. For example, a recent study has demonstrated that single cell RNA sequencing of human tumors can be leveraged to identify distinct intratumor subpopulations and characterize the gene expression profile of these differing cohorts (75).
A similar analysis in head and neck squamous cell carcinoma would informative: Not only would it allow a detailed characterization of the subpopulations present and their gene expression profiles, it would provide insight into the distinct contributions of each of these populations to tumor pathogenesis. For example, a uniquely expressed gene among the tumor stroma might be predictive of patient outcomes or serve as a novel therapeutic target following tumor resection. Similarly, if matched lymph nodes could also be sequenced, then theoretically one might identify differences in cellular subpopulations as well as unique genetic programs upregulated within the context of regional metastases. Clearly, single cell analyses of HNSCC hold great promise in improving diagnosis, prognosis, and treatment approaches – this work should proceed expeditiously as it will have important implications for clinical management of HNSCC.
Additional frontiers in HNSCC diagnostics and therapeutics
Despite advances in our understanding of HNSCC, the challenges discussed above have restricted the management of HNSCC to the same tools utilized decades ago, namely surgery, radiation therapy, and chemotherapy. However, there is growing interest into several areas of promise with ongoing clinical trials that may yield new biologic agents and therapeutics.
Biomarkers remain elusive but are worthy of pursuit
The difficulty of HNSCC surveillance due to limited recurrent tumor size, occult growth and progression, and localization in a variety of anatomical subsites emphasizes the need for reliable biomarkers in order to improve clinical management. This is especially critical considering the importance of early detection prior to the development of locoregional or distant metastases. To date, there is no usable serum or tissue biomarker that can be utilized for diagnosis of HNSCC. Hypermethylated circulating tumor DNA has been proposed as one potential candidate: A recent study examined serum from 100 HNSCC and 50 healthy controls to evaluate methylation status of EDNRB, p16, and DCC using quantitative methylation specific PCR methods (76). EDNRB hypermethylation was found in 10% of HNSCC patients (compared to 0% in controls), while DCC hypermethylation and p16 hypermethylation were rarely detected. Thus, EDNRB hypermethylation may be a highly specific albeit insensitive biomarker for HNSCC. Nevertheless, this approach may be used to identify a methylation profile for multiple genes, which could improve the sensitivity of testing and aid in the diagnosis and early detection of HNSCC (77).
To date, HPV is the only example of a biomarker in HNSCC that is utilized to predict prognosis. However, there is a still a real need to further stratify HPV+ patients given that a portion of these patients still have locoregional recurrences. Kumar and colleagues have suggested that HPV status be combined with measures of EGFR, p16, p53, and Bcl-XL expression to predict prognosis (78). Based on their analyses, patients with low EGFR, high HPV/p16, or low p53 with low Bcl-XL have improved overall survival and disease-free survival. Similar work has shown that increased Bcl-2 expression is an independent predictor of overall and disease free survival in oropharyngeal SCC (79). Interestingly, Bcl-2 expression was specifically associated with distant metastases rather than locoregional recurrences.
Genetic intratumor heterogeneity based on next generation sequencing may also serve as a potential biomarker. In the validating the TCGA HNSCC data set, both MATH and HPV status were significantly related to overall survival in bivariate Cox proportional hazards analysis, and a substantial portion of the relationship of MATH with outcome was independent of its relationship with HPV status (80). Importantly, MATH analysis improved HNSCC patient outcome prognostication beyond what was provided by HPV status alone. Ongoing prospective analysis in an oropharyngeal cohort will ultimately determine the role of MATH as a biomarker. Thus, additional biomarkers may help to further stratify HPV+ patients and provide insight into prognosis, thereby guiding the aggressiveness of clinical therapy a priori and facilitating deintensification of chemoradiation therapy.
MicroRNAs in HNSCC offer an additional, poorly understood layer of regulation in tumor biology
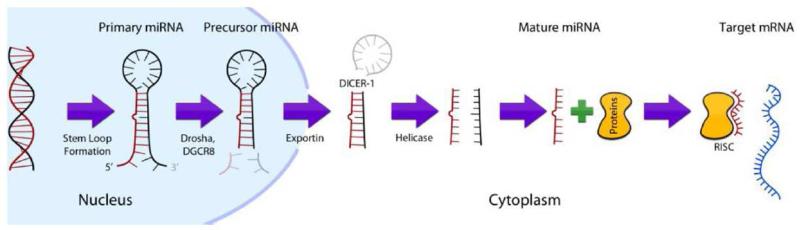
While microRNAs (miRNAs) have been well studied within the context of developmental biology and other tumor sites, within head and neck oncology, a role for microRNAs has been poorly characterized. MiRNAs are short 18-25 bp non-coding RNAs which modulate gene expression by binding to messenger RNA (mRNA) after transcription. miRNAs bind to the 3’ untranslated region (UTR), coding sequences, or the 5’ UTR of mRNA to inhibit translation or silence target transcripts by binding to the RNA-induced silencing complex (Figure 5). There is now growing evidence that dysregulation of miRNAs through direct genetic mutation, epigenetic changes, modifications in biogenesis, altered transcription factor expression, or changes in target sites may contribute to tumor progression (81). Several studies have cataloged the miRNAs present in HNSCC, identifying both oncogenic and tumor suppressor transcripts (81). For example miR-130b is upregulated in HNSCC, with a presumed role in regulating the epithelial-mesenchymal transition (82, 83). In contrast, miR-99 dysregulation and downregulation promotes OSCC survival through likely regulation of mTOR signaling, consistent with a tumor suppressor function (84). The Let-7 family of miRNAS, which is the largest family of miRNAs, is also downregulated in HNSCC (85-88).
Figure 5.
Schematic demonstrating the mechanism for miRNA based mRNA silencing.
From Mroz EA, Tward AM, Hammon RJ et al. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015; 12(2): e10001786; with permission.
Recent genome wide analyses of HNSCC have focused attention on miRNAs and convey the importance of miRNA-mRNA networks in HNSCC (17). These analyses identified decreased expression of Let-7c-5p and miR-100-5p in tumors compared to normal tissue, with a corresponding association with increased target gene expression of the cell cycle protein CDK6, transcription elongation factor E2F1, mitosis protein PLK1, and transcription factor HMGA2 (17). These critical observations raise the possibility of potentially novel therapeutics for HNSCC. Because miRNAs can be easily expressed through genetic constructs, an understanding of the role of miRNAs may allow for unique treatments or serum/tissue biomarkers. In addition, because many miRNAs have a more global regulatory role whereby they influence the activity of a multiple signaling cascades, modulation of miRNAs offers the possibility of more extensive inhibition or activation of relevant pathways than may be possible with small molecule agents.
Networking analyses can identify transcriptional and regulatory cassettes
Based on recent whole exome sequencing analyses, there are cohorts of tumor with characteristic signaling pathways, beginning the difficult task of capturing signaling networks within HNSCC. The Cancer Genome Atlas has utilized their whole exome sequencing data to define four molecular subtypes of HNSCC (atypical, mesenchymal, basal, and classical). These subtypes are defined by characteristic signaling cassettes and mutations. For example, p53 mutations, CDKN2A loss-of-function mutations, chr 3q amplification, changes in oxidative stress genes, and heavy smoking were associated with the classical subtype, while NOTCH1 mutations and HRAS-CASP8 co-mutations were associated with the basal subtype, suggesting that disrupted cell death is a common findings in this latter subtype (17). The atypical subtype lacked chr 7 amplifications, and the mesenchymal subtype had mutations in genes involved in innate immunity such as CD56 (17).
In addition, TCGA has also leveraged unsupervised clustering analysis of copy number alterations (CNAs) to delineate additional HNSCC cohorts. For example, this group describes an “M” class of tumors, which is driven primarily by mutations rather than CNAs (17). This cohort consists of a subset of oral cavity tumors with a characteristic three gene pattern of HRAS activating mutations, inactivating CASP8 mutations, and wild type p53. This cohort had a more favorable clinical outcome with what appears to be an alternative tumorigenesis pathway involving Ras and alterations in cell death/NFκB.
Recent work has focused on the biologic importance of NFκB in HNSCC by exploring the utility of bortezomib, a proteasome inhibitor which inactivates NFκB, in patients receiving concurrent chemoradiation therapy for recurrent or metastatic HNSCC (89). Preliminary data reveals a reduction of > 50% tumor size in two of six treated patients. NFκB regulated cytokines were decreased in patients with clinical responses, and bortezomib was show to induce apoptotic effects based on TUNEL analyses. This same group is now completing a phase I clinical trial investigating the safety profile of bortezomib when combined with cetuximab with or without cisplatin in patients with Stage IV head and neck cancer undergoing radiation therapy (90).
In the future, further bioinformatic analyses of existing larger tumor data sets (e.g. TCGA) will allow additional transcriptional and regulatory cassettes to be identified, perhaps setting up additional opportunities for translational therapeutics. Single cell analyses of HNSCC will be of great help in this regard, allowing similar analyses to be completed for tumor subpopulations and enabling a comprehensive characterization of tumor-stroma interactions and the signaling pathways governing cellular cross-talk.
Epigenetic changes and histone modification may enable tumor resistance
There has been recent interest in epigenetic changes, including histone modification, as a driver of tumorigenesis. Epigenetic changes have been touted as a major method for tumor resistance to chemotherapy, with cancer stem cells serving as a depot of self-renewing and self-propagating cells that may underlie treatment resistance. Epigenetic modifications may allow these cells to adapt to treatment regimens without requiring the acquisition of new mutations (91). For example, NFκB has been shown to localize to the nucleus in HNSCC where it modifies chromatin organization by influencing histone 3 acetylation and thereby condenses chromatin and desensitizes tumor cells to chemotherapy (92). Accordingly, treatment of cells with a histone deacetylase (HDAC) inhibitor mimics the effect of NFκB inhibition, providing a potential avenue for sensitizing tumor cells to chemotherapy (91). Interestingly, the same HDAC inhibitors appear to be required for the maintenance of cancer stem cells based on tumor sphere assays (93), suggesting that HNSCC progression and growth may critically depend on dynamic changes in chromatic organization through histone acetylation. Unfortunately, early trials with HDAC inhibitors have been disappointing: A phase II trial with romdepsin, a potent HDAC inhibitor derived from Chromobacterium vioaceum, effectively inhibited HDAC in vivo but yielded no objective clinical response (94). However, there are additional ongoing trials exploring other promising HDAC inhibitors. For example, Ohio State University has an ongoing clinical trial exploring the role of the HDAC inhibitor vorinostat in combination with chemoradiation for the treatment of stage III and IVA oropharyngeal SCC (95) based on promising preclinical data in cell line and animal models of HNSCC (96, 97). These additional studies should help clarify if HDAC inhibitors hold therapeutic promise.
Tumor-immune interactions and immunotherapy offer novel methods for cancer surveillance
There has been substantial interest in the interaction between immune infiltrates and tumor cells (see Head and Neck immuno-oncology section). Briefly, these immune cells, known as tumor infiltrating lymphocytes (TILs), have been characterized in several solid malignancies including HNSCC (98). The majority of studies in HNSCC have focused on the prognostic impact of TILs. For example, Rajjoub et al. used tissue microarrays to catalog 48 oropharyngeal SCC and determined the CD3+ T cells that were present (99). They found that tumors with more CD3 cells tended to have decreased rates of metastasis; however, multivariate analyses suggested that the difference was only statistically significant for HPV+ tumors. Similar analyses of CD8+ T cells have yielded mixed conclusions (98). Unfortunately, a detailed study of the biologic characteristics of HNSCC TILs has been lacking. Single cell sequencing of HNSCC including the CD45+ immune infiltrate may be one method of accomplishing this goal.
If these cells can be better characterized, then it may be possible to develop immunomodulatory therapies that can harness a patient's own immune surveillance mechanisms to target HNSCC. In addition, it may be possible to manipulate patient immune responses through ex vivo alterations in immune cells followed by re-implantation. Although there is significant progress to be made, biologic agents leveraging TILs may be a useful adjunct to current therapies, providing a method for cellular surveillance of cancer cells that escape traditional treatment modalities.
There are several ongoing clinical trials related to immunotherapy in HNSCC. For example, Merck has sponsored an open Phase II trial investigating the role of MK-3475 (Pembrolizumab), a potent inhibitor of PD-1 receptor, in recurrent or metastatic HNSCC after treatment with platinum-based and cetuximab therapy (100). It would be informative to biologically characterize these tumors before and after treatment with pembrolizumab to better understand how molecular signaling is altered with immunotherapy.
A Darwinian view of head and neck oncology offers creative perspectives into cancer biology and treatment
The past decade of research has seen an explosion of interest in the in situ evolution of tumorigenesis and the adaptation of cancer cells to their environment, not unlike Darwin's theory of evolution as it applies to species fitness (101). In the case of HNSCC, a Darwinian perspective emphasizes new angles of basic research that remain poorly characterized: What is the role of the “environment” (i.e. tumor niche) in carcinogenesis? What are the environmental “perturbations” (i.e. therapeutic silver bullets), if any, that might yield clonal extinction and eliminate the cancer so swiftly that none of its constituent members (individual cells) may adapt and survive? Is there a role for metronomic therapy in yielding better long-term control on HNSCC? (102). How can we slow clonal evolution to advance the efficacy of therapy? While abstract, this new understanding and perspective has begun to trickle down and influence how HNSCC may ultimately be successfully targeted. Certainly, collaboration with developmental and systems biologists is likely to maximize strides in HNSCC, providing new insight ranging from phylogenetic analysis of tumor clones to novel methods of targeting tumor resistance.
Summary
Our molecular understanding of HNSCC has undergone tremendous growth and evolution over the past several decades. We now know there are two clear cohorts of HNSCC, HPV+ which is caused by HPV infection and viral integration and HPV− which is driven by acquired mutations and alterations from environmental exposures such as tobacco and alcohol. Recent whole exome sequencing studies of HNSCC have provided dramatic insights into HNSCC tumor biology, demonstrating that tumor suppressors are the primary regulators of HNSCC with oncogenes rarely driving tumorigenesis.
In parallel a host of new observations have demonstrated the challenge and complexity of developing targeted therapies for HNSCC. Intratumor heterogeneity poses a major challenge to a “silver bullet” for HNSCC, providing genetic diversity that may allow clonal escape and tumor resistance. In addition, new insights from epigenetic studies, the identification of novel miRNA regulators, and tumor-immune system interactions suggest additional layers of regulation and control. Future work should leverage single cell sequencing and transcriptional network analyses to better characterized molecular cross-talk and cellular interactions, thereby providing a more complete and comprehensive view of HNSCC signaling.
While we have much to learn about HNSCC, we must not limit the lesson learned to those obtained directly from studying this tumor. Sequencing analyses have conveyed the point, more than ever, that dysregulation of differentiation is a major contribution to head and neck oncology. It is not gone unnoticed that similar pathways are aberrantly regulated in other squamous cell carcinomas of the cervix, skin, lung, and esophagus. In the end, we may find that the biology of these tumors is more of a reflection of their underlying tissue of origin than their anatomical subsite. By combining findings from HNSCC with these other tumors, we may be able to identify more dramatic insights that synergistically improve the treatment of other SCCs as well.
Although numerous clinical trials are ongoing, there is a necessity for greater translation research into areas of promise such as synthetic lethality and immunotherapy. While the mainstays of head and neck cancer treatment including surgery, radiation, and chemotherapy are likely to persist, the development of useful biological adjuncts could dramatically influence the way conventional therapy is utilized, offering the possibility of descalation and de-intensification of treatment, while simultaneously improving patient outcomes and quality of life.
Key Points.
Head and neck squamous cell carcinoma (HNSCC) is driven by numerous mutations, with HPV− cancers caused by more traditional risks factors (tobacco use/alcohol) tending to harbor more mutations, greater intratumor heterogeneity, and extensive copy number variation.
Recent genomic insights suggest that targeted therapy of HNSCC will remain a significant challenge. Most mutations identified based on sequencing analyses are loss of function mutations in known and putative tumor suppressor genes that may require novel approaches such as synthetic lethality.
Oncogenic drivers are few and far between and often are present at low mutant allele frequencies, suggesting they may be poor choices for targeted therapy.
One exception for targeted therapy may be activating Ras or PI3K mutations which occur at high frequency in HPV+ cancers, offering a potential avenue for therapy that may facilitate deintensification of chemoradiation therapy.
Identification of genes implicated in tumor-immune interactions as well as loss of function mutations suggest that immunotherapy and modulation of immune surveillance may be a valuable therapeutic approach, supporting ongoing immunotherapy clinical trials.
Abbreviations/Acronyms
- ATM
ataxia telangiectasia mutated
- BCR-Abl
breakpoint cluster region-Abelson
- Bcl-2
B-cell lymphoma-2
- Bcl-XL
B-cell lymphoma-extra long
- CASP8
caspase 8
- CDK6
cell division protein kinase 6
- CDKN2A
cyclin-dependent kinase inhibitor 2A
- chr
chromosome
- CML
chronic myelogenous leukemia
- CNA
copy number alterations
- DCC
deleted in colorectal carcinoma
- DDX3X
DEAD/H box 3, X-linked
- DNA
deoxyribonucleic acid
- E2F1
E2F transcription factor 1
- EDNRB
endothelin receptor type B
- EGFR
epidermal growth factor receptor
- EZH2
enhancer of zeste 2 polycomb repressive complex 2
- FAT1
FAT atypical cadherin 1
- HDAC
histone deacetylase
- HMGA2
high mobility group AT-hook 2
- HNSCC
head and neck squamous cell carcinoma
- HPV
human papilloma virus
- HRAS
Harvey rat sarcoma viral oncogene homolog
- INK4A
inhibitor of cyclin-dependent kinase type 4
- IRF6
interferon regulatory factor 6
- KRAS
V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
- LOH
loss of heterozygosity
- MATH
mutant allele tumor heterogeneity
- miRNA
microRNA
- MK2
stress-activated p38 mitogen-activated protein kinase MAPKAP kinase 2
- mRNA
messenger RNA
- NFκB
nuclear factor kappa B
- NSD1
nuclear receptor binding SET domain protein 1
- PAK3
serine/threonine protein kinase 3
- PCLO
piccolo presynaptic cytomatrix protein
- PD-1
programmed cell death 1
- PI3K
phosphoinositide 3-kinase
- PLK1
polo-like kinase 1
- PRDM9
PR domain zinc finger protein 9
- RIMS2
regulating synaptic membrane exocytosis 2
- RNA
ribonucleic acid
- SCC
squamous cell carcinoma
- SGK2
serum/glucocorticoid regulated kinase 2
- SMAD
Sma and Mad related family
- SYNE1/2
spectrin-repeat nuclear envelope ½
- TCGA
The Cancer Genome Atlas
- TGBβ
transforming growth factor
- TIL
tumor infiltrating lymphocytes
- TNF
tumor necrosis factor
- TRAF3
TNF receptor-associated factor 3
- TNM
tumor, node, metastasis
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- UTR
untranslated region
- WEE1
Wee1 –like protein kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures/Conflict of Interest: The authors have nothing to disclose.
References
- 1.International Agency for Research on Cancer . World Health Organisation. Estimated cancer incidence, mortality and prevalence worldwide in 2012. Globocan; 2014. http://globoscan.iarc.fr. [Google Scholar]
- 2.Rothenberg SM, Ellisen LW. The molecular pathogenesis of head and neck squamous cell carcinoma. Jnl of Clin Invest. 2012;122(6):1951–1957. doi: 10.1172/JCI59889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machiels JP, Lambrecht M, Hanin FX, et al. Advances in the management of squamous cell carcinoma of the head and neck. F1000 Prime Rep. 2014;6(44) doi: 10.12703/P6-44. eCollection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curado MP, Hashibe M. Recent changes in the epidemiology of head and neck cancer. Current Opinion in Oncology. 2009;21(3):194–200. doi: 10.1097/CCO.0b013e32832a68ca. [DOI] [PubMed] [Google Scholar]
- 5.Gillison ML. Human papillomavirus-associated head and neck cancer is a distinct epidemiologic, clinical, and molecular entity. Semin Oncol. 2004;31:744–754. doi: 10.1053/j.seminoncol.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Chaturvedi AK, Anderson WF, Lortet-Tieulent J, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31:4550–4559. doi: 10.1200/JCO.2013.50.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonilla-Velez J, Mroz EA, Hammon RJ, et al. Impact of human papillomavirus on oropharyngeal cancer biology and response to therapy: implications for treatment. Otolaryngol Clin North Am. 2013;46(4):521–43. doi: 10.1016/j.otc.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joseph AW, D'Souza G. Epidemiology of human papillomavirus-related head and neck cancer. Otolaryngol Clin North Am. 2012;45(4):739–764. doi: 10.1016/j.otc.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Marur S, D'Souza G, Westra WH, et al. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11(8):781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Cancer Institute Head and Neck Cancer. 2014 http://www.cancer.gov/cancertopics/types/head-and-neck.
- 11.Lin HW, Bhattacharyya N. Contemporary assessment of medical morbidity and mortality in head and neck surgery. Otolaryngol Head Neck Surg. 2012;146(3):385–389. doi: 10.1177/0194599811428557. [DOI] [PubMed] [Google Scholar]
- 12.Nigro JM, Baker SJ, Preisinger AC, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342(6250):705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- 13.Gasco M, Crook T. The p53 network in head and neck cancer. Oral Oncol. 2003;39(3):222–231. doi: 10.1016/s1368-8375(02)00163-x. [DOI] [PubMed] [Google Scholar]
- 14.Somers KD, Merrick MA, Lopez ME, et al. Frequent p53 mutations in head and neck cancer. Cancer Res. 1992;52(21):5997–6000. [PubMed] [Google Scholar]
- 15.Ohnishi K, Ota I, Takahashi A, Yane K, et al. Transfection of mutant p53 gene depresses X-ray- or CDDP-induced apoptosis in a human squamous cell carcinoma of the head and neck. Apoptosis. 2002;7(4):367–372. doi: 10.1023/a:1016131614856. [DOI] [PubMed] [Google Scholar]
- 16.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tassone P, Old M, Teknos TN, et al. p53-based therapeutics for head and neck squamous cell carcinoma. Oral Oncol. 2013;49(8):733–737. doi: 10.1016/j.oraloncology.2013.03.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, Song H, Evans JA, et al. Mutation and downregulation of the transforming growth factor beta type II receptor gene in primary squamous cell carcinomas of the head and neck. Carcinogenesis. 1997;18(11):2285–2290. doi: 10.1093/carcin/18.11.2285. [DOI] [PubMed] [Google Scholar]
- 20.Qiu W, Schonleben F, Li X, et al. Disruption of transforming growth factor beta–Smad signaling pathway in head and neck squamous cell carcinoma as evidenced by mutations of SMAD2 and SMAD4. Cancer Lett. 2007;245(1–2):163–170. doi: 10.1016/j.canlet.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han G, Lu SL, Li AG, et al. Distinct mechanisms of TGF-beta1-mediated epithelial-to-mesenchymal transition and metastasis during skin carcinogenesis. J Clin Invest. 2005;115(7):1714–1723. doi: 10.1172/JCI24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bornstein S, White R, Malkoski S, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119(11):3408–3419. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu SL, Herrington H, Reh D, et al. Loss of transforming growth factorbeta type II receptor promotes metastatic headand-neck squamous cell carcinoma. Genes Dev. 2006;20(10):1331–1342. doi: 10.1101/gad.1413306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagaraj NS, Datta PK. Targeting the transforming growth factor-beta signaling pathway in human cancer. Expert Opin Investig Drugs. 2010;19(1):77–91. doi: 10.1517/13543780903382609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doody RS, Raman R, Farlow M, et al. A phase 3 trial of semagacestat for treatment of Alzheimer's disease. N Engl J Med. 2013;369(4):341–350. doi: 10.1056/NEJMoa1210951. [DOI] [PubMed] [Google Scholar]
- 26.Morris LG1, Kaufman AM, Gong Y, et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45(3):253–261. doi: 10.1038/ng.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishikawa Y, Miyazaki T, Nakashiro K, et al. Human FAT1 cadherin controls cell migration and invasion of oral squamous cell carcinoma through the localization of β-catenin. Oncol Rep. 2011;26(3):587–92. doi: 10.3892/or.2011.1324. [DOI] [PubMed] [Google Scholar]
- 28.Jerby-Arnon L, et al. Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell. 2014;158(5):1199–209. doi: 10.1016/j.cell.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 29.McLornan DP1, List A, Mufti GJ. Applying synthetic lethality for the selective targeting of cancer. N Engl J Med. 2014;371(18):1725–35. doi: 10.1056/NEJMra1407390. [DOI] [PubMed] [Google Scholar]
- 30.Martin SA, McCabe N, Mullarkey M, et al. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH or MLH1. Cancer Cell. 2010;17:235–248. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin SA, McCarthy A, Barber LJ, et al. Methotrexate induces oxidative DNA damage and is selectively lethal to tumour cells with defects in the DNA mismatch repair gene MSH2. EMBO Mol Med. 2009;1:323–337. doi: 10.1002/emmm.200900040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morandell S, Reinhardt HC, Cannell IG, et al. A reversible gene-targeting strategy identifies synthetic lethal interactions between MK2 and p53 in the DNA damage response in vivo. Cell Rep. 2013;5:868–877. doi: 10.1016/j.celrep.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang H, Reinhardt HC, Bartkova J, et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–1909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baldwin A, Grueneberg DA, Hellner K, et al. Kinase requirements in human cells: V. Synthetic lethal interactions between p53 and the protein kinases SGK2 and PAK3. Proc Natl Acad Sci U S A. 2010;107:12463–12468. doi: 10.1073/pnas.1007462107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Simon R. Identification of potential synthetic lethal genes to p53 using a computational biology approach. BMC Med Genomics. 2013;6:30–30. doi: 10.1186/1755-8794-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moser R, Xu C, Kao M, et al. Functional kinomics identifies candidate therapeutic targets in head and neck cancer. Clin Cancer Res. 2014;20(16):4274–4288. doi: 10.1158/1078-0432.CCR-13-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalyankrishna S, Grandis JR. Epidermal growth factor biology in head and neck cancer. J Clin Oncol. 2006;24:2666–2672. doi: 10.1200/JCO.2005.04.8306. [DOI] [PubMed] [Google Scholar]
- 38.Temam S, Kawaguchi H, El-Naggar AK, et al. Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J Clin Oncol. 2007;25:2164–70. doi: 10.1200/JCO.2006.06.6605. [DOI] [PubMed] [Google Scholar]
- 39.Chung CH, Ely K, McGavran L, et al. Increased epidermal growth factor receptor gene copy number is associated with poor progonis in head and neck squamous cell carcinomas. J Clin Oncol. 2006;24:4170–4176. doi: 10.1200/JCO.2006.07.2587. [DOI] [PubMed] [Google Scholar]
- 40.Chiang WF, Liu SY, Yen CY, et al. Association of epidermal growth factor receptor (EGFR) gene copy number amplification with neck lymph node metastasis in areca-associated oral carcinoma. Oral Oncol. 2008;44:270–276. doi: 10.1016/j.oraloncology.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 41.Vermorken J, Mesia R, Rivera F, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Eng J Med. 2008;11:1116–27. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 42.Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010;11:21–28. doi: 10.1016/S1470-2045(09)70311-0. [DOI] [PubMed] [Google Scholar]
- 43.Ang KK, Zhang Q, Rosenthal DI, Nguyen-Tan PF, et al. Randomized phase III trial of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage III to IV head and neck carcinoma: RTOG 0522. J Clin Oncol. Sep 20. 2014;32(27):2940–2950. doi: 10.1200/JCO.2013.53.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okami K, Wu L, Riggins G, et al. Analysis of PTEN/MMAC1 alterations in aerodigestive tract tumors. Cancer Res. 1998;58(3):509–511. [PubMed] [Google Scholar]
- 45.Shao X, Tandon R, Samara G, et al. Mutational analysis of the PTEN gene in head and neck squamous cell carcinoma. Int J Cancer. 1998;77(5):684–688. doi: 10.1002/(sici)1097-0215(19980831)77:5<684::aid-ijc4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 46.Qiu W, Schönleben F, Li X, et al. PIK3CA mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12(5):1441–1446. doi: 10.1158/1078-0432.CCR-05-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu W, Tong GX, Manolidis S, et al. Novel mutant-enriched sequencing identified high frequency of PIK3CA mutations in pharyngeal cancer. Int J Cancer. 2008;122(5):1189–1194. doi: 10.1002/ijc.23217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476(7359):163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henken FE Banerjee NS, Snijders PJ, et al. PIK3CA-mediated PI3-kinase signalling is essential for HPV-induced transformation in vitro. Mol Cancer. 2011;10:71. doi: 10.1186/1476-4598-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saranath D, Chang SE, Bhoite LT, et al. High frequency mutation in codons 12 and 61 of H-ras oncogene in chewing tobacco-related human oral carcinoma in India. Br J Cancer. 1991;63(4):573–578. doi: 10.1038/bjc.1991.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson JA, Irish JC, Ngan BY. Prevalence of RAS oncogene mutation in head and neck carcinomas. J Otolaryngol. 1992;21(5):321–326. [PubMed] [Google Scholar]
- 52.Anderson JA, Irish JC, McLachlin CM, et al. H-ras oncogene mutation and human papillomavirus infection in oral carcinomas. Arch Otolaryngol Head Neck Surg. 1994;120(7):755–760. doi: 10.1001/archotol.1994.01880310059011. [DOI] [PubMed] [Google Scholar]
- 53.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9(8):550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 54.Slaughter DP, Southwick HW. ‘Field cancerization’ in oral stratified squamous epithelium. Clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 55.Califano J, van der Riet P, Westra W, et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56(11):2488–2492. [PubMed] [Google Scholar]
- 56.Liggett WH, Jr, Sewell DA, Rocco J, et al. p16 and p16 beta are potent growth suppressors of head and neck squamous carcinoma cells in vitro. Cancer Res. Sep 15. 1996;56(18):4119–4123. [PubMed] [Google Scholar]
- 57.Rocco JW, Li D, Liggett WH, Jr, et al. p16INK4A adenovirus-mediated gene therapy for human head and neck squamous cell cancer. Clin Cancer Res. 1998;4(7):1697–1704. [PubMed] [Google Scholar]
- 58.Grønhøj Larsen C, Gyldenløve M, Jensen DH, et al. Correlation between human papillomavirus and p16 overexpression in oropharyngeal tumours: a systematic review. Br J Cancer. 2014;110(6):1587–94. doi: 10.1038/bjc.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis JS., Jr p16 Immunohistochemistry as a standalone test for risk stratification in oropharyngeal squamous cell carcinoma. Head Neck Pathol. 2012;6:S75–82. doi: 10.1007/s12105-012-0369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sailasree R, Abhilash A, Sathyan KM, et al. Differential roles of p16INK4A and p14ARF genes in prognosis of oral carcinoma. Cancer Epidemiology Biomarkers and Prevention. 2008;17(2):414–420. doi: 10.1158/1055-9965.EPI-07-0284. [DOI] [PubMed] [Google Scholar]
- 61.Ogi K, Toyota M, Ohe-Toyota M, et al. Aberrant methylation of multiple genes and clinicopathological features in oral squamous cell carcinoma. Clinical Cancer Research. 2002;8(10):3164–3171. [PubMed] [Google Scholar]
- 62.Licitra L, Perrone F, Bossi P, et al. High-risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J Clin Oncol. 2006;24(36):5630–5636. doi: 10.1200/JCO.2005.04.6136. [DOI] [PubMed] [Google Scholar]
- 63.Butz K, Geisen C, Ullmann A, et al. Cellular responses of HPV-positive cancer cells to genotoxic anti-cancer agents: repression of E6/E7-oncogene expression and induction of apoptosis. Int J Cancer. 1996;68(4):506–513. doi: 10.1002/(SICI)1097-0215(19961115)68:4<506::AID-IJC17>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 64.Vu HL, Sikora AG, Fu S, et al. HPV-induced oropharyngeal cancer, immune response and response to therapy. Cancer Lett. 2010;288(2):149–155. doi: 10.1016/j.canlet.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 65.Wansom D, Light E, Worden F, et al. Correlation of cellular immunity with human papillomavirus 16 status and outcome in patients with advanced oropharyngeal cancer. Arch Otolaryngol Head Neck Surg. 2010;136(12):1267–1273. doi: 10.1001/archoto.2010.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wansom D, Light E, Thomas D, et al. Infiltrating lymphocytes and human papillomavirus-16--associated oropharyngeal cancer. Laryngoscope. 2012;122(1):121–127. doi: 10.1002/lary.22133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spanos WC, Nowicki P, Lee DW, et al. Immune response during therapy with cisplatin or radiation for human papillomavirus-related head and neck cancer. Arch Otolaryngol Head Neck Surg. 2009;135(11):1137–1146. doi: 10.1001/archoto.2009.159. [DOI] [PubMed] [Google Scholar]
- 68.Akagi K1, Li J, Broutian TR, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014;24(2):185–199. doi: 10.1101/gr.164806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ni CZ, Welsh K, Leo E, et al. Molecular basis for CD40 signaling mediated by TRAF3. Proc Natl Acad Sci U S A. 2000;97(19):10395–10399. doi: 10.1073/pnas.97.19.10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.el-Naggar AK1, Hurr K, Luna MA, Goepfert H, Hong WK, Batsakis JG. Intratumoral genetic heterogeneity in primary head and neck squamous carcinoma using microsatellite markers. Diagn Mol Pathol. 1997;6(6):305–308. doi: 10.1097/00019606-199712000-00001. [DOI] [PubMed] [Google Scholar]
- 71.Götte K1, Schäfer C, Riedel F, et al. Intratumoral genomic heterogeneity in primary head and neck cancer and corresponding metastases detected by dual-FISH. Oncol Rep. 2004;11(1):17–23. [PubMed] [Google Scholar]
- 72.Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49(3):211–215. doi: 10.1016/j.oraloncology.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mroz EA, Tward AD, Pickering CR, et al. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer. 2013;119(16):3034–3042. doi: 10.1002/cncr.28150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mroz EA, Tward AM, Hammon RJ, et al. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12(2) doi: 10.1371/journal.pmed.1001786. eCollection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mydlarz WK, Hennessey PT, Wang H, et al. Serum biomarkers for detection of head and neck squamous cell carcinoma. Head Neck. 2014 doi: 10.1002/hed.23842. ePub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arantes LM, de Carvalho AC, Melendez ME, et al. Methylation as a biomarker for head and neck cancer. Oral Oncol. 2014;50(6):587–592. doi: 10.1016/j.oraloncology.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 78.Kumar B, Cordell KG, Lee JS, et al. EGFR, p16, HPV Titer, Bcl-xL and p53, sex, and smoking as indicators of response to therapy and survival in oropharyngeal cancer. J Clin Oncol. 2008;26(19):3128–3137. doi: 10.1200/JCO.2007.12.7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nichols AC, Finkelstein DM, Faquin WC, et al. Bcl2 and human papilloma virus 16 as predictors of outcome following concurrent chemoradiation for advanced oropharyngeal cancer. Clin Cancer Res. 2010;16(7):2138–2146. doi: 10.1158/1078-0432.CCR-09-3185. [DOI] [PubMed] [Google Scholar]
- 80.Mroz EA, Tward AM, Hammon RJ, et al. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12(2):e10001786. doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sethi N, Wright A, Wood H, et al. MicroRNAs and head and neck cancer: reviewing the first decade of research. Eur J Cancer. 2014;50(15):2619–2635. doi: 10.1016/j.ejca.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 82.Cao P, Zhou L, Zhang J, et al. Comprehensive expression profiling of microRNAs in laryngeal squamous cell carcinoma. Head Neck. 2013;35:720–728. doi: 10.1002/hed.23011. [DOI] [PubMed] [Google Scholar]
- 83.Avissar M, Christensen BC, Kelsey KT, et al. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yan B, Fu Q, Lai L, et al. Downregulation of microRNA 99a in oral squamous cell carcinomas contributes to the growth and survival of oral cancer cells. Mol Med Rep. 2012;6:675–681. doi: 10.3892/mmr.2012.971. [DOI] [PubMed] [Google Scholar]
- 85.Hui AB, Lenarduzzi M, Krushel T, et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010;16:1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 86.Childs G, Fazzari M, Kung G, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maclellan SA, Lawson J, Baik J, et al. Differential expression of miRNAs in the serum of patients with high-risk oral lesions. Cancer Med. 2012;1:268–274. doi: 10.1002/cam4.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang WH, Lan HY, Tai SK, et al. Repression of bone morphogenetic protein 4 by let-7i attenuates mesenchymal migration of head and neck cancer cells. Biochem Biophys Res Commun. 2013;433:24–30. doi: 10.1016/j.bbrc.2013.02.076. [DOI] [PubMed] [Google Scholar]
- 89.Chang AA, Conley BA, Lebowitz PF, et al. Bortezomib with concurrent radiation therapy in head and neck squamous cell carcinoma. Otolaryngology Head and Neck Surgery. 2004;131(2):117. [Google Scholar]
- 90.National Cancer Institute . Radiation therapy and bortezomib and cetuximab with or without cisplatin to treat head and neck cancer. Clinicaltrials.gov; 2013. https://clinicaltrials.gov/ct2/show/NCT01445405. [Google Scholar]
- 91.Le JM, Squarize CH, Castilho RM. Histone modifications: Targeting head and neck cancer stem cells. World J Stem Cells. 2014;6(5):511–525. doi: 10.4252/wjsc.v6.i5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Almeida LO, Abrahao AC, Rosselli-Murai LK, et al. NFκB mediates cisplatin resistance through histone modifications in head and neck squamous cell carcinoma (HNSCC). FEBS Open Bio. 2014;4:96–104. doi: 10.1016/j.fob.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Giudice FS1, Pinto DS, Jr, Nör JE, et al. Inhibition of histone deacetylase impacts cancer stem cells and induces epithelial-mesenchyme transition of head and neck cancer. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0058672. ePub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haigentz M, Jr1, Kim M, Sarta C, et al. Phase II trial of the histone deacetylase inhibitor romidepsin in patients with recurrent/metastatic head and neck cancer. Oral Oncol. 2012;48(12):1281–1288. doi: 10.1016/j.oraloncology.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ohio State University Comprehensive Cancer Center . Ph I Vorinostat in the Treatment of Advanced Staged Oropharyngeal Squamous Cell Carcinoma. Clinicaltrials.gov; 2015. https://www.clinicaltrials.gov/ct2/show/NCT01064921. [Google Scholar]
- 96.Erlich RB, Kherrouche Z, Rickwood D, et al. Preclinical evaluation of dual PI3K-mTOR inhibitors and histone deacetylase inhibitors in head and neck squamous cell carcinoma. Br J Cancer. 2012;106(1):107–115. doi: 10.1038/bjc.2011.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang Y, Jung M, Dritschilo A, et al. Enhancement of radiation sensitivity of human squamous carcinoma cells by histone deacetylase inhibitors. Radiat Res. 2004;161(6):667–674. doi: 10.1667/rr3192. [DOI] [PubMed] [Google Scholar]
- 98.Uppaluri R, Dunn GP, Lewis JS., Jr Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in head and neck cancers. Cancer Immun. 2008;8:16. [PMC free article] [PubMed] [Google Scholar]
- 99.Rajjoub S, Basha SR, Einhorn E, et al. Prognostic significance of tumor-infiltrating lymphocytes in oropharyngeal cancer. Ear Nose Throat J. 2007;86(8):506–11. [PubMed] [Google Scholar]
- 100.Merck Sharpe and Dohme Corpo. Study of MK-3475 (Pembrolizumab) in Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma After Treatment With Platinum-based and Cetuximab Therapy (MK-3475-055/KEYNOTE-055) Clinicaltrials.gov; 2015. https://clinicaltrials.gov/ct2/show/NCT02255097. [Google Scholar]
- 101.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scharovsky OG, Mainetti LE, Rozados VR. Metronomic chemotherapy: changing the paradigm that more is better. Curr Oncol. 2009;16(2):7–15. doi: 10.3747/co.v16i2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rocco JW. Mutant Allele Tumor Heterogeneity (MATH) and Head and Neck Squamous Cell Carcinoma. Head Neck Pathol. 2015;9(1):1–5. doi: 10.1007/s12105-015-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]