SUMMARY
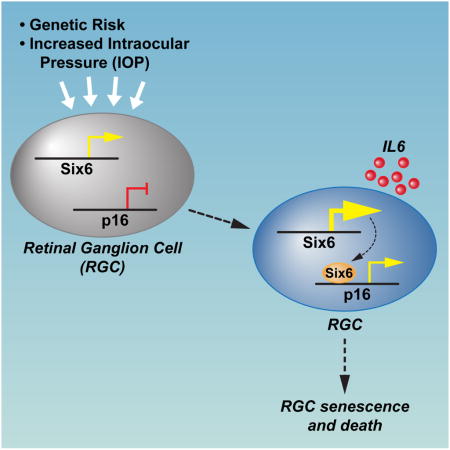
Glaucoma, a blinding neurodegenerative disease, whose risk factors include elevated intraocular pressure (IOP), age and genetics, is characterized by accelerated and progressive retinal ganglion cell (RGC) death. Despite decades of research, the mechanism of RGC death in glaucoma is still unknown. Here, we demonstrate that the genetic effect of the SIX6 risk-variant (rs33912345, His141Asn) is enhanced by another major POAG risk gene P16/INK4A (cyclin-dependent kinase inhibitor 2A). We further show that the upregulation of homozygous SIX6 risk alleles (CC) leads to an increase in P16/INK4A expression with a subsequent cellular senescence, as evidenced in a mouse model of elevated IOP and in human POAG eyes. Our data indicate that SIX6 and/or IOP promotes POAG by directly increasing P16/INK4A expression, leading to RGC senescence in adult human retinas. Our study provides important insights linking genetic susceptibility to the underlying mechanism of RGC death and provides a unified theory of glaucoma pathogenesis.
Graphical abstract
INTRODUCTION
Primary open-angle glaucoma (POAG) is a group of progressive optic neuropathies characterized by a slow and progressive degeneration of retinal ganglion cells (RGCs) and their axons, resulting in a distinct appearance of the optic disc and a concomitant pattern of vision loss (Zhang et al., 2012). POAG is the most frequent type of glaucoma in the western world, one of world’s leading causes of blindness, and the leading cause of blindness among African Americans (Kwon et al., 2009). Intraocular pressure (IOP) and age are the leading risk factors for both the development and progression of POAG.
Recent GWAS studies have indicated that the SIX1-SIX6 and P16/INK4A loci are among the strongest risk genes associated with POAG (Burdon et al., 2011; Osman et al., 2012; Wiggs et al., 2012). SIX6 is a member of the homeodomain transcription factor family that has been shown to be required for proper eye development (Anderson et al., 2012). While several common variants have been identified within this locus, the newly described SIX6 risk variant SIX6-rs33912345 (His141Asn) is strongly associated with POAG (Carnes et al., 2014; Iglesias et al., 2014; Osman et al., 2012), raising an intriguing question about the role of SIX6 in adult retina and in the pathology of POAG.
Expression of P16/INK4A (CDKN2A, cyclin-dependent kinase inhibitor 2A) is an indicator of irreversible growth arrest (senescence) in cultured cells and tissues (Campisi, 2013; Krishnamurthy et al., 2004; Naylor et al., 2013). Senescent cells have been implicated in many age-associated degenerative phenotypes, often through secreted proteins, which are components of Senescence Associated Secretory Phenotype (SASP)(Campisi, 2013). Interestingly, it has been reported that selective elimination of p16/Ink4A-positive senescent cells can prevent or delay age-related deterioration of mouse tissue (Baker et al., 2011). p16/Ink4A expression was found to be significantly elevated in a rat model of glaucoma (Burdon et al., 2011), suggesting a role for p16-dependent pathway in the progression of the disease.
Here, using a combination of genetic association and functional studies, we show that SIX6 risk variant increases P16/INK4A expression and leads to RGC senescence in cell culture, animal models, and human glaucoma retinas. This study provides insights into the mechanism of RGC death in glaucoma and suggests potential avenues for therapeutic intervention in glaucoma patients.
RESULTS
Association of SIX6-rs33912345 with risk of POAG
We performed genetic association studies and identified one missense variant of SIX6 - rs33912345 (NM_007374.2:c.421C>A; NP_031400.2: p.His141Asn) to be strongly associated with POAG. To further investigate the genetic association between SIX6-rs33912345 and POAG in Caucasian population, rs33912345 (C/A) in the SIX6 gene was genotyped using a single-nucleotide primer extension assay in 1130 POAG patients and 4036 controls. The C risk allele frequency of rs33912345 in SIX6 was significantly higher in POAG patients (0.46) than in controls (0.38), allelic P = 4.49E-12, odds ratio (OR) 1.39, 95%, CI 1.27–1.53 (Fig. 1a). We proceeded to perform a replication study using a Mexican cohort with POAG and found a significant association (P=0.11) (Table S1). We then performed meta-analysis using these two cohorts and another cohort reported in the literature (Carnes et al., 2014). Together, the combined results indicate a significant association (allelic P = 4.84E-16, Table S1).
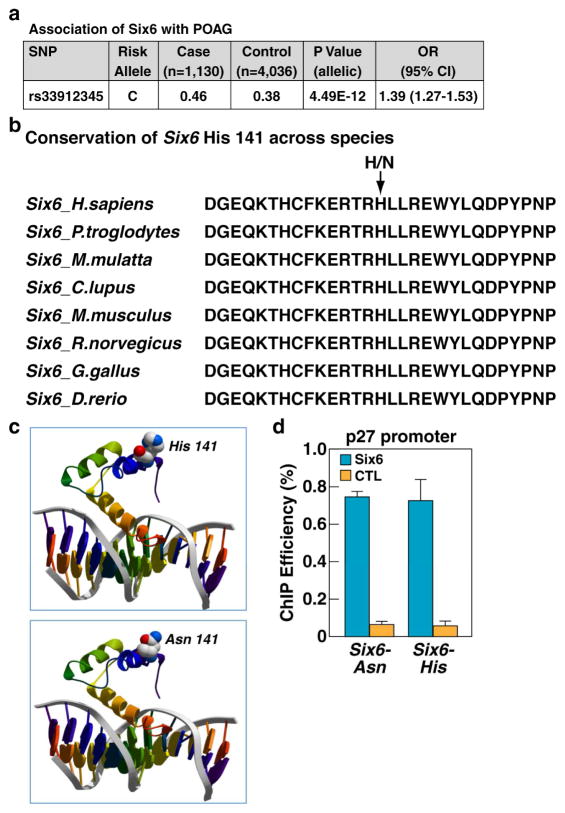
Figure 1. SIX6 protein residue 141 variants bind to DNA with similar efficiency.
a, Association of SIX6 risk variant with POAG. Shown are the frequencies of SIX6 141 amino acid variants in Caucasian population in correlation with POAG. b, Conservation of SIX6 His141 (risk) variant across the species, Ans141 (protective) variant is present only in human lineage. c, Computer modeling of SIX6 structure. Upper panel: model of SIX6 with histidine at position 141; lower panel: model of SIX6 with asparagine at position 141. d, ChIP-qPCR analysis of SIX6 binding to p27 regulatory element in patient-derived lymphoblastoid cells shows similar efficiency of binding of both SIX6 variants. Experiments repeated 3 times, +/− SD. CTL, negative control; See also Figure S1.
The human risk allele His141 in SIX6 is conserved across species (Fig. 1b) and amino acid 141 is located in the homeodomain of the SIX6 protein. 3D modeling indicated that this residue is positioned outside of the DNA-binding surface, and thus is predicted to have no impact on DNA binding (Fig. 1c), but could rather influence the ability of SIX6 to interact with other transcription factors and co-factors. To assess both SIX6 variants in vivo for their efficiency of binding to DNA regulatory elements, we first tested the specificity of our SIX6 antibody by chromatin immunoprecipitation (ChIP) assay on chromatins isolated from wild type (WT) and SIX6 knockout (KO) retinas. We observed that SIX6 protein could efficiently bind to the p27 regulatory element (Fig. S1a), previously reported to be directly recognized by SIX6 (Li et al., 2002). Using the same approach, we observed that both the His141 and Asn141 variants of the SIX6 protein can bind the p27 regulatory element in patient-derived lymphoblastoid cells with similar efficiencies (Fig. 1d). Additionally, we overexpressed HA-tagged versions of SIX6 in HEK293T cells and tested their ability to bind to the p27 regulatory element. ChIP experiments with antibodies specific to SIX6 protein or to the HA-tag demonstrated that both forms of the SIX6 protein (His141 and Asn141) bind efficiently to the p27 regulatory region (Fig. S1b, c). We therefore concluded that the presence of neither variant of residue 141 alters SIX6 binding efficiency to this known DNA-regulatory element, confirming our protein modeling prediction.
Joint effect of SIX6 and P16/INK4A on POAG risk
Since SIX6 and P16 are the two genes showing strongest genetic association with POAG risk, we investigated the joint effect of SIX6-rs33912345 and P16/INK4A-rs3731239 on POAG risk using a logistic regression model and calculated odds ratios (OR). A global two locus (9’2) contingency table, enumerating all 9 two-locus genotype combinations, was constructed. Compared to that of a baseline of non-risk alleles SIX6-rs33912345 AA and P16/INK4A-rs3731239 GG, the OR of the risk alleles SIX6-rs33912345 CC and P16/INK4A- AA is 2.73 (P<0.05, CI (1.63–4.62), Fig. 2a), suggesting their joint effect on POAG risk.
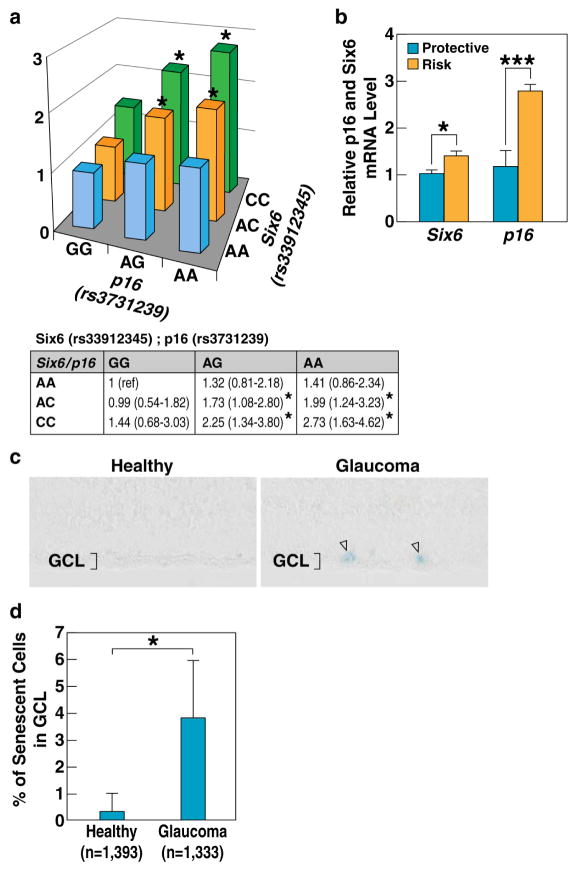
Figure 2. Joint effect of specific alleles of SIX6 (rs33912345) and P16/INK4A (rs3731239) suggest functional interaction between the two genes.
a, Results of the logistic regression analysis, plotted as Z-axis by odds ratios. b, RT-qPCR analysis of mRNA expression of SIX6 and P16/INK4A in human lymphocytes stratified by their SIX6 (rs33912345) genotypes. Four cell lines with rs33912345-AA (non-risk alleles) and four cell lines with rs33912345-CC (risk alleles) were analyzed. Relative mRNA levels were calculated by normalizing results with GAPDH and expressed relative to the AA genotype. p-values were calculated using two-tailed Student’s t-test. (+/− SD; *p<0.05, ***p<0.001). c, SA-βgal staining of human retinas indicating higher numbers of senescent cells in retinas with POAG. d, Quantification of senescent cells in healthy and POAG retinas (*p<0.05); See also Figure S2.
To investigate the correlation between SIX6-rs33912345 and P16/INK4A-rs3731239 genotypes and the expression of SIX6 and P16/INK4A, mRNA levels of both genes were measured in patient-derived human lymphoblastoid cells using reverse transcription followed by quantitative PCR (RT-qPCR). We detected higher SIX6 levels (1.4 fold) in cell lines carrying the SIX6 risk allele. In addition, the expression of P16/INK4A mRNA was 2.3-fold higher in cells with risk genotype as compared to the cells with protective genotype (Fig. 2b). We further measured the levels of P16/INK4A mRNA in retinas from healthy and glaucoma patients and observed significantly elevated expression in glaucoma eyes (Fig. S2b). To investigate whether the elevated expression of P16/INK4A correlated with increased cellular senescence, a senescence associated β-galactosidase (SA-βgal) assay was performed on healthy and glaucoma human retinas. Consistently, retinas from glaucoma patients exhibited elevated senescence as indicated by significantly higher number of blue-positive cells in the ganglion cell layer (GCL) (Fig. 2c, d and Fig. S2a).
Transcriptional regulation of P16/INK4A by Six6
To investigate whether SIX6 is involved in transcriptional regulation of P16/INK4A, we transiently expressed SIX6-His141 or SIX6-Asn141 variants in human fetal retinal progenitor cells (fRPCs) and quantified P16/INK4A mRNA levels using RT-qPCR. Significantly higher expression of P16/INK4A mRNA was observed in SIX6-His141 transfected cells (Fig. 3a). Similar trends were observed when SIX6-His141 and SIX6-Asn141 were overexpressed at similar levels in HEK293T cells (Fig. 3b, c), which suggests that the effect of the variation in SIX6 is not cell-type specific. To test whether both HA-tagged SIX6 protein variants can bind directly to P16/INK4A promoter, we performed ChIP assays using anti-SIX6 and anti-HA antibodies. We observed that both protein variants bind to P16/INK4A promoter with similar efficiency (Fig. 3d and not shown). Taken together, this data suggests that SIX6 can act as a direct activator of P16/INK4A gene, and that the SIX6-His141 variant has more potential to activate P16/INK4A expression.
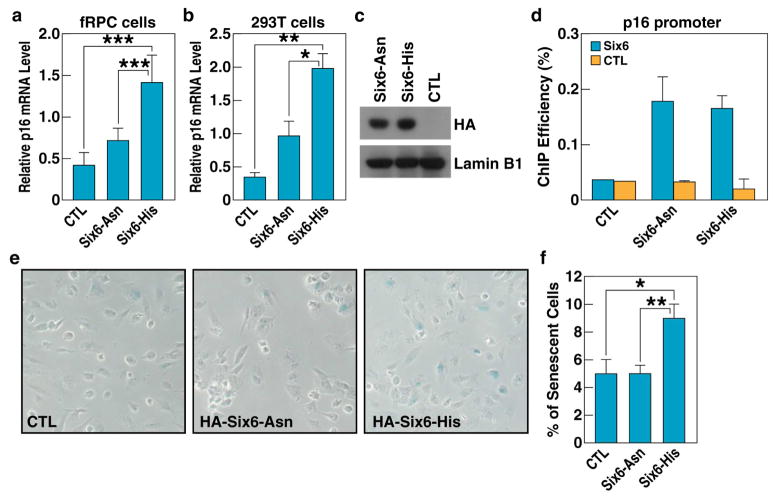
Figure 3. Increased expression of SIX6-risk variant correlates with a higher senescence rate.
a, RT-qPCR analysis shows that overexpression of SIX6-His variant increased p16/INK4A expression in fRPCs. Experiments repeated 3 times, p-values were calculated using two-tailed Student’s t-test. (+/− SD, ***p<0.001). CTL, negative control b, RT-qPCR analysis shows that the overexpression of the SIX6-His variant increased P16/INK4A expression in 293T cells. Experiments repeated 3 times, p-values calculated using a two-tailed Student’s t-test. (+/− SD; *p<0.05, **p<0.01). CTL, negative control c, Western-blot confirming similar levels of expression of both SIX6 variants in transient transfections experiments. CTL, negative control d, ChIP-qPCR analysis of SIX6 variant association with P16/INK4A promoter, showing similar level of binding. CTL, negative control e, SA-βgal staining of the fRPCs transfected with either of the two SIX6 variants showing higher ratio of senescence in cells transfected with SIX6-141His risk variant. CTL, negative control f, Quantification of β-galactosidase-positive cells in fRPCs transfected with SIX6 variants. p-values calculated using a two-tailed Student’s t-test. (+/− SD; *p<0.05, **p<0.01). CTL, negative control; See also Figure S3.
Given that elevated expression of P16/INK4A is a hallmark of cellular senescence (Campisi, 2013; Naylor et al., 2013), we tested whether transient overexpression of either variant of SIX6 (SIX6-His141 and SIX6-Asn141) could induce senescence in fRPCs. Interestingly, merely 24h post-transfection, fRPC cells expressing SIX6-His141 variant underwent senescence twice as readily as those under control conditions or those transfected with SIX6-Asn141 variant, as assessed by a SA-β-gal assay and by upregulation of IL6, a secretory marker of senescence (Fig. 3e, f and Fig. S3). Taken together, these data indicate that SIX6-His141 risk variant increased senescence in fRPC by direct induction of P16/INK4A expression.
Up-regulation of SIX6 and P16/INK4A in a mouse model of acute glaucoma
Experimental ocular hypertension mouse models have been used extensively to study the relationship between IOP and the mechanism of glaucomatous optic neuropathy (Gross et al., 2003). To investigate the association of glaucoma in the context of SIX6-His risk variant and P16/INK4A expression, we used a mouse glaucoma model in which the IOP is increased acutely. First, we analyzed the sequence of mouse Six6 gene and noticed that mouse genome encodes only the Six6-His variant (Fig. 1b). Western blot analysis confirmed that SIX6 was expressed in the adult retina at comparable levels to those observed in the embryonic stages (Fig. 4a). When both Six6 and P16/INK4A mRNA levels were measured 5 days post IOP elevation, both Six6 and P16/INK4A mRNA expression levels were significantly higher in retinas from IOP-treated eyes as compared to those from control eyes (Fig. 4b). We also noted increased expression of p15/CDKN2B and p19/ARF upon IOP elevation (Fig. S4a, b).
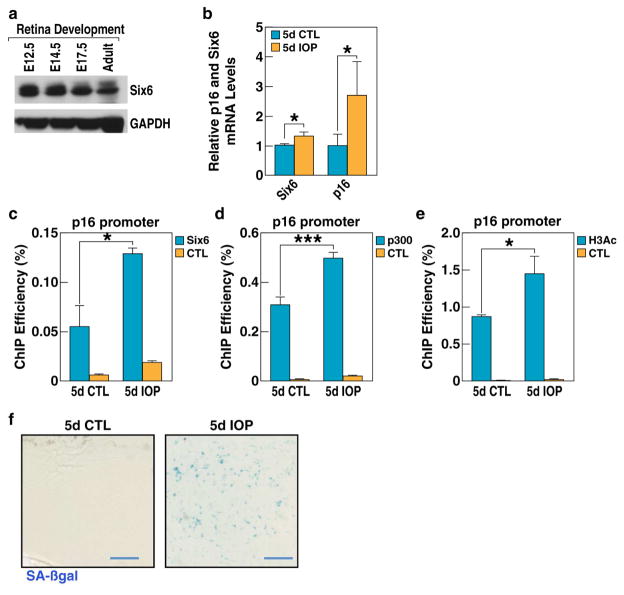
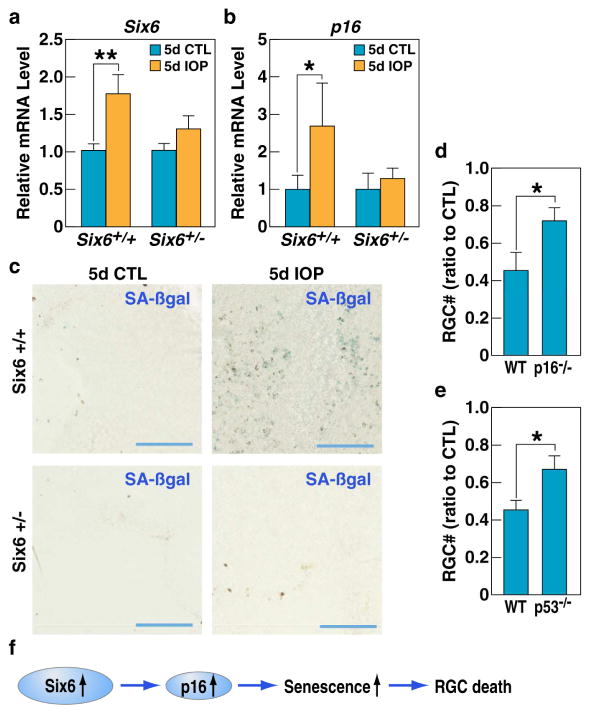
Figure 4. Increased expression of SIX6 and induction of cell senescence in retinas upon IOP elevation.
a, The expression of SIX6 protein in mouse retina during development and in the adult stage analyzed by Western blotting. b, RT-qPCR analysis of Six6 and P16/INK4A mRNA levels shows elevated expression of SIX6 and P16/INK4A in IOP-elevated mouse retinas 5 days after induction of acute experimental glaucoma (5d IOP) as compared to non-treated retina (5d CTL). Experiments were repeated in 8 animals, p-values calculated using a two-tailed Student’s t-test. (+/− SD; *p<0.05). c, ChIP-qPCR analysis of SIX6 protein binding shows its higher association with the P16/INK4A promoter in retinas subjected to acute intraocular pressure increase (5d IOP) as compared to non-treated retina (5d CTL). Experiments repeated 3 times, p-values calculated using a two-tailed Student’s t-test. (+/− SD; *p<0.05). CTL, negative control. d, ChIP-qPCR analysis shows higher levels of p300 association with P16/INK4A promoter upon experimental glaucoma (5d IOP) as compared to non-treated retina (5d CTL). Experiments repeated 3 times, p-values calculated using a two-tailed Student’s t-test. (+/− SD; ***p<0.001). CTL, negative control. e, ChIP-qPCR shows higher level of H3 acetylation at P16/INK4A promoter after acute intraocular pressure increase (5d IOP) as compared to non-treated retina (5d CTL). Experiments repeated 3 times, p-values calculated using a two-tailed Student’s t-test (+/− SD; *p<0.05). CTL, negative control. f, SA-βgal staining of flat mount retinas isolated from treated and non-treated eyes. High number of senescent cells is evident in IOP treated tissue. See also Figure S4.
Since SIX6 binds to P16/INK4A promoter upon engineered expression in cell culture (Fig. 3d), we asked if Six6 could also bind to this promoter in adult mouse retina in vivo. To test this, we compared ChIP efficiency in chromatins isolated from wild-type and Six6 KO retinas. We found that detectable signal could only be observed in wild-type retinas, and not in Six6 −/− retinas (Fig. S4c). Further, we investigated whether Six6 binding to p16/Ink4A promoter in the retina was altered by IOP elevation. We performed a ChIP-qPCR assay in chromatins isolated from IOP-elevated and control retinas and observed that binding of Six6 to the p16/Ink4A promoter was significantly elevated upon increased IOP (Fig. 4c). This phenomenon correlated with elevated recruitment of histone acetyltransferase p300, a known co-activator of p16/Ink4A expression (Fig. 4d) (Wang et al., 2008), and increased pan-acetylation of histone H3, a modification that is associated with active regulatory elements (Fig. 4e) (Strahl and Allis, 2000). Importantly, there was no recruitment of Six6 to p19ARF or p15/Cdkn2B promoters upon IOP elevation (Fig. S4d), suggesting that the effect of Six6 on p16/Ink4A is gene-specific. We then used a SA-β-gal assay to test if the cells in the treated retinas had undergone senescence. As expected, in 5 of 5 mice tested, we observed a dramatic accumulation of senescent cells in the IOP treated retinas, as compared to only a few senescent cells observed in untreated retinas (Fig 4f, Fig. S4e).
We further investigated whether it was the RGCs that undergo senescence in IOP treated retinas. As expected, images of the retinal cross-sections showed that most of the senescent cells were localized in the GCL (Fig. 5a). We performed immunohistochemistry using an anti-Brn3a antibody (an RGC marker) on IOP-treated and non-treated flat-mount retinas and detected that most of the β-galactosidase positive cells were also BRN3a positive (Fig. 5b). We further confirmed these findings by comparing the β-gal staining pattern in Thy1-CFP transgenic mice, in which the RGCs are specifically marked by CFP fluorescence (Fig. 5c and Fig. S5a) (Lindsey et al., 2013).
Figure 5. IOP elevation primarily affects retinal ganglion cells.
a, SA-βgal staining of cross-sections in IOP treated (5d IOP) or non-treated (5d CTL) retinas. Senescent cells are localized in the ganglion cell layer (GCL). b, Double staining of IOP treated (5d IOP) or non-treated (5d CTL) flat mount retinas with SA-βgal and BRN3a antibodies. Most senescent cells are also BRN3a positive. c, Immunostaining of IOP-treated Thy1-CFP retinas using anti-GFP antibody. Majority of SA-βgal positive cells are also Thy1-CFP positive. d, Schematic diagram of immunopanning, e, His but not the Asn version of Six6 significantly upregulates p16/INK4A expression as compared to non-transfected (RGC-CTL) or GFP-transfected (RGC-GFP) purified rat retinal ganglion cells. p-values calculated using a two-tailed Student’s t-test (+/− SD; *p<0.05). See also Figure S5.
We next investigated the effects of engineered expression of SIX6 variants on the expression of p16/Ink4A and senescence associated secretory phenotype marker, IL6, in cultured RGCs. RGCs from rat retina were isolated by immunopanning using anti-Thy-1 antibody (Winzeler and Wang, 2013) (Fig. 5d). The efficacy of this procedure was verified by cell morphology and Brn3a expression levels in isolated RGCs as compared to whole retina cells (Fig. S5d, e). Significantly, we observed increased expression of p16 and IL6 only in the RGCs in which we introduced the His variant of Six6 (Fig. 5e, Fig. S5f, g). Consistently, there were remarkably more IL6-positive RGCs in the IOP-treated retinas than in controls (Fig. S5b, c). Taken together, these data suggest that RGCs are the primary cells affected in this glaucoma model.
Lack of either Six6 or p16 protects against RGC death in glaucoma
The homozygous Six6 knockout adult retinas display numerous abnormalities (Clark et al., 2013), especially in the RGC layer. Therefore, we decided to use heterozygous mice to test the role of SIX6 in induced senescence and in the regulation of p16/Ink4A expression. As before, acute intraocular pressure elevation was applied and retinas were collected and assayed 5 days post IOP. In contrast to the wild-type littermates, reduced Six6 expression was accompanied by decreased P16/INK4A expression upon IOP elevation in Six6+/− retinas (Fig. 6a, b). Interestingly, in Six6+/− mice no β-gal positive cells could be observed (Fig. 6c, Fig. S6a), suggesting that haploinsufficiency of Six6 was sufficiently protective against senescence in RGCs.
Figure 6. Absence of either Six6 or P16 protects against RGC death in glaucoma.
a, b, RT-qPCR analysis of Six6 (a) and P16/INK4A (b) mRNA levels upon IOP treatment (5d IOP) shows elevated expression of Six6 and P16/INK4A only in wild-type (Six6+/+) retinas and not in Six6+/− retinas as compared to non-treated (5d CTL) retinas. Experiments repeated in 8 animals, p-values calculated using a two-tailed Student’s t-test. (+/− SD; *p<0.05, **p<0.01). c, SA-β-galactosidase staining of flat mount retinas isolated from IOP-treated and non-treated eyes of Six6+/+ and Six6+/− mice shows a lack of senescent cells in the treated tissue isolated from heterozygous mice (blue bar). d, Quantification of the RGC ratio in treated and non-treated retinas in WT and P16 −/− mice. e, Quantification of the RGC ratio in IOP treated and non-treated retinas in WT and p53 KO mice. f, Model of the sequence of events leading to RGC death upon Six6 upregulation in glaucoma. See also Figure S6.
Together, these results suggest a model in which increased p16/Ink4A expression is a major cause of cellular senescence in glaucoma. We then test whether the lack of p16/Ink4A expression in p16/Ink4A−/− mice could prevent RGC death in our acute glaucoma model. Indeed, consistent with the above data, absence of p16/Ink4a expression protected against RGC death (Fig. 6d).
Since P53 has been reported to play an important role in senescence (Campisi, 2013), we asked whether the lack of P53 can prevent RGC death caused by IOP elevation. To test this, we compared the numbers of RGCs in IOP-treated mouse retinas from p53−/− mice to IOP treated wild-type mice. We found that the lack of p53 significantly attenuated RGC death upon IOP when compared to wild type mice (Fig. 6e). We also observed a genetic association of a missense variant in P53 (rs1042522 (NM_000546.5:c.215C>G; NP_000537.3: p.Pro72Arg) with POAG, consistent with its role in glaucoma pathogenesis (Fig. S6b).
Taken together, these findings lead us to propose a model (Fig. 6f) in which the IOP elevation causes the upregulation of P16/INK4A through increased expression of SIX6 (in particular the His variant) and it’s binding to the p16/INK4A promoter. Increased p16/INK4A expression causes RGCs to enter cellular senescence. Prolonged senescence can cause increased retinal ganglion cell death and consequent blindness.
DISCUSSION
Glaucoma is the leading cause of blindness affecting tens of millions of people worldwide. Despite its prevalence, its etiology and pathogenesis are poorly understood and treatment is limited to lowering IOP. Despite aggressive IOP lowering therapies, most patients have progressive loss of visual function and some will eventually become legally blind. The relationship of raised IOP and RGC death is poorly understood.
Cellular senescence is a state of irreversible growth arrest. When senescent cells accumulate in the tissue, their impaired function can result in a predisposition to disease development and/or progression (Baker et al., 2011; Burton, 2009; Campisi, 2013). In the current study, we show that SIX6 directly regulates expression of P16/INK4A, an indicator of cell senescence and aging. Further, we show that upon acute IOP elevation, P16/INK4A expression is up-regulated, which, in turn, can be a cause of RGC death. Therefore P16/INK4A up-regulation appears to be a downstream integrator of diverse signals such as inherited genetic risk, age and other factors, such as raised IOP. Our hypothesis can help explain how IOP, the most common risk factor, can cause glaucoma. Moreover, it provides a molecular link between genetic susceptibility and other factors to the pathogenesis of glaucoma.
Our study suggests that cellular senescence plays a critical role in the pathogenesis of glaucoma. Consistent with this notion, we show that another key player in cellular senescence, P53 (Campisi, 2013) also contributes to RGC death, as evidenced by protection of RGC from IOP induced damage in mice lacking p53. Additionally, we provide data showing increased expression of secretory molecules, components of SASP, upon IOP-induced retinal damage. Moreover, we recently demonstrated that induction of IL1 is an early response to IOP induced RGC damage (Chi et al., 2014). Induction of IL1, in turn, is known to cause activation of NF-kB dependent senescence associated expression of IL6 and IL8 (Orjalo et al., 2009). Our data thus suggests that the senescence-associated cytokine network is activated in IOP-treated retinas.
P16 is a cyclin-dependent kinase inhibitor and a potent negative regulator of cell cycle progression. Consequently, upregulated P16/INK4A expression and senescence phenotype, as measured by SAβ-gal assay and SASP, usually indicate the irreversible cell cycle arrest (Campisi, 2013). In our study however, elevated p16 expression and senescence was observed in retinal ganglion cells (post-mitotic neurons), which are believed not to be replication-competent. Therefore, one possibility is that RGCs may contain a replication-competent, stem cell-like population; alternatively, p16 may play a previously unrecognized role in these post mitotic cells.
Although the primary focus of the current study is on the role of P16/INK4A on RGC death, our data do not exclude a potential role for other genes located in the 9p21 locus in the pathology of glaucoma. For example, p14ARF may contribute to glaucoma pathogenesis independently by influencing eye vasculature development (McKeller et al., 2002). Further studies are required to elucidate role of p14ARF and p15 in the pathogenesis of glaucoma.
SIX6 is a member of the SIX/Sine oculis family of homeobox transcription factors involved in the development of retina (Gallardo et al., 1999; Lopez-Rios et al., 1999). It has been shown to directly regulate expression of cyclin-dependent kinase inhibitor genes (Li et al., 2002) as well as GnRH (Larder et al., 2011) during mouse development. Although the role of SIX6 during retina development is being investigated by a number of laboratories, there are very few reports exploring its molecular role in adult retina or in glaucoma pathogenesis. Several SIX6 mutations and single nucleotide polymorphisms (SNPs) have been shown to correlate with developmental eye defects in human (Aldahmesh et al., 2013; Gallardo et al., 1999; Gallardo et al., 2004). Additionally, several SNPs have been associated with an increased risk of glaucoma (Carnes et al., 2014; Iglesias et al., 2014). The effects of SIX6 variants had always been assessed after late identification of the disease and this approach does not allow for correct dissociation of developmental defects from the genetic components that cause the disease.
The SIX6 variant referred to as “risk” (histidine encoding SIX-His) is actually evolutionary conserved across the phyla. The Asn variant (the protective allele for glaucoma) is detected only in the human branch; however, it is unknown why it would be advantageous for humans to have a protective variant. Several groups performed rescue experiments by overexpressing human His variant in zebrafish and found that it did not rescue eye defects induced by the removal of endogenous SIX6 proteins. Interestingly, both zSix6a and zSix6b carry the His amino acid at the orthologous position (Carnes et al., 2014; Iglesias et al., 2014). These results suggest that the inability to rescue eye phenotypes by human His version of SIX6 is likely the result of species-specific differences in other residues and not the sole effect of His/Asn variant.
Taken together, our study shows that SIX6 His variant increases P16/INK4A expression upon increased IOP, which in turn causes RGCs to enter into a senescent state and which may lead to increased RGC death in glaucoma. Our study provides important insights into the pathogenesis of glaucoma and suggests future therapeutic strategies based on targeted inhibition of P16/INK4A-induced cell senescence to prevent and treat glaucoma.
Experimental Procedures
Subjects and test procedure
This study was approved by the institutional review boards of the University of California, San Diego, the University of Southern California, and West China Hospital. All participants signed informed consent statements prior to participation. Blood was drawn from a vein in the patient’s arm into blood collection tubes containing the anticoagulant acid citrate dextrose. Genomic DNA was extracted from peripheral blood leukocytes with the Qiagen kit, according to the manufacturer’s instructions (Qiagen Inc., Chatsworth, CA, USA). For the initial cohort, 1130 POAG patients and 4036 Controls were enrolled in this study. All participants were of European descent. For the Mexican cohort, 105 POAG patients and 188 Controls were enrolled in this study.
Clinical definitions
Clinical assessment was performed using previously described methods (Wiggs et al., 2012). POAG cases were defined as individuals for whom reliable visual field (VF) tests showed characteristic VF defects consistent with glaucomatous optic neuropathy. Individuals were classified as affected if the VF defects were reproduced on a subsequent test or if a single qualifying VF was accompanied by a cup-disc ratio (CDR) of 0.7 or more in at least one eye. The examination of the ocular anterior segment did not show signs of secondary causes for elevated IOP such as exfoliation syndrome or pigment dispersion syndrome and the filtration structures were deemed to be open based on clinical measures. Controls had normal optic nerves (cup-disc ratios ≤ 0.6) and normal intraocular pressure (Wiggs et al., 2012).
Genotyping and statistical analysis
Genomic DNA was extracted from peripheral blood leukocytes with the Qiagen kit (Qiagen Inc., Chatsworth, CA, USA), according to the manufacturer’s instructions. rs33912345 (C/A) in SIX6, rs3731239 (A/G) in P16/INK4A and rs1042522 (C/G) in TP53 were genotyped using single-nucleotide primer extension assay (ABI Prism SNaPShot Multiplex Kit, Applied Biosystems) on an ABI 3130xl genetic analyzer as previously described (Yang et al., 2006). The primers used for genotyping are listed in Table S2. SNP genotyping results were screened for deviation from Hardy-Weinberg equilibrium using Chi-squared tests. Allele association was calculated via Chi-squared test performed using the program PLINK (Purcell et al., 2007) (http://pngu.mgh.harvard.edu/purcell/plink/). Asymptomatic p-values were also generated for this test.
Joint effects of SIX6-rs33912345 and P16/INK4A-rs3731239
Joint effects of SIX6-rs33912345 (C/A) and P16/INK4A-rs3731239 (A/G) were calculated by a logistic regression model (Chen et al., 2010; Chen et al., 2011). A global two locus (9’2) contingency table, enumerating all 9 two-locus genotype combinations, was constructed. Odds ratios and 95% confidence intervals, comparing each genotypic combination to the baseline of homozygosity for the common allele at both loci, were calculated, according to the previously described methods (Chen et al., 2010; Chen et al., 2011).
3D modeling of the SIX6 H141N variation
We performed a computer modeling analysis to investigate the effects of the SIX6 His141Asn variation on the three-dimensional structure and function of SIX6 with the ICM software (Cardozo et al., 1995). The SIX6 transcription factor has never been crystallized for X-ray analysis or studied by NMR. To build a structural model of the DNA-binding domain around the H141N variation, we searched the Protein Data Bank for homologous transcription factors. The search identified the following entries with fragments of related transcription factors: 2dmu, 1qry, 1vnd, 1yrn, 2lkx, and 2 solved by crystallography or NMR. Then we attempted a structural superposition of the identified domains. All had a structurally (topologically) similar DNA-binding motif, covering the area from residues around 134 to 189. The structural homology model of SIX6 was built for residues from 134 to 189 with the ICM software (Cardozo et al., 1995) by threading the SIX6 sequence onto the consensus structure.
Lymphoblastoid cell lines culture
The human lymphoblastoid cell lines were generated by the method “General protocol for the immortalization of human B-lymphocytes using EBV” (http://www.unclineberger.org/tissueculture/protocols/b-lymphocytesprotocol). Four lymphoblastoid cell lines with the genotypes of SIX6 rs33912345-CC and another four with the genotypes of SIX6 rs33912345-AA were used. The cells were cultured in RPMI-1640 base medium (61870127, Invitrogen) supplemented with 20% FBS and 1×Penicillin-Streptomycin (P0781, Sigma). RNA was extracted and the total mRNA levels of SIX6 and P16/INK4A were measured by qRT-PCR.
Acute IOP mouse model
Unilateral elevation of IOP in 1 month old C57BL/6J, p16 −/−, Thy1-CFP (Lindsey et al., 2013), Six6+/− (Larder et al., 2011), p53−/− (129-Trp53tm1Tyj/J, The Jackson Laboratory) mice was achieved through instilling the anterior chamber with saline solution and maintained at an elevated pressure of 90 mm Hg. The p16−/− line was engineered to remove the common exon of E2 and E3, therefore the line is a null allele for both p16 and p19/Arf genes (Serrano et al., 1996). IOP was measured by a tonometer. All procedures were conducted with the approval and under the supervision of the Institutional Animal Care Committee at the University of California San Diego and adherence to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. C57BL/6J male mice were purchased from The Jackson Laboratories (Bar Harbor, ME). To elevate IOP experimentally, animals were first anesthetized with a weight-based intraperitoneal injection of ketamine (80 mg/kg), xylazine (16 mg/kg). Additional anesthesia was provided via the same route at 45-minute intervals. Corneal anesthesia was achieved with a single drop of 0.5% proparacaine hydrochloride (Alcon Laboratories, Inc.). A drop of 0.5% proparacaine hydrochloride and 0.5% tropicamide (Alcon Laboratories, Inc., Fort Worth, TX) was then applied to the right eye. Body temperature was maintained between 37°C and 38°C, with a water-heat pad (TP500T/Pump; Gaymar Industries, Orchard Park, NY). A 30-gauge needle was used to puncture the mid-peripheral cornea of right eye; the anterior chambers were cannulated with sterile physiologic saline (balanced salt solution; BSS; Alcon Laboratories, Inc.) and IOP was manometrically controlled by adjusting the saline height. IOP was monitored with an indentation tonometer (Tonolab; Icare, Espoo, Finland) and maintained at 90 mm Hg pressure for one hour. Assessment of the senescence of RGCs was made in retinal flat mounts harvested 5 days after experiments. In brief, mice were euthanized with CO2, and eye balls were dissected and fixed in ice-cold phosphate buffered 4% paraformaldehyde, pH7.4, for 30 minutes, followed by flat mounting of the retinas.
SA-β-galacosidase assay to test senescence on retinas from human and IOP mouse eyes
Senescence assays were performed using the Senescence β-Galactosidase Staining Kit (Cell Signaling) according to the manufacturer’s protocol. For double staining, senescence assays were followed by anti-GFP antibody staining to reveal full fluorescence of Thy1-CFP retinas.
RNA extraction and real-time PCR
Total RNA extraction from mouse tissues or human lymphoblastoid cells, cDNA synthesis and qRT-PCR experiments were performed as previously described (Luo et al., 2013). Assays were performed in triplicate. Relative mRNA levels were calculated by normalizing results using GAPDH. The primers used for qRT-PCR are listed in Table S2. The differences in quantitative PCR data were analyzed with independent two-sample t-test.
Human fRPC isolation and expansion
All work with human material was performed with IRB approval. fRPCs were isolated from human fetal neural retina at 16 weeks gestational age as previously described (Luo et al., 2014). Whole neuroretina was separated from the RPE layer, minced, and digested with collagenase I (Sigma-Aldrich). Cells and cell clusters were plated onto human fibronectin (Akron)-coated flasks (Nunclon Delta) in Ultraculture Media (Lonza), supplemented with 2 mM L-glutamine (Invitrogen), 10 ng/ml rhbFGF (Peprotech), and 20 ng/ml rhEGF (Peprotech) in a low-oxygen incubator (37°C, 3% O2, 5% CO2, 100% humidity). Cells were passaged at 80% confluency using TrypZean (Sigma-Aldrich), benzonase (EMD Chemicals), and Defined Trypsin Inhibitor (Invitrogen).
Immunopanning
The immunopanning procedure and retinal ganglion cell culture was performed according to the published detailed protocol (Winzeler and Wang, 2013).
Plasmid transfection into HEK293 and fRPC cells
Overexpression was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer standard protocol. Empty or GFP containing vector was used as a negative control. For ChIP and expression analysis experiments, cells were harvested 48h post-transfection. For senescence analysis in fRPCs, cells were collected after 24h.
Chromatin immunoprecipitation (ChIP)
Chromatin immuno-precipitation was performed as previously described (Skowronska-Krawczyk et al., 2004; Skowronska-Krawczyk et al., 2009). Briefly, cells or dissected retinas were fixed for 10min with 1% formaldehyde; chromatin was isolated and sonicated to obtain fragments 300–700 bp in length. After adding antibodies, immunoprecipitation was carried out overnight and protein/DNA complexes were pulled out using protein G coated magnetic beads (Invitrogen). After 3 washes, DNA cross-linking was reversed. The DNA was then purified and subjected to qPCR. All experiments were repeated at least 3 times. P-values were calculated using student’s t-tests.
Antibodies
Antibodies used in this study include: guinea pig anti-SIX6 was a gift from Dr. Xue Li’s lab, rabbit anti-SIX6 (ab64995, Abcam), anti-H3Ac (06-599, Millipore), anti-HA (1867431 Roche), anti p-300 (C-20, Santa Cruz), anti-Brn3a (MAB1585, Millipore), anti-GFP (GFP-1020, Aves) and anti-IL6 (ab83339, Abcam).
Supplementary Material
Highlights.
SIX6 and P16/INK4A contribute to POAG genetic risk synergistically.
SIX6 directly regulates P16/INK4A expression.
SIX6 and P16/INK4A promote retinal ganglion cell senescence and death.
Acknowledgments
This study was supported in part by 973 program (2015CB94600 and 2013CB967504), NIH/NEI, State Key laboratory of Ophthalmology; and NIHR01 HD072754 (P.L.M.). We thank J. Hightower for figure preparation. D.S.-K. was supported by EMBO Long-Term Fellowship, The Swiss National Science Foundation, and The San Diego Foundation.
Footnotes
Authors Contribution
K.Z., D.S-K. and L.Z. designed the study. D.S.-K., L.Z., J.Z., R.N.W., J.L., K.F., S.P., C.W., M.K., H.L., H.O., D.L., W.W., G.L., Y.X., G.C., O.L., C.C., E.Y., M.J., M.A., Z.Z., W.S., L.Z., M.K., D.C., C.S., C.Z., J.Z., P.L.M., W.G., L.Z., X.S., S.Z., Y.Z., M.G.R., Y.L., K.Z. performed experiments, provided reagents and patients cohorts, analyzed results. K.Z., D.S.-K., L.Z. and R.N.W. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldahmesh MA, Khan AO, Hijazi H, Alkuraya FS. Homozygous truncation of SIX6 causes complex microphthalmia in humans. Clinical genetics. 2013;84:198–199. doi: 10.1111/cge.12046. [DOI] [PubMed] [Google Scholar]
- Anderson AM, Weasner BM, Weasner BP, Kumar JP. Dual transcriptional activities of SIX proteins define their roles in normal and ectopic eye development. Development. 2012;139:991–1000. doi: 10.1242/dev.077255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon KP, Macgregor S, Hewitt AW, Sharma S, Chidlow G, Mills RA, Danoy P, Casson R, Viswanathan AC, Liu JZ, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nature genetics. 2011;43:574–578. doi: 10.1038/ng.824. [DOI] [PubMed] [Google Scholar]
- Burton DG. Cellular senescence, ageing and disease. Age (Dordr) 2009;31:1–9. doi: 10.1007/s11357-008-9075-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. Aging, cellular senescence, and cancer. Annual review of physiology. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Totrov M, Abagyan R. Homology modeling by the ICM method. Proteins. 1995;23:403–414. doi: 10.1002/prot.340230314. [DOI] [PubMed] [Google Scholar]
- Carnes MU, Liu YP, Allingham RR, Whigham BT, Havens S, Garrett ME, Qiao C, Katsanis N, Wiggs JL, Pasquale LR, et al. Discovery and functional annotation of SIX6 variants in primary open-angle glaucoma. PLoS genetics. 2014;10:e1004372. doi: 10.1371/journal.pgen.1004372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Bedell M, Zhang K. Age-related macular degeneration: genetic and environmental factors of disease. Mol Interv. 2010;10:271–281. doi: 10.1124/mi.10.5.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zeng J, Zhao C, Wang K, Trood E, Buehler J, Weed M, Kasuga D, Bernstein PS, Hughes G, et al. Assessing susceptibility to age-related macular degeneration with genetic markers and environmental factors. Archives of ophthalmology. 2011;129:344–351. doi: 10.1001/archophthalmol.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Li F, Chen H, Wang Y, Zhu Y, Yang X, Zhu J, Wu F, Ouyang H, Ge J, et al. Caspase-8 promotes NLRP1/NLRP3 inflammasome activation and IL-1beta production in acute glaucoma. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11181–11186. doi: 10.1073/pnas.1402819111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DD, Gorman MR, Hatori M, Meadows JD, Panda S, Mellon PL. Aberrant development of the suprachiasmatic nucleus and circadian rhythms in mice lacking the homeodomain protein Six6. Journal of biological rhythms. 2013;28:15–25. doi: 10.1177/0748730412468084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo ME, Lopez-Rios J, Fernaud-Espinosa I, Granadino B, Sanz R, Ramos C, Ayuso C, Seller MJ, Brunner HG, Bovolenta P, et al. Genomic cloning and characterization of the human homeobox gene SIX6 reveals a cluster of SIX genes in chromosome 14 and associates SIX6 hemizygosity with bilateral anophthalmia and pituitary anomalies. Genomics. 1999;61:82–91. doi: 10.1006/geno.1999.5916. [DOI] [PubMed] [Google Scholar]
- Gallardo ME, Rodriguez De Cordoba S, Schneider AS, Dwyer MA, Ayuso C, Bovolenta P. Analysis of the developmental SIX6 homeobox gene in patients with anophthalmia/microphthalmia. American journal of medical genetics Part A. 2004;129A:92–94. doi: 10.1002/ajmg.a.30126. [DOI] [PubMed] [Google Scholar]
- Gross RL, Ji J, Chang P, Pennesi ME, Yang Z, Zhang J, Wu SM. A mouse model of elevated intraocular pressure: retina and optic nerve findings. Transactions of the American Ophthalmological Society. 2003;101:163–169. discussion 169–171. [PMC free article] [PubMed] [Google Scholar]
- Iglesias AI, Springelkamp H, van der Linde H, Severijnen LA, Amin N, Oostra B, Kockx CE, van den Hout MC, van Ijcken WF, Hofman A, et al. Exome sequencing and functional analyses suggest that SIX6 is a gene involved in an altered proliferation-differentiation balance early in life and optic nerve degeneration at old age. Human molecular genetics. 2014;23:1320–1332. doi: 10.1093/hmg/ddt522. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. The Journal of clinical investigation. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YH, Fingert JH, Kuehn MH, Alward WL. Primary open-angle glaucoma. The New England journal of medicine. 2009;360:1113–1124. doi: 10.1056/NEJMra0804630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder R, Clark DD, Miller NL, Mellon PL. Hypothalamic dysregulation and infertility in mice lacking the homeodomain protein Six6. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:426–438. doi: 10.1523/JNEUROSCI.1688-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science. 2002;297:1180–1183. doi: 10.1126/science.1073263. [DOI] [PubMed] [Google Scholar]
- Lindsey JD, Duong-Polk KX, Dai Y, Nguyen DH, Leung CK, Weinreb RN. Protection by an oral disubstituted hydroxylamine derivative against loss of retinal ganglion cell differentiation following optic nerve crush. PloS one. 2013;8:e65966. doi: 10.1371/journal.pone.0065966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rios J, Gallardo ME, Rodriguez de Cordoba S, Bovolenta P. Six9 (Optx2), a new member of the six gene family of transcription factors, is expressed at early stages of vertebrate ocular and pituitary development. Mechanisms of development. 1999;83:155–159. doi: 10.1016/s0925-4773(99)00017-9. [DOI] [PubMed] [Google Scholar]
- Luo J, Baranov P, Patel S, Ouyang H, Quach J, Wu F, Qiu A, Luo H, Hicks C, Zeng J, et al. Human retinal progenitor cell transplantation preserves vision. The Journal of biological chemistry. 2014;289:6362–6371. doi: 10.1074/jbc.M113.513713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Zhao L, Chen AY, Zhang X, Zhu J, Zhao J, Ouyang H, Luo H, Song Y, Lee J, et al. TCF7L2 variation and proliferative diabetic retinopathy. Diabetes. 2013;62:2613–2617. doi: 10.2337/db12-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeller RN, Fowler JL, Cunningham JJ, Warner N, Smeyne RJ, Zindy F, Skapek SX. The Arf tumor suppressor gene promotes hyaloid vascular regression during mouse eye development. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3848–3853. doi: 10.1073/pnas.052484199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor RM, Baker DJ, van Deursen JM. Senescent cells: a novel therapeutic target for aging and age-related diseases. Clinical pharmacology and therapeutics. 2013;93:105–116. doi: 10.1038/clpt.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J. Cell surface-bound IL-1alpha is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17031–17036. doi: 10.1073/pnas.0905299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman W, Low SK, Takahashi A, Kubo M, Nakamura Y. A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Human molecular genetics. 2012;21:2836–2842. doi: 10.1093/hmg/dds103. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D, Ballivet M, Dynlacht BD, Matter JM. Highly specific interactions between bHLH transcription factors and chromatin during retina development. Development. 2004;131:4447–4454. doi: 10.1242/dev.01302. [DOI] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D, Chiodini F, Ebeling M, Alliod C, Kundzewicz A, Castro D, Ballivet M, Guillemot F, Matter-Sadzinski L, Matter JM. Conserved regulatory sequences in Atoh7 mediate non-conserved regulatory responses in retina ontogenesis. Development. 2009;136:3767–3777. doi: 10.1242/dev.033449. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Wang X, Pan L, Feng Y, Wang Y, Han Q, Han L, Han S, Guo J, Huang B, Lu J. P300 plays a role in p16(INK4a) expression and cell cycle arrest. Oncogene. 2008;27:1894–1904. doi: 10.1038/sj.onc.1210821. [DOI] [PubMed] [Google Scholar]
- Wiggs JL, Yaspan BL, Hauser MA, Kang JH, Allingham RR, Olson LM, Abdrabou W, Fan BJ, Wang DY, Brodeur W, et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS genetics. 2012;8:e1002654. doi: 10.1371/journal.pgen.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler A, Wang JT. Purification and culture of retinal ganglion cells from rodents. Cold Spring Harbor protocols. 2013;2013:643–652. doi: 10.1101/pdb.prot074906. [DOI] [PubMed] [Google Scholar]
- Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- Zhang K, Zhang L, Weinreb RN. Ophthalmic drug discovery: novel targets and mechanisms for retinal diseases and glaucoma. Nature reviews Drug discovery. 2012;11:541–559. doi: 10.1038/nrd3745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.