Abstract
C-reactive protein (CRP), an acute-phase plasma protein, is a major component of inflammatory reactions functioning as a mediator of innate immunity. It has been widely used as a validated clinical biomarker of the inflammatory state in trauma, infection, and age-associated chronic diseases, including cancer and cardiovascular disease (CVD). Despite this, the molecular mechanisms that regulate CRP expression are not well understood. Given that the CRP 3′ untranslated region (UTR) is long and AU rich, we hypothesized that CRP may be regulated posttranscriptionally by RNA-binding proteins (RBPs) and by microRNAs. Here, we found that the RBP HuR bound directly to the CRP 3′ UTR and affected CRP mRNA levels. Through this interaction, HuR selectively increased CRP mRNA stability and promoted CRP translation. Interestingly, treatment with the age-associated inflammatory cytokine interleukin-6 (IL-6) increased binding of HuR to CRP mRNA, and conversely, HuR was required for IL-6-mediated upregulation of CRP expression. In addition, we identified microRNA 637 (miR-637) as a microRNA that potently inhibited CRP expression in competition with HuR. Taken together, we have uncovered an important posttranscriptional mechanism that modulates the expression of the inflammatory marker CRP, which may be utilized in the development of treatments for inflammatory processes that cause CVD and age-related diseases.
INTRODUCTION
Inflammatory processes and their inherent regulatory controls are critical for the immune response to injury and pathogens throughout the life span. However, inflammation has now been identified as an important underlying factor in many chronic diseases, including cardiovascular disease (CVD), diabetes mellitus, cancer, and metabolic disorders. Age itself is a critical factor in the development of the inflammatory state and risk for these conditions. This age-associated inflammatory state, known as inflammaging, is defined as a state of low-grade, sterile inflammation that occurs with age and is characterized by elevations of serum concentrations of proinflammatory cytokines, including interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α), as well as the acute-phase reactant C-reactive protein (CRP) (1). Given the incidence, morbidity, and mortality of inflammation-based chronic disease, proinflammatory molecules, including CRP, are avidly studied.
CRP, a pentraxin protein, is an established marker of acute-phase reactions (2). It is an important inflammatory biomarker that is influenced by the action of numerous activated cytokines, such as IL-6, IL-1β, and TNF-α (3, 4). It is well established that circulating levels of CRP and IL-6 are correlated in humans (5, 6). CRP has been widely used as a validated clinical biomarker of the inflammatory state and an independent predictor of cardiovascular disease. There is some evidence that CRP is not only a biomarker of cardiovascular and metabolic disease but also a specific risk factor for disease, with some data supporting the idea that CRP is an active participant in atherogenesis and events at the endothelium (7). In diabetes, CRP contributes to the development of insulin resistance and may thus be an etiologic factor in diabetes mellitus type 2, especially in the elderly (8). In cancer, CRP may play any of three possible roles: as a marker of cancer susceptibility in the setting of chronic inflammation, as a marker of occult cancer, or as a causal factor (9). Currently, anti-inflammatory clinical trials in the setting of cardiovascular disease and other inflammatory conditions focus on modulating CRP production by inhibiting TNF-α and IL-6, thereby reducing hepatic production of the protein (10). Even though CRP plays a central role in aging and age-related disease, most of the molecular mechanisms that regulate CRP expression are not known.
Several studies have focused on the transcriptional regulation of CRP. The CRP promoter contains consensus sequences for the transcription factors STAT3 and C/EBPβ, which activate CRP transcription downstream of IL-6 signaling (11–14). In addition, it has been shown that NF-κB p50 and Oct-1 bind to the CRP gene promoter via a nonconsensus κB site, which also overlaps the proximal C/EBP site (15, 16). IL-1β, which alone does not induce CRP expression in human hepatoma Hep3B cells, synergistically enhances the effects of IL-6 by activating the transcription factor NF-κB (17, 18). The HNF-1 and HNF-3 transcription factors are also involved in regulating CRP expression via IL-6 (12, 19). Although the transcriptional modulation of CRP has been explored, we have limited knowledge of the posttranscriptional mechanisms that regulate CRP expression.
Mammalian cells regulate gene expression robustly via posttranscriptional mechanisms controlled by RNA-binding proteins (RBPs) and microRNAs (miRNAs), two types of major etiologic factors in disease (20, 21). In addition, these factors are being evaluated for the diagnosis and management of disease. Human antigen R (HuR) is a ubiquitously expressed RBP belonging to the Hu/Elav family that modulates the stability, translation, and localization of subsets of mRNAs by interacting with uridylate (U)-rich or adenylate-uridylate (AU)-rich elements in their 3′ untranslated regions (UTRs) (22, 23). By regulating the expression of specific sets of proteins, HuR critically influences a variety of processes, such as cell proliferation and survival as well as immune and stress responses (24, 25). HuR modulates inflammatory responses by promoting the expression of proinflammatory proteins such as COX-2, IL-8, transforming growth factor β (TGF-β), and TNF-α and by inhibiting the expression of anti-inflammatory proteins such as IL-10 (26, 27). HuR is implicated in inflammatory diseases, including rheumatoid arthritis, asthma, and inflammatory bowel disease, as well as in cardiovascular disease, cancer, and neurodegenerative diseases. HuR also plays a role in cellular senescence and vascular aging by controlling the turnover and/or translation of mRNAs encoding SIRT1, TNF-α, ICAM1, and VCAM1 (28–30).
miRNAs are small noncoding RNAs that modulate gene expression posttranscriptionally by binding to target mRNAs bearing regions of partial complementarity to the miRNA (31). By affecting the expression of target mRNAs, miRNAs regulate a variety of important processes, including cell proliferation, cellular senescence, and inflammation (32, 33), and various age-related diseases such as CVD, neurodegeneration, and metabolic diseases (34). Given the fact that the CRP 3′ UTR contains AU-rich elements, we hypothesized that posttranscriptional regulatory factors bind and regulate CRP expression. Here, we present evidence that CRP expression is regulated via a competitive interaction between the RBP HuR and miRNA 637 (miR-637) with the CRP 3′ UTR.
MATERIALS AND METHODS
Cell culture and small RNA transfection.
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS). HepG2 cells were maintained in minimal essential medium (MEM) containing 10% FBS. Where indicated, cells were treated with 50 ng/ml IL-6 (R&D Systems), 2 μg/ml actinomycin D (EMD Millipore), or the vehicle control (phosphate-buffered saline [PBS] and dimethyl sulfoxide [DMSO], respectively). Control small interfering RNA (siRNA) and HuR siRNA (5′-AAGAGGCAATTACCAGTTTCA-3′) were obtained from Qiagen. The pre-miRNA precursor control, the miR-637 and anti-miRNA inhibitor controls, and miR-637 were purchased from Ambion. All siRNAs, miRNAs, and plasmids were transfected by using either Lipofectamine-2000 (Invitrogen) or Lipofectamine-RNAiMAX (Invitrogen). RNA and protein were isolated from the cells 48 h after transfection.
Ribonucleoprotein immunoprecipitation assay.
For ribonucleoprotein (RNP) immunoprecipitation (RIP), HepG2 cells or HeLa cells (24 h after transfection with miR-637) (see Fig. 5B) were lysed in a solution containing 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, and 0.5% NP-40 for 10 min on ice and centrifuged at 10,000 × g for 15 min at 4°C. The supernatants were incubated with mouse IgG-agarose beads (Sigma) and coated with anti-HuR (Santa Cruz Biotechnology) or with normal mouse IgG (Santa Cruz Biotechnology) antibodies for 1 h at 4°C. After repeated washing with ice-cold NT2 buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40), the complexes were incubated with RNase-free DNase I for 10 min at 30°C and subsequently incubated with 0.1% SDS–0.5 mg/ml proteinase K for 15 min at 55°C. The RNA from the immunoprecipitation (IP) samples was extracted by using acidic phenol, precipitated in isopropanol, and analyzed by reverse transcription and quantitative real-time PCR (RT-qPCR).
FIG 5.
HuR and miR-637 regulate CRP expression competitively. (A) Control siRNA or HuR siRNA and either biotinylated miR-637 (bio-miR-637) or the control (bio-miR-Ctrl) were transfected into HeLa cells for 24 h. Biotinylated miRNAs were precipitated by using streptavidin beads. Enrichment of CRP mRNA was assessed by RT-qPCR. (B) Twenty-four hours after transfection of HeLa cells with miR-Ctrl or miR-637, RIP followed by RT-qPCR analysis was used to measure the enrichment of CRP mRNA in HuR IP compared to IgG IP. (C) Effects of silencing of HuR together with miR-637 on CRP expression. HeLa cells were cotransfected with HuR siRNA and miR-637; 48 h later, lysates were assessed for the level of CRP mRNA by RT-qPCR analysis (left) and the levels of HuR, CRP, and the loading control β-actin by immunoblot analysis (right). CRP protein levels were quantified from immunoblots and normalized to β-actin levels. (D) Forty-eight hours after HeLa cells were cotransfected with Flag-HuR and miR-637, CRP mRNA levels were measured by RT-qPCR and normalized to GAPDH mRNA levels. (E) HepG2 cells were transfected with HuR siRNA or miR-637 and then treated with IL-6 (50 ng/ml) for 24 h or left untreated. The CRP mRNA levels were examined by RT-qPCR. The histograms show the means and standard errors of the means from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant (as determined by Student's t test [A to D] and by one-way analysis of variance and Tukey's post hoc test [E]).
RNA isolation and RT-qPCR analysis.
Total RNA was isolated from cells by using TRIzol (Invitrogen) according to the manufacturer's instructions. After RT using random hexamers (Invitrogen) and SuperScript II reverse transcriptase (Invitrogen), the abundance of transcripts was assessed by qPCR analysis using 2× SYBR green master mix (Applied Biosystems). A QuantiMir RT kit (System Biosciences) was used for cDNA synthesis for miRNAs, U6 snRNA, and snoRNAs. RT-qPCR analysis was performed on an Applied Biosystems model 7500 real-time PCR machine. The following primer pairs were used (forward and reverse, respectively, in each case): AGACATGTCGAGGAAGGCTTTT and TCGAGGACAGTTCCGTGTAGAA for CRP, GTGACATCGGGAGAACGAAT and GCGGTCACGTAGTTCACAAA for ELAVL1 (HuR), GCTCCTCCTGTTCGACAGTCA and ACCTTCCCCATGGTGTCTGA for the glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH), CCCTATCAACTTTCGATGGTAGTCG and CCAATGGATCCTCGTTAAAGGATTT for 18S, AGATGGTCAAGGTCGCAAGCT and GGGCATATCCTACAACAAACTTGTC for HPRT1, ATTTGGGTCGCGGTTCTTG and TGCCTTGACATTCTCGATGGT for UBC, TACAAGTACCTCACCGCTTGGT and TGATCTTGTCTTGGTGCTCGTA for the Renilla luciferase gene (RL), and CTAAGAAGGGCCTGCAGAAGAT and AAGCCCTGGTAGTCGGTCTTAG for the firefly luciferase gene (FL). To measure the abundance of miRNAs, the following forward primers were used: GCGATAACTGACGAAGACTAC for RNU49, TTAAACCACCAAGATCGCTGA for RNU24, GATATCACTGTAAAACCGTTCC for U47, ACTGGGGGCTTTCGGGCTCTGCGT for miR-637, and CACCACGTTTATACGCCGGTG for U6. The universal primer supplied by the QuantMir RT kit was used as the reverse primer. For peripheral blood mononuclear cell (PBMC) analysis, HuR expression was normalized to the average HPRT and UBC expression levels by using gene-specific primers. The miR-637 expression levels were normalized to the average expression levels of snoRNAs RNU24, RNU49, and U47.
Biotin pulldown analysis.
CRP 3′-UTR fragments a, b, c, and d were amplified from the psiCHECK2 luciferase vectors containing the respective 3′ UTRs by using primers that contained the T7 RNA polymerase promoter sequence (5′-CCAAGCTTCTAATACGACTCACTATAGGGAGA-3′) (35). After purification of the template PCR products, biotinylated transcripts were synthesized by using the MaxiScript T7 kit (Ambion). Whole-cell lysates were incubated with 1 μg of purified biotinylated transcripts for 1 h at room temperature, and complexes were then isolated with streptavidin-coupled Dynabeads M-280 (Invitrogen). The proteins present in the pulldown material were analyzed by immunoblotting with anti-HuR antibodies (Santa Cruz Biotech). The biotinylated GAPDH 3′ UTR was used as a negative control. The following primer pairs were used: (T7)AGCTGTGGGTCCTGAAGGTA and AAGTAAACAGGGGCTTTATT for fragment a, (T7)AGCTGTGGGTCCTGAAGGTA and AGAAATTATCTCCAAGATCT for fragment b, (T7)GATAATTTCTTACCTCACAT and ATTTATACCTAGTGCTTCAT for fragment c, (T7)ATGAAGCACTAGGTATAAAT and AAGTAAACAGGGGCTTTATT for fragment d, and (T7)CCTCAACGACCACTTTGTCA and GGTTGAGCACAGGGTACTTTATT for the GAPDH 3′-UTR fragment.
3′-UTR luciferase reporter assays.
The cDNA fragments corresponding to the entire 3′ UTR and partial 3′ UTR of human CRP mRNAs were amplified by PCR using specific primers. After XhoI and NotI digestion, the PCR product was cloned downstream of the Renilla open reading frame of the psiCHECK2 reporter plasmid. The psiCHECK2-CRP (3′-UTR-Mut) construct containing specific point mutations (CC to GG) in the miR-637-binding site was generated by using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. For Fig. 1E, 24 h after transfection with 200 ng of the reporter constructs, RIP assays were performed as described above, with the exception that complexes were incubated with RNase-free DNase I for 1 h at 30°C. HuR binding to the ectopic luciferase transcripts was determined by RT-qPCR analysis using primers specific for the RL and FL mRNAs. HeLa cells were transfected with 100 ng of the indicated 3′-UTR luciferase reporter constructs and transfected again 24 h later with the control and HuR siRNAs or miR-637. Twenty-four hours later, RL and FL activities were measured by using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's instructions, or RL mRNA and FL mRNA levels were determined by RT-qPCR analysis.
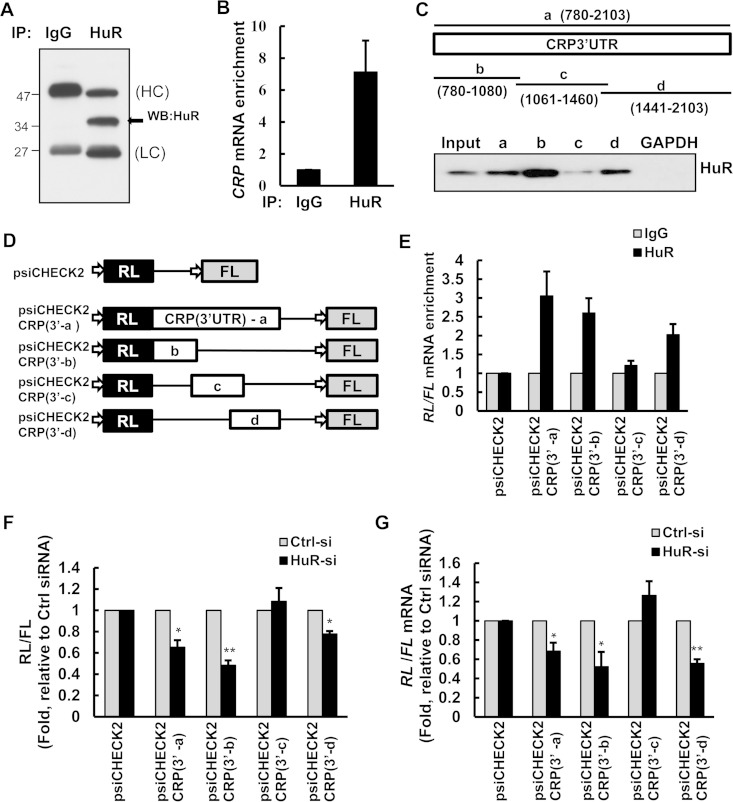
FIG 1.
HuR binds to the CRP 3′ UTR. (A) HuR immunoprecipitates from HepG2 cells were analyzed by immunoblotting. HuR (arrowhead), the immunoglobulin heavy chain (HC) and light chain (LC), and molecular weight markers are indicated. WB, Western blotting. (B) RIP followed by RT-qPCR analysis was used to measure the enrichment of CRP mRNA in HuR IP compared to IgG IP in HepG2 cell lysates, normalized to GAPDH mRNA levels for each IP reaction. (C, top) Schematic of the CRP 3′ UTR and the different biotinylated RNAs synthesized for use in biotin pulldown analyses (full length and fragments a, b, c, and d). (Bottom) The indicated biotinylated CRP RNAs or the control GAPDH 3′ UTR was incubated with HeLa cell lysates, and HuR was detected by immunoblotting with anti-HuR antibodies. (D) Schematic of the CRP 3′-UTR dual-luciferase reporters. (E) HeLa cells transfected with the luciferase constructs were analyzed by RIP and RT-qPCR to measure the enrichment of Renilla luciferase (RL) mRNA in HuR IPs compared to that in IgG IPs. RL mRNA levels in each IP were normalized to firefly luciferase (FL) mRNA levels and to GAPDH mRNA levels. (F) After transfection of HeLa cells with control (Ctrl) or HuR siRNAs, cells were transfected with either psiCHECK2 or the psiCHECK2-CRP (3′-UTR) reporters, as indicated. The ratio of RL to FL was normalized to the value for the parent vector (psiCHECK2) and then normalized to luciferase activity in control siRNA-transfected cells. (G) Cells were transfected as described for panel E, and the ratio of RL mRNA/FL mRNA for each reporter construct was quantified by RT-qPCR. Histograms represent the means and standard errors of the means from three independent experiments. *, P < 0.05; **, P < 0.01 (compared to control siRNA, as determined by Student's t test).
Polysome analysis.
For polysome analysis, 48 h after transfection of HeLa cells with control siRNA or HuR siRNA, cells were incubated with 100 μg/ml cycloheximide for 10 min and lysed with PEB (polysome extraction buffer) containing 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, and 0.5% NP-40. Cytoplasmic lysates were fractionated by centrifugation through 10 to 50% linear sucrose gradients and divided into 12 fractions. The total RNA in each fraction was extracted with TRIzol (Invitrogen) and analyzed by reverse transcription followed by RT-qPCR analysis.
Biotinylated miR-637 pulldown assays.
Twenty-four hours after transfection of HeLa cells with biotinylated miR-637 or with biotinylated control miRNA from Caenorhabditis elegans cel-miR-67 (biot-miR-637 and biot-Ctrl-miR, respectively; both from GE Healthcare Dharmacon), HeLa cells were lysed in a solution containing 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.3% NP-40, 50 U of RNase Out (Invitrogen), and a protease cocktail inhibitor (Roche Applied Science) for 10 min on ice and centrifuged at 10,000 × g for 10 min at 4°C. Streptavidin Dynabeads (Invitrogen) were preincubated in lysis buffer with yeast tRNA (1 mg/ml) and bovine serum albumin (BSA) (1 mg/ml) overnight at 4°C. Cytoplasmic lysates were added to the beads, and the beads were incubated for 4 h at 4°C. After the beads were washed with lysis buffer, the RNA bound to the beads and the RNA in cytoplasmic extracts were isolated by using TRIzol (Invitrogen) as described above and analyzed by RT-qPCR.
Western blot analysis.
Cells were washed twice with 1× cold PBS and lysed by using 2× Laemmli sample buffer. The cell lysates were fractionated by SDS-PAGE, transferred onto membranes, and analyzed by using primary antibodies that recognized HuR (Santa Cruz Biotechnology), CRP (Millipore), or the loading control β-actin (Santa Cruz Biotechnology). Following incubation with appropriate secondary antibodies, signals were detected by using enhanced chemiluminescence (ECL).
Clinical study participants.
A subcohort of women with either low (≤3 mg/liter) or high (≥20 mg/liter) high-sensitivity CRP (hsCRP) levels (n = 15/group) were chosen from the Healthy Aging in Neighborhoods of Diversity across the Life Span (HANDLS) study of the National Institute on Aging Intramural Research Program (NIA IRP), National Institutes of Health (NIH). This study has been approved by the NIEHS Institutional Review Board (IRB). All study participants signed informed-consent documents approved by the IRB. Previously, we examined CRP levels in a larger cohort containing these women (36), and here, we analyzed a subset of these participants from whom we had also obtained and stored PBMCs. Fifteen white and 15 African American females with an average age (± standard deviation) of 49.7 ± 8.1 years were used for this study. RNA was isolated from PBMCs by using TRIzol (Invitrogen) according to the manufacturer's instructions. Serum IL-6 levels were quantified by using Searchlight protein arrays from Aushon Biosystems (Billerica, MA).
RESULTS
The RNA-binding protein HuR interacts with the 3′ UTR of CRP mRNA.
To investigate whether CRP mRNA was associated with HuR, we performed ribonucleoprotein (RNP) immunoprecipitation (RIP) assays with anti-HuR antibodies in parallel with control IgG antibodies using the human hepatoma cell line HepG2, chosen because hepatic cells are a major source of circulating CRP in vivo. The interaction of HuR with CRP mRNA was assessed by the isolation of RNA in IP material (Fig. 1A), followed by reverse transcription (RT) and quantitative real-time PCR (qPCR) analysis to measure the levels of CRP mRNA in the HuR IP relative to the control IgG IP. As shown in Fig. 1B, CRP mRNA was highly enriched (∼>7-fold) in HuR IP samples compared with IgG IP samples. To identify the area(s) of interaction of endogenous HuR with ectopic CRP mRNA, partial in vitro-transcribed biotinylated RNAs spanning the CRP 3′ UTR (Fig. 1C) were incubated with HeLa cell lysates. After pulldown using streptavidin-coated beads, the association of HuR with each biotinylated transcript was assessed by immunoblotting. As shown, HuR associated with CRP 3′-UTR fragments a, b, and d and most strongly with fragment b. HuR did not bind to a negative-control transcript, the biotinylated GAPDH 3′ UTR (Fig. 1C).
To confirm that HuR binds to the CRP 3′ UTR in vivo, we used heterologous luciferase reporter plasmids (Fig. 1D) that express Renilla luciferase (RL) from constructs lacking or containing the CRP 3′ UTR (psiCHECK2 and psiCHECK2-CRP 3′UTR, respectively). These plasmids also contain firefly luciferase (FL), which served as an internal transfection control. We used RIP assays to test the binding of HuR to the ectopically expressed luciferase transcripts. As shown, HuR selectively associated with RL mRNAs containing CRP 3′-UTR fragments a, b, and d (Fig. 1E), further confirming our in vitro binding data.
To investigate if HuR regulates CRP mRNA stability and translation via the specific regions where it binds preferentially within the CRP 3′ UTR (regions b and d) (Fig. 1C), we first analyzed the luciferase reporter constructs. The ratio of RL activity/FL activity from each transfected reporter plasmid indicated that silencing of HuR significantly decreased the levels of psiCHECK2-CRP 3′-a, -b, and -d luciferase reporters, while it did not affect the activity of the psiCHECK2-CRP 3′-c reporter (Fig. 1F). Consistent with the strong HuR binding in the biotin precipitation experiments (Fig. 1C), the most robust regulation of luciferase activity by HuR was mapped to CRP segment 3′-b (Fig. 1F). To verify if the individual HuR binding sites (a,b, and d) regulate the stability and/or translation of the CRP mRNA, we examined the ratio of the reporter mRNA levels after silencing HuR. HuR silencing decreased the levels of RL mRNA expressed from psiCHECK2-CRP 3′-a, -b, and -d, indicating that HuR primarily affects the mRNA stability of these individual binding sites (Fig. 1G). Taken together, these data strongly suggest that HuR enhances CRP expression by associating with specific positive regulatory elements in the CRP 3′ UTR to regulate CRP mRNA stability.
HuR promotes CRP expression by regulating CRP mRNA stability and translation.
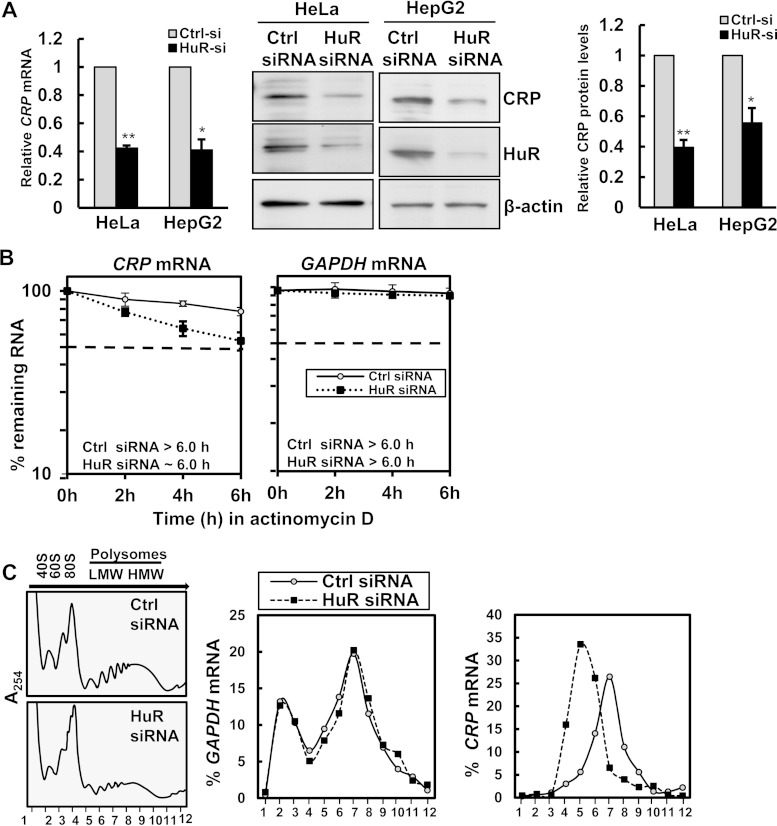
We tested whether HuR directly affected CRP expression by silencing HuR using specific HuR-directed small interfering RNAs (siRNAs). Silencing of HuR decreased CRP mRNA and protein levels (Fig. 2A), indicating that HuR positively regulates CRP expression. Because HuR was previously shown to stabilize several target mRNAs (27) and the CRP 3′-UTR reporter constructs shown in Fig. 1G, we investigated whether HuR regulates endogenous CRP mRNA turnover. After silencing HuR in HeLa cells, actinomycin D was used to inhibit de novo transcription, and the time needed for CRP mRNA to be reduced to 50% of its initial abundance (its half-life [t1/2]) was then calculated by measuring CRP mRNA levels by RT-qPCR, using 18S rRNA levels for normalization. We found that silencing of HuR selectively decreased the t1/2 of CRP mRNA to ∼6 h, compared to its half-life in control siRNA-transfected cells, which was far higher than 6 h. The half-life of GAPDH mRNA, encoding a housekeeping protein, was unaffected by HuR silencing (Fig. 2B). These data suggest that HuR enhances CRP expression at least in part by stabilizing CRP mRNA.
FIG 2.
HuR promotes CRP mRNA stability and translation. (A, left) Forty-eight hours after transfection of HeLa and HepG2 cells with either control siRNA or HuR siRNA, CRP mRNA levels were measured by RT-qPCR analysis. (Right) Transfected HepG2 cells or HeLa cells were assessed by immunoblotting using anti-CRP, anti-HuR, and anti-β-actin antibodies as a loading control. CRP protein levels were quantified from immunoblots and normalized to the levels of actin. Histograms represent the means and standard errors of the means from three independent experiments. *, P < 0.05; **, P < 0.01 (compared to control siRNA, as determined by Student's t test). (B) HeLa cells transfected as described for panel A were treated with actinomycin D (2 μg/ml), and RNA was isolated at the times indicated. The CRP mRNA and GAPDH mRNA levels were assessed by RT-qPCR and normalized to 18S rRNA levels. The t1/2 values for CRP and GAPDH mRNAs were quantified by measuring the time needed for the transcript levels to reach 50% of their original abundance at time zero. (C) HeLa cells transfected with either control siRNA or HuR siRNA were fractionated into cytoplasmic extracts through sucrose gradients. The arrow indicates the direction of sedimentation. Small (40S) and large (60S) ribosomal subunits and monosomes (80S) in fractions 2 to 4 and progressively larger polysomes from low to high molecular weights in fractions 6 to 12 are shown at the left. The distribution (percent) of CRP and GAPDH mRNAs was quantified by RT-qPCR analysis of RNA isolated from 12 gradient fractions.
In addition to stabilizing some target mRNAs, HuR also modulates the translation of several mRNAs (22). To investigate if HuR also affects CRP translation, we performed polysome analysis in HeLa cells expressing normal HuR levels or HuR levels reduced by silencing. The cytosolic fractions were centrifuged through sucrose gradients in order to separate ribosome components (40S and 60S [fractions 2 and 3]), monosomes (single ribosomes) (80S [fraction 4]), as well as low-molecular-weight (LMW) (fractions 5 to 7) and high-molecular-weight (HMW) (fractions 8 to 10) polysomes. RNA was extracted from each of the 12 fractions obtained, and the levels of CRP and GAPDH mRNAs were quantified by RT-qPCR analysis. The results showed that in control cells, CRP mRNA levels peaked at fraction 7, while HuR silencing showed a distinct leftward shift, peaking at fraction 5, indicating that CRP mRNA formed on average smaller polysomes after silencing of HuR (Fig. 2C). The distribution of the housekeeping GAPDH mRNA was the same between the two groups, indicating that silencing of HuR specifically affected CRP mRNA translation. In sum, HuR increases CRP expression levels by both stabilizing CRP mRNA and enhancing its translation.
IL-6 upregulation of CRP is dependent on HuR.
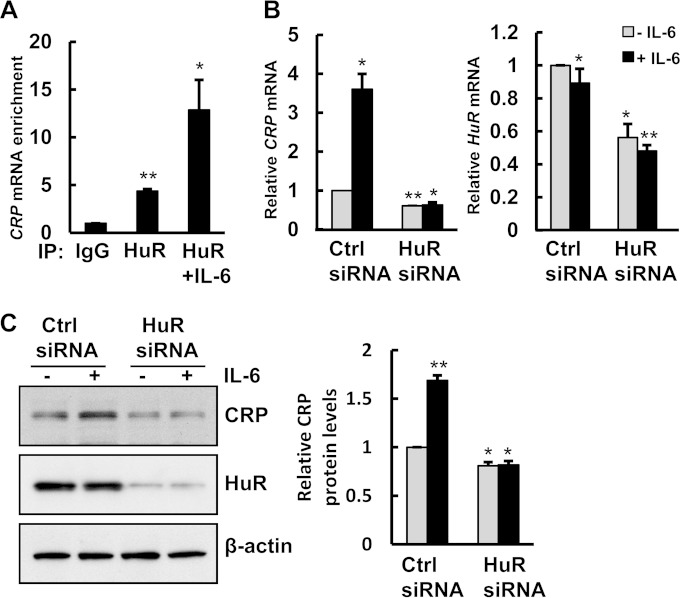
IL-6 is a potent mediator of inflammatory processes, and it has been proposed that the age-associated increase in IL-6 levels accounts for some of the phenotypic changes observed with advancing age. IL-6 and CRP levels increase with age and are highly correlated (37). In addition, previously reported data showed that IL-6 can upregulate CRP levels (14). Given this relationship, we hypothesized that HuR may contribute to the increased CRP mRNA expression levels in response to IL-6. To test this idea, we treated HepG2 cells with IL-6 and then performed a HuR RIP assay. This analysis revealed that more HuR was associated with CRP mRNA after treatment with IL-6 (Fig. 3A). To address whether HuR is required for IL-6-mediated upregulation of CRP, we silenced HuR and treated cells with IL-6 (Fig. 3B and C). HuR silencing significantly decreased CRP mRNA and protein levels in the presence of IL-6. In contrast, HuR mRNA and protein levels were not affected by IL-6 treatment. These findings indicate that IL-6-mediated upregulation of CRP is dependent on HuR.
FIG 3.
IL-6-mediated upregulation of CRP is dependent on HuR. (A) After IL-6 (50 ng/ml) treatment for 15 min, RIP analysis was performed by using HuR or IgG antibodies. CRP mRNA levels were measured by RT-qPCR analysis. (B and C) HepG2 cells were transfected with either control siRNA or HuR siRNA and treated with or without IL-6 for 24 h. The mRNA and protein levels of CRP and HuR were examined by RT-qPCR (B) and immunoblotting (C). CRP protein levels were quantified from immunoblots and normalized to values for the β-actin control. The histograms represent the means and standard errors of the means from three independent experiments. *, P < 0.05; **, P < 0.01 (by Student's t test).
miR-637 interacts with the CRP mRNA.
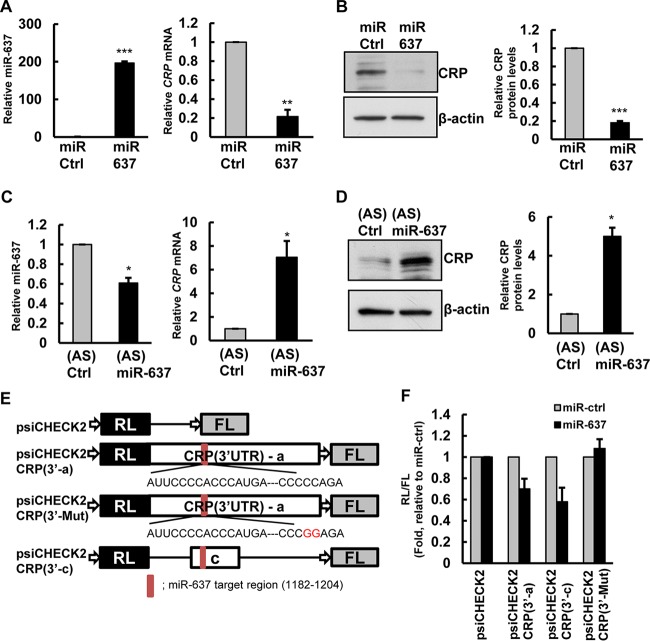
Recently, several studies showed that RBPs and miRNAs bind and functionally regulate shared target mRNAs (21, 25, 35). Therefore, in addition to HuR, it is likely that miRNAs also regulate CRP. Using TargetScan to test this possibility, we found that the CRP mRNA contains one predicted miR-637 site in its 3′ UTR. To investigate whether miR-637 regulates CRP expression, we overexpressed miR-637 by transfecting the miR-637 precursor and found that this intervention decreased CRP mRNA and protein abundances (Fig. 4A and B), indicating that CRP levels can be modulated by miR-637. In contrast, decreasing the levels of endogenous miR-637 by transfection with an miR-637 antagomir (anti-miR-637) increased both CRP mRNA and protein expression levels (Fig. 4C and D). Similar results were also observed by using a locked nucleic acid (LNA) (LNA-anti-miR-637) (data not shown). For further analysis of the effects of miR-637 on CRP expression, we performed luciferase assays using miR-637 target reporter constructs bearing CRP 3′ UTRs (Fig. 4E). We observed that miR-637 repressed luciferase activity from the psiCHECK2-CRP 3′ UTR and the CRP 3′-c fragment, which contains the predicted miR-637 site (Fig. 4E and F), but this effect was abolished when the miR-637 site was mutated (Fig. 4F). These results suggest that miR-637 reduces the expression levels of CRP by interacting with the CRP 3′ UTR.
FIG 4.
miR-637 interacts with CRP mRNA. (A and B) Forty-eight hours after transfection with control miRNA (miR-Ctrl) or with miR-637, the levels of miR-637 (left) and CRP mRNA (right) were measured by RT-qPCR analysis (A), or protein levels were assessed by immunoblotting with anti-CRP and anti-β-actin antibodies, as a loading control (B). CRP protein levels were quantified from immunoblots and normalized to values for β-actin. (C and D) HeLa cells were transfected with antisense (AS) miR-637 or control miRNA. After 72 h, the levels of miR-637 (left) and CRP mRNA (right) were measured by RT-qPCR analysis (C), or the protein levels of CRP and control β-actin were analyzed by immunoblotting (D). CRP protein levels were quantified from immunoblots and normalized to values for β-actin. (E) Schematic of psiCHECK2-CRP dual-luciferase reporters. Red bar, predicted miR-637 binding site. (F) Twenty-four hours after transfection of HeLa cells with miR-637, cells were transfected with different CRP 3′-UTR luciferase reporter plasmids, and luciferase activity was measured (RL/FL ratio). The histograms represent the means and standard errors of the means from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by Student's t test).
To confirm that miR-637 binds to CRP mRNA, we performed precipitations with biotin-labeled miR-637 followed by RNA isolation and RT-qPCR analysis. Indeed, CRP mRNA was enriched in the biotin miR-637 pulldown (Fig. 5A). Since RBPs can cooperate or compete with miRNAs to control gene expression either cooperatively or antagonistically (38, 39), and since both miR-637 and HuR interact with the CRP 3′ UTR, we tested whether miR-637 and HuR might compete or work cooperatively to modulate expression of CRP. First, we examined whether silencing of HuR affects miR-637 binding to CRP mRNA. Decreasing HuR levels significantly increased the binding of biotin miR-637 to the CRP transcript (Fig. 5A). Second, we performed the reverse experiment, overexpressing miR-637 and examining HuR binding to CRP mRNA by RIP analysis. miR-637 overexpression significantly decreased HuR binding to CRP mRNA (Fig. 5B), indicating that HuR and miR-637 bind competitively to CRP mRNA.
Next, we examined the effect of HuR silencing on miR-637-induced repression of CRP expression. Silencing of HuR additively repressed the miR-637-mediated repression of CRP expression, as indicated by decreases in the levels of CRP mRNA and protein in miR-637-overexpressing cells (Fig. 5C). In contrast, ectopic HuR overexpression inhibited the miR-637-mediated repression of CRP expression (Fig. 5D). In addition, we investigated whether HuR and miR-637 competed to regulate CRP expression in response to IL-6 treatment. As shown in Fig. 5E, overexpression of miR-637 or silencing of HuR decreased CRP mRNA levels following IL-6 treatment. Simultaneous miR-637 overexpression and HuR silencing in cells had an additive effect on decreasing the CRP abundance in response to IL-6 (Fig. 5E). These findings suggest that HuR and miR-637 competitively regulate CRP mRNA.
Individuals with high CRP levels have high levels of HuR and low levels of miR-637.
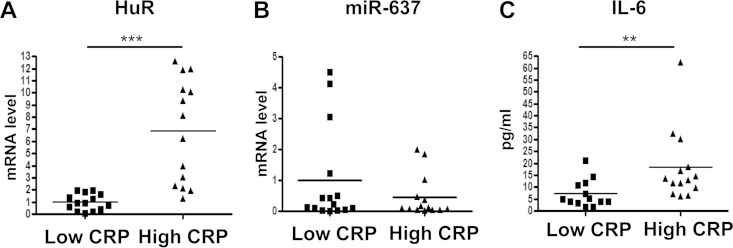
In order to test whether HuR and miR-637 may affect CRP levels in vivo, we obtained peripheral blood mononuclear cells from participants in the HANDLS study who have either low (≤3 mg/liter) or high (≥20 mg/liter) circulating levels of hsCRP. HuR mRNA levels were higher and miR-637 levels were lower in individuals with high hsCRP levels, consistent with our in vitro data showing that HuR positively and miR-637 negatively regulated CRP expression (Fig. 6A and B). As previously reported, we found a positive relationship between circulating IL-6 levels and hsCRP levels (Fig. 6C).
FIG 6.
Individuals with high CRP levels have high levels of HuR and low levels of miR-637. (A and B) RNA was isolated from PBMCs of individuals with either low (≤3 mg/liter) or high (≥20 mg/liter) circulating CRP levels (n = 15/group). The levels of HuR mRNA and miR-637 were quantified by using RT-qPCR. HuR mRNA levels were normalized to HPRT1 mRNA and UBC mRNA levels. miR-637 levels were normalized to snoRNA levels (SNU24, SNU49, and U47). (C) Serum protein levels of IL-6 from the same individuals as those for panels A and B were quantified. Means in the graph are indicated by bars. **, P < 0.01; ***, P < 0.001 (by a Mann-Whitney U test).
DISCUSSION
CRP has been well studied as an acute-phase marker during infection and inflammation, and it is now also well known to have clinical and pathological significance as a marker of age-associated disease, most prominently for CVD. Although some mechanisms that regulate CRP transcription have been explored, whether CRP expression is regulated by posttranscriptional mechanisms has not been reported. Here, we found that HuR and miR-637 play key roles in the posttranscriptional regulation of CRP through the AU-rich or U-rich elements in the CRP 3′ UTR.
The RBP HuR, a well-established RNA-binding protein and posttranscriptional regulator, affects the expression of its target mRNAs by binding to specific AU- or U-rich elements in their 3′ UTRs (22). Our results indicate that HuR interacts with CRP mRNA via the CRP 3′ UTR and regulates CRP mRNA stability and translation. Notably, HuR binds to CRP mRNA after IL-6 treatment, indicating the importance of HuR in mediating IL-6-induced CRP expression. The fact that HuR regulates other proinflammatory proteins, such as COX-2, IL-8, TGF-β, and TNF-α, suggests that HuR can serve as a master upstream modulator of inflammation (26, 27). These data indicate that HuR might be valuable as a therapeutic target for acute or chronic inflammatory diseases. Currently, anti-inflammatory agents under investigation are directed at the IL-1, TNF-α, and IL-6 pathways. Canakinumab and low-dose methotrexate lead to reductions in IL-6 and CRP levels (10); however, our findings that HuR is essential for IL-6-mediated upregulation of CRP suggest that HuR may be a promising target for the design of therapeutic agents to reduce CRP levels in CVD, diabetes mellitus, and other inflammatory diseases. Nonetheless, given the untoward and unintended serious side effects (heart failure and vascular complications) observed with anti-inflammatory agents that inhibit TNF-α and cyclooxygenase-2, it will be important to take into account the potential adverse events that may accompany the modulation of HuR function.
We also found that miR-637 is able to inhibit the expression of CRP. miRNAs play a significant role in posttranscriptional gene regulation predominately by acting as repressors (40). miR-637, a primate-specific miRNA, was first identified in colorectal tumor tissue (41). While the biological function of miR-637 is still not fully understood, several targets, such as osterix and collagen type IV alpha 1 (COL4A1), have been reported (42, 43). Decreased miR-637 expression levels were reported for four hepatocellular carcinoma cell lines; overexpression of miR-637 inhibited growth and enhanced apoptosis by suppressing the expression of the autocrine leukemia-inhibitory factor (LIF) (44), suggesting a tumor suppressor function for miR-637 in hepatocytes. Importantly, we found that miR-637 levels were lower in individuals with high circulating levels of hsCRP. These data indicate that miR-637 may have a direct role in modulating CRP levels in vivo and that miR-637 may have an anti-inflammatory effect by decreasing circulating CRP levels. Future research is needed in order to fully investigate whether miR-637 targets other inflammatory pathways. It is possible that miR-637 can be used to decrease the levels of CRP in at-risk individuals.
Here, we report the binding of HuR and miR-637 to the CRP 3′ UTR. Through the use of ectopic reporters, we found that the predominant sites where HuR interacted with and regulated CRP mRNA were the regions spanning positions 780 to 1080 and 1441 to 2103 of the CRP 3′ UTR, while the target site of miR-637 was located at the sequence spanning positions 1182 to 1204, directly between these two segments. These findings suggest that HuR and miR-637 interact with different binding sites of the CRP 3′ UTR. HuR and miR-637 modulated CRP mRNA in a functionally competitive manner: HuR promoted while miR-637 inhibited CRP expression. This result is consistent with data from other reports showing that HuR and miRNAs operate in opposite directions (23). For example, HuR inhibits miR-331-3p-mediated repression of ERBB2 expression (45) and miR-494-mediated repression of NCL expression (35). Also, HuR competes with miR-195 to regulate STIM1 mRNA (39) and suppresses miR-1192 to modulate HMGB1 mRNA (46). Although the specific mechanisms whereby HuR and miRNAs compete to regulate joint target mRNAs are not well understood, it is interesting to note that several competing miRNAs have binding sites near the HuR sites (47, 48), suggesting that HuR and miRNAs may compete via steric hindrance or local conformational changes of the mRNA.
IL-6 and CRP levels increase with age and are highly correlated (5, 6). Consistent with these findings, we found that IL-6 levels were higher in individuals with high hsCRP levels, regardless of age. Most significantly, in the small human cohort tested here, individuals with high levels of hsCRP also had high levels of HuR and low levels of miR-637, mirroring the correlations that we found with cultured cells.
As inflammation is now understood to be an underlying etiologic factor in a number of different diseases, it is important to fully understand the mechanisms by which these inflammatory markers are regulated. Unraveling the factors that modulate CRP provides insight for understanding the factors that contribute to the low-grade inflammatory state of aging and will undoubtedly shed light on age-related chronic diseases. The design of primary and secondary prevention therapies and treatment modalities aimed at the process of inflammation is already successfully under way. However, therapeutic agents target mainly upstream biomarkers. The role of CRP as both a risk factor and a biomarker warrants investigation of its regulation by other molecular factors. We have discovered a posttranscriptional mechanism by which the RBP HuR and the miRNA miR-637 modulate CRP expression downstream of IL-6 signaling. We have identified HuR and miR-637 as new factors that may play an integral role in the development of the inflammatory state and may represent new valuable targets in the diagnosis, treatment, or prevention of inflammation and diseases with a major inflammatory component.
ACKNOWLEDGMENTS
This study was supported by the Intramural Research Program of the National Institutes of Health National Institute on Aging.
We thank Kotb Abdelmohsen for critical reading of the manuscript and Althaf Lohani for technical assistance. We also thank Ngozi Ejiogu and the HANDLS medical staff for their careful evaluation and management of study participants.
REFERENCES
- 1.Franceschi C, Bonafe M, Valensin S, Olivieri F, De Luca M, Ottaviani E, De Benedictis G. 2000. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci 908:244–254. [DOI] [PubMed] [Google Scholar]
- 2.Pepys MB, Hirschfield GM. 2003. C-reactive protein: a critical update. J Clin Invest 111:1805–1812. doi: 10.1172/JCI200318921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volanakis JE. 2001. Human C-reactive protein: expression, structure, and function. Mol Immunol 38:189–197. doi: 10.1016/S0161-5890(01)00042-6. [DOI] [PubMed] [Google Scholar]
- 4.Ebersole JL, Cappelli D. 2000. Acute-phase reactants in infections and inflammatory diseases. Periodontol 2000 23:19–49. doi: 10.1034/j.1600-0757.2000.2230103.x. [DOI] [PubMed] [Google Scholar]
- 5.Harris TB, Ferrucci L, Tracy RP, Corti MC, Wacholder S, Ettinger WH Jr, Heimovitz H, Cohen HJ, Wallace R. 1999. Associations of elevated interleukin-6 and C-reactive protein levels with mortality in the elderly. Am J Med 106:506–512. doi: 10.1016/S0002-9343(99)00066-2. [DOI] [PubMed] [Google Scholar]
- 6.Bermudez EA, Rifai N, Buring J, Manson JE, Ridker PM. 2002. Interrelationships among circulating interleukin-6, C-reactive protein, and traditional cardiovascular risk factors in women. Arterioscler Thromb Vasc Biol 22:1668–1673. doi: 10.1161/01.ATV.0000029781.31325.66. [DOI] [PubMed] [Google Scholar]
- 7.Salazar J, Martinez MS, Chavez M, Toledo A, Anez R, Torres Y, Apruzzese V, Silva C, Rojas J, Bermudez V. 2014. C-reactive protein: clinical and epidemiological perspectives. Cardiol Res Pract 2014:605810. doi: 10.1155/2014/605810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh T, Newman AB. 2011. Inflammatory markers in population studies of aging. Ageing Res Rev 10:319–329. doi: 10.1016/j.arr.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allin KH, Nordestgaard BG. 2011. Elevated C-reactive protein in the diagnosis, prognosis, and cause of cancer. Crit Rev Clin Lab Sci 48:155–170. doi: 10.3109/10408363.2011.599831. [DOI] [PubMed] [Google Scholar]
- 10.Ridker PM, Luscher TF. 2014. Anti-inflammatory therapies for cardiovascular disease. Eur Heart J 35:1782–1791. doi: 10.1093/eurheartj/ehu203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D, Sun M, Samols D, Kushner I. 1996. STAT3 participates in transcriptional activation of the C-reactive protein gene by interleukin-6. J Biol Chem 271:9503–9509. doi: 10.1074/jbc.271.16.9503. [DOI] [PubMed] [Google Scholar]
- 12.Li SP, Goldman ND. 1996. Regulation of human C-reactive protein gene expression by two synergistic IL-6 responsive elements. Biochemistry 35:9060–9068. doi: 10.1021/bi953033d. [DOI] [PubMed] [Google Scholar]
- 13.Nishikawa T, Hagihara K, Serada S, Isobe T, Matsumura A, Song J, Tanaka T, Kawase I, Naka T, Yoshizaki K. 2008. Transcriptional complex formation of c-Fos, STAT3, and hepatocyte NF-1 is essential for cytokine-driven C-reactive protein gene expression. J Immunol 180:3492–3501. doi: 10.4049/jimmunol.180.5.3492. [DOI] [PubMed] [Google Scholar]
- 14.Sun H, Zhang Y, Gao P, Li Q, Sun Y, Zhang J, Xu C. 2011. Adiponectin reduces C-reactive protein expression and downregulates STAT3 phosphorylation induced by IL-6 in HepG2 cells. Mol Cell Biochem 347:183–189. doi: 10.1007/s11010-010-0627-y. [DOI] [PubMed] [Google Scholar]
- 15.Voleti B, Agrawal A. 2005. Regulation of basal and induced expression of C-reactive protein through an overlapping element for OCT-1 and NF-κB on the proximal promoter. J Immunol 175:3386–3390. doi: 10.4049/jimmunol.175.5.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cha-Molstad H, Young DP, Kushner I, Samols D. 2007. The interaction of C-Rel with C/EBPbeta enhances C/EBPbeta binding to the C-reactive protein gene promoter. Mol Immunol 44:2933–2942. doi: 10.1016/j.molimm.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Agrawal A, Cha-Molstad H, Samols D, Kushner I. 2003. Overexpressed nuclear factor-kappaB can participate in endogenous C-reactive protein induction, and enhances the effects of C/EBPbeta and signal transducer and activator of transcription-3. Immunology 108:539–547. doi: 10.1046/j.1365-2567.2003.01608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer F, Torzewski J, Kamenz J, Veit K, Hombach V, Dedio J, Ivashchenko Y. 2008. Interleukin-1beta stimulates acute phase response and C-reactive protein synthesis by inducing an NFkappaB- and C/EBPbeta-dependent autocrine interleukin-6 loop. Mol Immunol 45:2678–2689. doi: 10.1016/j.molimm.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Toniatti C, Demartis A, Monaci P, Nicosia A, Ciliberto G. 1990. Synergistic trans-activation of the human C-reactive protein promoter by transcription factor HNF-1 binding at two distinct sites. EMBO J 9:4467–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keene JD. 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 21.Ciafre SA, Galardi S. 2013. MicroRNAs and RNA-binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol 10:935–942. doi: 10.4161/rna.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. 2008. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem 389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srikantan S, Tominaga K, Gorospe M. 2012. Functional interplay between RNA-binding protein HuR and microRNAs. Curr Protein Pept Sci 13:372–379. doi: 10.2174/138920312801619394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 25.Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. 2008. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci U S A 105:20297–20302. doi: 10.1073/pnas.0809376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nabors LB, Gillespie GY, Harkins L, King PH. 2001. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res 61:2154–2161. [PubMed] [Google Scholar]
- 27.Srikantan S, Gorospe M. 2012. HuR function in disease. Front Biosci 17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhee WJ, Ni CW, Zheng Z, Chang K, Jo H, Bao G. 2010. HuR regulates the expression of stress-sensitive genes and mediates inflammatory response in human umbilical vein endothelial cells. Proc Natl Acad Sci U S A 107:6858–6863. doi: 10.1073/pnas.1000444107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yi J, Chang N, Liu X, Guo G, Xue L, Tong T, Gorospe M, Wang W. 2010. Reduced nuclear export of HuR mRNA by HuR is linked to the loss of HuR in replicative senescence. Nucleic Acids Res 38:1547–1558. doi: 10.1093/nar/gkp1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W. 2014. HuR and post-transcriptional regulation in vascular aging. Sci China Life Sci 57:863–866. doi: 10.1007/s11427-014-4706-2. [DOI] [PubMed] [Google Scholar]
- 31.Krol J, Loedige I, Filipowicz W. 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 32.Noren Hooten N, Fitzpatrick M, Wood WH III, De S, Ejiogu N, Zhang Y, Mattison JA, Becker KG, Zonderman AB, Evans MK. 2013. Age-related changes in microRNA levels in serum. Aging (Albany NY) 5:725–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olivieri F, Rippo MR, Monsurro V, Salvioli S, Capri M, Procopio AD, Franceschi C. 2013. MicroRNAs linking inflamm-aging, cellular senescence and cancer. Ageing Res Rev 12:1056–1068. doi: 10.1016/j.arr.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Dimmeler S, Nicotera P. 2013. MicroRNAs in age-related diseases. EMBO Mol Med 5:180–190. doi: 10.1002/emmm.201201986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tominaga K, Srikantan S, Lee EK, Subaran SS, Martindale JL, Abdelmohsen K, Gorospe M. 2011. Competitive regulation of nucleolin expression by HuR and miR-494. Mol Cell Biol 31:4219–4231. doi: 10.1128/MCB.05955-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noren Hooten N, Ejiogu N, Zonderman AB, Evans MK. 2012. Association of oxidative DNA damage and C-reactive protein in women at risk for cardiovascular disease. Arterioscler Thromb Vasc Biol 32:2776–2784. doi: 10.1161/ATVBAHA.112.300276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sesso HD, Wang L, Buring JE, Ridker PM, Gaziano JM. 2007. Comparison of interleukin-6 and C-reactive protein for the risk of developing hypertension in women. Hypertension 49:304–310. doi: 10.1161/01.HYP.0000252664.24294.ff. [DOI] [PubMed] [Google Scholar]
- 38.Xiao L, Cui YH, Rao JN, Zou T, Liu L, Smith A, Turner DJ, Gorospe M, Wang JY. 2011. Regulation of cyclin-dependent kinase 4 translation through CUG-binding protein 1 and microRNA-222 by polyamines. Mol Biol Cell 22:3055–3069. doi: 10.1091/mbc.E11-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhuang R, Rao JN, Zou T, Liu L, Xiao L, Cao S, Hansraj NZ, Gorospe M, Wang JY. 2013. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res 41:7905–7919. doi: 10.1093/nar/gkt565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breving K, Esquela-Kerscher A. 2010. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol 42:1316–1329. doi: 10.1016/j.biocel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, Raymond CK, Roberts BS, Juhl H, Kinzler KW, Vogelstein B, Velculescu VE. 2006. The colorectal microRNAome. Proc Natl Acad Sci U S A 103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang JF, Fu WM, He ML, Wang H, Wang WM, Yu SC, Bian XW, Zhou J, Lin MC, Lu G, Poon WS, Kung HF. 2011. miR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol Biol Cell 22:3955–3961. doi: 10.1091/mbc.E11-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lisse TS, Chun RF, Rieger S, Adams JS, Hewison M. 2013. Vitamin D activation of functionally distinct regulatory miRNAs in primary human osteoblasts. J Bone Miner Res 28:1478–1488. doi: 10.1002/jbmr.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang JF, He ML, Fu WM, Wang H, Chen LZ, Zhu X, Chen Y, Xie D, Lai P, Chen G, Lu G, Lin MC, Kung HF. 2011. Primate-specific microRNA-637 inhibits tumorigenesis in hepatocellular carcinoma by disrupting signal transducer and activator of transcription 3 signaling. Hepatology 54:2137–2148. doi: 10.1002/hep.24595. [DOI] [PubMed] [Google Scholar]
- 45.Epis MR, Barker A, Giles KM, Beveridge DJ, Leedman PJ. 2011. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J Biol Chem 286:41442–41454. doi: 10.1074/jbc.M111.301481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dormoy-Raclet V, Cammas A, Celona B, Lian XJ, van der Giessen K, Zivojnovic M, Brunelli S, Riuzzi F, Sorci G, Wilhelm BT, Di Marco S, Donato R, Bianchi ME, Gallouzi IE. 2013. HuR and miR-1192 regulate myogenesis by modulating the translation of HMGB1 mRNA. Nat Commun 4:2388. doi: 10.1038/ncomms3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. 2011. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell 43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M Jr, Tuschl T, Ohler U, Keene JD. 2011. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell 43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]