Abstract
During mouse preimplantation development, the generation of the inner cell mass (ICM) and trophoblast lineages comprises upregulation of Nanog expression in the ICM and its silencing in the trophoblast. However, the underlying epigenetic mechanisms that differentially regulate Nanog in the first cell lineages are poorly understood. Here, we report that BRG1 (Brahma-related gene 1) cooperates with histone deacetylase 1 (HDAC1) to regulate Nanog expression. BRG1 depletion in preimplantation embryos and Cdx2-inducible embryonic stem cells (ESCs) revealed that BRG1 is necessary for Nanog silencing in the trophoblast lineage. Conversely, in undifferentiated ESCs, loss of BRG1 augmented Nanog expression. Analysis of histone H3 within the Nanog proximal enhancer revealed that H3 lysine 9/14 (H3K9/14) acetylation increased in BRG1-depleted embryos and ESCs. Biochemical studies demonstrated that HDAC1 was present in BRG1-BAF155 complexes and BRG1-HDAC1 interactions were enriched in the trophoblast lineage. HDAC1 inhibition triggered an increase in H3K9/14 acetylation and a corresponding rise in Nanog mRNA and protein, phenocopying BRG1 knockdown embryos and ESCs. Lastly, nucleosome-mapping experiments revealed that BRG1 is indispensable for nucleosome remodeling at the Nanog enhancer during trophoblast development. In summary, our data suggest that BRG1 governs Nanog expression via a dual mechanism involving histone deacetylation and nucleosome remodeling.
INTRODUCTION
Cell fate decisions are crucial for the development of multicellular organisms. In higher animals, such as placental mammals, the first cell fate decision occurs during preimplantation development, when the totipotent blastomeres differentiate into the blastocyst inner cell mass (ICM) and trophoblast lineages (1). The proper development of the blastocyst ICM and trophoblast lineages is critical for embryo implantation, placentation, gastrulation, and full-term development. Abnormal development of the ICM and trophoblast lineages may contribute to pregnancy loss, reproductive disorders, and birth defects.
Early lineage formation in preimplantation embryos is mediated by a combination of transcriptional and epigenetic mechanisms (2, 3). During blastocyst formation, the expression of key transcription factors, such as octamer-binding transcription factor 4 (OCT4), Nanog homeobox (NANOG), and sex-determining region Y box 2 (SOX2), becomes restricted to the pluripotent ICM, while transcription factor AP-2 gamma (TFAP2C), GATA binding protein 3 (GATA3), and caudal type homeobox 2 (CDX2) are expressed exclusively in the trophoblast lineage (4–10). The spatial and temporal expression of these lineage-specific factors is controlled by position-dependent HIPPO signaling, transcription factor regulatory loops, and chromatin modifications (2, 3, 10–12). For example, the HIPPO signaling pathway differentially regulates lineage formation via the downregulation of CDX2 expression in the ICM and SOX2 expression in the trophoblast (10, 12). In conjunction with the HIPPO pathway, OCT4 and CDX2 negatively regulate one another's expression in the ICM and trophoblast lineages via binding to each other's promoters and cooperating with ERG-associated protein with SET domain (ESET) and BRG1 (Brahma-related gene 1), a chromatin-remodeling protein, to block transcription (13–15). Other epigenetic modifiers, such as embryonic ectoderm development (EED) and lysine (K)-specific demethylase 6B (KDM6B), work in opposition to restrict Cdx2 and Gata3 expression to the trophoblast lineage (16). Altogether, these studies demonstrate that ICM and trophoblast lineage development is regulated by overlapping transcriptional and epigenetic mechanisms.
Despite our current understanding of the mechanisms that regulate the spatial and temporal expression of Oct4, Gata3, and Cdx2, less is known about the epigenetic mechanisms that govern Nanog expression during the first cell fate decision in preimplantation embryos. Recent studies in mice revealed that Nanog expression is controlled by epigenetic modifications, such as Tet methylcytosine dioxygenase 1 (Tet1)-dependent 5-hydroxymethylcytosine (17) and coactivator-associated arginine methyltransferase 1 (CARM1)-dependent arginine methylation (18). Previous work in our laboratory demonstrated that Nanog expression is upregulated in BRG1 knockdown (KD) blastocysts (19). Furthermore, we and others established that BRG1 occupies the Nanog promoter in mouse embryonic stem cells (ESCs) (19, 20). Combined, these findings suggest that BRG1 may act as key regulator of Nanog expression during early lineage formation.
Here, we report that BRG1 functions as a major regulator of Nanog expression during early embryogenesis. In blastocysts, BRG1 is required for downregulation of Nanog in the trophoblast lineage. Conversely, in pluripotent ESCs, BRG1 regulates Nanog expression by fine-tuning the transcriptional outcome at the Nanog locus. We show that this mode of regulation depends on histone deacetylase 1 (HDAC1); in ESCs and preimplantation embryos, BRG1 interacts with HDAC1 to antagonize histone H3 lysine 9 and 14 (H3K9/14) acetylation at the Nanog proximal enhancer. Disruption of BRG1 and/or HDAC1 augments H3K9/14 acetylation and Nanog transcription. Finally, we show that, during ESC differentiation into trophoblast-like cells, BRG1 is required for nucleosome occupancy at the Nanog proximal enhancer and transcriptional start site (TSS). These findings demonstrate that during early embryonic development, BRG1 governs Nanog expression via a dual mechanism involving histone deacetylation and nucleosome remodeling.
MATERIALS AND METHODS
Embryo collection, embryo manipulation, and inhibitor treatment.
Mouse embryo collection, in vitro culture, and small interfering RNA (siRNA) microinjection were performed as previously described (7, 15, 21). Briefly, embryos were collected from either superovulated CF1 or B6D2/F1 females mated with B6D2/F1 males (Charles River Laboratories, Wilmington, MA). One-cell embryos were microinjected with 5 to 10 pl of 50 to 100 μM Brg1, Hdac1, or nontargeting control siRNA (siGenome SMARTpool; GE Dharmacon, Lafayette, CO). Following injection, the embryos were cultured in modified KSOM medium (EMD Millipore, Billerica, MA) for 3 to 4 days. For cell fate-mapping experiments, chimeric embryos were constructed as previously described (7). GFP mRNA was coinjected with Brg1 siRNA to track the fate of BRG1 KD blastomeres. At the 8-cell stage, acid Tyrode solution (Sigma, St. Louis, MO) was used to remove the zona pellucida. Two zona pellucida-free embryos were paired in a microwell and cultured until the blastocyst stage. Green fluorescent protein (GFP) expression was evaluated by epifluorescence on a Nikon Eclipse Ti inverted microscope equipped with LED illumination and a fluorescein isothiocyanate (FITC) filter. In a subset of experiments, compacted morulae were cultured in the presence of a histone deacetylase inhibitor. A 1 M stock solution of sodium butyrate (NaB) (Sigma) was prepared in water. NaB was then added to modified KSOM medium to achieve the desired concentration. Embryos were cultured in the presence of the inhibitor for 24 h until blastocyst formation. All animal work in this study was approved by the Institutional Animal Care and Use Committee (IACUC) at Michigan State University and conformed to the institutional guidelines and regulatory standards.
ESC culture, lentivirus transduction, and inhibitor treatment.
Cdx2-inducible mouse ESCs (Coriell Institute, Camden, NJ) were cultured as previously described, with slight modifications for lentiviral transduction of short hairpin RNA (shRNA) constructs (22). ESCs were initially propagated on mitomycin-treated, puromycin- and doxycycline-resistant mouse embryonic fibroblasts (MEFs) and then switched to gelatinized dishes for growth under feeder-free conditions. The growth medium consisted of high-glucose Dulbecco's modified Eagle's medium (DMEM) with l-glutamine and sodium pyruvate (Gibco, Life Technologies, Carlsbad, CA) supplemented with 20% fetal bovine serum (FBS) (HyClone, GE Healthcare Life Sciences, South Logan, UT), 0.1 mM nonessential amino acids, 0.1 mM β-mercaptoethanol, 100 U/ml leukemia-inhibitory factor (LIF) (EMD-Millipore), 1 μg/ml puromycin, and 0.2 μg/ml doxycycline. For lentiviral transduction of shRNA constructs, feeder-free ESCs were passaged and plated in doxycycline- and puromycin-free medium supplemented with 8 μg/ml Polybrene (Sigma) and a 1.25× concentration of lentiviral stock. Eight hours following viral transduction, doxycycline was returned to the medium to prevent expression of the transgene. After 24 h, puromycin was returned to the growth medium, and during subsequent medium changes, the level of puromycin was gradually increased to 6 μg/ml by 72 h after transduction. Cdx2 expression was induced by the removal of doxycycline from the medium. Mouse R1 ESCs (ATCC, Manassas, VA) were cultured in growth medium under conditions similar to those for the Cdx2-inducible cells, but without supplementation with doxycycline or puromycin. For the histone deacetylase inhibitor experiment, NaB from a 1 M NaB stock was added to the growth medium to the desired concentration. The cells were grown in the presence of inhibitor for 48 h.
RNA interference (RNAi) targeting sequences.
Lentiviral pLKO.1 vectors carrying shRNA sequences were used to knock down BRG1 in ESCs. The sequences of the hairpins were as follows: Brg1 shRNA, CCGGCGGCTCAAGAAGGAAGTTGAACTCGAGTTCAACTTCCTTCTTGAGCCGTTTTTG (Open Biosystems; TRCN0000071385), and scrambled shRNA, CCGGTCCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGGTTTTTG (Addgene plasmid number 1864). These plasmids were a kind gift from Gerald Crabtree for the Brg1 shRNA construct and David Sabatini for the scrambled shRNA construct (23, 24). Lentiviral particles were prepared at the University of Michigan Vector Core.
qRT-PCR and Western blot analysis.
Total RNA was extracted from ESCs using the RNeasy minikit (Qiagen, Valencia, CA) or from pools of 10 embryos using the PicoPure RNA isolation kit (Arcturus, Mountain View, CA). First-strand cDNA synthesis was performed using SuperScript II reverse transcriptase (Life Technologies, Carlsbad, CA). Quantitative reverse transcriptase (qRT)-PCR analysis was performed with TaqMan probes or gene-specific primers using SYBR green detection on a StepOne Plus thermocycler (Applied Biosystems, Foster City, CA). Data were analyzed by the ΔΔCT method and normalized to Ubtf for embryos or Eef1a1 for ESCs. Western blot analysis was performed as previously described (15, 22). In brief, whole ESC lysates were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. The antibodies against proteins detected by Western blotting were BRG1 (Santa Cruz Biotechnology, Dallas, TX; catalog no. sc-10768), M2 FLAG (Sigma; catalog no. F1802), NANOG (Cosmo Bio, Carlsbad, CA; catalog no. RCAB002P-F), β-actin (Sigma; catalog no. A5441), BAF155 (Santa Cruz Biotechnology; catalog no. sc-10756), HDAC1 (Santa Cruz Biotechnology; catalog no. sc-81598), and HDAC2 (Santa Cruz Biotechnology; catalog no. sc-7899). Intensity quantification of BRG1 and NANOG was performed using ImageJ (NIH).
Immunofluorescence, PLA, and confocal microscopy.
Immunofluorescent staining of preimplantation embryos was performed as previously described (7). In brief, morulae and blastocysts were fixed in 3.7% paraformaldehyde for 20 min, permeabilized, washed, blocked, and then incubated overnight in primary antibodies diluted in blocking solution. The following antibodies were used: NANOG (Cosmo Bio; catalog no. RCAB002P-F), TFAP2A (Santa Cruz Biotechnology; catalog no. sc-12726), BRG1 (Santa Cruz Biotechnology; catalog no. sc-10768), HDAC1 (Santa Cruz Biotechnology; catalog no. sc-81598), and acetyl-histone H3 (EMD Millipore; catalog no. 06-599). After washing, the embryos were incubated with secondary antibodies coupled to Alexa Fluor 488 or Alexa Fluor 594 (Molecular Probes, Eugene, OR). The embryos were mounted in Vectashield containing DAPI (4′,6-diamidino-2-phenylindole). For proximity ligation assay (PLA), the protocol was identical to our immunofluorescence procedure through the primary antibody incubation step. Washing steps, incubation with PLA probes, ligation, and amplification were performed using the Duolink in situ PLA kit according to the manufacturer's protocol (Olink Bioscience, Uppsala, Sweden). Imaging was performed on an inverted Olympus IX81 microscope equipped with an Olympus Fluoview 1000 confocal system (Olympus America, Center Valley, PA) or a spinning-disc confocal module (CARV; Atto Bioscience, Rockville, MD) with MetaMorph software (Molecular Devices, Sunnyvale, CA). Fluorescence intensities were quantified using ImageJ version 1.47 (National Institutes of Health). PLA quantification was performed using Blobfinder v3.2 (Center for Image Analysis, Uppsala University, Uppsala, Sweden).
ChIP assay.
Quantitative chromatin immunoprecipitation (ChIP) in ESCs and microscale ChIP (microChIP) coupled with whole-genome amplification for mouse blastocysts were performed as previously described (9, 15, 22). ESCs and mouse blastocysts (pools of 30 embryos per replicate) were fixed with 1% formaldehyde, quenched with glycine, washed in phosphate-buffered saline (PBS), and flash frozen. Sonicated chromatin extracts were incubated with antibodies specific for histone H3K9/K14 acetylation (EMD-Millipore; catalog no. 06-599), HDAC1 (Abcam, Boston, MA; catalog no. ab7028), BRG1 antiserum (obtained from Anthony Imbalzano, University of Massachusetts Medical School, Worcester, MA), or an isotype control (EMD-Millipore; catalog no. 12-370). Immunocomplexes were washed, eluted, de-cross-linked, and purified. Purified ChIP DNA from ESCs was directly ready for analysis. ChIP DNA from mouse blastocysts was amplified using the Genomeplex single-cell genome amplification kit (Sigma; WGA4). ChIP data were analyzed by real-time quantitative PCR (qPCR) on a StepOne Plus thermocycler with SYBR green reagents (Applied Biosystems) using the percent input method. The following primers were used to analyze the Nanog proximal enhancer: 5′-CTGGGTGCCTGGGAGAATAG-3′ and 5′-CCAACGGCTCAAGGCGATAG-3′. The intergenic control region was amplified with 5′-TTTTCAGTTCACACATATAAAGCAGA-3′ and 5′-TGTTGTTGTTGTTGCTTCACTG-3′.
Co-IP of nuclear complexes.
Nuclear coimmunoprecipitation (co-IP) was performed using the Nuclear Complex Co-IP kit (Active Motif, Carlsbad, CA) according to the manufacturer's protocol with some modifications. Nuclear extracts were prepared fresh from R1 ESCs. Following a two-step nuclear-lysis protocol, extracts were treated with nuclease enzyme included with the kit. Two hundred to 250 μg of nuclear extract was used per immunoprecipitation, as quantified by bicinchoninic acid (BCA) assay (Life Technologies). Dilution of nuclear extract and further washing steps were performed using 1× low-stringency co-IP buffer containing protease inhibitors and 150 mM NaCl. No dithiothreitol (DTT) was added to the co-IP buffer, contrary to the manufacturer's recommendation of 1 mM DTT. Prepared and diluted nuclear extracts were incubated with BRG1 antiserum (obtained from Anthony Imbalzano, University of Massachusetts Medical School, Worcester, MA) or HDAC1 antibody (Abcam, Boston, MA; catalog no. ab7028) or rabbit IgG (EMD-Millipore; catalog no. 12-370). Complexes were collected on magnetic protein G Dynabeads (Life Technologies). The beads were washed with co-IP buffer, and during the final wash step, the beads were divided for either direct analysis by Western blotting or HDAC enzymatic activity assay. For Western blot analysis of co-IP material, proteins were directly eluted from beads with 2× Laemmli buffer (130 mM Tris-HCl, pH 6.8, 4% SDS, 0.02% bromophenol blue, 100 mM DTT), boiled for 5 min. and loaded onto SDS-PAGE gels.
HDAC activity assay.
Enzymatic histone deacetylase activity was assayed using the Fluorescent HDAC Assay kit (Active Motif). Preparation of standards and control was performed according to the manufacturer's protocol. To assay co-IP material, beads bound with immunoprecipitated complexes were resuspended in 40 μl of HDAC assay buffer. Ten microliters of HDAC substrate was added, and samples were incubated at 37°C for 1 h. Following this incubation, a magnet was used to collect the beads, and the supernatant was transferred to a 96-well half-volume black microplate (Greiner Bio-One, Monroe, NC). Fifty microliters of developing solution was then added, following a 10-min incubation, fluorescence was measured using 360-nm excitation and 460-nm emission on a Gemini EM fluorescence microplate reader (Molecular Devices, Sunnyvale, CA).
Nucleosome-mapping assay.
The micrococcal nuclease (MNase) digestion conditions and mapping approach were adapted from a method previously described (25). Cells were formaldehyde fixed under conditions similar to those for the preparation of ChIP chromatin. The cells were lysed in a buffer composed of 10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 0.1% Triton X-100, and 1× protease inhibitor cocktail (Sigma; catalog no. P8340). Following centrifugation, the pellets were resuspended in MNase digestion buffer composed of 50 mM Tris-HCl, pH 7.4, 25 mM KCl, 4 mM MgCl2, 12.5% glycerol, and 1 mM CaCl2. Resuspended cells from each experimental group were split into several aliquots and then subjected to various concentrations of MNase (USB, Cleveland, OH) or left untreated with nuclease as a genomic control. Samples were then incubated at 37°C for 10 min, and digestion was stopped by addition of SDS-EDTA-EGTA buffer. Samples underwent DNA purification and then gel purification on a 1.8% agarose gel for the mononucleosome-size fragment. DNA from the 100-U/ml MNase treatment contained the most mononucleosome-size DNA and was used for downstream analyses. A tiling primer approach was used to map nucleosome density along an ∼1-kb region centered at the Nanog transcriptional start site. Primers were designed to obtain products around 100 bp in length, and each primer pair amplified a region located approximately 30 bp away from the region amplified by the adjacent primer pair. Control primers were used to amplify a regulatory region of the interleukin 12b gene (Il12b) and the RNA binding protein fox-1 homolog 3 gene (Rbfox3); these genes are repressed in the early cell lineages and do not undergo nucleosome remodeling. Primer sequences will be provided upon request. Relative protection from digestion by MNase was calculated by comparing amplification of MNase-digested DNA to that of an undigested control by a ΔCT method. The data were then normalized to the highest protection calculated from a known heterochromatic or repressed region (26).
Statistical analyses.
Student's t test was used to determine statistical significance between control and treatment groups, where appropriate. Analyses of variance (ANOVA) were performed using SAS 9.4 (SAS, Cary, NC). P values of <0.05 were considered statistically significant unless otherwise stated.
RESULTS
BRG1 regulates Nanog transcription during blastocyst formation.
Previously, we demonstrated that BRG1 is required for normal blastocyst development in mice (19). BRG1-depleted blastocysts exhibit defects in the trophoblast lineage and express increased levels of pluripotency genes. However, the precise role of BRG1 in trophoblast lineage specification and Nanog regulation was not investigated. To examine the biological role of BRG1 in early lineage formation, we first evaluated the expression and localization of Nanog mRNA and protein in BRG1 KD and control blastocysts (Fig. 1). To accomplish this, we microinjected Brg1 siRNA or control siRNA into fertilized 1-cell embryos. Using this approach, we can obtain a greater than 85% reduction in Brg1 transcripts (15, 19) and can phenocopy Brg1-null embryos (27). At the blastocyst stage, treated and control embryos were subjected to real-time qPCR and immunofluorescence analysis. To distinguish between the ICM and trophoblast lineages, blastocysts were costained with an antibody for transcription factor AP-2α (TFAP2A), a trophoblast-specific marker. These analyses demonstrated that Nanog transcripts were increased approximately 100% in BRG1 KD blastocysts compared to controls (P < 0.05) (Fig. 1A). Immunofluorescence staining showed that in control blastocysts the NANOG protein was restricted to cells in the ICM and was absent in the TFAP2A-positive trophoblasts (Fig. 1B). In contrast, in BRG1 KD blastocysts, the NANOG protein was expressed in both the ICM and trophoblast lineages. These results demonstrate that BRG1 is necessary for downregulation of Nanog expression in the emerging trophoblast lineage.
FIG 1.

BRG1 negatively regulates Nanog expression in blastocysts and is required for trophoblast lineage development. (A) qRT-PCR analysis of Nanog and Brg1 transcripts in BRG1 KD blastocysts relative to control blastocysts; the error bars represent standard errors of the mean (SEM) from 3 replicates. The dashed line denotes control expression, set at 1. The asterisks indicate a significant difference between BRG1 KD and control embryos (P < 0.05). (B) Confocal immunofluorescence analysis of NANOG expression and localization in BRG1 KD and control blastocysts. Embryos were costained with a TFAP2A antibody to highlight the ectopic expression of NANOG in the trophoblast lineage. Nuclei were counterstained with DAPI (blue) (scale bars = 20 μm). (C) Lineage-tracing experiments using chimeric embryos reveal that GFP-positive BRG1 KD blastomeres give rise to the ICM and fail to efficiently contribute to the trophoblast lineage. Shown are Hoffman modulation contrast (HMC) (top row) and GFP fluorescence (bottom row) in the respective group.
Because loss of BRG1 is associated with increased levels of NANOG, we tested whether BRG1-deficient blastomeres exhibit a preferential commitment to the ICM lineage. We generated chimeric embryos using blastomeres from 8-cell stage control embryos and GFP-labeled (GFP+) BRG1 KD embryos. Control-control and BRG1 KD-BRG1 KD chimeric embryos were used as additional controls. These experiments revealed that the vast majority (85%) of control-BRG1 KD blastocysts contained GFP-positive cells localized predominantly in the ICM (Fig. 1C). Altogether, these results demonstrate that BRG1 negatively regulates Nanog expression during blastocyst formation and suggest that BRG1 is important for establishment of the trophoblast lineage.
BRG1 modulates Nanog expression in ESCs and represses Nanog during differentiation into trophoblast-like cells.
To elucidate the underlying molecular mechanisms by which BRG1 regulates Nanog expression during trophoblast lineage formation, we utilized a Cdx2-inducible ESC model in tandem with preimplantation embryos. Cdx2-inducible ESCs differentiate into trophoblast-like cells that resemble native trophoblast stem (TS) cells in terms of gene expression and function (13, 28). Recently, we used this ESC-based model to characterize the transcriptional and epigenetic changes associated with pluripotency gene silencing during trophoblast differentiation (22). To determine the biological role of BRG1 in this context, a lentiviral-mediated RNAi approach was employed to deplete BRG1 (23). Seventy-two hours after Cdx2 induction, subsequent experiments were performed. An overview of the experimental design can be found in Fig. S1 in the supplemental material. The 72-h time point is based on a previous study in which we showed that the majority of transcriptional and epigenetic changes occur between 48 and 96 h after Cdx2 induction (22).
In the first set of experiments Nanog expression was evaluated in BRG1 KD and control ESCs with and without Cdx2 induction. Real-time qPCR analysis revealed that Nanog transcripts were upregulated approximately 50 and 100% in uninduced and Cdx2-induced BRG1 KD ESCs, respectively, compared to ESCs expressing scrambled shRNA (P < 0.05) (Fig. 2A). Consistent with this observation, Western blot analysis demonstrated that NANOG protein was increased in BRG1 KD Cdx2-induced ESCs (P < 0.05) (Fig. 2B and C). To examine the differentiation status of these BRG1 KD Cdx2-induced ESCs, several trophoblast and epithelial cell markers were evaluated (Fig. 2D). This analysis showed that E74-like factor 5 gene (Elf5), fibroblast growth factor receptor 2 gene (Fgfr2), eomesodermin gene (Eomes), claudin 4 gene (Cldn4), and Tfap2c gene transcripts were expressed at lower levels in BRG1 KD Cdx2-induced ESCs than in control Cdx2-induced ESCs (P < 0.05). Altogether, these results demonstrate that BRG1 functions as a negative regulator of Nanog expression and that downregulation of BRG1 in Cdx2-induced ESCs impairs trophoblast lineage differentiation. Importantly, the phenotype of BRG1 KD Cdx2-induced ESCs closely resembles that of BRG1-depleted blastocysts, providing a powerful cell-based model to investigate the molecular mechanisms of BRG1-dependent gene regulation during early embryonic development.
FIG 2.
BRG1 modulates Nanog in ESCs and represses Nanog during differentiation of ESCs into trophoblast-like cells. (A) qRT-PCR analysis of Nanog and Brg1 transcripts in BRG1 KD ESCs versus control ESCs at 72 h postinduction. The asterisks indicate a significant difference between BRG1 KD and control cells (P < 0.05). (B) Western blot analysis of BRG1, Flag-CDX2, and NANOG in uninduced and Cdx2-induced ESCs (72 h postinduction) infected with either a Brg1 or scrambled shRNA lentivirus. β-Actin was used as a loading control. (C) Quantification of BRG1 and NANOG protein levels in BRG1 KD uninduced and Cdx2-induced ESCs. BRG1 protein was reduced by 69% and 65% in the BRG1 KD uninduced and Cdx2-induced ESCs, respectively. NANOG was increased by 13% and 283% in the BRG1 KD uninduced and Cdx2-induced ESCs, respectively. A total of three Western blots were used for quantitation. (D) qRT-PCR analysis of trophoblast lineage markers in BRG1 KD and control Cdx2-induced ESCs relative to uninduced ESCs (72 h postinduction). The asterisks indicate a significant difference between control and BRG1 KD cells in Cdx2-induced groups (P < 0.05). A total of 3 replicates were performed. The error bars indicate SEM.
BRG1 antagonizes histone H3 acetylation at the Nanog proximal enhancer.
Since depletion of BRG1 causes an increase in Nanog transcripts, we postulated that BRG1 controls Nanog transcription via epigenetic modifications. One such modification is acetylated H3K9/14 (acH3K9/14). In eukaryotic cells, acH3K9/14 is tightly associated with transcriptionally active genes (29, 30), and in ESCs and trophoblast stem cells (TSCs), acH3K9/14 is enriched in regulatory regions of active genes (22, 31–33). To determine whether histone H3 acetylation is altered at the Nanog proximal enhancer, ChIP experiments were performed in BRG1 KD embryos and ESCs (Fig. 3). Real-time qPCR analysis revealed that acH3K9/14 was significantly increased at the Nanog proximal enhancer in BRG1 KD blastocysts and BRG1 KD Cdx2-induced ESCs (P < 0.05) (Fig. 3A and B). In uninduced BRG1 KD ESCs, acH3K9/14 was also increased (P < 0.05) (Fig. 3B), suggesting that BRG1 may modulate Nanog transcription in undifferentiated ESCs via histone acetylation. Control experiments revealed that the effects of BRG1 depletion on histone acetylation were specific to the Nanog enhancer; no changes in acH3K9/14 were observed in an intergenic region outside the Nanog locus (P > 0.05). These results strongly suggest that BRG1 regulates Nanog transcription by antagonizing acH3K9/14 at its proximal enhancer.
FIG 3.
BRG1 antagonizes histone H3K9/14 acetylation at the Nanog proximal enhancer in blastocysts and ESCs. (A) MicroChIP analysis of histone H3K9/14 acetylation at the OCT4-SOX2 binding motif (PE, proximal enhancer) of Nanog in control and BRG1 KD blastocysts. Preimmune rabbit IgG was used as a negative control. An intergenic control region was analyzed for specificity of the acetylation mark. (B) ChIP analyses of histone H3K9/14 acetylation in BRG1 KD and control ESCs. The same promoter regions were analyzed as in the embryo. ChIP was performed with both uninduced and Cdx2-induced ESCs (72 h postinduction) infected with either a Brg1 or a scrambled shRNA lentivirus. The asterisks indicate a significant difference in enrichment for the acetylation mark between the indicated groups (P < 0.05). The error bars indicate SEM.
BRG1 and HDAC1 form a functional complex during early embryogenesis.
BRG1 can activate and repress gene expression through different mechanisms (34). In some cell types, BRG1 can modulate gene expression independently of its chromatin-remodeling activity. For instance, BRG1 can act as a scaffold to recruit various coactivators and corepressors to target genes (35). Recently, we showed via ChIP that BRG1 and HDAC1 cooccupy the Nanog proximal enhancer in mouse ESCs (22). Hence, we hypothesized that BRG1 antagonizes acH3K9/14 by interacting with HDAC1. To test this, we immunoprecipitated BRG1 from ESC nuclear extracts using a rabbit polyclonal antibody (Fig. 4A). To substantiate that BRG1 and HDAC1 interactions were direct and not mediated by DNA bridging, co-IPs were carried out in ESC extracts treated with a nuclease. We confirmed that BAF155, a major component of BRG1/BAF complexes in mouse ESCs (23), coimmunoprecipitated with BRG1 in these extracts. Western blot analysis revealed that HDAC1, but not HDAC2, was present in these co-IPs. Conversely, immunoprecipitation of HDAC1 from ESC nuclear extracts pulled out BRG1 and BAF155. The interaction between BRG1 and HDAC1 was found to persist under a variety of co-IP conditions, including ESC extracts that were not nuclease treated (data not shown). To test whether BRG1 immunoprecipitates contained inherent HDAC activity, fluorescence-based HDAC activity assays were carried out in ESC nuclear extracts (Fig. 4B). HDAC1 and IgG immunoprecipitates were used as positive and negative controls, respectively. BRG1 immunoprecipitates contained intrinsic HDAC activity that was significantly greater than that of the IgG control (P < 0.05), suggesting that BRG1 may antagonize acH3K9/14 via HDAC1. Treatment of the ESC extract or a control HeLa cell extract with trichostatin A abolished the observed HDAC activity (Fig. 4C).
FIG 4.
BRG1 forms a histone deacetylation complex in ESCs. (A) BRG1 and HDAC1 protein complexes were coimmunoprecipitated from ESC nuclear lysates with antibodies for BRG1, HDAC1, or preimmune IgG and analyzed by Western blotting (WB). (B) Histone deacetylase activity of BRG1-immunoprecipitated material was measured by a fluorescence assay. HDAC1 and IgG immunoprecipitates were used as positive and negative controls, respectively. The asterisks indicate a significant difference in deacetylase activity between the indicated groups (P < 0.05). (C) Treatment of HeLa cell or ESC nuclear extracts with trichostatin A, a potent HDAC inhibitor, abolished the HDAC activity in the extracts. The error bars indicate SEM.
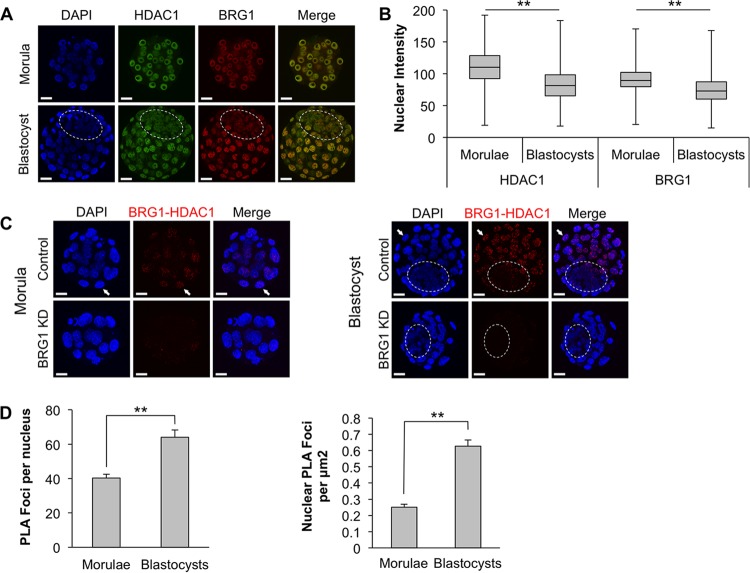
To test whether BRG1 interacts with HDAC1 during embryonic development, immunofluorescence assays and PLAs were carried out in preimplantation embryos. In the first set of experiments, the global expression of BRG1 and HDAC1 was evaluated during the morula-to-blastocyst transition, when Nanog expression is normally downregulated in the emerging trophoblast lineage (Fig. 5). In preliminary experiments, the specificity of each antibody was verified in knockdown experiments by microinjecting Brg1 and Hdac1 siRNAs into zygotes (19 and data not shown). Immunofluorescence analysis revealed that both BRG1 and HDAC1 were broadly expressed in the nuclei of morulae and blastocysts (Fig. 5A). During the morula-to-blastocyst transition, their overall expression was moderately reduced (P < 0.05) (Fig. 5B). Next, PLAs were performed to examine protein-protein interactions between BRG1 and HDAC1. This assay allows the visualization of proteins that are located within 30 nm of each other. To confirm that the observed interactions were specific, control experiments were carried out using BRG1 KD embryos. These experiments demonstrated that BRG1 and HDAC1 interact in morulae and that during blastocyst formation BRG1-HDAC1 interactions are enriched in the trophoblast lineage (P < 0.05) (Fig. 5C and D). Collectively, these experiments demonstrate that BRG1 and HDAC1 form a functional complex during early embryonic development that might antagonize histone acH3K9/14 at the Nanog proximal enhancer to govern its transcriptional activity.
FIG 5.
BRG1 and HDAC1 interactions are enriched in the trophoblast lineage during the morula-to-blastocyst transition. (A) Representative z-stack projections of morulae and blastocysts stained with BRG1 and HDAC1 antibodies. The position of the blastocyst ICM is indicated by the dashed oval (scale bars = 20 μm). (B) Summary of nuclear staining intensity calculated using ImageJ. The box plot represents the distribution of the intensities for all nuclei analyzed in each group. The top and bottom horizontal lines indicate the maximum and minimum values. The line within each box is the median. The asterisks indicate a significant difference in the average intensities of the two groups (P < 0.01). Centrally located z-sections from 23 morulae and 24 blastocysts were used to calculate the intensities. (C) Specific interactions between BRG1 and HDAC1 in morulae and blastocysts were determined using PLA, which detects interacting proteins (<30 nm apart). The red staining is representative of the BRG1-HDAC1 interaction; a greater number of foci is indicative of increased interactions. BRG1 KD embryos were used as a negative control. The arrows indicate representative nuclei containing BRG1-HDAC1 interactions. Nuclei were counterstained with DAPI. The location of the blastocyst ICM is indicated by the dashed oval (scale bar = 20 μm). (D) Quantification of PLA foci acquired using Blobfinder software. Centrally located z-sections from 19 morulae and 20 blastocysts, pooled from three independent PLA experiments, were analyzed for quantification of the PLA foci. Nuclei located along the outside edges of morulae and from the trophectoderms of blastocysts were used for the quantitative analysis. The error bars indicate SEM. The asterisks indicate a significant difference between the comparisons indicated (P < 0.01).
A BRG1-HDAC1 complex mediates Nanog repression during early embryonic development.
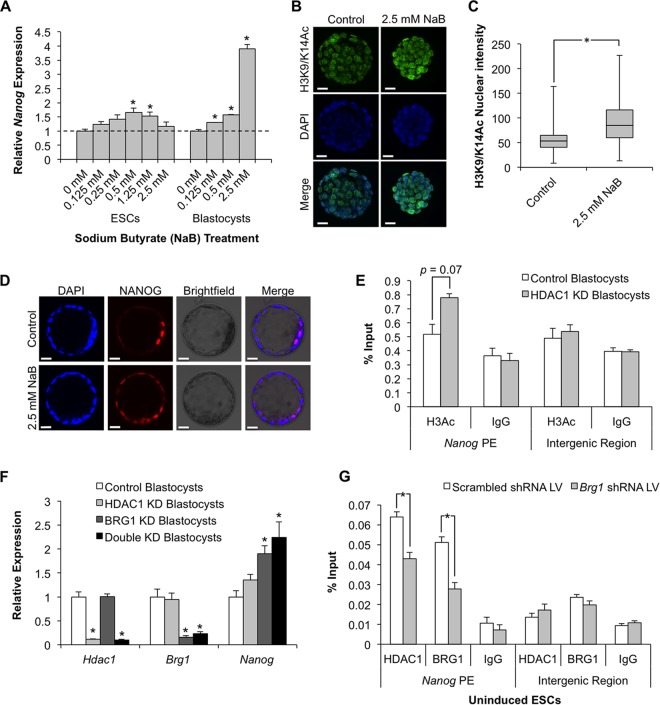
To further explore the potential relationship between BRG1, HDAC1, and Nanog transcription, several HDAC1 and/or BRG1 loss-of-function experiments were performed in ESCs and preimplantation embryos. Previous studies in mouse and human ESCs demonstrated that lower concentrations of NaB promote ES cell pluripotency (36). Furthermore, HDAC1 knockout ESCs exhibit increased levels of Nanog mRNA (32). Thus, we hypothesized that HDAC1 cooperates with BRG1 to negatively regulate Nanog expression during early embryonic development. In the first set of experiments, ESCs and preimplantation embryos at the morula stage were cultured in the presence of increased concentrations of the HDAC inhibitor NaB (Fig. 6). ESCs cultured in the presence of 0, 0.125, 0.25, 0.5, 1.25, and 2.5 mM NaB exhibited a bell-shaped dose response. At low (0.125 mM) and high (2.5 mM) concentrations, the levels of Nanog transcripts were similar to those in ESCs treated with the vehicle, whereas at intermediate doses (0.5 mM), Nanog transcripts were significantly increased (P < 0.05) (Fig. 6A). Morulae cultured in the presence of 0, 0.125, 0.5, and 2.5 mM NaB for 24 h exhibited an increase in histone acH3K9/14 and a dose-dependent rise in Nanog transcripts at the blastocyst stage compared to vehicle-treated controls (P < 0.05) (Fig. 6A to C). Similar to the phenotype of BRG1 KD blastocysts (15, 19), embryos cultured in the presence of NaB ectopically expressed NANOG in the trophoblast lineage (Fig. 6D). To confirm that HDAC1 is responsible for the observed changes in histone acetylation at the Nanog enhancer, microChIP experiments were performed on HDAC1-deficient blastocysts (Fig. 6E). To accomplish this, Hdac1 siRNA was microinjected into fertilized 1-cell embryos, and then the embryos were cultured to the blastocyst stage. qPCR analysis confirmed that there was an 88% reduction in Hdac1 transcripts. Analysis of histone acH3K9/14 revealed that there was an increase in histone acetylation (P = 0.0748) at the Nanog proximal enhancer that was similar to what was observed in BRG1 KD blastocysts. No changes were detected in an intergenic region outside the Nanog locus.
FIG 6.
HDAC1 cooperates with BRG1 to negatively regulate Nanog expression during early embryogenesis. (A) qRT-PCR analysis of Nanog transcripts following treatment of ESCs and embryos with various concentrations of NaB. Embryos at the morula stage were cultured in the presence of NaB for 24 h. Nanog expression was evaluated at the blastocyst stage. The asterisks indicate doses of NaB that elicited a significant increase in expression versus vehicle-treated samples. The dashed line denotes vehicle-treated control expression, set at 1. (B) Representative z-stack projection of immunofluorescent staining for histone H3K9/K14 acetylation (green) in NaB-treated embryos. Scale bars = 20 μm. (C) Box plot summary of nuclear staining intensities of H3K9/K14 acetylation calculated using ImageJ. The asterisk indicates a significant difference between the indicated samples (P < 0.05). (D) Representative immunofluorescence images of control and treated embryos stained for NANOG (red). Nuclei were counterstained with DAPI, and transmitted light images were also collected at the same focal plane to assess embryo morphology (scale bars = 20 μm). (E) MicroChIP analysis of histone H3K9/14 acetylation at the proximal enhancer (PE) of Nanog in control and HDAC1 KD blastocysts. Preimmune rabbit IgG was used as a negative control. An intergenic control region was analyzed for specificity of the acetylation mark. (F) qRT-PCR analysis of Nanog transcripts in blastocysts obtained following injection of siRNAs targeting Hdac1, Brg1, or both Hdac1 and Brg1. Gene expression is shown relative to embryos injected with a nontargeting control; the asterisks indicate a significant difference compared to the control (P < 0.05). (G) ChIP analysis of HDAC1 binding to the Nanog proximal enhancer in BRG1 KD and control ESCs. ChIP was also used to quantitate the degree of BRG1 enrichment at the Nanog proximal enhancer in each group. Preimmune rabbit IgG was used as a negative control. The asterisks indicate a significant difference in HDAC1 and BRG1 binding between the indicated groups (P < 0.05). The error bars indicate SEM.
To test whether manipulation of BRG1 and HDAC1 levels exerts an additive or synergistic effect on Nanog transcription, BRG1 and/or HDAC1 was downregulated in preimplantation embryos by microinjection of Brg1 and Hdac1 siRNAs alone or in combination. Manipulated embryos were cultured to the blastocyst stage, and Nanog transcripts were evaluated. Depletion of BRG1 or HDAC1 alone triggered 90 and 40% increases in Nanog transcripts, respectively (Fig. 6F). Moreover, combined depletion of BRG1 and HDAC1 resulted in a 124% increase in Nanog transcripts, which was greater than that with downregulation of BRG1 or HDAC1 alone (P < 0.05) (Fig. 6F).
To decipher the mechanism by which BRG1 cooperates with HDAC1 at the Nanog proximal enhancer, we performed ChIP assays for HDAC1 in BRG1 KD and control uninduced ESCs (Fig. 6G). We hypothesized that BRG1 is required for HDAC1 binding to the Nanog proximal enhancer. As a quality control, BRG1 ChIP was carried out in the same chromatin extracts. BRG1 occupancy was reduced by 46% in BRG1 KD ESCs compared to the control. This amount of reduction was consistent with the efficiency of the shRNA-mediated silencing of BRG1 in these extracts. Remarkably, we observed a 33% decrease in HDAC1 binding at the Nanog proximal enhancer in BRG1 KD ESCs compared to the control (P < 0.05), suggesting that BRG1 may be necessary for the recruitment and/or tethering of HDAC1 in the enhancer region. Altogether, these results indicate that BRG1 cooperates with HDAC1 to regulate Nanog transcription in the first cell lineages.
BRG1 is required for remodeling nucleosomes at the Nanog proximal enhancer during differentiation of ESCs into trophoblast-like cells.
The acquisition of nucleosomes in gene-regulatory regions serves as one mechanism for attenuating transcription in eukaryotic cells (37, 38). Recent work in our laboratory established that downregulation of Oct4 and Nanog expression in ESCs is associated with dynamic changes in chromatin structure at core enhancers (22). To test whether BRG1 is required for chromatin remodeling at the Nanog proximal promoter, nucleosome-mapping experiments were performed in ESCs (Fig. 7). BRG1 KD and control cells were treated with MNase, and isolated mononucleosome DNA was subjected to real-time qPCR analysis using overlapping primer sets that span the Nanog proximal enhancer and TSS. In addition, chromatin from MEFs was used as a positive control for increased nucleosome occupancy; in this cell type, Nanog is epigenetically silenced. In BRG1 KD and control uninduced ESCs, the Nanog proximal promoter region was largely devoid of nucleosomes (Fig. 7A). In control Cdx2-induced ESCs, two prominent nucleosomes that had intermediate density compared to the nucleosomes in MEFs were established at the Nanog proximal enhancer and TSS (P < 0.05) (Fig. 7B). Interestingly, in BRG1 KD Cdx2-induced ESCs, nucleosome remodeling was compromised. Nucleosome occupancy was low and resembled that in control and BRG1 KD uninduced ESCs (P > 0.05) (Fig. 7A and B). To rule out the possibility that there was a global change in nucleosome occupancy during differentiation of control ESCs into trophoblast-like cells, we performed qPCR on the regulatory regions of Il12b and Rbfox3. In ESCs and TSCs, these genes are repressed, and nucleosome occupancy should remain constant. The nucleosome densities at the regulatory elements of Il12b and Rbfox3 were similar in BRG1 KD and control ESCs before and during differentiation into trophoblast-like cells (Fig. 7C). Altogether, these results demonstrate that BRG1 is required for nucleosome remodeling at the Nanog proximal promoter during trophoblast lineage development.
FIG 7.

BRG1 is required for nucleosome remodeling at the Nanog proximal promoter during differentiation of ESCs into trophoblast-like cells. (A and B) MNase protection assays were performed on control and BRG1 KD ESCs (uninduced and Cdx2 induced) to determine nucleosome occupancy at the Nanog proximal promoter. MNase protection was determined by qRT-PCR using 23 primer pairs covering an ∼1-kb region around the Nanog TSS. MEFs were used as a control. (C) Control qPCRs for genomic regions that do not exhibit a change in nucleosome occupancy during Cdx2 induction or BRG1 KD were performed using primers for Il12b and Rbfox3. The error bars indicate SEM. One-way ANOVA revealed no change in the relative MNase protection at Il12b and Rbfox3 (P = 0.489 and P = 0.800, respectively).
DISCUSSION
Previous work from our laboratory and others established that Oct4, Nanog, and Sox2 are direct targets of BRG1 in mouse ESCs (15, 19, 20, 22). Notably, these studies demonstrated that BRG1 binding is enriched at key regulatory elements, such as enhancers and TSSs, within the Oct4, Nanog, and Sox2 genes. The results of the present study expand on these observations and provide new insights into the BRG1-dependent mechanisms that govern Nanog expression during early embryonic development in mice. We found that during early embryogenesis, (i) BRG1 is required for transcriptional silencing of Nanog, (ii) BRG1 regulates Nanog expression via interactions with HDAC1 and antagonism of histone acH3K9/14, and (iii) BRG1-dependent chromatin-remodeling activity is required for nucleosome remodeling at the Nanog proximal enhancer. Collectively, our data demonstrate that during mouse early embryogenesis, a BRG1-HDAC1 complex negatively controls Nanog transcription via a combination of histone H3K9/14 deacetylation and nucleosome remodeling. These overlapping modes of regulation are summarized in a model shown in Fig. 8.
FIG 8.
Model for BRG1-dependent regulation of Nanog transcription. Shown is a working model proposing a dual mechanism by which BRG1 regulates Nanog expression during trophoblast lineage development. BRG1/BAF155 modulates Nanog expression in pluripotent cells and the emerging trophoblast lineage by antagonizing H3K9/14 acetylation at the Nanog proximal enhancer. BRG1 achieves this by interacting with HDAC1. Upon differentiation, BRG1's chromatin-remodeling activity acts in conjunction with histone deacetylation to silence Nanog expression in the trophoblast lineage. The dashed circle on the left represents low nucleosome occupancy or the absence of a nucleosome. The gray circles represent the presence of a nucleosome or high nucleosome occupancy. Ac, acetyl.
During preimplantation development, proper cell fate decisions are vital for establishment of the pluripotent ICM and multipotent extraembryonic lineages. NANOG is a key regulator of pluripotency during early development (5, 39). In preimplantation embryos, Nanog is widely expressed initially, and then, during the morula-to-blastocyst transition, it becomes restricted to the ICM and is silenced in the extraembryonic lineages (5, 6). Functional studies in mice demonstrated that NANOG plays a pivotal role in the establishment of pluripotency during preimplantation development. Genetic ablation of Nanog results in failure to form the embryonic epiblast, an excess of extraembryonic endoderm, and lethality between days 3.5 and 5.5 of development (5). Moreover, Nanog-deficient ESCs differentiate into extraembryonic lineages, whereas overexpression of Nanog in ESCs promotes self-renewal (5, 39). Thus, Nanog is an essential gene required for establishment of pluripotency, and its precise regulation is critical for normal embryonic development.
In the current study, we show that BRG1-dependent regulation of Nanog may be important for trophoblast development in preimplantation embryos and Cdx2-inducible ESCs. Proper development of the trophoblast lineage requires downregulation of pluripotency genes (4) and upregulation of trophoblast-specific transcription factors, such as TFAP2C, CDX2, ELF5, and EOMES (4, 7, 40). Interestingly, in BRG1-deficient ESCs undergoing differentiation into trophoblast-like cells, there were significantly higher levels of Nanog mRNA and protein and lower levels of Elf5, Tfap2c, and Eomes mRNAs. Consistent with these findings, BRG1-deficient blastomeres in chimeric embryos exhibited a bias and preferentially contributed to the ICM. Since NANOG is a known suppressor of the extraembryonic lineages (5, 41–43), we envisage that BRG1-dependent repression of Nanog may serve as a mechanism to facilitate trophoblast lineage specification. Ongoing experiments in our laboratory are elucidating the precise role of BRG1 in trophoblast lineage development.
Although BRG1 was originally identified as an activator of gene expression in Saccharomyces cerevisiae (44, 45), work in mammalian cells over the last 15 years has shown that BRG1 can function as either an activator or a repressor of gene transcription (35). Its role as a transcriptional activator or repressor depends on both the cellular context and which coactivators or corepressors are present (46–48). In the present study, we show that BRG1 can differentially regulate Nanog transcription in ESCs and trophoblasts by two mechanisms. The first mode of regulation occurs in ESCs, where BRG1 interacts with the corepressor HDAC1 to antagonize histone acH3K9/14 at the Nanog proximal enhancer. Disruption of BRG1 and/or HDAC1 augmented acH3K9/14 and caused an increase in Nanog expression. This type of regulation appears to be important for fine-tuning Nanog expression in pluripotent cells, where a specific amount of NANOG is important for maintaining an ESC identity (5, 39, 49). The second mechanism operates during formation of the trophoblast lineage, when Nanog expression is normally silenced. Disruption of BRG1 and/or HDAC1 blocked Nanog repression by maintaining an open chromatin structure and sustaining higher levels of acH3K9/14 at the Nanog proximal enhancer. This type of regulation is likely critical for silencing Nanog expression in the trophoblast lineage. In future studies, it will be exciting to determine whether interactions between HDAC1 and BRG1 stimulate one another's activity to regulate Nanog expression. Moreover, it will be interesting to test whether OCT4 and CDX2 are involved in recruiting and/or regulating the activities of BRG1 and HDAC1 at the Nanog proximal enhancer. In this regard, we demonstrated that BRG1 can physically interact and/or colocalize with OCT4 and CDX2 at pluripotency gene enhancers (15, 22).
Our most novel and intriguing finding from the current study is the observation that BRG1 antagonizes histone H3K9/14 acetylation to modulate Nanog expression in undifferentiated ESCs. In support of this finding, a recent study in human ESCs showed that BRG1 can regulate lineage-specific genes via inhibition of histone H3K27 acetylation at their enhancers (50). Interestingly, genome-wide ChIP studies in mouse ESCs and T lymphocytes unexpectedly revealed that HDACs are enriched at regulatory elements of highly expressed genes (32, 51). This raises the question of how HDAC1 is targeted to the regulatory elements of active genes. In the present study, we observed reduced amounts of HDAC1 protein at the Nanog proximal enhancer in BRG1-depleted ESCs, indicating that BRG1 may be required for the recruitment and/or tethering of HDAC1 to gene-regulatory elements. Future experiments are necessary to establish the exact mechanism by which BRG1 cooperates with HDAC1 to modulate histone acetylation at gene enhancers. Modulation of histone acetylation levels via a BRG1-HDAC complex may serve as a much larger regulatory mechanism to control transcription of key pluripotency and lineage-specific genes to ensure proper development.
In summary, the results reported here demonstrate that BRG1 cooperates with HDAC1 to regulate Nanog expression in the early cell lineages. Disruption of BRG1 or HDAC1 activity perturbed Nanog expression and blocked embryonic development. Such information may be pertinent to understanding some causes of early embryonic failure in humans. Furthermore, our findings have broader implications in ESCs and induced pluripotency stem cells, where NANOG plays a crucial role in maintenance of self-renewal and pluripotency, as well as acquisition of pluripotency during nuclear reprogramming. Manipulation of BRG1 and/or HDAC1 expression in some cell types could serve as a tool to alter cell identity.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by a grant from the National Institute of General Medical Sciences to J.G.K. (GM095347) and grants from the National Institute of Child Health and Human Development to S.P. (HD079363, HD062546, and HD075233).
We thank Gerald Crabtree (Stanford University) for generously providing the Brg1 shRNA plasmids and Anthony Imbalzano (University of Massachusetts Medical School) for providing the rabbit BRG1 antiserum. We are grateful for Monique Floer's (Michigan State University) critical reading of the manuscript. We thank Michael McAndrew from the Floer laboratory for providing us with PCR primers for Il12b and Rbfox3.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00546-15.
REFERENCES
- 1.Cockburn K, Rossant J. 2010. Making the blastocyst: lessons from the mouse. J Clin Invest 120:995–1003. doi: 10.1172/JCI41229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul S, Knott JG. 2014. Epigenetic control of cell fate in mouse blastocysts: the role of covalent histone modifications and chromatin remodeling. Mol Reprod Dev 81:171–182. doi: 10.1002/mrd.22219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oron E., Ivanova N. 2012. Cell fate regulation in early mammalian development. Phys Biol 9:045002. doi: 10.1088/1478-3975/9/4/045002. [DOI] [PubMed] [Google Scholar]
- 4.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. 2005. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 5.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. 2003. The Homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113:631–642. doi: 10.1016/S0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 6.Dietrich JE, Hiiragi T. 2007. Stochastic patterning in the mouse pre-implantation embryo. Development 134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- 7.Choi I, Carey TS, Wilson CA, Knott JG. 2012. Transcription factor AP-2gamma is a core regulator of tight junction biogenesis and cavity formation during mouse early embryogenesis. Development 139:4623–4632. doi: 10.1242/dev.086645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuckenberg P, Buhl S, Woynecki T, van Furden B, Tolkunova E, Seiffe F, Moser M, Tomilin A, Winterhager E, Schorle H. 2010. The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol Cell Biol 30:3310–3320. doi: 10.1128/MCB.01215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S. 2009. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem 284:28729–28737. doi: 10.1074/jbc.M109.016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wicklow E, Blij S, Frum T, Hirate Y, Lang RA, Sasaki H, Ralston A. 2014. HIPPO pathway members restrict SOX2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst. PLoS Genet 10:e1004618. doi: 10.1371/journal.pgen.1004618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Z, Carey TS, Ganguly A, Wilson CA, Paul S, Knott JG. 2015. Transcription factor AP-2gamma induces early Cdx2 expression and represses HIPPO signaling to specify the trophectoderm lineage. Development 142:1606–1615. doi: 10.1242/dev.120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, Makita R, Kurihara H, Morin-Kensicki EM, Nojima H, Rossant J, Nakao K, Niwa H, Sasaki H. 2009. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 14.Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B, Ng HH. 2009. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev 23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Sengupta S, Magnani L, Wilson CA, Henry RW, Knott JG. 2010. Brg1 is required for Cdx2-mediated repression of Oct4 expression in mouse blastocysts. PLoS One 5:e10622. doi: 10.1371/journal.pone.0010622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha B, Home P, Ray S, Larson M, Paul A, Rajendran G, Behr B, Paul S. 2013. EED and KDM6B coordinate the first mammalian cell lineage commitment to ensure embryo implantation. Mol Cell Biol 33:2691–2705. doi: 10.1128/MCB.00069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. 2007. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kidder BL, Palmer S, Knott JG. 2009. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 20.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. 2009. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A 106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi I, Carey TS, Wilson CA, Knott JG. 2013. Evidence that transcription factor AP-2gamma is not required for Oct4 repression in mouse blastocysts. PLoS One 8:e65771. doi: 10.1371/journal.pone.0065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carey TS, Choi I, Wilson CA, Floer M, Knott JG. 2014. Transcriptional reprogramming and chromatin remodeling accompanies Oct4 and Nanog silencing in mouse trophoblast lineage. Stem Cells Dev 23:219–229. doi: 10.1089/scd.2013.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. 2009. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A 106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 25.Gévry N, Svotelis A, Larochelle M, Gaudreau L. 2009. Nucleosome mapping. Methods Mol Biol 543:281–291. doi: 10.1007/978-1-60327-015-1_19. [DOI] [PubMed] [Google Scholar]
- 26.Infante JJ, Law GL, Young ET. 2012. Analysis of nucleosome positioning using a nucleosome-scanning assay. Methods Mol Biol 833:63–87. doi: 10.1007/978-1-61779-477-3_5. [DOI] [PubMed] [Google Scholar]
- 27.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell 6:1287–1295. doi: 10.1016/S1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 28.Tolkunova E, Cavaleri F, Eckardt S, Reinbold R, Christenson LK, Scholer HR, Tomilin A. 2006. The caudal-related protein cdx2 promotes trophoblast differentiation of mouse embryonic stem cells. Stem Cells 24:139–144. doi: 10.1634/stemcells.2005-0240. [DOI] [PubMed] [Google Scholar]
- 29.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. 2007. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res 17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roh TY, Cuddapah S, Zhao K. 2005. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev 19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L. 2012. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13:424. doi: 10.1186/1471-2164-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kidder BL, Palmer S. 2012. HDAC1 regulates pluripotency and lineage specific transcriptional networks in embryonic and trophoblast stem cells. Nucleic Acids Res 40:2925–2939. doi: 10.1093/nar/gkr1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de la Serna IL, Ohkawa Y, Imbalzano AN. 2006. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet 7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 35.Trotter KW, Archer TK. 2008. The BRG1 transcriptional coregulator. Nucl Recept Signal 6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ware CB, Wang L, Mecham BH, Shen L, Nelson AM, Bar M, Lamba DA, Dauphin DS, Buckingham B, Askari B, Lim R, Tewari M, Gartler SM, Issa JP, Pavlidis P, Duan Z, Blau CA. 2009. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell 4:359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai L, Morozov AV. 2010. Gene regulation by nucleosome positioning. Trends Genet 26:476–483. doi: 10.1016/j.tig.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Voss TC, Hager GL. 2014. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet 15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113:643–655. doi: 10.1016/S0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 40.Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M. 2008. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol 10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hyslop L, Stojkovic M, Armstrong L, Walter T, Stojkovic P, Przyborski S, Herbert M, Murdoch A, Strachan T, Lako M. 2005. Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells 23:1035–1043. doi: 10.1634/stemcells.2005-0080. [DOI] [PubMed] [Google Scholar]
- 42.Chen L, Yabuuchi A, Eminli S, Takeuchi A, Lu CW, Hochedlinger K, Daley GQ. 2009. Cross-regulation of the Nanog and Cdx2 promoters. Cell Res 19:1052–1061. doi: 10.1038/cr.2009.79. [DOI] [PubMed] [Google Scholar]
- 43.Cho LT, Wamaitha SE, Tsai IJ, Artus J, Sherwood RI, Pedersen RA, Hadjantonakis AK, Niakan KK. 2012. Conversion from mouse embryonic to extra-embryonic endoderm stem cells reveals distinct differentiation capacities of pluripotent stem cell states. Development 139:2866–2877. doi: 10.1242/dev.078519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirschhorn JN, Brown SA, Clark CD, Winston F. 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev 6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 45.Winston F, Carlson M. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet 8:387–391. [DOI] [PubMed] [Google Scholar]
- 46.Zhang B, Chambers KJ, Faller DV, Wang S. 2007. Reprogramming of the SWI/SNF complex for co-activation or co-repression in prohibitin-mediated estrogen receptor regulation. Oncogene 26:7153–7157. doi: 10.1038/sj.onc.1210509. [DOI] [PubMed] [Google Scholar]
- 47.Xu Z, Meng X, Cai Y, Koury MJ, Brandt SJ. 2006. Recruitment of the SWI/SNF protein Brg1 by a multiprotein complex effects transcriptional repression in murine erythroid progenitors. Biochem J 399:297–304. doi: 10.1042/BJ20060873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu W, Cho H, Kadam S, Banayo EM, Anderson S, Yates JR 3rd, Emerson BM, Evans RM. 2004. A methylation-mediator complex in hormone signaling. Genes Dev 18:144–156. doi: 10.1101/gad.1141704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Darr H, Mayshar Y, Benvenisty N. 2006. Overexpression of NANOG in human ES cells enables feeder-free growth while inducing primitive ectoderm features. Development 133:1193–1201. doi: 10.1242/dev.02286. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X, Li B, Li W, Ma L, Zheng D, Li L, Yang W, Chu M, Chen W, Mailman RB, Zhu J, Fan G, Archer TK, Wang Y. 2014. Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Reports 3:460–474. doi: 10.1016/j.stemcr.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.