Abstract
BHLHE40 and BHLHE41 (BHLHE40/41) are basic helix-loop-helix type transcription factors that play key roles in multiple cell behaviors. BHLHE40/41 were recently shown to be involved in an epithelial-to-mesenchymal transition (EMT). However, the precise mechanism of EMT control by BHLHE40/41 remains unclear. In the present study, we demonstrated that BHLHE40/41 expression was controlled in a pathological stage-dependent manner in human endometrial cancer (HEC). Our in vitro assays showed that BHLHE40/41 suppressed tumor cell invasion. BHLHE40/41 also suppressed the transcription of the EMT effectors SNAI1, SNAI2, and TWIST1. We identified the critical promoter regions of TWIST1 for its basal transcriptional activity. We elucidated that the transcription factor SP1 was involved in the basal transcriptional activity of TWIST1 and that BHLHE40/41 competed with SP1 for DNA binding to regulate gene transcription. This study is the first to report the detailed functions of BHLHE40 and BHLHE41 in the suppression of EMT effectors in vitro. Our results suggest that BHLHE40/41 suppress tumor cell invasion by inhibiting EMT in tumor cells. We propose that BHLHE40/41 are promising markers to predict the aggressiveness of each HEC case and that molecular targeting strategies involving BHLHE40/41 and SP1 may effectively regulate HEC progression.
INTRODUCTION
Basic helix-loop-helix (bHLH) type transcription factors play key roles in cell differentiation, proliferation, apoptosis, and metabolism. BHLHE40 (basic helix-loop-helix family member e40 gene) and BHLHE41 are members of the Hairy/E(spl)/HES family. BHLHE40 and BHLHE41 (BHLHE40/41) exhibit more than 90% similarity in the bHLH region and approximately 50% in total.
BHLHE40/41 have been shown to function as transcriptional repressors by binding to the class B E-box. BHLHE40/41 interact with TF2B, TBP, or TF2D or recruit a histone deacetylase at the E-box site (1–5). On the other hand, BHLHE40/41 were previously reported to modulate the expression of some genes in an E-box-independent manner. BHLHE40 has been shown to associate with SP1 binding sites in the BIRC5 promoter to activate its transcription (6) and with STAT3 to regulate the transcription of STAT3-dependent target genes (7). BHLHE41 suppressed VEGF transcription by interacting with HIF1A (8). BHLHE40 and BHLHE41 were reported to associate with retinoid X receptor (RXR), MYOD1, or CEBP in order to regulate the transcription of their target genes (9–12).
In diverse types of cancer species, such as colon, oral, and liver cancer or brain tumors, BHLHE40 expression levels were found to be higher in tumors than in adjacent normal tissues (13–15). On the other hand, in human endometrial cancer (HEC) and non-small-cell lung cancer, no changes in BHLHE40 expression were reported between cancer and normal tissues (16, 17). Regarding expression profiles with the development of cancer, studies on oral, lung, liver, and esophageal cancer showed that BHLHE40 expression inversely correlated with the tumor stage or differentiation grade (18–21). These findings suggest that patients who strongly expressed BHLHE40 had better prognoses (19). An in vitro analysis also revealed the tumor-suppressive effects of BHLHE40. The overexpression of BHLHE40 in multiple cell types has been shown to inhibit cell proliferation, migration, or invasion and to induce cellular senescence (3, 4, 13, 18, 19, 22). Among the mechanisms involved, BHLHE40 was demonstrated to directly inhibit CCND1 or ID1 transcription (18, 20, 23). Fewer studies have examined the expression of BHLHE41 in cancer. The higher expression of BHLHE41 in HEC than in normal adjacent endometrial tissue has been reported (17). Although some studies have found a positive correlation between BHLHE41 expression levels and tumor progression, others reported an inverse correlation (24–26). In triple-negative breast cancer (TNBC), the stronger expression of BHLHE41 correlated with better prognoses, including metastasis-free survival (26). This study demonstrated that BHLHE41 promoted the degradation of HIF1A and EPAS1 in order to suppress TNBC metastasis by direct binding (26). In breast cancer, BHLHE41 has also been shown to directly suppress CCND1 transcription (27).
Epithelial-to-mesenchymal transition (EMT) is an essential mechanism to explain the properties of tumors that allow them to invade adjacent stromal tissue. In pancreatic cancer cells, the transforming growth factor β1 (TGFβ1)-induced expression of BHLHE40 enhanced EMT, whereas that of BHLHE41 inhibited EMT by directly suppressing SNAI2 expression (28, 29). TWIST1 transcription was very recently reported to be suppressed by BHLHE41 through an E-box in the TWIST1 promoter (30). These are the only studies to have described the influence of BHLHE40 and BHLHE41 on EMT; therefore, further studies are needed in order to obtain more detailed information. In addition to these findings, several transcription factors, including SP1, have been shown to regulate EMT effector genes (31–33).
In the present study, we examined the impact of BHLHE40/41 on EMT and cell invasion by HEC. BHLHE40 and BHLHE41 expression levels both correlated with the pathological stages of HEC patients. BHLHE40/41 directly regulated the transcription of the EMT effector gene TWIST1 by affecting its promoter regions. An intimate analysis revealed that BHLHE40/41 regulated TWIST1 transcription by associating with an SP1 binding site in its promoter. This regulation was independent of E-boxes. This is the first study to have elucidated a novel mechanism by which BHLHE40/41 influenced EMT in HEC. We propose that BHLHE40/41 are promising markers for predicting the prognosis of HEC, and our results may lead to a new strategy to control HEC development.
MATERIALS AND METHODS
Cell culture and reagents.
HEC-1, HEC-6, Ishikawa, HHUA, hEM, and 293T cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, penicillin, and streptomycin. HEC-1 and HEC-6 cells were purchased from the Japanese Collection of Research Bioresources (Tokyo, Japan). HHUA cells were from the RIKEN BioResource Center (Ibaraki, Japan). Ishikawa cells were from Sigma-Aldrich (St. Louis, MO). 293T cells were from Invitrogen (Carlsbad, CA). hEM cells were a kind gift from Satoru Kyo. hEM cells were established from human endometrial glandular cells immortalized using human papillomavirus 16 E6/7 and human telomerase reverse transcriptase (34). 293T and Ishikawa cells were used within 6 months of receipt. The identities of HEC-1, HEC-6, and HHUA cells were confirmed by the Japanese Collection of Research Bioresources (JCRB) Cell Bank using DNA profiling (short tandem repeat [STR]) in March 2015. WP631 methanesulfonate was purchased from Sigma-Aldrich and dissolved in dimethyl sulfoxide.
Patients and tissue samples.
Eight-six HEC patients who underwent surgery at the Department of Obstetrics and Gynecology of Kyushu University Hospital between 2005 and 2010 were recruited for this study. Normal control uterine endometrial tissues were removed from 20 patients undergoing hysterectomy for benign reasons such as uterine myoma or endometriosis. The 20 normal endometrial tissue specimens and 37 HEC primary specimens (30 endometrioid adenocarcinoma specimens, including 16 grade 1, 10 grade 2, and 4 grade 3, and 7 serous adenocarcinoma specimens) out of 86 HEC cases (77 cases of endometrioid adenocarcinoma, including 37 grade 1, 27 grade 2, and 13 grade 3, and 9 cases of serous adenocarcinoma) were used in mRNA assays. Eighty-six HEC primary specimens were used for immunohistochemistry. All patients involved in this study provided their written informed consent. This study was approved by the Ethical Committee of Kyushu University.
Real-time RT-PCR.
Total RNA from tissue samples and cultured cells was extracted using an RNeasy minikit (Qiagen, Germantown, MD) or Isogen (Nippon Gene, Tokyo, Japan). cDNA was synthesized by a ReverTra Ace kit (Toyobo, Osaka, Japan). Real-time reverse transcription (RT)-PCR was carried using Brilliant II SYBR master mixes (Agilent Technologies, Santa Clara, CA). The relative expression levels of target genes were calculated using a ΔΔCT method after normalization using those of the housekeeping gene, ACTB (35). The sequence information used is shown in Table 1. All primers were designed to be located across an intron.
TABLE 1.
Primers used for the qRT-PCR analysis
| Target gene | Accession no. | Sequence |
Amplicon (bp) | |
|---|---|---|---|---|
| Forward primer | Reverse primer | |||
| BHLHE40 | NM_003670 | 5′-GACCGGATTAACGAGTGCAT-3′ | 5′-TGCTTTCACATGCTTCAAGG-3′ | 123 |
| BHLHE41 | NM_030762 | 5′-GCATGAAACGAGACGACACC-3′ | 5′-ATTTCAGATGTTCAGGCAGT-3′ | 126 |
| SNAI1 | NM_005985 | 5′-AAGGCCTTCTCTAGGCCCT-3′ | 5′-CGCAGGTTGGAGCGGTCAG-3′ | 113 |
| SNAI2 | NM_003068 | 5′-TTCGGACCCACACATTACCT-3′ | 5′-GCAGTGAGGGCAAGAAAAAG-3′ | 122 |
| TWIST1 | NM_000474 | 5′-CAGCTACGCCTTCTCGGTCT-3′ | 5′-CTGTCCATTTTCTCCTTCTCTGGA-3′ | 138 |
| ACTB | NM_001101 | 5′-TTGCCGACAGGATGCAGAAG-3′ | 5′-CAGCGAGGCCAGGATGGAGC-3′ | 122 |
Immunohistochemistry.
Surgical tissue samples were freshly frozen until examined immunohistochemically. Four-micrometer-thick sections were made with a cryostat, air dried, and fixed in a 4% paraformaldehyde–phosphate-buffered saline (PBS) solution. Endogenous peroxidase was inactivated in a 3% hydrogen peroxide-PBS solution. After the reaction was blocked with protein block (Agilent Technologies), the sections were incubated overnight at 4°C with a diluted anti-BHLHE40 (HPA028921; Atlas Antibodies, Stockholm, Sweden) or anti-BHLHE41 (E-4; Santa Cruz Biotechnology, Santa Cruz, CA) antibody. The sections were washed, and EnVision+ dual link system-horseradish peroxidase (HRP) (Agilent Technologies) was used for the addition of a secondary antibody and color development. Mayer's hematoxylin was used for nuclear counterstaining.
Immunoblotting and immunoprecipitation.
In immunoblotting, cell lysates were separated on 10% SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). The primary antibodies used were anti-BHLHE40 (S-8), -BHLHE41 (E-4), -SNAI1 (H-130), -SNAI2 (D-19), -SP1 (PEP2), -TWIST1 (H-81), -VIM (V-9), -FN1 (EP5), -CDH1 (H-108), -CDH2 (H-63), -CCLD1 (DCS-6), and -GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase; FL-335) (Santa Cruz Biotechnology) antibodies. Antihemagglutinin (anti-HA; HA-7) and -FLAG (M5) antibodies were from Sigma-Aldrich (St. Louis, MO).
After the membranes were incubated with horseradish peroxidase-linked secondary antibodies (Promega, Madison, WI), blots were detected using an enhanced chemiluminescence (ECL) system. The intensities of the bands were quantified with NIH Image software (http://rsb.info.nih.gov/nih-image/).
Whole-cell lysates from 293T cells expressing MYC-SP1 and either a combination of HA-BHLHE40 and FLAG-BHLHE40, HA-BHLHE41 and FLAG-BHLHE41, or HA-BHLHE40 and FLAG-BHLHE41 were prepared for immunoprecipitation. The lysates were immunoprecipitated overnight at 4°C with either anti-MYC, -HA, or -FLAG antibody bound to protein G PLUS-agarose (Santa Cruz Biotechnology) and separated by SDS-PAGE. The immunoprecipitated proteins were analyzed by Western blotting using either an anti-MYC, -HA, or -FLAG antibody.
Plasmid transfection, lentivirus vector, and luciferase assay.
HA- or FLAG-tagged human BHLHE40 and BHLHE41 open reading frames (ORFs) were amplified by PCR using cDNA from HHUA cells. MYC-tagged human SP1 was also constructed by PCR using cDNA from human normal endometrial tissue. The DNA sequence of each construct was confirmed by a sequence reaction using an ABI PRISM BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Carlsbad, CA). They were ligated into a pCDNA3 expression vector. HA-tagged BHLHE40 and FLAG-tagged BHLHE41 were also inserted into a pENTR4 vector and transferred into pLX302 and pLX303 vectors by recombination reaction (Gateway Technology by Invitrogen). Lentivirus vectors were produced as described previously (36). In order to construct stable cell lines, puromycin and blasticidin were used for pLX302 and pLX303, respectively.
Several regions upstream of SNAI1 (spanning from bp −902 from the transcription start site [TSS]), SNAI2 (from bp −1650 from the TSS), and TWIST1 (from bp −1876, −1587, −170, +34, and +116 from the TSS) were amplified by PCR and ligated into the pGL4.22 basic luciferase vector (Promega). Site-directed mutagenesis was performed to generate the mutated luciferase reporter constructs. The sequences of the mutants generated are shown in Table 2. The DNA sequence of each construct was confirmed by a sequencing reaction. In reporter assays, cells (1 × 105) were transfected with 200 ng of each luciferase reporter, 100 ng of an expressing vector, and 25 ng of pCDNA3.1-His-LacZ using Lipofectamine 2000 reagent (Invitrogen). Twenty-four hours after transfection, cell lysates were collected and assayed using a luciferase assay kit (Promega) and a beta-galactosidase assay kit (Clontech Laboratories, Mountain View, CA). Luciferase activity values were normalized using those of beta-galactosidase activity.
TABLE 2.
Sequences of reporter constructs
| Construct | Sense oligonucleotidea |
|---|---|
| pTWIST1-wtSP1BS | +151 GTCCCCTCCCCCTCCCGCCTCCCTCCCCGCCTCC +184 |
| pTWIST1-mtSP1BS-a | +151 GTCCACTACACATCACGCCTCCCTCCCCGCCTCC +184 |
| pTWIST1-mtSP1BS-b | +151 GTCCCCTCCCCCTCCCGCATACATACACGACTCC +184 |
| pTWIST1-mtSP1BS-ab | +151 GTCCACTACACATCACGCATACATACACGACTCC +184 |
| pTWIST1-wtSP1BS-c | +193 CCCTCCCC +200 |
| pTWIST1-mtSP1BS-c | +193 ACATACAC +200 |
Mutated nucleotides are underlined.
Knockdown by siRNA and shRNA.
Double-stranded small interfering RNA (siRNA) for SP1 (siSP1) (sc-29487) was purchased with control siRNA (sc-37007) from Santa Cruz Biotechnology. We also established HHUA cells with the stable knockdown of BHLHE40 and BHLHE41 by a lentivirus system (36). The transduced cells were selected by puromycin. Short hairpin RNA (shRNA) target sites and sequences are shown in Table 3. shRNA sequences for BHLHE40 and BHLHE41 were from Sigma-Aldrich (Mission shRNA validated sequence). Two shRNAs for each of the BHLHE40 and BHLHE41 genes were made, and both showed similar efficiencies (see Fig. S4A and B in the supplemental material), and shBHLHE40S2 for BHLHE40 and shBHLHE41S1 for BHLHE41 were used in most cases.
TABLE 3.
Sequences of shRNA constructs
| shRNA name | Target | Target site | shRNA sequence (cloned into AgeI and EcoRI)a |
|---|---|---|---|
| Control shRNA (shCtrl) | None | CCTAAGGTTAAGTCGCCCTCG | 5′-accggtCCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGGTTTTTTTgaattc-3′ |
| shBHLHE40S1 (TRCN0000013249) | BHLHE40 exon 4 | GCACTAACAAACCTAATTGAT | 5′-accggtGCACTAACAAACCTAATTGATCTCGAGATCAATTAGGTTTGTTAGTGCTTTTTTgaattc-3′ |
| shBHLHE40S2 (TRCN0000232187) | BHLHE40 exon 4 | CATGTGAAAGCACTAACAAAC | 5′-accggtGCATGTGAAAGCACTAACAAACCTCGAGGTTTGTTAGTGCTTTCACATGCTTTTTTgaatttc-3′ |
| shBHLHE41S1 (TRCN0000086556) | BHLHE41 exon 2 | CTGGACTATTCCTCTTTGTAT | 5′-accggtGCTGGACTATTCCTCTTTGTATCTCGAGATACAAAGAGGAATAGTCCAGCTTTTTTgaattc-3′ |
| shBHLHE41S2 (TRCN0000016945) | BHLHE41 exon 1 | AGAGACAGTTACTGGAACATA | 5′-accggtAGAGACAGTTACTGGAACATACTCGAGTATGTTCCAGTAACTGTCTCTTTTTTTgaattc-3′ |
Cloning sites are shown in lowercase letters.
EMSA.
An electrophoretic mobility shift assay (EMSA) was performed using nuclear extracts from 293T cells expressing HA-BHLHE40 and/or FLAG-BHLHE41 as described elsewhere (36). The sequences of the probes used are as follows: pTWIST1-wtSP1BS, 5′-CCGTCCCCTCCCCCTCCCGCCTCCCTCCCCGCCTCCCC-3′; pTWIST1-mtSP1BS, 5′-CCGTCCACTACACATCACGCATACATACACGACTCCCC-3′. The mutated nucleotides are underlined. In order to determine whether SP1, BHLHE40, and BHLHE41 were involved in protein-nucleotide complexes, an anti-HA (HA-7; Sigma-Aldrich), -FLAG (M5; Sigma-Aldrich), -SP1 (PEP2; Santa Cruz Biotechnology), or -CEBPA (14AA; Santa Cruz Biotechnology) antibody was added to the incubation mixture.
Chromatin immunoprecipitation assay.
A chromatin immunoprecipitation assay was performed as described elsewhere (36). The DNA-protein complex was immunoprecipitated using anti-SP1 (PEP-2; Santa Cruz Biotechnology), -acetylated histone H3 (Millipore), -HDAC1 (ab7028; Abcam), and -KAT2B (ab12188; Abcam) antibodies and protein G PLUS-agarose (sc-2002; Santa Cruz Biotechnology). Precipitated DNA samples were used to amplify the SP1 binding site of the TWIST1 promoter with the primers as follows: forward primer, 5′-CCTCCAAGTCTGCAGCTCTC-3′; reverse primer, 5′-CCCGAGGTCCAAAAAGAAAG-3′. Specific DNA fragments were semiquantified by real-time PCR. The fold enrichment of immunoprecipitated DNA fragments was normalized based on 10% input.
Transwell chamber assay.
Cell motility and invasion were evaluated by the transwell chamber assay as described previously (35). A total of 4.0 × 104 or 5.0 × 104 cells were plated in upper wells without serum. Complete growth medium with 10% fetal bovine serum was placed in the lower wells. After 48 h for HEC-1, HHUA, and Ishikawa cells and 72 h for HEC-6 cells, the membranes were collected for analysis. The membranes were stained with Diff-Quik staining solutions (Sysmex Corp., Kobe, Japan).
Statistics.
Data are represented as the mean ± standard deviation (SD). Reporter assay data were analyzed with the Student t test. Case-control data were analyzed with the Mann-Whitney U test. The relationships between BHLHE40 and BHLHE41, BHLHE40 and TWIST1, and BHLHE41 and TWIST1 were evaluated by Pearson's product-moment correlation coefficient. The significance of these relationships was determined by the F-test. A P value of <0.05 was considered significant.
RESULTS
Clinical evaluation of BHLHE40 and BHLHE41 expression in HEC.
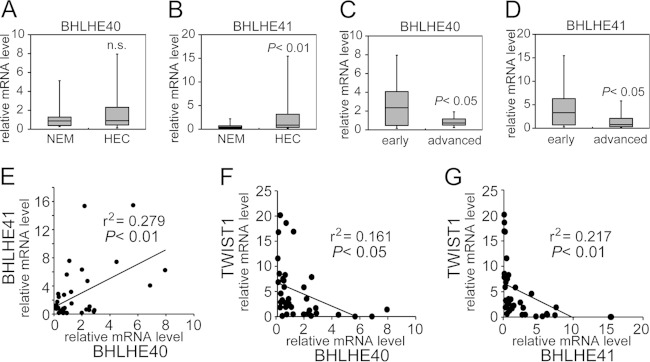
In order to elucidate the involvement of BHLHE40 and BHLHE41 (BHLHE40/41) in human endometrial cancer (HEC), we first examined their expression levels in cancer specimens. Thirty-seven cases of surgically removed specimens from primary cancer sites were used for mRNA assays (endometrioid adenocarcinoma, 30 cases; serous adenocarcinoma, 7 cases; stage IA, 20 cases; more than stage IB, 17 cases [based on the surgical staging system of FIGO 2008 {55}]). Normal endometrial tissues from benign tumor cases were also examined as controls. Only slight differences were observed in BHLHE40 mRNA levels between cancer tissues and normal endometrial tissues, whereas BHLHE41 mRNA levels were markedly higher in cancer tissues than in normal endometrial tissues (Fig. 1A and B). In order to determine the relationship between BHLHE40/41 expression and the clinical features of cancer, cases at stage IA with no or less than 50% invasion into the adjacent myometrium were compared with cases at more than stage IB showing more than 50% invasion into the myometrium. The mRNA levels of BHLHE40 and BHLHE41 were both significantly higher in cases at stage IA than in cases at more than stage IB (Fig. 1C and D). A positive correlation was found between BHLHE40 and BHLHE41mRNA levels (Fig. 1E). No significant differences in BHLHE40/41 mRNA levels were observed between the group of grade 1 or 2 endometrioid adenocarcinoma (type I) and that of grade 3 endometrioid adenocarcinoma or serous adenocarcinoma (type II) (see Fig. S1A and B in the supplemental material).
FIG 1.
mRNA analysis of endometrial cancer specimens by real-time RT-PCR. (A and B) Twenty normal endometrial tissue specimens (NEM) and 37 primary HEC specimens were used to analyze their mRNA levels of BHLHE40 (A) and BHLHE41 (B). (C and D) BHLHE40/41 mRNA levels of the HEC group at the early stage (stage IA based on FIGO 2008 system criteria) were compared with those of the group at the advanced stage (more than stage IB). (E) The relationship between BHLHE40 and BHLHE 41 mRNA levels from 37 specimens was analyzed by Pearson's product-moment correlation coefficient. r values are correlation coefficients. The significance of these relationships was determined by the F-test. (F and G) The relationship between TWIST1 and BHLHE40 or TWIST1 and BHLHE41 mRNA levels was also analyzed by Pearson's product-moment correlation coefficient. A P value of <0.05 was considered significant. n.s., not significant.
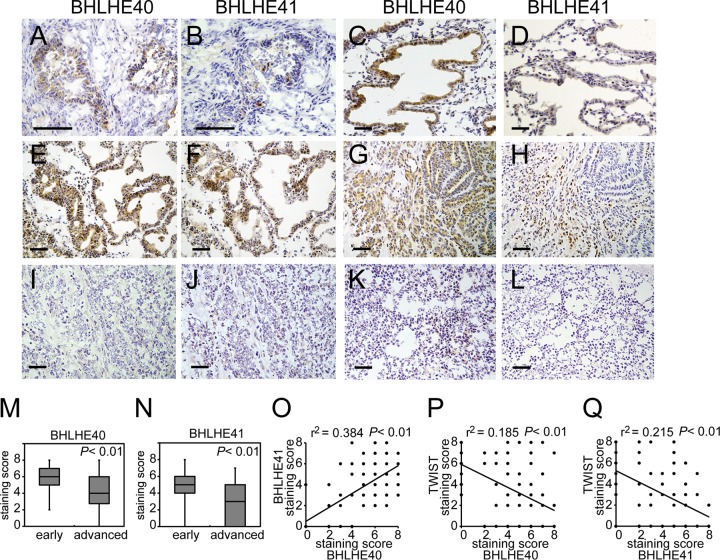
BHLHE40/41 protein levels were also analyzed by immunohistochemistry. In normal endometrial tissues, the BHLHE40 protein was exclusively detected in glandular epithelial cells, not in the surrounding stromal cells (Fig. 2A and C). In contrast, the BHLHE41 protein was almost absent in normal endometrial tissues (Fig. 2B and D). BHLHE40/41 protein expression was examined in 86 cases of HEC tissue samples (endometrioid adenocarcinoma, 77 cases; serous adenocarcinoma, 9 cases; stage IA, 37 cases; more than stage IB, 49 cases [based on the surgical staging system of FIGO 2008]). The nuclear expression of BHLHE40/41 was evaluated by a staining scoring system described by Allred et al. (37). Representative samples tested positive for BHLHE40/41 (Fig. 2E to H), while others were negative (Fig. 2I to L). Consistent with the results obtained in the mRNA assay, BHLHE40/41 protein levels were both higher in cases at stage IA than in cases at more than stage IB (Fig. 2M and N). A positive correlation was detected between BHLHE40 and BHLHE41 protein levels (Fig. 2O). In contrast to the results from the mRNA assay, a significant difference was observed for BHLHE40 but not for BHLHE41 between type I and type II HEC cases (see Fig. S1C and D in the supplemental material).
FIG 2.
Immunohistochemistry of endometrial cancer specimens. Surgical samples from 86 HEC patients were analyzed for BHLHE40/41expression by immunohistochemistry. Representative results are shown. (A to D) Results are for the late proliferative phase (A and B) and the secretory phase (C and D) of normal endometrial tissue. (E and F) A grade 1 endometrioid adenocarcinoma (EAC) case at stage IA; (G and H) a grade 2 EAC case at stage IA; (I and J) a grade 3 EAC case at stage IB; (K and L) a grade 2 EAC case at stage IIIA. Immunohistochemical images with an anti-BHLHE40 antibody (A, C, E, G, I, K) and anti-BHLHE41 antibody (B, D, F, H, J, L) are shown. The scale bars represent 200 μm. (M and N) The staining scores of immunohistochemical images were analyzed. Eighty-six cases were divided into a group at the early stage (stage IA) and that at the advanced stage (more than stage IB). (O) The relationship between BHLHE40 and BHLHE41 staining levels from 86 specimens was analyzed by Pearson's product-moment correlation coefficient. r values are correlation coefficients. The significance of coefficients was determined by the F-test. (P and Q) The relationship between TWIST1 and BHLHE40 or TWIST1 and BHLHE41 staining levels was also analyzed by Pearson's product-moment correlation coefficient. A P value of <0.05 was considered significant.
Forced BHLHE40/41 expression suppressed tumor properties of HEC cells.
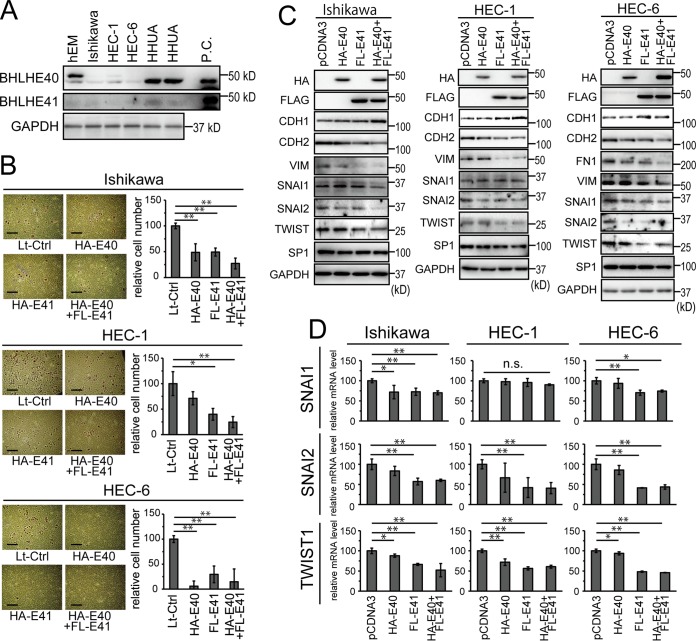
In order to investigate the impact of BHLHE40/41 expression in HEC cells, we induced the exogenous expression of BHLHE40/41 or knocked down the endogenous expression of BHLHE40/41 in HEC cells. BHLHE40/41 protein expression was initially examined in four HEC cell lines. The expression of BHLHE40/41 was observed in HHUA cells but was absent in Ishikawa, HEC-1, and HEC-6 cells (Fig. 3A). The exclusive expression of BHLHE40 was detected in an immortalized endometrial glandular cell line, hEM cells (34) (Fig. 3A). We used a lentivirus transduction system to induce the stable expression of BHLHE40/41. Plasmid transfection was also performed to examine the transient expression of BHLHE40/41. Four types of cells were obtained by infection with an empty lentivirus vector, either the vector to express HA-tagged BHLHE40 or FLAG-tagged BHLHE41, and both vectors to express HA-BHLHE40 and FLAG-BHLHE41 (see Fig. S2B in the supplemental material). The forced expression of BHLHE40/41 in Ishikawa, HEC-1, and HEC-6 cells resulted in the greater suppression of cell invasion than that in control cells (Fig. 3B). BHLHE40/41 expression did not affect the motility of HEC-1 or Ishikawa cells (see Fig. S3A in the supplemental material). A protein analysis was performed by immunoblotting of Ishikawa, HEC-1 and HEC-6 cells. We focused on the expression of EMT-related molecules. The transient expression of BHLHE40/41 resulted in the downregulation of VIM, CDH2, and FN1 and induction of CDH1 (Fig. 3C). The intensities of the bands were quantified and are shown in Fig. S5A in the supplemental material. The mRNA and protein levels of the EMT effector genes SNAI1, SNAI2, and TWIST1 were suppressed in most cell lines (Fig. 3C and D; see Fig. S5A in the supplemental material). SNAI1 expression was not altered in HEC-1 cells, and some differences were observed in gene expression patterns that depended on the cellular context. In most experiments, we used an HA tag for BHLHE40 and a FLAG tag for BHLHE41 in order to distinguish between the expression of their genes because the molecular weights of their two gene products were similar. Therefore, in order to confirm that exogenous BHLHE40 and BHLHE41 were expressed at similar levels, HA-BHLHE40 and HA-BHLHE41 or FLAG-BHLHE40 and FLAG-BHLHE41 were transfected in HEC-6 cells and the cell lysates were immunoblotted with anti-HA or anti-FLAG antibodies. The expression levels of exogenous BHLHE40 and BHLHE41 were similar regardless of the tags (see Fig. S2A in the supplemental material).
FIG 3.
Impact of forced BHLHE40/41 expression in HEC cells. (A) Expression status of BHLHE40/41 in HEC cell lines analyzed by immunoblotting. (B) In vitro invasion of Ishikawa, HEC-1, and HEC-6 cells infected with lentivirus vectors to express HA-BHLHE40, FLAG-BHLHE41, or both. The scale bars represent 200 μm. The right graphs show quantification data of the results. (C) Protein expression of EMT markers in Ishikawa, HEC-1, and HEC-6 cells transfected with vectors to express HA-BHLHE40, FLAG-BHLHE41, or both. (D) mRNA levels of SNAI1, SNAI2, and TWIST1 in each transfectant were analyzed by real-time RT-PCR. E40, BHLHE40; E41, BHLHE41; n.s., not significant; P.C., positive control; Lt-Ctrl, control lentivirus vector; *, P < 0.05; **, P < 0.01.
Knockdown of BHLHE40/41 in HEC cells enhanced cell invasion and EMT.
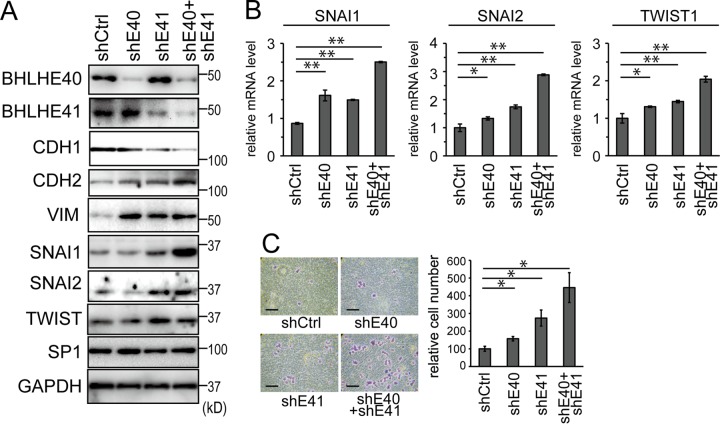
The knockdown of BHLHE40/41 using shRNA transduced by a lentivirus was performed on HHUA cells, which originally expressed BHLHE40/41. Within the extent of our screening, HHUA cells were the only cell line that expressed BHLHE40/41. The successful knockdown of BHLHE40/41 was confirmed at the protein level (Fig. 4A). The shRNAs used for BHLHE40 and BHLHE41 were validated by the vendor. Since the two shRNAs for BHLHE40 and BHLHE41 showed similar efficiencies (see Fig. S4 in the supplemental material), shBHLHE40S2 for BHLHE40 and shBHLHE41S1 for BHLHE41 were used in most cases. In vitro assays showed that the knockdown of BHLHE40 and/or BHLHE41 enhanced cell invasion and motility (Fig. 4C; see also Fig. S3B). Double knockdown led to the most prominent changes. The knockdown of BHLHE40 and/or BHLHE41 upregulated the expression of VIM and CDH2, while it downregulated that of CDH1 (Fig. 4A). The intensities of the bands were quantified and are shown in Fig. S5B in the supplemental material. Furthermore, the knockdown of BHLHE40 and/or BHLHE41 upregulated the expression of SNAI1, SNAI2, and TWIST1 at the mRNA and protein levels (Fig. 4A and B; see also Fig. S5B).
FIG 4.
Impact of BHLHE40/41 in HEC cells analyzed by knockdown. (A) Protein expression levels of EMT markers in HHUA cells infected with lentivirus vectors to knock down BHLHE40, BHLHE41, or both were determined by immunoblotting. (B) A mRNA analysis of SNAI1, SNAI2, and TWIST1 in each knocked-down cell type was performed by real-time RT-PCR. (C) In vitro invasion of each knocked-down cell type. The right graph shows quantification data of the results. The scale bars represent 200 μm. shE40, shBHLHE40; shE41, shBHLHE41; *, P < 0.05; **, P < 0.01.
BHLHE40/41 regulated promoter activities of SNAI1, SNAI2, and TWIST1.
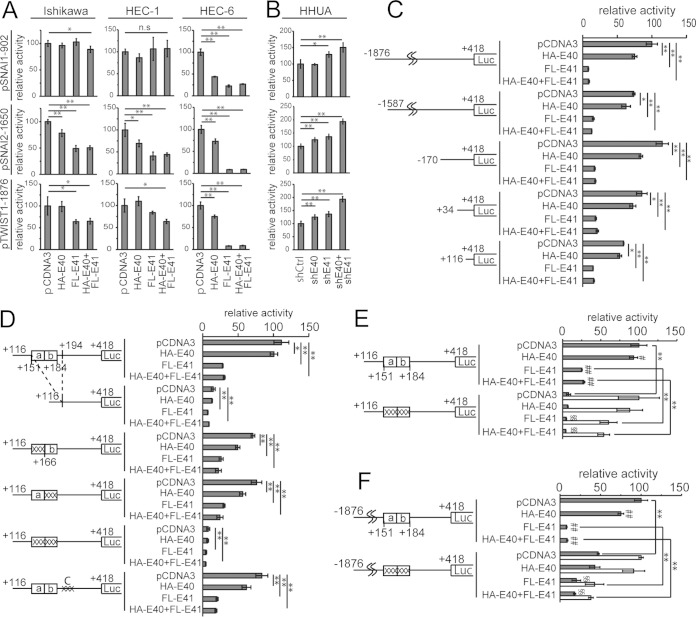
In order to explore the mechanism underlying the transcriptional regulation of SNAI1, SNAI2, and TWIST1 by BHLHE40/41, a reporter assay using their promoters was performed. The promoter regions spanning from bp −902 to +88 of SNAI1 (pSNAI1-902), bp −1650 to +215 of SNAI2 (pSNAI2-1650), and bp −1876 to +418 of TWIST1 (pTWIST1-1876), which are highly conserved between human and mouse, were used for this assay. Ishikawa, HEC-1, and HEC-6 cells were transfected with each reporter gene, HA-BHLHE40, and/or FLAG-BHLHE41. BHLHE40/41 expression suppressed the reporter activities of SNAI1, SNAI2, and TWIST1 in most cases, except for the SNAI1 reporter in HEC-1 cells (Fig. 5A). Since BHLHE40/41 expression showed the greatest suppression of all three luciferase activities in HEC-6 cells, these cells were subsequently used in reporter assays. In contrast, the knockdown of BHLHE40/41 in HHUA cells upregulated the three reporter activities (Fig. 5B).
FIG 5.
Identification of the BHLHE40/41 responsible site in the TWIST1 promoter. (A) Reporter analysis of the SNAI1, SNAI2, and TWIST1 promoters in Ishikawa, HEC-1, and HEC-6 cells transfected with vectors to express HA-BHLHE40 and/or FLAG-BHLHE41. (B) Reporter analysis of the SNAI1, SNAI2, and TWIST1 promoters in HHUA cells transfected with the vectors to knock down BHLHE40 and/or BHLHE41. (C) Truncation assay of TWIST1 reporters. Five kinds of reporters possessing upstream regions from bp −1876, −1587, −170, +34, and +116 from the transcription start site were analyzed for their activity. (D) The pTWIST1+116 reporters possessing a deletion or mutations at the SP1 binding site (SBS) were used to analyze their activity. (E) The control activity of the mutant pTWIST1+116 reporter was adjusted to the same value as that of the wild-type reporter to evaluate the effects of BHLHE40/41 expression (white bars). (F) Impact of the SBS in the full-length TWIST1 reporter was evaluated using the pTWIST1-1876 reporter possessing a mutation at the SBS. The control activity of the mutant pTWIST1-1876 reporter was adjusted to the same value as that of the wild-type reporter to evaluate the effects of BHLHE40/41 expression (white bars). n.s., not significant; *, P < 0.05; **, P < 0.01. #, P < 0.05; ##, P < 0.01 (significantly different from the pCDNA3 control). §, P < 0.05; §§, P < 0.01 (significantly different from the pCDNA3 control).
Identification of critical promoter regions regulating TWIST1 transcription.
A previous study reported that TWIST expression was associated with the depth of myometrial invasion and was an independent predictive factor of patient survival (38); therefore, we focused on the transcriptional regulation of TWIST1 by BHLHE40/41. The TWIST1 promoter was examined to identify the critical region for its activity and responsiveness to BHLHE40/41 expression. Truncated reporters were constructed as shown in Fig. 5C. The pTWIST1+116 reporter still exhibited high activity and also showed responsiveness to BHLHE40/41 expression (Fig. 5C). A search of the region from bp +116 to +418 by rVista 2.0 (http://rvista.dcode.org/) revealed that there were no conserved E-boxes, which is a well-known consensus motif for BHLHE40/41 to bind. We instead found that a consensus SP1 binding site (SBS) in the region from bp +151 to +184 was conserved between human and mouse. We focused on the SBS because BHLHE40 was previously reported to associate with the SBSs of the BIRC5 promoter (6). We generated a mutant reporter possessing a deletion in the SBS. Deletion of the region from bp +151 to +184 resulted in a marked decrease in basal activity. Responsiveness to BHLHE40/41 expression was also diminished (Fig. 5D). Based on these results, mutant reporters were generated by base substitutions (Table 2). While a mutation in any half of the region from bp +151 to +184 resulted in a partial decrease in basal reporter activity, a mutation in the entire region from bp +151 to +184 markedly diminished basal activity. Responsiveness to BHLHE40/41 expression also declined (Fig. 5D and E). The impact of the region from bp +151 to +184 was confirmed by analyzing a mutant reporter generated using the pTWIST1-1876 reporter. We observed similar results to that in the mutant pTWIST1+116 reporter (Fig. 5F).
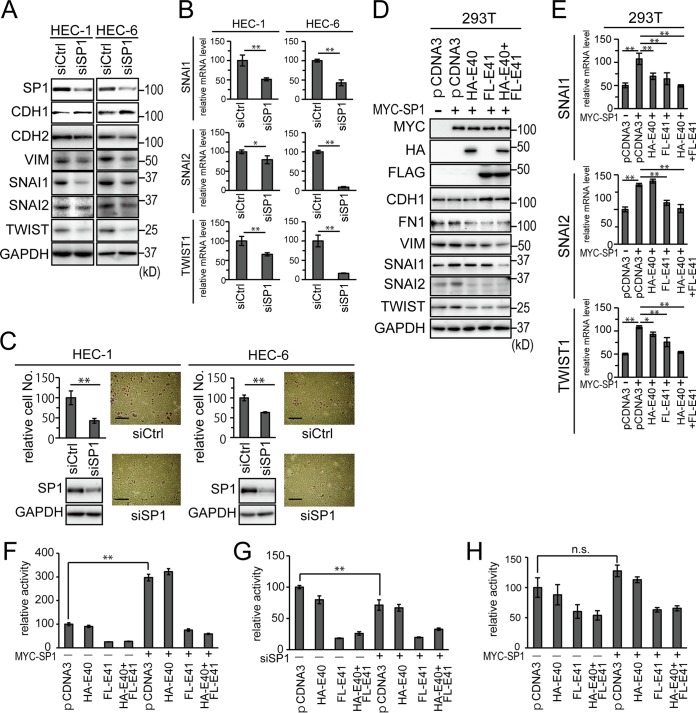
SP1 regulated SNAI and TWIST1 transcription.
The results obtained above suggested that the SBS was required for the basal transcriptional activity of the TWIST1 promoter and also that the SBS was required for the transcriptional suppression by BHLHE40/41. In order to verify whether SP1 was involved in the transcriptional regulation of TWIST1, HEC-1 and HEC-6 cells were used to knock down SP1 expression. The successful knockdown of SP1 by siRNA transfection resulted in lower SNAI1, SNAI2, and TWIST1 mRNA and protein levels (Fig. 6A and B). The upregulation of CDH1, downregulation of CDH2 and VIM, and suppression of in vitro cell invasion were observed in HEC-1 and HEC-6 cells with SP1 knocked down (Fig. 6A and C). The intensities of the bands in Fig. 6A were quantified and are shown in Fig. S6A in the supplemental material.
FIG 6.
SP1 is critical for TWIST1 transcription. (A and B) Immunoblotting analysis (A) and mRNA analysis (B) of EMT marker expression in HEC-1 and HEC-6 cells with SP1 knockdown. (C) HEC-1 and HEC-6 cells showing SP1 knockdown were used for the in vitro cell invasion assay. The graphs show quantification of the data. The scale bars represent 200 μm. (D and E) 293T cells transfected with MYC-SP1 alone or with a combination of MYC-SP1 and HA-BHLHE40 and/or FLAG-BHLHE41 were analyzed for their EMT marker expression at the protein (D) and mRNA (E) levels. (F) The wild-type pTWIST+116 reporter was transfected into HEC-6 cells with MYC-SP1 alone or with a combination of MYC-SP1 and HA-BHLHE40 and/or FLAG-BHLHE41, and its activity was compared with that of control transfectants without MYC-SP1. (G) Same as panel F, but siSP1 was used instead of MYC-SP1. (H) Same as panel F, but the pTWIST+116 reporter possessing mutations at the SBS was used instead of the wild-type reporter. n.s., not significant; *, P < 0.05; **, P < 0.01.
We also examined the forced expression of SP1 in cells. SP1 is a large protein (∼106 kDa) with a low transfection efficiency in HEC cell lines; therefore, 293T cells were used to obtain high expression levels of SP1. MYC-tagged SP1 was expressed with empty pCDNA3 or HA-BHLHE40 and/or FLAG-BHLHE41. The upregulation of SNAI1, SNAI2, and TWIST1 was demonstrated by SP1 transfection. The cotransfection of SP1 and BHLHE40/41 resulted in the resuppression of SNAI1, SNAI2, and TWIST1 (Fig. 6D and E). The intensities of the bands in Fig. 6D were quantified and shown in Fig. S6B in the supplemental material.
A reporter assay was performed to show that SP1 regulated TWIST1 transcription by associating with the SBS of the TWIST1 promoter. SP1 expression enhanced TWIST1 reporter activity, whereas BHLHE40/41 still suppressed it (Fig. 6F). The enhancement in reporter activity disappeared following the introduction of a mutation in the SBS in the TWIST1 reporter (Fig. 6H). The suppression of activity by BHLHE40/41expression was markedly weaker in the mutant reporter than in the wild type (Fig. 6H). As expected, basal reporter activity was markedly suppressed when SP1 expression was knocked down by siRNA (Fig. 6G). The protein expression of each gene construct transfected with the reporter genes was confirmed by immunoblotting (see Fig. S2D and E in the supplemental material). An examination of clinical samples revealed an inverse correlation between TWIST1 and BHLHE40/41 (Fig. 1F and G and 2P and Q).
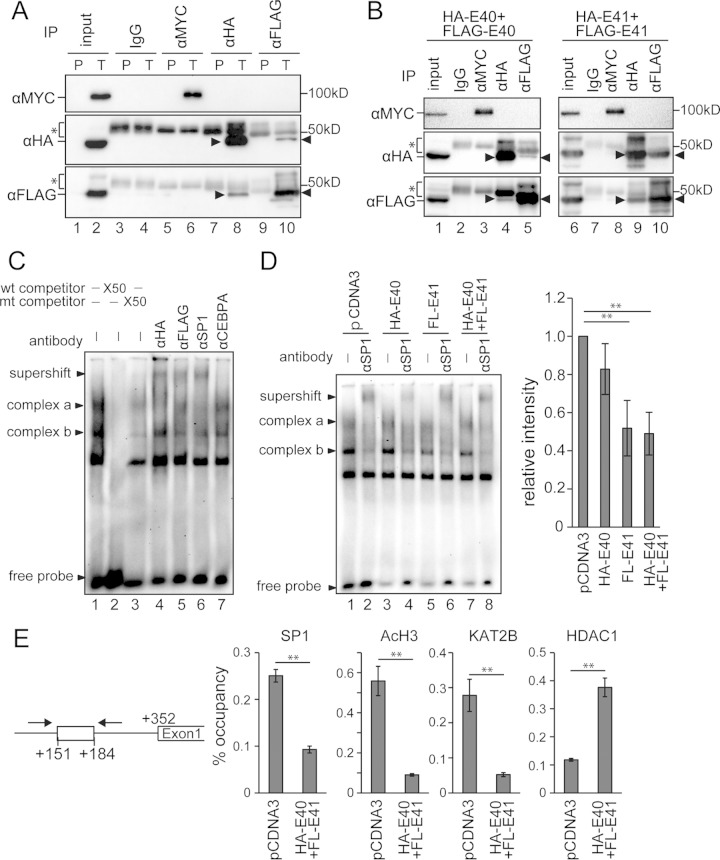
BHLHE40/41 competed with SP1 for binding to the SP1 binding site of the TWIST1 promoter.
The results described above suggested that BHLHE40/41 and SP1 both affected the same region of the TWIST1 promoter to regulate its activity. In order to elucidate the mechanism underlying SP1 inhibition by BHLHE40/41, we raised three possibilities: (i) BHLHE40/41 suppress SP1 expression, (ii) a physical interaction between SP1 and BHLHE40/41 inhibits SP1 function, and (iii) BHLHE40/41 compete with SP1 for binding to the same DNA site. In order to examine the first possibility, the protein levels of SP1 were assayed in HEC cells in which BHLHE40/41 expression had been modified. SP1 protein levels were not altered in HEC cells showing the forced expression of BHLHE40/41 (Fig. 3C; see Fig. S5A in the supplemental material). The knockdown of BHLHE40/41 in HHUA cells also did not alter SP1 expression (Fig. 4A; see also Fig. S5B).
Regarding the second possibility, we investigated whether BHLHE40/41 and SP1 associated with each other by use of an immunoprecipitation assay. Cell lysates from 293T cells transfected with MYC-SP1, HA-BHLHE40, and FLAG-BHLHE41 were used for immunoprecipitation with an anti-MYC, -HA, or -FLAG antibody. The results obtained suggest that SP1 associated with neither BHLHE40 nor BHLHE41 (Fig. 7A, lanes 6, 8, and 10). Consistent with previous findings (39), our results also demonstrated that BHLHE40 associated with BHLHE41 (Fig. 7A, lanes 8 and 10). In order to carefully exclude the possibility that SP1 associated with a homodimer of BHLHE40-BHLHE40 or BHLHE41-BHLHE41, 293T cells transfected with a combination of MYC-SP1, HA-BHLHE40, and FLAG-BHLHE40 or a combination of MYC-SP1, HA-BHLHE41, and FLAG-BHLHE41 were used for immunoprecipitation. The results obtained show that SP1 associated with neither the BHLHE40-BHLHE40 dimer nor the BHLHE41-BHLHE41 dimer (Fig. 7B, lanes 3 to 5 and 8 to 10). By a relative comparison, the amount of the BHLHE41-BHLHE41 dimer was markedly larger than that of the BHLHE40-BHLHE40 or BHLHE40-BHLHE41 dimer (Fig. 7A, compare lanes 8 and 10; Fig. 7B, lanes 4 and 5; Fig. 7B, lanes 9 and 10).
FIG 7.
BHLHE40/41 suppressed SP1 function by a competitive mechanism for binding to the SBS of the TWIST1 promoter. (A) Immunoprecipitation (IP) assay using 293T cells transfected with MYC-SP1, HA-BHLHE40, and FLAG-BHLHE41. The cell lysate was immunoprecipitated with the antibodies shown at the top and immunoblotted with the antibodies indicated on the left (α, anti-). P, cells transfected with pCDNA3 alone; T, cells transfected with all three constructs. (B) IP assay using 293T cells transfected with MYC-SP1, HA-BHLHE40, and FLAG-BHLHE40 (left panels) or 293T cells transfected with MYC-SP1, HA-BHLHE41, and FLAG-BHLHE41 (right panels). The arrowheads indicate the target bands. The bands indicated by asterisks are IgG heavy chains. (C) Electrophoretic mobility shift assay using the nuclear extract of 293T cells transfected with HA-BHLHE40 and FLAG-BHLHE41. Two main SP1-DNA complexes (complexes a and b) were formed. Supershift bands were formed by incubation with anti-HA, -FLAG, and -SP1 antibodies. An anti-CEBPA antibody was used as a negative control. wt, wild type; mt, mutant. (D) Nuclear extracts from 293T cells transfected with HA-BHLHE40 and/or FLAG-BHLHE41 were incubated with the labeled SBS probe. An anti-SP1 antibody was used to form supershift bands. The right graph shows the quantified band intensities of complex b in lanes 1, 3, 5, and 7 from four independent experiments. (E) Chromatin immunoprecipitation assay using 293T cells transfected with HA-BHLHE40 and FLAG-BHLHE41. Protein-DNA complexes immunoprecipitated with each of the anti-SP1, -acetylated H3, -KAT2B, and -HDAC1 antibodies were used to amplify the SBS site by PCR. The 10% input samples were used to calculate the occupancy ratio (%) from the values measured by real-time PCR. **, P < 0.01.
Concerning the third possibility, we initially determined whether BHLHE40/41 and SP1 directly associated with the SBS of the TWIST1 promoter using an EMSA. The nuclear extract from 293T cells transfected with HA-BHLHE40 and FLAG-BHLHE41 was used to form protein-DNA complexes with the digoxigenin-labeled oligonucleotide probe. Antibodies against HA, FLAG, and SP1 were used to demonstrate that the protein-DNA complexes specifically contained BHLHE40, BHLHE41, and SP1 (Fig. 7C, lanes 4 to 6, supershift). These results strengthened the possibility that BHLHE40/41 competed with SP1 for binding to the SBS of the TWIST1 promoter. In order to examine whether BHLHE40/41 expression inhibited SP1 binding to the SBS, nuclear extracts from 293T cells transfected with either HA-BHLHE40, FLAG-BHLHE41, or both were used to form complexes with the SBS probe. The EMSA showed that BHLHE41 expression alone or the combination of BHLHE40 and BHLHE41 expression markedly diminished the SP1-DNA complexes (Fig. 7D, complex b, compare lanes 1, 3, 5, and 7). BHLHE40 alone had a modest inhibitory effect on the SP1-DNA complexes. These results were consistent with those from the reporter assays showing that BHLHE41 had stronger effects than BHLHE40. Complex b was confirmed to contain SP1 because of the disappearance of the band and supershift formation after incubation with an anti-SP1 antibody (Fig. 7D, complex b, lanes 2, 4, 6, and 8). The inhibition of SP1 binding to the SBS by BHLHE40/41 expression was also demonstrated using a chromatin immunoprecipitation assay. 293T cells expressing HA-BHLHE40 and FLAG-BHLHE41 were compared with control cells for the level of SP1-DNA binding. BHLHE40/41 expression markedly diminished SP1 binding to the SBS of the TWIST1 promoter (Fig. 7E). As reported previously, BHLHE40/41 expression recruited HDAC1 and excluded KAT2B and acetylated histone H3 from the SBS (11, 40, 41) (Fig. 7E).
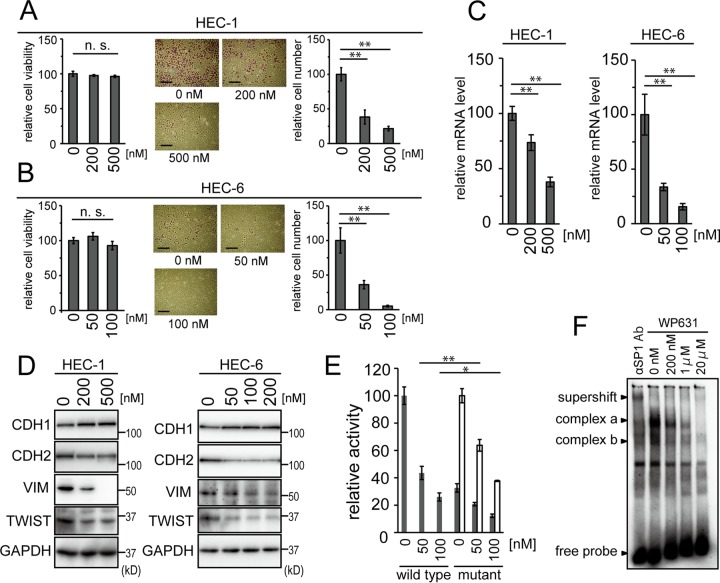
Inhibition of SP1 binding to the SBS of the TWIST1 promoter by WP631.
In order to confirm that SP1 binding to the SBS was critical for the transcriptional activation of the TWIST1 promoter, a chemical compound, WP631, was introduced. WP631 is a fluorescent bisintercalating anthracycline antibiotic which has been shown to inhibit SP1 binding to DNA by its extremely high DNA binding affinity (42). We searched for noncytotoxic concentrations of WP631 for HEC-1 and HEC-6 (Fig. 8A and B, left graphs). In vitro cell invasion of HEC-1 and HEC-6 cells was inhibited in a dose-dependent manner (Fig. 8A and B, middle panels and right graphs). The upregulation of CDH1 and downregulation of CDH2 and VIM were observed to occur in a dose-dependent manner (Fig. 8D). The intensities of the bands were quantified and are shown in Fig. S6C in the supplemental material. TWIST1 expression in cells was also dose dependently suppressed at the mRNA and protein levels (Fig. 8C and D; see also Fig. S6C). As expected, wild-type TWIST1 reporter activity was suppressed in a dose-dependent manner, and the TWIST1 reporter possessing a mutation in the SBS was less responsive to the addition of WP631 (Fig. 8E). Our EMSA directly demonstrated that WP631 inhibited SP1-DNA complexes in a dose-dependent manner (Fig. 8F).
FIG 8.
The SP1 inhibitor WP631 was used to suppress EMT in HEC cells. (A and B) Noncytotoxic concentrations of WP631 were used in HEC-1 (A) and HEC-6 (B) cells in order to examine in vitro cell invasion. The left graphs show cell viability assayed at each concentration of WP631. The right graphs show quantification of the data. The scale bars represent 200 μm. (C) The mRNA levels of TWIST1 in HEC-1 and HEC-6 cells treated with each concentration of WP631 were examined by real-time RT-PCR. (D) Protein levels of EMT markers, including TWIST, in HEC-1 and HEC-6 cells were analyzed by immunoblotting. (E) The wild-type and mutant pTWIST1+116 reporters were used to examine their responses to WP631 in HEC-6 cells. The control activities of the mutant reporter were adjusted to the same values as those of the wild-type reporter and are shown as white bars. (F) Electrophoretic mobility shift assay showing the inhibition of SP1-DNA complexes by WP631 by use of the nuclear extract from 293T cells. An anti-SP1 antibody was used to confirm that the complexes contained SP1 (supershift). n.s., not significant; *, P < 0.05; **, P < 0.01.
DISCUSSION
BHLHE40 and BHLHE41 (BHLHE40/41) have been suggested to play roles in carcinogenesis, cancer development, invasion, and metastasis. However, the detailed functions and mechanisms of BHLHE40/41 in cancer invasion have not yet been clarified. In the present study, we used human uterine endometrial cancer (HEC) cells to examine the influence of BHLHE40/41 on the epithelial-to-mesenchymal transition (EMT). TWIST expression is known to be crucial for invasion and is an independent factor that predicts the prognosis of HEC patients (38). In this study, we demonstrated for the first time that BHLHE40 and BHLHE41 suppressed the transcription of TWIST1 by competing with SP1 for binding to the SP1 binding site (SBS) of the TWIST1 promoter. In addition to TWIST1, our results showed that BHLHE40/41 suppressed the transcription of SNAI1 and SNAI2 activated by SP1 (Fig. 6E). These results suggest that a transcriptional mechanism that includes SP1 and BHLHE40/41 was also involved in the transcriptional regulation of SNAI1 and SNAI2.
Although several studies have reported the overexpression of BHLHE40 in diverse types of cancer species, no such changes have been found in BHLHE40 expression between cancer and normal tissues in non-small-cell lung cancer or endometrial cancer (16, 17). Our results showed that BHLHE40 expression was already present in normal endometrial tissues and that there were only modest differences in BHLHE40 mRNA levels between HEC and normal endometrial tissues (Fig. 1A, 2A, and 2C). A previous study reported that the normal rat uterus abundantly expressed BHLHE40 mRNA but not BHLHE41 mRNA (43). Our results showed that in contrast to BHLHE40, BHLHE41 expression was absent in normal endometrial tissues and also that the expression of BHLHE41 mRNA was higher in HEC specimens than in normal endometrial tissues (Fig. 1B and 2B and 2D). This is also consistent with previous findings (17). Although the mechanism underlying the upregulated expression of BHLHE41 in the process of carcinogenesis has not been elucidated, several studies have indicated that BHLHE41 suppresses apoptosis and enhances genomic instability (25, 44–46). These findings suggest that the upregulated expression of BHLHE41 is expedient for carcinogenesis in endometrial epithelial cells.
Although some studies have found a positive correlation between the overexpression of BHLHE40/41 and tumor progression or a poor prognosis, others reported an inverse correlation (18–21, 24–26, 47, 48). Our results showed that the expression of BHLHE40 and BHLHE41 inversely correlated with clinicopathological stages (Fig. 1C and D and 2M and N). Since the exogenous expression of BHLHE40/41 is known to inhibit cell proliferation, migration, and invasion in multiple cell types, they may be tumor suppressors (3, 4, 18, 26–28, 30). While previous studies demonstrated that BHLHE40 and BHLHE41 mutually suppressed the transcription of each other in vitro (1, 2), our results clearly showed a positive relationship between BHLHE40 and BHLHE41 expression levels (Fig. 1E and 2O). BHLHE40/41 expression is regulated in multiple ways, and the regulation mechanism of BHLHE40/41in the normal endometrium and HEC remains to be elucidated (49, 50).
Although BHLHE40 and BHLHE41 have been suggested to play opposite roles (28, 45), BHLHE40 may have effects similar to those of BHLHE41 on some target genes (9, 44, 51, 52). Previous studies reported similar effects between BHLHE40 and BHLHE41, whereas others showed that BHLHE40 had weaker effects on target genes than BHLHE41 in vitro (51, 52). Our results were consistent with the latter findings. Since the expression levels of BHLHE40 and BHLHE41 were similar (see Fig. S2A in the supplemental material), structural differences may determine their impact. Our immunoprecipitation results suggested that the BHLHE41-BHLHE41 homodimer was the more preferred form to the BHLHE40-BHLHE40 homodimer or the BHLHE40-BHLHE41 heterodimer (Fig. 7A and B). These results may explain why BHLHE41 suppressed EMT effectors more effectively than BHLHE40.
Transcriptional suppression by BHLHE40/41 is known to mediate E-boxes on target promoters (1–3). On the other hand, BHLHE40/41 were previously shown to modulate the expression of some genes in an E-box-independent manner. BHLHE40 in particular was found to associate with SBSs in order to regulate a target gene (6). Our results also suggest that BHLHE40/41 regulated the transcription of TWIST1 by suppressing SP1 binding to their promoters. Whereas Li et al. did not exclude the possibility that BHLHE40 affected SP1 expression or that BHLHE40 binds to SP1 (6), we examined both possibilities and demonstrated that BHLHE40 did not alter SP1 expression and that SP1 did not bind to BHLHE40 or BHLHE41. Therefore, we concluded that SP1 and BHLHE40/41 competed for binding to the same proximal region of the TWIST1 promoter. Since BHLHE41 in particular had higher affinity for the SBS, BHLHE41 efficiently extruded SP1 from the SBS (Fig. 7D). In the present study, we were unable to provide evidence to show that BHLHE40/41 suppressed promoter activity mediated by E-boxes. This was unexpected because a previous study reported that BHLHE41 suppressed TWIST1 in an E-box-dependent manner (30). This inconsistency remains to be clarified, and a cellular context may explain it.
Our results indicated that a mutation or deletion in the SBS did not completely abolish the suppression of reporter activity induced by the expression of BHLHE40/41 (Fig. 5D and E). Similar findings were observed by Li et al., who made a mutant reporter using a pGL3 basic luciferase vector that still responded to the expression of BHLHE41 (1). We used a pGL4.22 basic luciferase vector for the reporter assay. Although many of the candidate binding sites for transcription factors present in a pGL3 basic backbone were previously reported to be eliminated in a pGL4 basic backbone, our results indicated that the pGL4.22 basic empty vector still responded to the expression of BHLHE41 (see Fig. S7A in the supplemental material). As we were unable to exclude the possibility that BHLHE40/41 associated with regions other than the SBS, this responsiveness may explain why our mutated reporter still responded to the expression of BHLHE41. On the other hand, the pGL4.22 basic empty vector did not respond to the expression of SP1 (see Fig. S7B in the supplemental material).
The bisanthracycline WP631 prefers to intercalate into G/C-rich DNA regions in a highly sequence-selective manner. WP631 is a potent inhibitor of SP1 and was previously demonstrated to compete with SP1 for binding to its consensus binding sites (42). Our results indicated that WP631 suppressed in vitro cell invasion by inhibiting TWIST1 transcription and EMT (Fig. 8). WP631 inhibited SP1 binding to the SBS of TWIST1 promoter in a dose-dependent manner (Fig. 8). Our results have provided a novel insight into the function of WP631 in the regulation of EMT, besides G2/M cell cycle arrest or the induction of apoptosis (53, 54). The application of small molecules, such as WP631, to control SP1 function may be a useful strategy to control EMT in cancer.
In conclusion, we clarified the impact of BHLHE40/41 expression in HEC cells. BHLHE40/41 suppressed the transcription of the EMT effectors SNAI1, SNAI2, and TWIST1. A detailed analysis of the TWIST1 promoter showed that SP1 was required for the basal transcription of TWIST1 and that BHLHE40/41 suppressed SP1 binding to the SBS of the TWIST1 promoter. Since SP1 is known to play a crucial role in carcinogenesis, exquisite modulation of the BHLHE40/41-SP1 axis may represent a strategy to control cancer invasion.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the technical support of Emiko Hori, Yumi Konno, and the Research Support Center, Graduate School of Medical Science, Kyushu University.
This work was supported by JSPS KAKENHI grant number 26462529, the Takeda Science Foundation, and the Foundation for the Advancement of Clinical Medicine (funds to K.A.).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00678-15.
REFERENCES
- 1.Li Y, Xie M, Song X, Gragen S, Sachdeva K, Wan Y, Yan B. 2003. DEC1 negatively regulates the expression of DEC2 through binding to the E-box in the proximal promoter. J Biol Chem 278:16899–16907. doi: 10.1074/jbc.M300596200. [DOI] [PubMed] [Google Scholar]
- 2.Azmi S, Sun H, Ozog A, Taneja R. 2003. mSharp-1/DEC2, a basic helix-loop-helix protein functions as a transcriptional repressor of E box activity and Stra13 expression. J Biol Chem 278:20098–20109. doi: 10.1074/jbc.M210427200. [DOI] [PubMed] [Google Scholar]
- 3.Zawel L, Yu J, Torrance CJ, Markowitz S, Kinzler KW, Vogelstein B, Zhou S. 2002. DEC1 is a downstream target of TGF-beta with sequence-specific transcriptional repressor activities. Proc Natl Acad Sci U S A 99:2848–2853. doi: 10.1073/pnas.261714999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun H, Taneja R. 2000. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc Natl Acad Sci U S A 97:4058–4063. doi: 10.1073/pnas.070526297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian Y, Zhang J, Jung YS, Chen X. 2014. DEC1 coordinates with HDAC8 to differentially regulate TAp73 and DeltaNp73 expression. PLoS One 9:e84015. doi: 10.1371/journal.pone.0084015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Xie M, Yang J, Yang D, Deng R, Wan Y, Yan B. 2006. The expression of antiapoptotic protein survivin is transcriptionally upregulated by DEC1 primarily through multiple sp1 binding sites in the proximal promoter. Oncogene 25:3296–3306. doi: 10.1038/sj.onc.1209363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanova AV, Ivanov SV, Zhang X, Ivanov VN, Timofeeva OA, Lerman MI. 2004. STRA13 interacts with STAT3 and modulates transcription of STAT3-dependent targets. J Mol Biol 340:641–653. doi: 10.1016/j.jmb.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 8.Sato F, Bhawal UK, Kawamoto T, Fujimoto K, Imaizumi T, Imanaka T, Kondo J, Koyanagi S, Noshiro M, Yoshida H, Kusumi T, Kato Y, Kijima H. 2008. Basic-helix-loop-helix (bHLH) transcription factor DEC2 negatively regulates vascular endothelial growth factor expression. Genes Cells 13:131–144. doi: 10.1111/j.1365-2443.2007.01153.x. [DOI] [PubMed] [Google Scholar]
- 9.Cho Y, Noshiro M, Choi M, Morita K, Kawamoto T, Fujimoto K, Kato Y, Makishima M. 2009. The basic helix-loop-helix proteins differentiated embryo chondrocyte (DEC) 1 and DEC2 function as corepressors of retinoid X receptors. Mol Pharmacol 76:1360–1369. doi: 10.1124/mol.109.057000. [DOI] [PubMed] [Google Scholar]
- 10.Azmi S, Ozog A, Taneja R. 2004. Sharp-1/DEC2 inhibits skeletal muscle differentiation through repression of myogenic transcription factors. J Biol Chem 279:52643–52652. doi: 10.1074/jbc.M409188200. [DOI] [PubMed] [Google Scholar]
- 11.Gulbagci NT, Li L, Ling B, Gopinadhan S, Walsh M, Rossner M, Nave KA, Taneja R. 2009. SHARP1/DEC2 inhibits adipogenic differentiation by regulating the activity of C/EBP. EMBO Rep 10:79–86. doi: 10.1038/embor.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park YK, Park H. 2012. Differentiated embryo chondrocyte 1 (DEC1) represses PPARgamma2 gene through interacting with CCAAT/enhancer binding protein beta (C/EBPbeta). Mol Cells 33:575–581. doi: 10.1007/s10059-012-0002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Zhang H, Xie M, Hu M, Ge S, Yang D, Wan Y, Yan B. 2002. Abundant expression of Dec1/stra13/sharp2 in colon carcinoma: its antagonizing role in serum deprivation-induced apoptosis and selective inhibition of procaspase activation. Biochem J 367:413–422. doi: 10.1042/bj20020514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Preusser M, Birner P, Ambros IM, Ambros PF, Budka H, Harris AL, Hainfellner JA. 2005. DEC1 expression in 1p-aberrant oligodendroglial neoplasms. Histol Histopathol 20:1173–1177. [DOI] [PubMed] [Google Scholar]
- 15.Shi XH, Zheng Y, Sun Q, Cui J, Liu QH, Qu F, Wang YS. 2011. DEC1 nuclear expression: a marker of differentiation grade in hepatocellular carcinoma. World J Gastroenterol 17:2037–2043. doi: 10.3748/wjg.v17.i15.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giatromanolaki A, Koukourakis MI, Sivridis E, Turley H, Wykoff CC, Gatter KC, Harris AL. 2003. DEC1 (STRA13) protein expression relates to hypoxia-inducible factor 1-alpha and carbonic anhydrase-9 overexpression in non-small cell lung cancer. J Pathol 200:222–228. doi: 10.1002/path.1330. [DOI] [PubMed] [Google Scholar]
- 17.Yunokawa M, Tanimoto K, Nakamura H, Nagai N, Kudo Y, Kawamoto T, Kato Y, Hiyama E, Hiyama K, Nishiyama M. 2007. Differential regulation of DEC2 among hypoxia-inducible genes in endometrial carcinomas. Oncol Rep 17:871–878. doi: 10.3892/or.17.4.871. [DOI] [PubMed] [Google Scholar]
- 18.Bhawal UK, Sato F, Arakawa Y, Fujimoto K, Kawamoto T, Tanimoto K, Ito Y, Sasahira T, Sakurai T, Kobayashi M, Kashima I, Kijima H, Kuniyasu H, Abiko Y, Kato Y, Sato S. 2011. Basic helix-loop-helix transcription factor DEC1 negatively regulates cyclin D1. J Pathol 224:420–429. doi: 10.1002/path.2878. [DOI] [PubMed] [Google Scholar]
- 19.Xu Q, Ma P, Hu C, Chen L, Xue L, Wang Z, Liu M, Zhu H, Xu N, Lu N. 2012. Overexpression of the DEC1 protein induces senescence in vitro and is related to better survival in esophageal squamous cell carcinoma. PLoS One 7:e41862. doi: 10.1371/journal.pone.0041862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Wang L, Lin XY, Wang J, Yu JH, Miao Y, Wang EH. 2013. The transcription factor DEC1 (BHLHE40/STRA13/SHARP-2) is negatively associated with TNM stage in non-small-cell lung cancer and inhibits the proliferation through cyclin D1 in A549 and BE1 cells. Tumour Biol 34:1641–1650. doi: 10.1007/s13277-013-0697-z. [DOI] [PubMed] [Google Scholar]
- 21.Wong VC, Ko JM, Qi RZ, Li PJ, Wang LD, Li JL, Chan YP, Chan KW, Stanbridge EJ, Lung ML. 2011. Abrogated expression of DEC1 during oesophageal squamous cell carcinoma progression is age- and family history-related and significantly associated with lymph node metastasis. Br J Cancer 104:841–849. doi: 10.1038/bjc.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, Xu L, Zhang J, Hu X, Liu Y, Yin H, Lv T, Zhang H, Liu L, An H, Liu H, Xu J, Lin Z. 2013. Sunitinib induces cellular senescence via p53/Dec1 activation in renal cell carcinoma cells. Cancer Sci 104:1052–1061. doi: 10.1111/cas.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian Y, Chen X. 2008. ID1, inhibitor of differentiation/DNA binding, is an effector of the p53-dependent DNA damage response pathway. J Biol Chem 283:22410–22416. doi: 10.1074/jbc.M800643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu T, He N, Yang Y, Yin C, Sang N, Yang Q. 2015. DEC2 expression is positively correlated with HIF-1 activation and the invasiveness of human osteosarcomas. J Exp Clin Cancer Res 34:22. doi: 10.1186/s13046-015-0135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inaguma S, Riku M, Hashimoto M, Murakami H, Saga S, Ikeda H, Kasai K. 2013. GLI1 interferes with the DNA mismatch repair system in pancreatic cancer through BHLHE41-mediated suppression of MLH1. Cancer Res 73:7313–7323. doi: 10.1158/0008-5472.CAN-13-2008. [DOI] [PubMed] [Google Scholar]
- 26.Montagner M, Enzo E, Forcato M, Zanconato F, Parenti A, Rampazzo E, Basso G, Leo G, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. 2012. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature 487:380–384. doi: 10.1038/nature11207. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Shen Q, Kim HT, Bissonnette RP, Lamph WW, Yan B, Brown PH. 2011. The rexinoid bexarotene represses cyclin D1 transcription by inducing the DEC2 transcriptional repressor. Breast Cancer Res Treat 128:667–677. doi: 10.1007/s10549-010-1083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato F, Kawamura H, Wu Y, Sato H, Jin D, Bhawal UK, Kawamoto T, Fujimoto K, Noshiro M, Seino H, Morohashi S, Kato Y, Kijima H. 2012. The basic helix-loop-helix transcription factor DEC2 inhibits TGF-beta-induced tumor progression in human pancreatic cancer BxPC-3 cells. Int J Mol Med 30:495–501. doi: 10.3892/ijmm.2012.1037. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y, Sato F, Yamada T, Bhawal UK, Kawamoto T, Fujimoto K, Noshiro M, Seino H, Morohashi S, Hakamada K, Abiko Y, Kato Y, Kijima H. 2012. The BHLH transcription factor DEC1 plays an important role in the epithelial-mesenchymal transition of pancreatic cancer. Int J Oncol 41:1337–1346. doi: 10.3892/ijo.2012.1559. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki M, Sato F, Bhawal UK. 2014. The basic helix-loop-helix (bHLH) transcription factor DEC2 negatively regulates Twist1 through an E-box element. Biochem Biophys Res Commun 455:390–395. doi: 10.1016/j.bbrc.2014.11.030. [DOI] [PubMed] [Google Scholar]
- 31.Jungert K, Buck A, von Wichert G, Adler G, Konig A, Buchholz M, Gress TM, Ellenrieder V. 2007. Sp1 is required for transforming growth factor-beta-induced mesenchymal transition and migration in pancreatic cancer cells. Cancer Res 67:1563–1570. doi: 10.1158/0008-5472.CAN-06-1670. [DOI] [PubMed] [Google Scholar]
- 32.Venkov C, Plieth D, Ni T, Karmaker A, Bian A, George AL Jr, Neilson EG. 2011. Transcriptional networks in epithelial-mesenchymal transition. PLoS One 6:e25354. doi: 10.1371/journal.pone.0025354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nam EH, Lee Y, Park YK, Lee JW, Kim S. 2012. ZEB2 upregulates integrin alpha5 expression through cooperation with Sp1 to induce invasion during epithelial-mesenchymal transition of human cancer cells. Carcinogenesis 33:563–571. doi: 10.1093/carcin/bgs005. [DOI] [PubMed] [Google Scholar]
- 34.Kyo S, Nakamura M, Kiyono T, Maida Y, Kanaya T, Tanaka M, Yatabe N, Inoue M. 2003. Successful immortalization of endometrial glandular cells with normal structural and functional characteristics. Am J Pathol 163:2259–2269. doi: 10.1016/S0002-9440(10)63583-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D, Takao T, Tsunematsu R, Morokuma S, Fukushima K, Kobayashi H, Saito T, Furue M, Wake N, Asanoma K. 2013. Inhibition of AHR transcription by NF1C is affected by a single-nucleotide polymorphism, and is involved in suppression of human uterine endometrial cancer. Oncogene 32:4950–4959. doi: 10.1038/onc.2012.509. [DOI] [PubMed] [Google Scholar]
- 36.Asanoma K, Kubota K, Chakraborty D, Renaud SJ, Wake N, Fukushima K, Soares MJ, Rumi MA. 2012. SATB homeobox proteins regulate trophoblast stem cell renewal and differentiation. J Biol Chem 287:2257–2268. doi: 10.1074/jbc.M111.287128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allred DC, Harvey JM, Berardo M, Clark GM. 1998. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol 11:155–168. [PubMed] [Google Scholar]
- 38.Kyo S, Sakaguchi J, Ohno S, Mizumoto Y, Maida Y, Hashimoto M, Nakamura M, Takakura M, Nakajima M, Masutomi K, Inoue M. 2006. High Twist expression is involved in infiltrative endometrial cancer and affects patient survival. Hum Pathol 37:431–438. doi: 10.1016/j.humpath.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Song X, Ma Y, Liu J, Yang D, Yan B. 2004. DNA binding, but not interaction with Bmal1, is responsible for DEC1-mediated transcription regulation of the circadian gene mPer1. Biochem J 382:895–904. doi: 10.1042/BJ20040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanov SV, Salnikow K, Ivanova AV, Bai L, Lerman MI. 2007. Hypoxic repression of STAT1 and its downstream genes by a pVHL/HIF-1 target DEC1/STRA13. Oncogene 26:802–812. doi: 10.1038/sj.onc.1209842. [DOI] [PubMed] [Google Scholar]
- 41.Hsiao SP, Huang KM, Chang HY, Chen SL. 2009. P/CAF rescues the Bhlhe40-mediated repression of MyoD transactivation. Biochem J 422:343–352. doi: 10.1042/BJ20090072. [DOI] [PubMed] [Google Scholar]
- 42.Martin B, Vaquero A, Priebe W, Portugal J. 1999. Bisanthracycline WP631 inhibits basal and Sp1-activated transcription initiation in vitro. Nucleic Acids Res 27:3402–3409. doi: 10.1093/nar/27.17.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossner MJ, Dorr J, Gass P, Schwab MH, Nave KA. 1997. SHARPs: mammalian enhancer-of-split- and hairy-related proteins coupled to neuronal stimulation. Mol Cell Neurosci 10:460–475. [PubMed] [Google Scholar]
- 44.Nakamura H, Tanimoto K, Hiyama K, Yunokawa M, Kawamoto T, Kato Y, Yoshiga K, Poellinger L, Hiyama E, Nishiyama M. 2008. Human mismatch repair gene, MLH1, is transcriptionally repressed by the hypoxia-inducible transcription factors, DEC1 and DEC2. Oncogene 27:4200–4209. doi: 10.1038/onc.2008.58. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Sato F, Kawamoto T, Fujimoto K, Morohashi S, Akasaka H, Kondo J, Wu Y, Noshiro M, Kato Y, Kijima H. 2010. Anti-apoptotic effect of the basic helix-loop-helix (bHLH) transcription factor DEC2 in human breast cancer cells. Genes Cells 15:315–325. doi: 10.1111/j.1365-2443.2010.01381.x. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y, Sato F, Bhawal UK, Kawamoto T, Fujimoto K, Noshiro M, Seino H, Morohashi S, Kato Y, Kijima H. 2012. BHLH transcription factor DEC2 regulates pro-apoptotic factor Bim in human oral cancer HSC-3 cells. Biomed Res 33:75–82. doi: 10.2220/biomedres.33.75. [DOI] [PubMed] [Google Scholar]
- 47.Chakrabarti J, Turley H, Campo L, Han C, Harris AL, Gatter KC, Fox SB. 2004. The transcription factor DEC1 (stra13, SHARP2) is associated with the hypoxic response and high tumour grade in human breast cancers. Br J Cancer 91:954–958. doi: 10.1038/sj.bjc.6602059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Miao Y, Wang J, Lin X, Wang L, Xu HT, Wang EH. 2013. DEC1 is positively associated with the malignant phenotype of invasive breast cancers and negatively correlated with the expression of claudin-1. Int J Mol Med 31:855–860. doi: 10.3892/ijmm.2013.1279. [DOI] [PubMed] [Google Scholar]
- 49.Yamada K, Miyamoto K. 2005. Basic helix-loop-helix transcription factors, BHLHB2 and BHLHB3; their gene expressions are regulated by multiple extracellular stimuli. Front Biosci 10:3151–3171. doi: 10.2741/1772. [DOI] [PubMed] [Google Scholar]
- 50.Kato Y, Kawamoto T, Fujimoto K, Noshiro M. 2014. DEC1/STRA13/SHARP2 and DEC2/SHARP1 coordinate physiological processes, including circadian rhythms in response to environmental stimuli. Curr Top Dev Biol 110:339–372. doi: 10.1016/B978-0-12-405943-6.00010-5. [DOI] [PubMed] [Google Scholar]
- 51.Choi SM, Cho HJ, Cho H, Kim KH, Kim JB, Park H. 2008. Stra13/DEC1 and DEC2 inhibit sterol regulatory element binding protein-1c in a hypoxia-inducible factor-dependent mechanism. Nucleic Acids Res 36:6372–6385. doi: 10.1093/nar/gkn620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamaguchi H, Fujimoto K, Kawamoto T, Noshiro M, Maemura K, Takeda N, Nagai R, Furukawa M, Honma S, Honma K, Kurihara H, Kato Y. 2004. Expression of the gene for Dec2, a basic helix-loop-helix transcription factor, is regulated by a molecular clock system. Biochem J 382:43–50. doi: 10.1042/BJ20031760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villamarin S, Ferrer-Miralles N, Mansilla S, Priebe W, Portugal J. 2002. Induction of G(2)/M arrest and inhibition of c-myc and p53 transcription by WP631 in Jurkat T lymphocytes. Biochem Pharmacol 63:1251–1258. doi: 10.1016/S0006-2952(02)00865-1. [DOI] [PubMed] [Google Scholar]
- 54.Mansilla S, Priebe W, Portugal J. 2006. Mitotic catastrophe results in cell death by caspase-dependent and caspase-independent mechanisms. Cell Cycle 5:53–60. doi: 10.4161/cc.5.1.2267. [DOI] [PubMed] [Google Scholar]
- 55.Pecorelli S. 2009. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol Obstet 105:103–104. doi: 10.1016/j.ijgo.2009.02.012 (Erratum, 108:176, 2010, .) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.