SUMMARY
Transcriptional cyclin-dependent kinases play important roles in eukaryotic gene expression. CDK7, CDK9 (P-TEFb) and CDK13 are also critical for HIV replication. However, the function of CDK11 remained enigmatic. In this report, we determined that CDK11 regulates the cleavage and polyadenylation (CPA) of all viral transcripts. CDK11 was found associated with the TREX/THOC, which recruited this kinase to DNA. Once at the viral genome, CDK11 phosphorylated serines at position 2 in the CTD of RNAPII, which increased levels of CPA factors at the HIV 3’ end. In its absence, cleavage of viral transcripts was greatly attenuated. In contrast, higher levels of CDK11 increased the length of HIV polyA tails and the stability of mature viral transcripts. We conclude that CDK11 plays a critical role for the co-transcriptional processing of all HIV mRNA species.
INTRODUCTION
The human immunodeficiency virus (HIV) is a retrovirus of the lentivirus family that integrates into the host genome, where it behaves similarly to other human genes. Its transcription and replication have been studied extensively and lessons learned from HIV have contributed greatly to the understanding of eukaryotic biology (Peterlin and Price, 2006). For example, studies of its transcriptional transactivator (Tat) revealed that the control of RNA polymerase II (RNAPII) elongation is an important step in gene expression (Kao et al., 1987). It is now known that most if not all genes have RNAPII already engaged at their promoters and that extracellular cues can release it thereby promoting diverse cellular processes, which include activation, proliferation, differentiation and reprogramming (Rahl et al., 2010). Further steps in co-transcriptional processing, 5’ capping, mRNA splicing and 3’ end formation, which include cleavage and polyadenylation (CPA), have also profited greatly from studies of HIV (Karn and Stoltzfus, 2012; Valente et al., 2009). Importantly, all these events involve RNAPII, especially its C-terminal domain (CTD). It contains 52 heptapeptide repeats (YSPTSPS), which contain 5 residues that can be phosphorylated by various kinases for specific functions (Adelman and Lis, 2012).
In association with their respective cyclins (Cycs), transcriptional cyclin-dependent kinases (CDKs) phosphorylate different serines (Ser2P, Ser5P or Ser7P) and the threonine (Thr4P) in the CTD (Drogat et al., 2012; Loyer et al., 2005). These modifications play critical roles for 5’ capping, mRNA splicing and CPA of primary transcripts (Blazek et al., 2011; Drogat et al., 2012; Loyer et al., 2005). Indeed, differentially phosphorylated forms of RNAPII provide binding platforms for complexes that execute these early and late events in transcription. They ensure that nascent RNA species mature into functional mRNAs, which can be exported into the cytoplasm for translation (Huang and Carmichael, 1996; Muller-McNicoll and Neugebauer, 2013; Proudfoot, 2011; Wahle and Ruegsegger, 1999).
For HIV, the involvement of CDK7, CDK9, CDK11 and CDK13 has been documented. Tat potentiates the role of CycH:CDK7 in 5’ capping of all viral transcripts (Zhou et al., 2003). Tat also recruits its co-activator, CycT1:CDK9, which forms the positive transcription elongation factor b (P-TEFb), for the transition from initiation to elongation of HIV transcription (Peterlin and Price, 2006). P-TEFb exists in two states, the active free form and the inactive 7SK snRNP, from which it must be released to target the HIV long terminal repeat (LTR) (Peterlin and Price, 2006). CycK:CDK13 affects mRNA splicing so that the over-expression and depletion of CDK13 decreases and increases the production of HIV Gag and Env proteins and the production of new viral particles, respectively (Berro et al., 2008). The role/s of CycL:CDK11 is/are less well defined, especially since the depletion and over-expression of CDK11 decreased levels of HIV replication, albeit in greatly different contexts (Valente et al., 2009; Yu et al., 2008).
The transcription/export (TREX) complex connects transcription and co-transcriptional processes. In yeast, TREX couples transcription, mRNA processing and mRNA nuclear export (Strasser et al., 2002). Suppressors of the Transcriptional defects of Hpr1Δ by Over-expression complex (THOC) is required for transcription elongation (Li et al., 2005; Rehwinkel et al., 2004). THO proteins are also found in the TREX complex, hence the name TREX/THOC (Masuda et al., 2005). TREX is found at the coding and 3’ ends of genes (Kim et al., 2004; Strasser et al., 2002). In addition to RNAPII and TREX/THOC, the human 3’ end processing complex contains Cleavage and Polyadenylation Specificity Factors (CPSFs), Cleavage stimulatory Factors (CstFs), the PolyA Polymerase (PAP) and over 50 other proteins (Shi and Manley, 2015). Indeed, the depletion of THOC5 from the TREX/THOC inhibited the expression of polyadenylated mRNA species (Katahira et al., 2013). However, the mechanism of these effects remained unknown.
In this study, we were able to address these conundra. First, we found that CDK11 binds to subunits of TREX/THOC, which we validated structurally and functionally. In addition, levels of CDK11 affected greatly HIV gene expression and replication. We also demonstrated that CDK11 is recruited to the HIV genome via TREX/THOC, where it increased levels of Ser2P and the assembly of 3’ end processing machinery. In cells depleted of CDK11, HIV replicated poorly, the CPA machinery was not recruited to the RNAPII and HIV transcripts were not cleaved or polyadenylated. In cells that over-expressed CDK11, just the opposite was observed, which led to longer polyA tails and greater stability of all HIV transcripts. We conclude that CDK11 mediates effects of TREX/THOC on transcription via critical modifications of RNAPII.
RESULTS
CDK11 associates with TREX/THOC in 293T cells
Humans have two genes, CDC2L1 and CDC2L2, which encode multiple isoforms of CDK11/PITSLRE. CDK11, referred as a p110 (110 kDa), is ubiquitously expressed in cells and its p58 (58 kDa) isoform is specific for the G2/M phase (Loyer et al., 2008). Another isoform, p46, is generated by caspase cleavage of larger precursors and promotes apoptosis in human cells (Mikolajczyk and Nelson, 2004). A tandem affinity purification (TAP) in yeast also identified Cyclin L as the partner for CDK11 (Drogat et al., 2012). Another integrative analysis of 3290 affinity-purified proteins found CDK11 within the complex-complex interaction network (CCI), which are type II co-regulators that do not have consistent steady-state stoichiometric partners (Malovannaya et al., 2011).
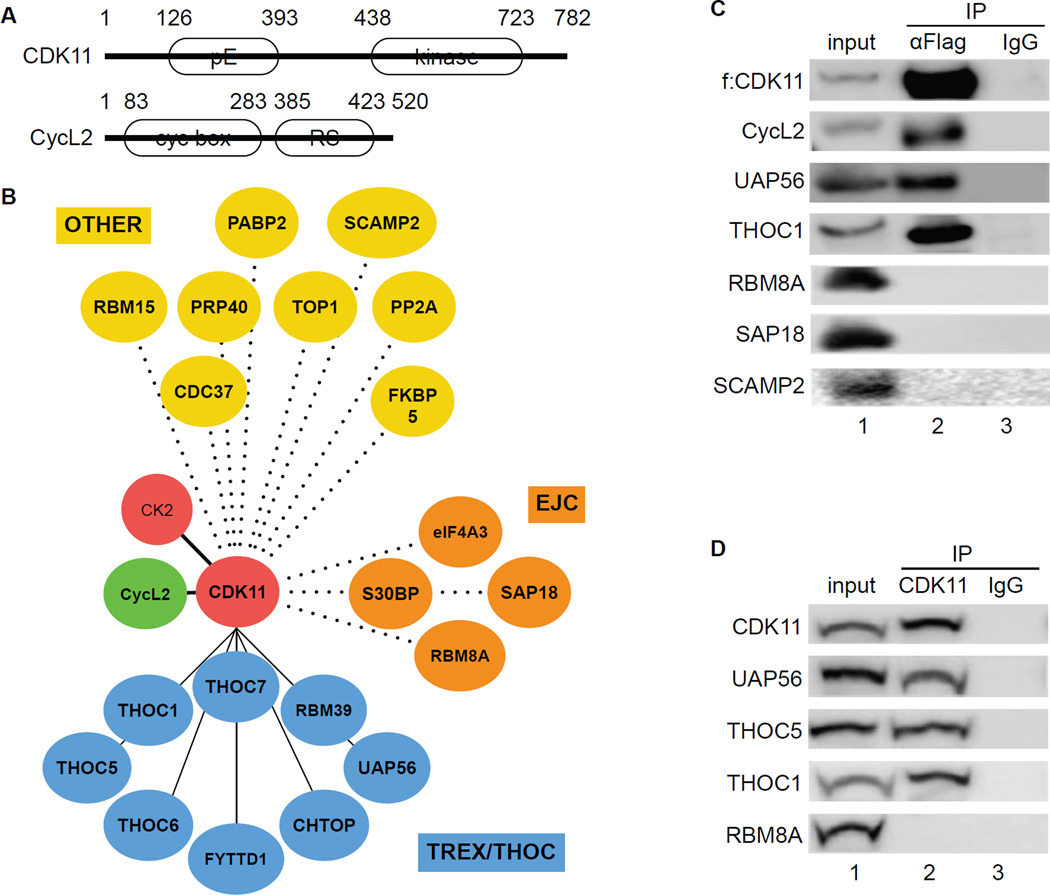
In this study, we examined the long form of CDK11 (p110), which displays extensive homology to other CDKs in its C-terminal catalytic domain (Figure 1A, top). Its N-terminus contains many glutamic acid residues (polyE, pE), which are encoded by an RNA that also serves as an Internal Ribosome Entry Site (IRES) (Loyer et al., 2005). CycL2 is the cyclin partner of CDK11 (Figure 1A, bottom). It contains two cyclin boxes near its N-terminus and an arginine/serine-rich domain (RS domain) near its C-terminus. The RS domain is a hallmark of many proteins involved in pre-mRNA processing (Figure 1A) (Loyer et al., 2005). To identify potential CDK11-binding partners, we used affinity purification and mass spectrometry (Jager et al., 2012). After confirming the expression of FLAG epitope-tagged CDK11 (f:CDK11) protein by western blotting, samples were processed further. We identified 23 proteins from f:CDK11 mass spectrometry using CompPASS (Sowa et al., 2009) and MiST quantitative scoring systems (Jager et al., 2012; Verschueren et al., 2015). Next, we organized proteins associated with CDK11 into functional categories (Figure 1B). Most associated peptides were from subunits of TREX/THOC. Also included were members of the exon junction complex (EJC) and other proteins, some of which bind to RNA (OTHER) (Figure 1B). In agreement with previous reports, casein kinase II (CK2) and CycL2 were present in our immunoprecipitations (de Graaf et al., 2004; Dickinson et al., 2002; Trembley et al., 2003). Further sequential f:CDK11 immunoprecipitations confirmed the binding between CDK11, CycL2 and subunits of TREX/THOC (Figure 1C). In contrast, RBM8A, SAP18 and SCAMP2 did not interact with f:CDK11 (Figure 1C). Co-immunoprecipitations with the endogenous CDK11 protein confirmed these associations (Figure 1D). Thus, CycL2:CDK11 bind to TREX/THOC in cells.
Figure 1. CDK11 associates with TREX/THOC in 293 T cells.
A. A schematic representation of CDK11 and CycL2. CDK11 contains 782 residues and migrates with an apparent molecular mass of 110 kDa. CycL2 contains 520 residues with an apparent molecular mass of 58 kDa. White ovals depict regions rich in glutamic acids (pE), the kinase domain of CDK11, cyclin boxes and the arginine-serine (RS)-rich domain in CycL2.
B. Dendrogram of the CDK11 proteome. Mass spectrometry of proteins that co-immunoprecipitated with CDK11 identified TREX/THOC (blue circles), EJC (brown circles) and various OTHER (yellow circles) proteins.
C. Co-immunoprecipitations of f:CDK11-interacting proteins. Immunoprecipitation of FLAG epitope-tagged CDK11 protein (f:CDK11) was followed by western blotting with anti-FLAG, UAP56, THOC1, CycL2, RBM8A, SAP18 and SCAMP2 antibodies. Lanes contain the following: lane 1, input; lane 2, co-immunoprecipitated proteins; lane 3, IgG control.
D. Co-immunoprecipitations between endogenous proteins. Immunoprecipitation of the endogenous CDK11 protein (CDK11) was followed by western blotting with anti-UAP56, THOC5, THOC1 and RBM8A antibodies (lanes 3–7). Lanes are as in panel C.
CDK11 increases HIV gene expression and replication in 293T and Jurkat cells
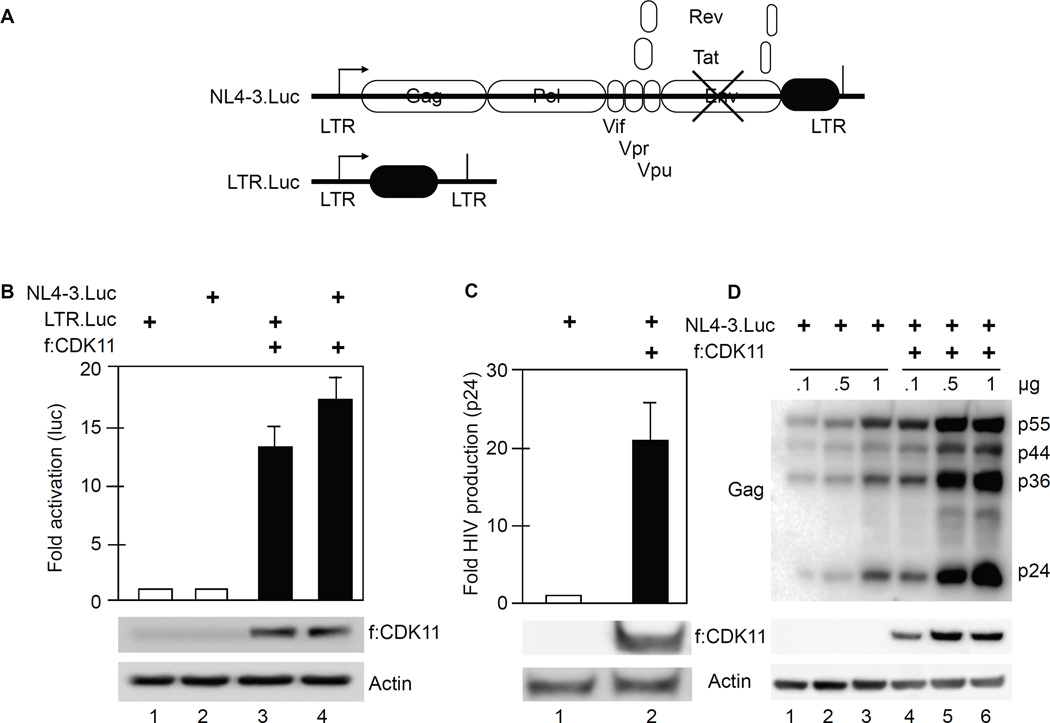
HIV gene expression requires cellular transcription factors (TFs), where the role of CDK11 remained enigmatic (Valente et al., 2009; Yu et al., 2008). To investigate the function of CDK11 in HIV transcription, f:CDK11 effector and NL4-3.Luc or LTR.Luc plasmid targets (Figure 2A) were co-expressed transiently in 293T (Figure 2B, 2C and 2D) and Jurkat (Figure S1) cells. Plasmid targets contained the luciferase reporter gene, which enables a precise quantification of small changes in transcription. Note that the NL4-3.Luc contained the luciferase reporter gene in place of Nef, which must undergo multiple mRNA splicing events for expression. Additionally, the plasmid target lacked the Env gene so that it did not infect other cells. For control purposes, the empty pcDNA3.1 plasmid vector was also co-expressed with these plasmid targets. Cells were lysed 48 hours after the transfection and luciferase activity was measured. Cells expressing f:CDK11 and NL4-3.Luc or LTR.Luc exhibited a 13- and 17-fold increased levels of luciferase, respectively, compared to those with pcDNA3.1 alone (Figure 2B, bars 1, 2, 3 and 4). Since both plasmid targets had very similar responses, this finding excludes increased mRNA splicing of NL4-3.Luc transcripts. To confirm that CDK11 increases HIV replication, we also collected supernatants from transfected 293T cells in triplicate. As presented in Figure 2C, levels of p24 (Gag) were also increased 22-fold in supernatants of cells that co-expressed f:CDK11 and NL4-3.Luc. This increase in viral proteins was confirmed by western blotting (Figure 2D). Next, f:CDK11 was co-expressed with increasing amounts of NL4-3.Luc. After 48 hours, cells were lysed and submitted to western blotting with anti-p24, FLAG or actin antibodies. Levels of HIV p55, p44 and p24 (Gag proteins) increased at least 10-fold in cells expressing f:CDK11 compared to controls (Figure 2D, compare lanes 1 and 4, 2 and 5, 3 and 6). Since f:CDK11 affected all HIV plasmid targets, we also examined its effects on several host cell and other viral promoters in Jurkat cells (Figure S1). In all these cases, f:CDK11 increased their activities from 7- to 30-fold (Figure S1). Thus, as was the case with P-TEFb, CDK11 activates not only HIV but affects other eukaryotic transcription units.
Figure 2. CDK11 increases HIV gene expression and replication in 293 T cells.
A. Schematic representation of NL4-3.Luc and LTR.Luc plasmid targets, respectively. NL4-3.Luc does not express Env (crossed lines) and contains the luciferase gene in the place of Nef.
B. CDK11 activates NL4-3.luc and LTR.Luc. Luciferase activity was measured in lysates of cells co-expressing f:CDK11 or the empty plasmid vector and LTR.Luc or pNL4-3.Luc. Error bars represent mean ± SE, n = 3. Western blots of f:CDK11 and actin as the loading control are presented below the bar graph. See also Figures S1 and S2.
C. CDK11 increases the production of new viral particles. Presented is the HIV p24 ELISA of supernatants from cells that co-expressed f:CDK11 and NL4-3.Luc. Error bars are as in panel B.
D. CDK11 increases the production of HIV structural proteins. Presented is a western blot of HIV Gag levels in cells expressing f:CDK11 and NL4-3.Luc or plasmid vector control. Amounts of plasmid effectors are presented above the western blot. Lanes 1 and 4, 2 and 5 and 3 and 6 should be compared. Below this panel are western blots of f:CDK11 and actin, as in panel C.
To validate our proteomic data, we also knocked down proteins from Figure 1 via short interfering RNA (siRNA) in the presence and absence of f:CDK11. All interacting proteins were subjected to RNAi screening. We found that the depletion of THOC1 or THOC5 inhibited effects of f:CDK11 (Figures S2A and S2B). Depletion of a subunit of the EJC (eIF4A3) and CK2 had no effect (Figures S2C and S2D). Knockdowns (KDs) were confirmed by western blotting (Figure S2, panels below the bar graphs). We conclude that CycL2:CDK11 interacts with subunits of TREX/THOC structurally and functionally. Importantly, this new complex plays a critical role in HIV gene expression and replication.
CDK11 regulates levels of all HIV transcripts in 293T and Jurkat cells
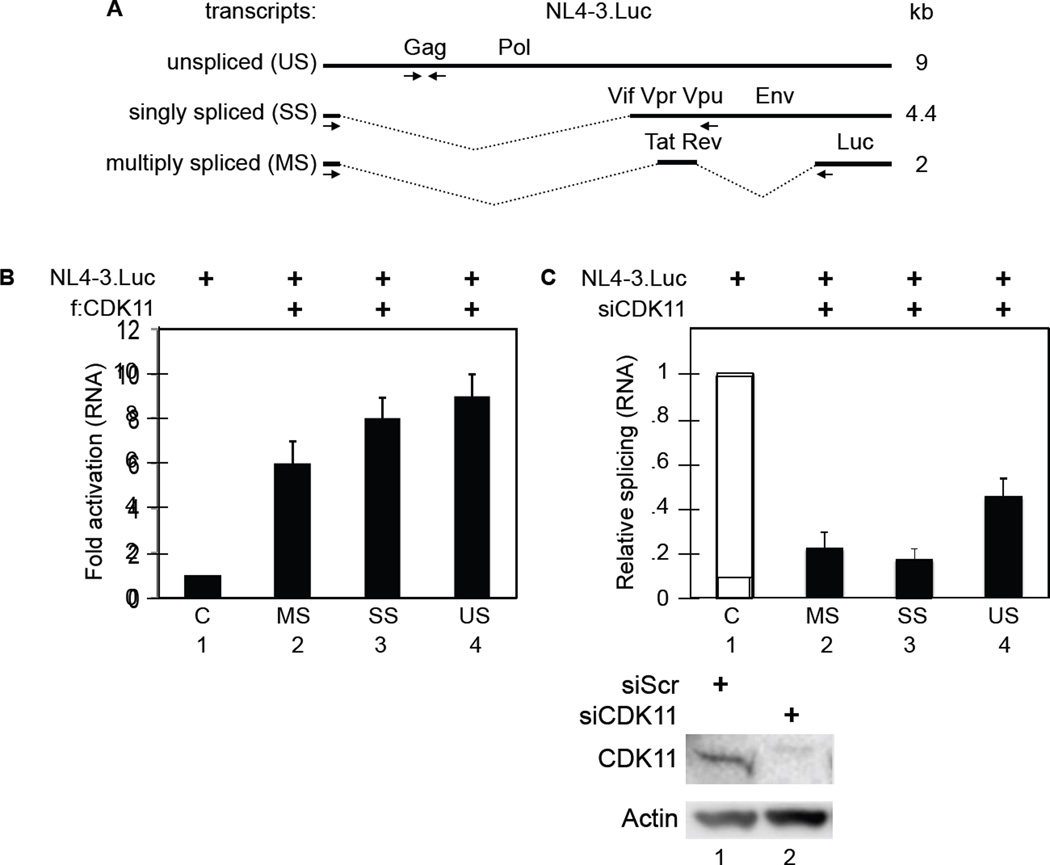
HIV transcripts undergo a series of mRNA splicing events that generate singly spliced (SS) 4kb mRNA species that code for Env, Vpr, Vpu and Vif and multiply spliced (MS) 2kb mRNA species that code for Tat, Rev and Nef. The unspliced (US) 9kb mRNA codes for Gag and Gag/Pol polyproteins that form new viral particles (Figure 3A). Balanced splicing of the viral mRNA is critical for HIV replication and infectivity (Ocwieja et al., 2012; Purcell and Martin, 1993). Primers for reverse transcription quantitative polymerase chain reactions (RT-qPCR) (Figure 3A, arrows below HIV RNA species) were designed to detect different mRNA species (Jablonski and Caputi, 2009). RT-qPCRs revealed that all three different viral transcripts increased 6- to 10-fold in f:CDK11-expressing 293T cells (Figure 3B, bars 1, 2, 3, 4). These data confirm the increased expression of viral particles and proteins observed in Figures 2C and 2D, respectively. In addition, the knockdown of CDK11 decreased US, SS and MS transcripts in 293T cells (Figure 3C, bars 1, 2, 3, 4). Whereas US transcripts were reduced 2-fold, those for spliced transcripts decreased 5-fold (Figure 3C, bars 2, 3, and 4). The ratio of spliced to US transcripts was thus reduced 2- to 3-fold. A similar effect on mRNA splicing was also observed in Jurkat cells (Figure S3). This finding agrees with other reports on effects of CDK11 depletion on mRNA splicing (Loyer et al., 2005). We conclude that CDK11 affects directly all HIV transcripts.
Figure 3. CDK11 regulates levels of all HIV transcripts in 293T cells.
A. Schematic representation of HIV transcripts. The three main viral mRNA species are represented: unspliced (US), singly spliced (SS) and multiply spliced (MS) HIV transcripts. Arrows indicate the direction and location of primers.
B. CDK11 increases levels of all HIV transcripts. RT-qPCR of HIV RNA from cells that co-expressed f:CDK11 or plasmid vector and pNL4-3.Luc. Primers were designed to detect unspliced (US) genomic, singly spliced (SS) and multiply spliced (MS) HIV transcripts. Error bars represent mean ± SE, n = 3.
C. CDK11 depletion decreases HIV mRNA splicing. CDK11-depleted cells co-expressed the same plasmids as in panel B. Presented are RT-qPCR data with errors as in panel B. Western blots of CDK11 and actin are presented below the bar graph. See also Figure S3.
CK2 does not contribute to CTD phosphorylation by CDK11
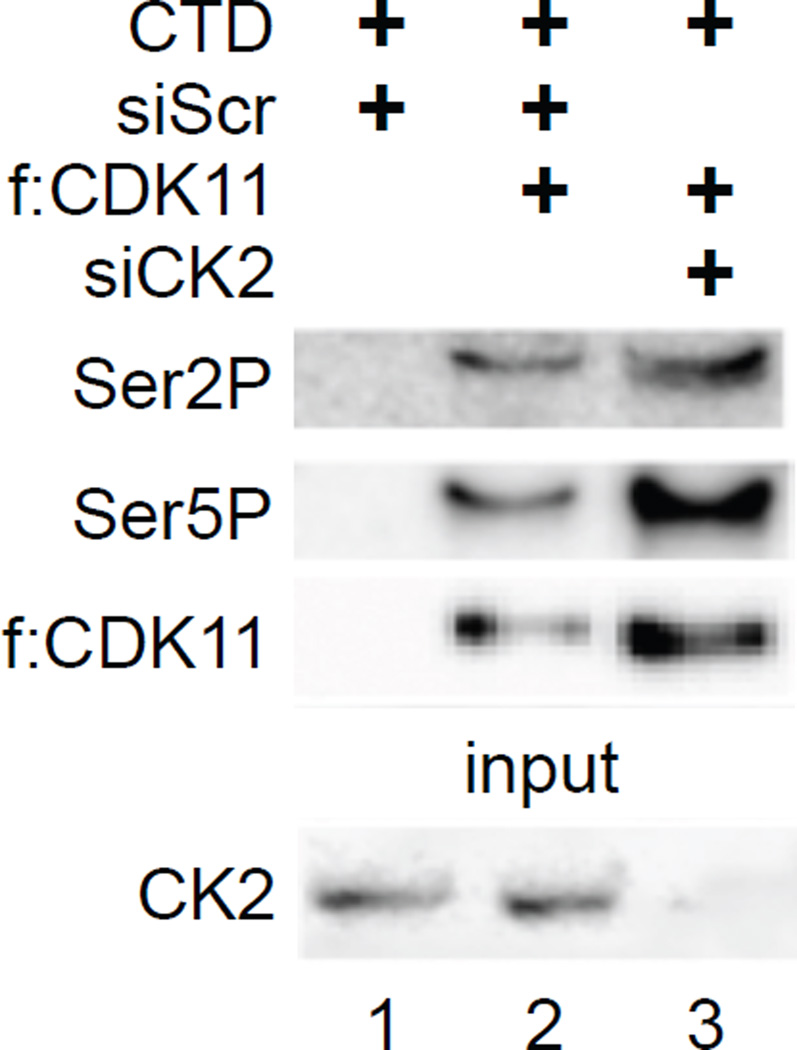
It was reported previously that CK2 phosphorylates CDK11 and the CTD (Trembley et al., 2003). To determine if CDK11 can also phosphorylate the CTD of RNAPII independently of CK2, we performed in vitro kinase assays with CDK11 in the presence and absence of CK2 and the CTD of RNAPII. CK2 was depleted using siCK2 RNA. f:CDK11 was immunoprecipitated from wild type (WT) and CK2-depleted 293T cells and combined with the glutathione S-transferase (GST)-CTD chimera, which was purified from E. coli (Peterson et al., 1992). Kinase reactions were performed in the presence of cold ATP. Residues in the CTD that were phosphorylated were detected with anti-Ser2P and Ser5P antibodies by western blotting (Czudnochowski et al., 2012). Importantly, CDK11 phosphorylated serines at positions 2 and 5 in the CTD independently of CK2 (Figure 4, lanes 2 and 3). This result extends our finding that CK2 depletion did not affect the increased HIV gene expression by f:CDK11 (Figure S2D). These data also suggest that CDK11 is a bona-fide CTD kinase.
Figure 4. CK2 does not contribute to CTD phosphorylation by CDK11.
Kinase assays were performed with the immunoprecipitated f:CDK11 protein and GST-CTD using cold ATP, followed by western blotting with anti-Ser2P and Ser5P antibodies. Lane 3 contains reactions with the immunoprecipitated f:CDK11 protein from CK2-depleted cells. The western blot for the depletion of CK2 is presented at the bottom.
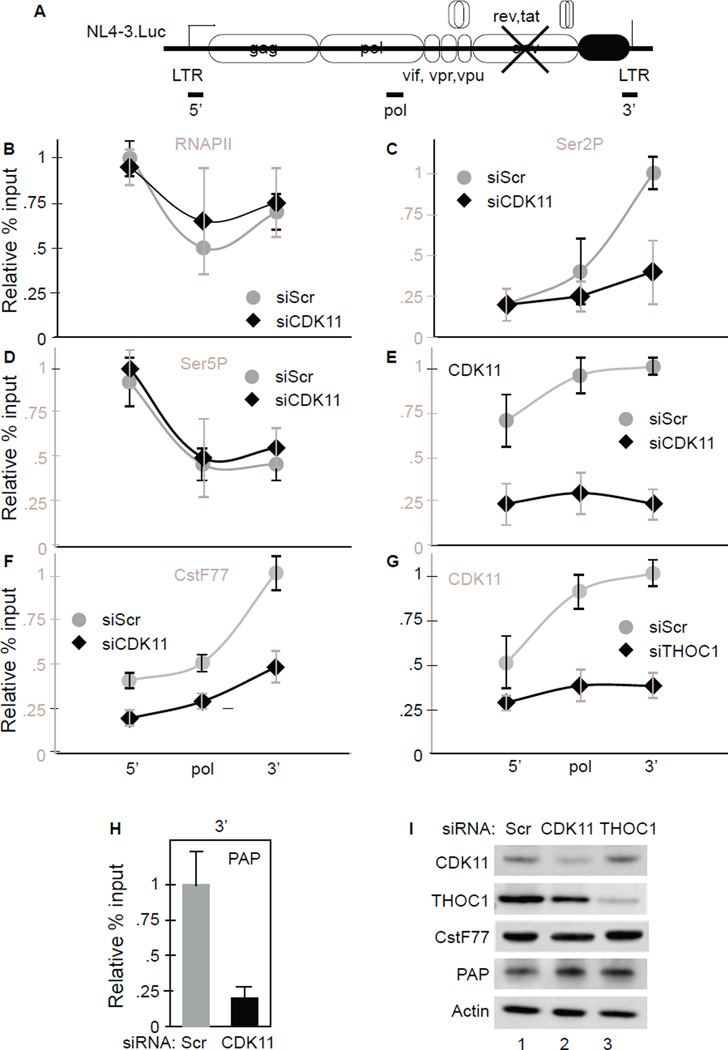
CDK11 depletion decreases levels of Ser2P, CstF77 and PAP at the HIV gene in HeLa P4 cells
RNAPII synthesized transcripts undergo extensive processing before they are transported to the cytoplasm and the CTD directs much of this activity (Barilla et al., 2001; McCracken et al., 1997). Whereas Ser5P plays a critical role in 5’ capping, increased levels of Ser2P during transcription recruit splicing and CPA complexes to RNAPII (Gu et al., 2013). We used chromatin immunoprecipitations (ChIPs) to investigate the effects of CDK11 and TREX/THOC on levels of RNAPII, Ser2P and Ser5P and the 3’ end formation machinery during HIV transcription. The schematic of the viral genome and primers used in ChIPs are presented in Figure 5A. Importantly, 5’ and 3’ primers do not share any sequences. In the rest of Figure 5, panels B to F contain ChIPs comparing WT (gray line) to CDK11-depleted (black line) cells. Panel G contains data from THOC1-depleted cells. Panels H and I contain PAP ChiPs and western blots for depleted proteins, respectively.
Figure 5. Knockdown of CDK11 decreases levels of Ser2P and CPA factors at the HIV 3’ end in HeLaP4 cells.
A. Schematic representation of the NL4-3.Luc. Horizontal lines beneath the gene represent regions amplified by qPCR after ChIPs.
B, C, D, E, F, H. Effects of CDK11 depletion and THOC1 depletion (G) on levels of RNAPII, Ser2P, Ser5P, CDK11, CstF77 and PAP at the HIV gene. ChIPs were performed using indicated antibodies in cells expressing the NL4-3.Luc and treated with siCDK11 or siTHOC1 (black diamonds and lines) and compared to those treated with siScr RNA (gray circles and lines). Sequences corresponding to the HIV 5’ end, Pol, and 3’ end were amplified by qPCR. Values represent percentage of input normalized to IgG levels. See also Figure S4.
I. Confirmation of depleted proteins. Presented are western blots of CDK11, THOC1, CstF77 and PAP in cell lysates, where actin represents the loading control.
As presented in Figure 5B, CDK11 depletion did not affect greatly levels of RNAPII at the HIV LTR, the Pol gene or the 3’ end. In contrast, levels of Ser2P at the 3’ end of HIV gene decreased 2-fold (Figure 5C). Effects on Ser5P were insignificant (Figure 5D). As expected, CDK11 depletion also reduced levels of CDK11 throughout the HIV coding region (Figure 5E). In these cells, levels of CstF77, an important component of the CPA, were also reduced, especially at the 3’ end (Figure 5F). Of note, depletion of THOC1 and CDK11 had similar effects on levels of CDK11 at the HIV gene (compare Figure 5E and 5G). CDK11 depletion also reduced levels of PAP 4-fold at the 3’ end (Figure 5H). Depletion of THOC1 mirrored that of CDK11 (Figure S4) except that the knock down of CDK11 did not reduce levels of THOC1 at the HIV gene (Figure S4F). Reduced levels of targeted proteins were confirmed by western blotting (Figure 5I). Importantly, CDK11 depletion did not affect steady state levels of THOC1, CstF77, PAP or actin proteins (Figure 5I, lanes 1, 2 and 3). In agreement with our data, levels of Ser2P are known to increase from the body to 3’ end of genes (Davidson et al., 2014; Eifler et al., 2015). We conclude that TREX/THOC recruits CDK11, which increases levels of Ser2P and CPA factors towards the HIV 3’ end.
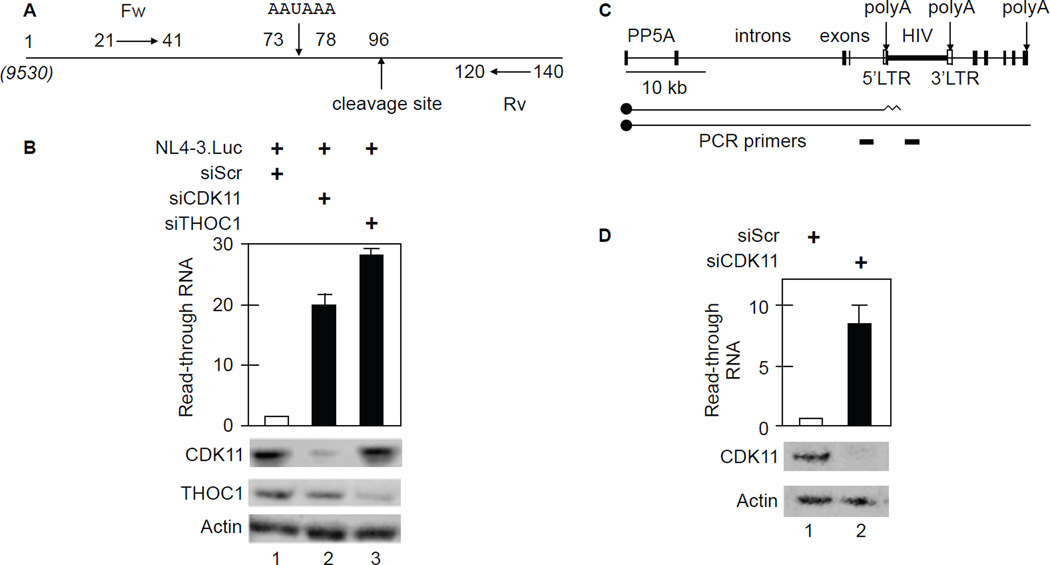
Knockdown of CDK11 or THOC1 increases HIV LTR read through transcription in HeLa P4 and J-Lat 9.2 cells
Since knockdowns of CDK11 and THOC1 affected levels of CPA factors at the 3’ end, we examined this aspect of HIV co-transcriptional processing. First, we interrogated the cleavage reaction. Read through transcripts and primers are depicted in Figure 6A. Note that position 1 corresponds to position 9530 with respect to the 5’ HIV LTR in full length transcripts of NL4-3.Luc (Figure 6A). Primers were designed based on HIV cleavage, which occurs 18 nucleotides downstream of the polyA site (AAUAAA). Read through transcription is the ratio of uncleaved RNA to total RNA 5’ to the cleavage site. RT-qPCR with the indicated primers (Fw and Rv) demonstrated that levels of uncleaved HIV RNA increased 20- to 30-fold in CDK11- and THOC1-depleted cells, respectively (Figure 6B, black bars 2 and 3). Thus, knockdown of CDK11 reduced levels of total HIV RNA (Figure 3B) and increased the proportion of extended 3’ ends (Figure 6B). All RT-qPCR samples were normalized to GAPDH levels. We conclude that CDK11 depletion decreases cleavage of HIV transcripts.
Figure 6. Knockdown of CDK11 or THOC1 increases HIV 3’end read through transcription in HeLa P4 and J-Lat 9.2 cells.
A. Schematic representation of primers used for the HIV 3’ end read through assay. Note that position 1 corresponds to position 9530 from the 5’ HIV LTR. Locations of polyA and cleavage sites are depicted by arrows. Fw and Rv are forward and reverse primers, respectively.
B. Knockdown of CDK11 or THOC1 increases read through of HIV transcripts. Black bars represent fold-increased read-through over WT cells. Error bars represent the mean +/− SE, n = 3. Western blots of CDK11 and THOC1 with actin as the loading control are presented below the bar graph.
C. Schematic representation of the PP5 gene and the integrated HIV in J-Lat 9.2 cells. Two different capped (black circles) transcripts are diagrammed below the gene, those that terminate in the 5’ HIV LTR and those that read through into the viral coding sequences. PCR primers for RT-qPCR are presented below primary transcripts.
D. CDK11 depletion increases read through transcription past the 5’ HIV LTR in J-Lat 9.2 cells. Black bar and error bars are as in panel B. Western blots of CDK11 with actin as the loading control are presented below the bar graph.
See also Figure S5.
To confirm these findings, we also examined J-Lat 9.2 cells, where HIV had integrated into the PP5 gene (Lenasi et al., 2008). In these cells, HIV is silenced because RNAPII from PP5 gene occludes and terminates in the 5’ HIV LTR and the 3’ HIV LTR initiates transcription into the rest of the PP5 gene. This integration is depicted in Figure 6C. With CDK11 depletion, 8-fold more transcripts read through the 5’ HIV LTR and into the coding region of the virus (Figure 6D). Again, all RT-qPCR samples were normalized to GAPDH levels and the knockdown of CDK11 was confirmed by western blotting. Thus, CDK11 depletion also increases read through transcription of the integrated provirus in a latently infected T cell line.
We also used a recently published dual fluorescence repoter assay for read through transcription (Ji et al., 2011). In this system, the CMV promoter drives the expression of RFP and EGFP genes, which are separated by polyA sites. In addition, EGFP 3’ to the first polyA site is translated via an IRES (Figure S5). With this plasmid target, when CPA occurs normally, cells turn red. However, when the first polyA site is ignored, then EGFP is also expressed and cells turn yellow. With CDK11 depletion, 10-fold more 293T cells turned yellow (Figure S5, panels B and C). CDK12 depletion had only a small effect. Knock down of CDK12 was an important control as it also affects 3’ end processing via the EJC, but mostly of longer genes with many exons and introns (Blazek et al., 2011). This dual fluorescence plasmid target contained no exon junctions. Thus, in all these situtations, with different polyA sites, CDK11 plays a critical role in CPA.
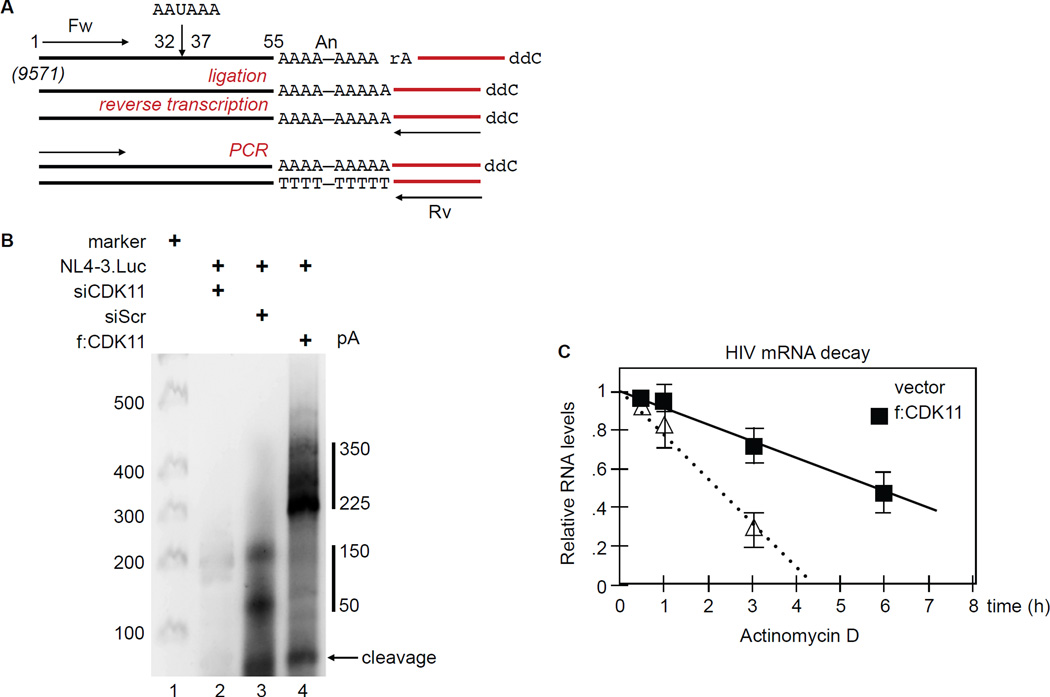
Levels of CDK11 affect polyadenylation and stability of HIV transcripts in HeLa P4 cells
RNAPII transcription and mRNA processing are coordinated within the nucleus. CPA occurs co-transcriptionally, suggesting that RNAPII regulates these events. Indeed, phosphorylation of the CTD is important for capping, mRNA splicing and CPA (Barilla et al., 2001; McCracken et al., 1997). Polyadenylation of the 3’ end of eukaryotic transcripts is associated with several aspects of mRNA metabolism, such as mRNA stability, export and translation (Shi and Manley, 2015). To determine if increased levels of HIV transcripts in Figure 3B could be due to greater stability and polyadenylation of HIV mRNA species, we measured their polyA tail lengths using the All Tail kit (Bioscientific). Figure 7A depicts the experimental scheme, where a linker oligonucleotide was ligated to the polyA tail at the 3’end of viral transcripts. Next, RNA was reverse transcribed with this linker primer (Rv) and amplified using an HIV-specific primer (Fw) by 2 rounds of PCR. Note that position 1 corresponds to position 9571 with respect to the 5’ HIV LTR in full length transcripts of NL4-3.Luc. In addition, the 3’ linker measured 20 nucleotides, which increases all PCR products by that amount.
Figure 7. CDK11 increases length of polyA tails and stability of HIV transcripts in HeLa P4 cells.
A. Schematic representation of extreme 3’ RACE using the All Tail kit for polyA tail length, with indicated primer sets. Position 1 was assigned arbitrarily to the beginning of the forward primer (Fw). It is 9571 nucleotides removed from the 5’ end of the unsplised (US) genomic NL4.3.Luc transcript. Note that the ligated primer (in red) measures 20 nucleotides. Steps in the reaction are depicted in red. Rv, reverse primer.
B. Length of HIV polyA tails. HIV polyA tails in HeLa P4 cells co-expressing siCDK11, vector or f:CDK11 and pNL4-3.Luc were amplified by RT-qPCR and separated by a 2.5% agarose gel electophoresis.
C. CDK11 increases the stability of HIV transcripts. Measured was the stability of HIV genomic transcripts in cells co-expressing f:CDK11 and pNL4-3.Luc. After 24 hours, cells were treated with Actinomycin D and RNA was extracted at different time points. RT-qPCR was performed in duplicate. Error bars represent mean ± SE, n =3.
As presented in Figure 7B, lane 3, polyA tails of viral transcripts in control samples measured 50 to 150 nucleotides. Cleavage occurred at position 55, which together with the 20 nucleotides linker measures 75 nucleotides in the agarose gel (Figure 7B, arrow). This cleavage site was confirmed by direct sequencing of PCR-amplified isolated fragments from agarose gels. In cells expressing f:CDK11, polyA tails measured between 225 to 350 nucleotides (Figure 7B, lane 4). CDK11 depletion not only reduced the general pool of HIV mRNA, but also resulted in insignificant cleavage and polyadenylation (Figure 7B, lane 2).
To determine if increased length of polyA tails in HIV transcripts enhances their stability, we analyzed their degradation kinetics. We used Actinomycin D mRNA stability assay as described previously (Zhang et al., 2011). We added Actinomycin D to block transcription 24 hours after the transfection, extracted mRNA at different time points, and measured levels of HIV transcripts by RT-qPCR. Results were normalized to 18S ribosomal RNA. We found that the half-life of HIV mRNA increased from 2.5 hours in control to 6 hours in f:CDK11-expressing cells (Figure 7C). We conclude that by affecting Ser2P near the 3’ end, CDK11 increases the recruitment of CPA factors, cleavage of primary transcripts and their polyadenylation. Combined effects of these changes increase the stability of HIV mRNA species, which results in greater levels of viral gene expression and replication.
DISCUSSION
In this study, we explored the relationship between CDK11 and HIV replication. We first defined the CDK11 proteome structurally and functionally and determined that CDK11 is an integral part of the TREX/THOC, which could inform all previous studies on this complex. Next, we found that levels of CDK11 affected directly HIV gene expression and replication. Importantly, CDK11 functioned as a CTD kinase even in the absence of CK2. Moreover, although it phosphorylated serines at positions 2 and 5 in the CTD in vitro, ChIPs revealed that CDK11 primarily affected levels of Ser2P at the HIV 3’ end in cells. Increased levels of Ser2P led to a greater recruitment of CPA factors to the RNAPII near the HIV 3’ end. In CDK11-depleted cells, this absence resulted in increased read through transcription at the HIV polyA site and diminished polyadenylation of all viral transcripts. Different lengths of HIV transcripts also affected their stability. We conclude that CDK11 is recruited to RNAPII via TREX/THOC, where it phosphorylates serines at position 2 in the CTD, which improves 3’ end processing of all viral transcripts and increases HIV replication.
Although we do not know which subunit of TREX/THOC binds to CycL2:CDK11, this binding appears to be direct and independent of RNA. If RNA were involved, CDK11 should have also co-immunoprecipitated with EJC proteins. Importantly, interactions between CDK11 and TREX/THOC were confirmed with endogenous proteins and validated by functional studies. Moreover, the over-expression of f:CDK11 increased the luciferase activity of LTR.Luc as well as NL4-3.Luc. In addition, we found that levels of Gag polyprotein and its processed subunits were increased. These findings argue against a major effect of CDK11 on mRNA splicing. Examination of all 3 major classes of HIV transcripts confirmed directly this conclusion. In contrast, CDK11 had major effects on HIV 3’ formation. First, depletion of CDK11 attenuated severely the cleavage of HIV transcripts 3’ to the polyA site. Second, HIV transcripts had much longer polyA tails in the presence of f:CDK11. They were also much more stable. These effects on CPA were due to increased levels of Ser2P and CPA factors near the 3’ end of viral transcripts. Although we did not examine all CstF or CPSF proteins, they and PAP co-aggregate and accumulate near the polyA site and are required for efficient termination of transcription.
Our findings build on the work of two previous reports on CDK11 and HIV replication. In one study, the CDK11 depletion attenuated greatly HIV replication (Yu et al., 2008). Another study found that the over-expression of CDK11 could also restrict the infection by a pseudotyped, puromycin-resistant HIV in colony forming assays (Valente et al., 2009). As mentioned previously, CDK11 has three isoforms: p110, p58 and p46. The apoptosis-inducing p46 isoform represents a caspase cleavage product of p110 (Mikolajczyk and Nelson, 2004). Since puromycin activates caspases (Koh et al., 2011; Naarmann-de Vries et al., 2013), such treatment of cells over-expressing CDK11 p110 (Valente et al., 2009) could have increased levels of p46, thus resulting in fewer HIV colonies. Indeed, we failed to establish stable f:CDK11-expressing cell lines in the presence of puromycin (data not presented). Importantly, we did not observe any diminution of HIV gene expression or replication with our over-expression of f:CDK11 in our transient assays. Moreover, we determined the mechanism by which CDK11 affected these changes in HIV gene expression. Another important observation concerns physiological levels of CDK11 in human cells. In all transformed cell lines, be they of lymphoid or epithelial origin, levels of CycL2:CDK11 are high. In contrast, they are vanishingly low in resting CD4+ T cells (data not presented). This observation prevented us from repeating our studies in primary cells. Nevertheless, our findings and those already published on CDK11 and HIV were obtained in cells that are fully permissive for HIV replication. Together with observed low levels of P-TEFb in resting lymphocytes, the lack of these additional transcriptional CDKs could represent major obstacles to the reactivation of HIV in latently infected cells in humans.
Another important issue concerns the uniqueness and redundancy of CTD kinases. Thus far, CDK7, CDK8, CDK9, CDK11, CDK12 and CDK13 can all phosphorylate the CTD of RNAPII. However, some, like CycH:CDK7, prefer serines at position 5, whereas P-TEFb, CDK11 and CycK complexes target serines at position 2 in cells. For this differential phosphorylation, it is possible that after serines at positions 5 and/or 7 had been phosphorylated by CDK7, the CDKs that are recruited later target the now better-exposed serines at position 2. Of interest, CDK11, CDK12 and CDK13 associate with RNAPII during elongation. There is also a hierarchy of importance, such that most if not all promoters require P-TEFb for transition from initiation to elongation of transcription. CDK12 affects only a small subset of acute response and longer genes, e.g. those that code for DNA damage response factors. CDK11 appears to have a broader range, especially since TREX/THOC is recruited to many coding genes. That these kinases are not redundant is also apparent from studies of HIV. Thus, Tat augments the ability of CycH:CDK7 to cap HIV transcripts (Zhou et al., 2003). P-TEFb is required for Tat transactivation and transition from initiation to elongation of viral transcription (Peterlin and Price, 2006). CDK13 affects HIV splicing, such that its over-expression favors multiply spliced rather than genomic RNA species (Berro et al., 2008). CDK11 now takes its place in HIV 3’ end formation. How these transcriptional CDKs imprint directions on the CTD, i.e. which heptapeptide repeats are targeted and when remain to be explored. Nevertheless, the CTD appears to be the central processing unit of transcription and co-transcriptional processing of eukaryotic and HIV genes.
CDK11 also plays an important role for other genes and appears to be the critical CTD kinase for their 3’ end formation. In addition to our work, in one study, where levels of only P-TEFb were increased due to the disruption of the 7SK snRNP, RNAPII elongated efficiently, but did not terminate and read through megabases of genomic DNA (Castelo-Branco et al., 2013). In this context, levels of other transcriptional CDKs or CDK11 must have been insufficient for proper co-transcriptional processing of these genes. Similarly, in our study, CDK11 depletion led to increased read through transcription of a transcriptionally interfered HIV, which had integrated into an active cell host gene in the sense orientation. Indeed, most HIV integrates into actively transcribed genes (Schroder et al., 2002). Thus, during attempts at HIV reactivation from latency, if P-TEFb levels increase but those of CDK11 lag behind, many apparent HIV-specific transcripts will represent read through transcripts from cell host genes. Thus, RNA levels would not translate to amounts of HIV proteins and numbers of new viral particles. This consideration is of importance in all studies of HIV reactivation from latency and attempts at purging the viral reservoir in infected individuals.
Our study also addresses the function of TREX/THOC, which is a highly conserved multimeric complex from yeast to humans and is required for mRNP biogenesis (Luna et al., 2012). This complex connects transcription elongation with 3’ end mRNA processing (Aguilera, 2005). For example, one member of the THO subcomplex, THOC5, is necessary for recruiting CPSF100 to immediate early genes (Tran et al., 2014). Thus, CPA of newly synthesized mRNA at 3’ end ensures that nascent RNA is protected from degradation (Proudfoot, 2011). Our studies address these processes and connect TREX/THOC, CDK11 and the CTD of RNAPII in transcription. Finally, CDK11 also plays an important role in cancer. Its expression is high and critical for growth and proliferation of breast cancer, osteosarcoma and liposarcoma (Duan et al., 2012; Jia et al., 2014). In these cells, the depletion of CDK11 resulted in the loss of cancer cell viability and led to their apoptosis (Kren et al., 2015). Taken together, these findings suggest that CDK11 is a promising target for therapeutic drug development to combat HIV/AIDS and cancer.
EXPERIMENTAL PROCEDURES
Plasmids
pUHD10-3CDK11FLAG p110 (f:CDK11) (Jill M. Lahti), PCMV( Bin Tian), NL4-3Luc (AIDS Reagents Program, #3418), LTR.Luc (Bartholomeeusen et al., 2013), CMV.luc, c-Jun.Luc, MCK.Luc, EIAV.Luc, and RSV.Luc (Clarke et al., 1998; Kowalska et al., 2012; Nojima et al., 2008; Taube et al., 2000; Vasanwala et al., 2002), pcDNA3.1 (V790-20, Invitrogen).
Antibodies
Anti-CDK11, SAP18, SCAMP2, THOC1, THOC5, UAP56, THOC7 (Abcam, 19393, 31748, 154181, 487, 47955, 86070, 72295), anti-CDK11, CycL1, CycL2, IgG, eiF4A3, RBM8A (Bethyl, A300–310A, A302-058A, A301–677A, MI10–102, A302–981A, A301-033A), anti-THOC6, THOC1, CK2, RNAPII N20, PAP (Santa Cruz, 101177, 136426, 6479, 899, 32915), anti-Ser2P, Ser5P (Covance, H5, H14), anti-FLAG, HA, IgM, IgG (Sigma-Aldrich, F3165, F7425, H3663, H6908, A4540, A5420), peroxidase-conjugated monoclonal mouse anti-IgG (Jackson ImmunoResearch, 211-032–171, 115-035–174), anti-CstF77 (David Bentley).
Transfections, luciferase reporter assays and p24 CA ELISA
293T and HeLa P4 were transfected with 0.1 µg of NL4-3.Luc or LTR.Luc plasmids with 2 µg of f:CDK11 or empty plasmid vector in 12-well plates using X-TremeGENE (Roche) reagent following the manufacturer’s protocol. Transfections of Jurkat and J-Lat 9.2 cells were carried out using the Neon® Transfection System (ThermoFisher Scientific) at 1150 V for 30 seconds with two pulses with 5 µg of NL4-3.Luc or LTR.Luc plasmids with 10 µg of f:CDK11 or empty plasmid vector. For siRNA Neon® transfection siRNA used in range 100nM −1 µM. After 48–72 hours, cells were lysed and a standard luciferase (Pak et al., 2011) assay was performed. Supernatants from cells co-transfected with NL4-3.Luc and f:CDK11 or empty plasmid vector were collected and passed through a 0.45 µm filter. Supernatants were diluted for standard ELISA protocol. HIV p24 CA ELISA kit (Perkin Elmer, NEK050B) was used in duplicates to determine levels of new viral particles in supernatants following the manufacturer’s protocol. Knockdown experiments with siRNA (IDT) were performed using RNAiMAx (Invitrogen) or Pepmute (SignaGen) reagents to transfect 293T and HeLa P4 cells while Jurkat were transfected using the Neon® Transfection System as described above. After 24 hours, 0.1 µg of NL4-3.Luc and 2 µg of f:CDK11 or empty plasmid vector were transfected in 12-well plates. After 48 hours, cells were lysed and processed for Luciferase assay.
Immunoprecipitation and western blotting
For immunoprecipitation, f:CDK11 was over-expressed in 293T cells. Cells were lysed in lysis buffer (HEPES pH 7.9, 100 mM KCl, 0.2 mM EDTA, 5 mM β-mercaptoethanol, 0.1% Nonidet P-40, 10% glycerol, and protease inhibitors (Roche Molecular Biochemicals)), sonicated and cleared by centrifugation at 20,000 g for 15 min at 4°C. Washed Sepharose 4B beads (Invitrogen) were incubated with lysates for 1 hour at 4 °C. Beads were then discarded and lysates were incubated overnight with rabbit anti-FLAG antibodies, protein specific or rabbit IgG antibodies at 4 °C. Pre-washed protein G-conjugated Sepharose 4B beads (Invitrogen) were added to the lysate and the lysate-bead mixtures were incubated for 1 hour at 4 °C. Beads-antibody conjugates were washed 5 times in lysis buffer and then heated at 95 °C for 5 minutes with SDS-PAGE sample buffer (Bio-Rad). Collected samples were processed for gel electrophoresis. For western blotting, samples were run on 4–18% SDS PAGE, transferred to nitrocellulose membranes, probed with specific primary antibodies, washed 5 times and incubated with peroxidase conjugated or IRDye fluorescent secondary antibodies (Licor). Membranes were visualized using the Odyssey Fc imaging system.
RT-qPCR
293T, HeLa P4 and Jurkat cells were co-transfected as described above. After 48 hours, RNA was extracted using TRIZOL reagent (Invitrogen) and treated with Turbo DNAse (Ambion). RNA samples were reverse transcribed using the Superscript® III First Strand System (Invitrogen) with the oligo dT primer, random hexamers or gene specific primers to produce cDNA. Primers used to detect different mRNA species are described previously (Jablonski and Caputi, 2009). qPCR was performed using SensiFAST SYBR Lo-ROX kit (Bioline) following the manufacturer’s protocol. Results were normalized to GAPDH.
Chromatin Immunoprecipitation Assays (ChIPs)
CDK11 or THOC1 were depleted in 293T cells or HeLa P4 cells by transfection of siRNA as described above. After 24 hours, siRNA treated cells were transfected with NL4-3.Luc. After 48 hours, chromatin was extracted from the cells as described by Carey and colleagues in Cold Spring Harbor Protocols (Carey et al., 2009) with modifications, where the total sonication time was increased to 5 min and performed in ice-cold ethanol or an ice-salt slurry.
GST-CTD purification and phosphorylation assay
GST-CTD purification and phosphorylation assays were performed as described previously (Czudnochowski et al., 2012).
Mass spectrometry
Was performed essentially as described previously for CDK12 (Eifler et al., 2015).
Determination of polyA tail length and 3’ end formation
PolyA tail was measured using the ALL-TAIL kit (Biooscientific). Briefly, HeLa P4 cells co-expressed 0.1 µg of NL4-3.Luc and 2 µg of f:CDK11 or empty plasmid vector in 12 well plates. After 48 hours, RNA was extracted using TRIZOL reagent (Invitrogen) and 1 µg RNA was treated with Turbo DNAse (Ambion). Isolated RNA was ligated to the AIR Adenylated Linker C and then reverse transcribed with Linker C universal primer. Primers: Fw (GGCAGCTGTAGATCTTAGCC) and Universal Linker C, Rv from ALL-TAIL kit (Biooscientific). polyA tails (or 3’ end) were resolved on 2.5% agarose gels, stained with SYBR Gold nucleic acid gel stain (Molecular probe, S11494) and visualized using Odyssey Fc imaging system (Licor).
Read through transcription
Read through transcription assays were performed as described (Eifler et al., 2015). cDNA was synthesized using random hexamers and subjected to qPCR analysis with primers: Fw, (TAGTGTGTGCCCGTCTGTT); Rv, (CCTCCTGGGTGCTAGAGATTT). For J-Lat 9.2 cells, primers were: Fw, (GGAAAAGTTATCTTGGTAGC); Rv, (GTTTGTATGTCTGTTGCTATT).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. David L. Bentley, Bin Tian and Jill M. Lahti for reagents. This work was supported by a fellowship from CHRP (California HIV/AIDS Research Program Award ID: F13-SF-302 to VP), by the National Institute of Health (U19 AI076113 and AI076113 to BMP; P50 GM082250 to BMP and NJK, PO1 AI090935 and P50 GM081879 to NJK) and the American Foundation of AIDS Research (108241-51-RGRL to KF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental data include supplemental experimental procedures and 5 figures.
AUTHOR CONTRIBUTIONS
VP, TTE, KF and BMP designed and performed the experiments. SJ and NJK performed mass spectrometry and analyzed proteomic data.
REFERENCES
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr Opin Cell Biol. 2005;17:242–250. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Barilla D, Lee BA, Proudfoot NJ. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98:445–450. doi: 10.1073/pnas.98.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomeeusen K, Fujinaga K, Xiang Y, Peterlin BM. Histone deacetylase inhibitors (HDACis) that release the positive transcription elongation factor b (P-TEFb) from its inhibitory complex also activate HIV transcription. J Biol Chem. 2013;288:14400–14407. doi: 10.1074/jbc.M113.464834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berro R, Pedati C, Kehn-Hall K, Wu W, Klase Z, Even Y, Geneviere AM, Ammosova T, Nekhai S, Kashanchi F. CDK13, a new potential human immunodeficiency virus type 1 inhibitory factor regulating viral mRNA splicing. J Virol. 2008;82:7155–7166. doi: 10.1128/JVI.02543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, Cimermancic P, Ule J, Peterlin BM. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25:2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey MF, Peterson CL, Smale ST. Chromatin immunoprecipitation (ChIP) Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5279. pdb prot5279. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco G, Amaral PP, Engstrom PG, Robson SC, Marques SC, Bertone P, Kouzarides T. The non-coding snRNA 7SK controls transcriptional termination, poising, and bidirectionality in embryonic stem cells. Genome Biol. 2013;14:R98. doi: 10.1186/gb-2013-14-9-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke N, Arenzana N, Hai T, Minden A, Prywes R. Epidermal growth factor induction of the c-jun promoter by a Rac pathway. Mol Cell Biol. 1998;18:1065–1073. doi: 10.1128/mcb.18.2.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czudnochowski N, Bosken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun. 2012;3:842. doi: 10.1038/ncomms1846. [DOI] [PubMed] [Google Scholar]
- Davidson L, Muniz L, West S. 3' end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 2014;28:342–356. doi: 10.1101/gad.231274.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf K, Hekerman P, Spelten O, Herrmann A, Packman LC, Bussow K, Muller-Newen G, Becker W. Characterization of cyclin L2, a novel cyclin with an arginine/serine-rich domain: phosphorylation by DYRK1A and colocalization with splicing factors. J Biol Chem. 2004;279:4612–4624. doi: 10.1074/jbc.M310794200. [DOI] [PubMed] [Google Scholar]
- Dickinson LA, Edgar AJ, Ehley J, Gottesfeld JM. Cyclin L is an RS domain protein involved in pre-mRNA splicing. J Biol Chem. 2002;277:25465–25473. doi: 10.1074/jbc.M202266200. [DOI] [PubMed] [Google Scholar]
- Drogat J, Migeot V, Mommaerts E, Mullier C, Dieu M, van Bakel H, Hermand D. Cdk11-cyclinL controls the assembly of the RNA polymerase II mediator complex. Cell Rep. 2012;2:1068–1076. doi: 10.1016/j.celrep.2012.09.027. [DOI] [PubMed] [Google Scholar]
- Duan Z, Zhang J, Choy E, Harmon D, Liu X, Nielsen P, Mankin H, Gray NS, Hornicek FJ. Systematic kinome shRNA screening identifies CDK11 (PITSLRE) kinase expression is critical for osteosarcoma cell growth and proliferation. Clin Cancer Res. 2012;18:4580–4588. doi: 10.1158/1078-0432.CCR-12-1157. [DOI] [PubMed] [Google Scholar]
- Eifler TT, Shao W, Bartholomeeusen K, Fujinaga K, Jager S, Johnson JR, Luo Z, Krogan NJ, Peterlin BM. Cyclin-dependent kinase 12 increases 3' end processing of growth factor-induced c-FOS transcripts. Mol Cell Biol. 2015;35:468–478. doi: 10.1128/MCB.01157-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Eick D, Bensaude O. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res. 2013;41:1591–1603. doi: 10.1093/nar/gks1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Carmichael GG. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski JA, Caputi M. Role of cellular RNA processing factors in human immunodeficiency virus type 1 mRNA metabolism, replication, and infectivity. J Virol. 2009;83:981–992. doi: 10.1128/JVI.01801-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Luo W, Li W, Hoque M, Pan Z, Zhao Y, Tian B. Transcriptional activity regulates alternative cleavage and polyadenylation. Mol Syst Biol. 2011;7:534. doi: 10.1038/msb.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B, Choy E, Cote G, Harmon D, Ye S, Kan Q, Mankin H, Hornicek F, Duan Z. Cyclin-dependent kinase 11 (CDK11) is crucial in the growth of liposarcoma cells. Cancer Lett. 2014;342:104–112. doi: 10.1016/j.canlet.2013.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Karn J, Stoltzfus CM. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med. 2012;2:a006916. doi: 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Okuzaki D, Inoue H, Yoneda Y, Maehara K, Ohkawa Y. Human TREX component Thoc5 affects alternative polyadenylation site choice by recruiting mammalian cleavage factor I. Nucleic Acids Res. 2013;41:7060–7072. doi: 10.1093/nar/gkt414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3' ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh W, Jeong SJ, Lee HJ, Ryu HG, Lee EO, Ahn KS, Bae H, Kim SH. Melatonin promotes puromycin-induced apoptosis with activation of caspase-3 and 5'-adenosine monophosphate-activated kinase-alpha in human leukemia HL-60 cells. J Pineal Res. 2011;50:367–373. doi: 10.1111/j.1600-079X.2010.00852.x. [DOI] [PubMed] [Google Scholar]
- Kowalska E, Ripperger JA, Muheim C, Maier B, Kurihara Y, Fox AH, Kramer A, Brown SA. Distinct roles of DBHS family members in the circadian transcriptional feedback loop. Mol Cell Biol. 2012;32:4585–4594. doi: 10.1128/MCB.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kren BT, Unger GM, Abedin MJ, Vogel RI, Henzler CM, Ahmed K, Trembley JH. Preclinical evaluation of cyclin dependent kinase 11 and casein kinase 2 survival kinases as RNA interference targets for triple negative breast cancer therapy. Breast Cancer Res. 2015;17:19. doi: 10.1186/s13058-015-0524-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenasi T, Contreras X, Peterlin BM. Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe. 2008;4:123–133. doi: 10.1016/j.chom.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang X, Zhang X, Goodrich DW. Human hHpr1/p84/Thoc1 regulates transcriptional elongation and physically links RNA polymerase II and RNA processing factors. Mol Cell Biol. 2005;25:4023–4033. doi: 10.1128/MCB.25.10.4023-4033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyer P, Trembley JH, Grenet JA, Busson A, Corlu A, Zhao W, Kocak M, Kidd VJ, Lahti JM. Characterization of cyclin L1 and L2 interactions with CDK11 and splicing factors: influence of cyclin L isoforms on splice site selection. J Biol Chem. 2008;283:7721–7732. doi: 10.1074/jbc.M708188200. [DOI] [PubMed] [Google Scholar]
- Loyer P, Trembley JH, Katona R, Kidd VJ, Lahti JM. Role of CDK/cyclin complexes in transcription and RNA splicing. Cell Signal. 2005;17:1033–1051. doi: 10.1016/j.cellsig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Luna R, Rondon AG, Aguilera A. New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochim Biophys Acta. 2012;1819:514–520. doi: 10.1016/j.bbagrm.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5'-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajczyk M, Nelson MA. Regulation of stability of cyclin-dependent kinase CDK11p110 and a caspase-processed form, CDK11p46, by Hsp90. Biochem J. 2004;384:461–467. doi: 10.1042/BJ20040848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- Naarmann-de Vries IS, Urlaub H, Ostareck DH, Ostareck-Lederer A. Caspase-3 cleaves hnRNP K in erythroid differentiation. Cell Death Dis. 2013;4:e548. doi: 10.1038/cddis.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima M, Huang Y, Tyagi M, Kao HY, Fujinaga K. The positive transcription elongation factor b is an essential cofactor for the activation of transcription by myocyte enhancer factor 2. J Mol Biol. 2008;382:275–287. doi: 10.1016/j.jmb.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocwieja KE, Sherrill-Mix S, Mukherjee R, Custers-Allen R, David P, Brown M, Wang S, Link DR, Olson J, Travers K, et al. Dynamic regulation of HIV-1 mRNA populations analyzed by single-molecule enrichment and long-read sequencing. Nucleic Acids Res. 2012;40:10345–10355. doi: 10.1093/nar/gks753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak V, Heidecker G, Pathak VK, Derse D. The role of amino-terminal sequences in cellular localization and antiviral activity of APOBEC3B. J Virol. 2011;85:8538–8547. doi: 10.1128/JVI.02645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Peterson SR, Dvir A, Anderson CW, Dynan WS. DNA binding provides a signal for phosphorylation of the RNA polymerase II heptapeptide repeats. Genes Dev. 1992;6:426–438. doi: 10.1101/gad.6.3.426. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell DF, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol. 2004;11:558–566. doi: 10.1038/nsmb759. [DOI] [PubMed] [Google Scholar]
- Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Shi Y, Manley JL. The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev. 2015;29:889–897. doi: 10.1101/gad.261974.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Taube R, Fujinaga K, Irwin D, Wimmer J, Geyer M, Peterlin BM. Interactions between equine cyclin T1, Tat, and TAR are disrupted by a leucine-to-valine substitution found in human cyclin T1. J Virol. 2000;74:892–898. doi: 10.1128/jvi.74.2.892-898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DD, Saran S, Williamson AJ, Pierce A, Dittrich-Breiholz O, Wiehlmann L, Koch A, Whetton AD, Tamura T. THOC5 controls 3'end-processing of immediate early genes via interaction with polyadenylation specific factor 100 (CPSF100) Nucleic Acids Res. 2014;42:12249–12260. doi: 10.1093/nar/gku911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trembley JH, Hu D, Slaughter CA, Lahti JM, Kidd VJ. Casein kinase 2 interacts with cyclin-dependent kinase 11 (CDK11) in vivo and phosphorylates both the RNA polymerase II carboxyl-terminal domain and CDK11 in vitro. J Biol Chem. 2003;278:2265–2270. doi: 10.1074/jbc.M207518200. [DOI] [PubMed] [Google Scholar]
- Valente ST, Gilmartin GM, Venkatarama K, Arriagada G, Goff SP. HIV-1 mRNA 3' end processing is distinctively regulated by eIF3f, CDK11, and splice factor 9G8. Mol Cell. 2009;36:279–289. doi: 10.1016/j.molcel.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J Immunol. 2002;169:1922–1929. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- Verschueren E, Von Dollen J, Cimermancic P, Gulbahce N, Sali A, Krogan NJ. Scoring Large-Scale Affinity Purification Mass Spectrometry Datasets with MiST. Curr Protoc Bioinformatics. 2015;49 doi: 10.1002/0471250953.bi0819s49. 8 19 11–18 19 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Ruegsegger U. 3'-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Yu W, Ramakrishnan R, Wang Y, Chiang K, Sung TL, Rice AP. Cyclin T1-dependent genes in activated CD4 T and macrophage cell lines appear enriched in HIV-1 co-factors. PLoS One. 2008;3:e3146. doi: 10.1371/journal.pone.0003146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, Stein GS. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011;108:9863–9868. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Deng L, Kashanchi F, Brady JN, Shatkin AJ, Kumar A. The Tat/TAR-dependent phosphorylation of RNA polymerase II C-terminal domain stimulates cotranscriptional capping of HIV-1 mRNA. Proc Natl Acad Sci U S A. 2003;100:12666–12671. doi: 10.1073/pnas.1835726100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.