Abstract
The epigenetic marks located throughout the genome exhibit great variation between normal and transformed cancer cells. While normal cells contain hypomethylated CpG islands near gene promoters and hypermethylated repetitive DNA, the opposite pattern is observed in cancer cells. Recently, it has been reported that alteration in the microenvironment of melanocyte cells, such as substrate adhesion blockade, results in the selection of anoikis-resistant cells, which have tumorigenic characteristics. Melanoma cells obtained through this model show an altered epigenetic pattern, which represents one of the first events during the melanocytes malignant transformation. Because microRNAs are involved in controlling components of the epigenetic machinery, the aim of this work was to evaluate the potential association between the expression of miR-203, miR-26, and miR-29 family members and the genes Dnmt3a, Dnmt3b, Mecp2, and Ezh2 during cells transformation. Our results show that microRNAs and their validated or predicted targets are inversely expressed, indicating that these molecules are involved in epigenetic reprogramming. We also show that miR-203 downregulates Dnmt3b in mouse melanocyte cells. In addition, treatment with 5-aza-CdR promotes the expression of miR-26 and miR-29 in a nonmetastatic melanoma cell line. Considering the occurrence of CpG islands near the miR-26 and miR-29 promoters, these data suggest that they might be epigenetically regulated in cancer.
1. Introduction
Epigenetic modifications play a pivotal role in modulating gene expression during development and tissue differentiation, in the establishment and maintenance of genomic imprinting, and in X chromosome inactivation in female mammals. The most commonly studied type of epigenetic modification is the DNA methylation of CpG dinucleotides. In normal human cells DNA methylation typically occurs throughout the genome and at repetitive DNA, preserving genomic stability and inhibiting unwanted transposon reactivation [1–3]. CpG-rich regions, known as CpG islands [4], which are localized near 60% of coding gene promoters, are hypomethylated in normal cells [5]. Only one-tenth of these CpG islands are methylated in a tissue-specific manner [6].
DNA methylation is regulated through a family of DNA methyltransferases (Dnmt) comprising 5 members. Three of these proteins mediate the addition of a methyl group predominantly to DNA cytosines of CpG dinucleotide sequences. Dnmt1 is ubiquitously expressed at high levels in proliferating cells and is involved in the maintenance of DNA methylation. Dnmt3a and Dnmt3b promote de novo DNA methylation and are highly expressed at the early stages of development and also in embryonic stem cells (ESC). Immediately after differentiation, both proteins are downregulated and then remain ubiquitously expressed at low levels in somatic cells [7].
Posttranslational histone modifications are also involved in gene regulation, as the acetylation, methylation, or phosphorylation of histones can influence their affinity for DNA. Depending on the specific combination of these marks, they can either facilitate (e.g., histone 3 lysine 4 trimethylation, H3K4me3; histone 3 lysine 9 acetylation, H3K9ac) or impede (e.g., H3K9me3; H3K27me3) the binding of transcriptional factors to promoter regions [8–10]. DNA methylation and histone modifications are associated through methyl-binding proteins (MBP). The best-known MBP, Mecp2, contains two domains: one that specifically recognizes methylated DNA, a methyl-binding domain (MBD) [11]; and another that binds to Sin3 recruiting histone deacetylases, a transcriptional repression domain (TRD) [12–14] promoting gene silencing. Despite being ubiquitously expressed throughout all human tissues, MECP2 is highly expressed in the brain [15].
Contrasting observations have been made in tumor cells, where repetitive DNA is globally demethylated, promoting genomic instability [16–18], and oncogene promoters are less frequently methylated, leading to aberrant expression [19]. However, a myriad of methylated CpG islands near promoters, many of which found in tumor suppressor genes [20], have been observed in cancer cells [21], and the hypermethylation profile depends on the tumor type [22–24]. The pattern of posttranslational histone modifications is also disrupted in tumor cells and might play an essential role in tumor initiation [25]. As an evidence of how important epigenetic alterations are during the early steps of malignant transformation, a recent study showed that murine melanocyte cells that are resistant to apoptosis induction by adhesion blockade (anoikis-resistant cells) could be selected when cultured in agarose-coated plates. The clonal colonies obtained after sequential cycles through the same process were reported as tumorigenic when injected in syngeneic mice [26]. However, when cells were treated with 5-aza-2′-deoxycytidine, a demethylating agent, before each cycle of anchorage blockade, the derived cells were not malignantly transformed. Therefore, this study shows that most epigenetic modifications occur as primordial and essential events during the establishment of malignant transformation in this murine melanoma model, rather than as a consequence of cell transformation [27]. Since microRNAs (miRNAs) are predicted to regulate the expression of 60% of all coding genes [28] including the epigenetic machinery (reviewed in [29]), they are good candidates for promoting aberrant expression of epigenetic machinery, contributing to the deregulation of epigenetic marks during cell transformation.
Here, using this murine melanoma model [26, 27], we demonstrate that Dnmt3a and Dnmt3b are downregulated in nonmetastatic (4C3−) and metastatic (4C3+) melanoma cell lines, concomitant with the overexpression of miR-29b and miR-29c, which are known Dnmt3a and Dnmt3b regulators [30]. miR-26a, which targets Ezh2 [31], a histone methyltransferase belonging to the Polycomb family, was found to be downregulated in a nonmetastatic melanoma cell line (4C3−) but upregulated in 4C3+. These data are consistent with the fluctuation of Ezh2 expression during the transition from a nonmetastatic to a metastatic melanoma phenotype. Moreover, the members of the miR-26 family potentially target Mecp2 and Dnmt3b mRNAs, as predicted by several in silico miRNA target prediction programs, are upregulated in 4C3+ cells, whereas Mecp2 and Dnmt3b were downregulated. Indeed, recent studies have shown that transfection of miR-26b in breast cancer cells showed diminished expression of DNMT3B. On the other hand, transfection of miR-26b antagomiR showed an overexpression of DNMT3B [32], suggesting that this miRNA is involved in DNMT3b regulation.
We have also seen an inverse correlation between this miR-203 and DNMT3b in nonmetastatic melanoma cells compared to normal melanocytes. This result is in agreement with the predicted target of miR-203 and with the findings of Coleman group that have reported that breast cancer cells with hypermethylated DNA phenotype have DNMT3b overexpression and a downregulation of miR-203 [32, 33]. In order to validate Dnmt3b as miR-203 target, we have transfected cell with miR-203 expressing vector, which promotes Dnmt3b downregulation. Additionally, miR-203 is also predicted to target Mecp2, showing an inverse correlation of expression between normal melanocytes and nonmetastatic melanoma.
Furthermore, to determine whether miR-26, miR-29 family members and miR-203 could be epigenetically regulated, cells were treated with the hypomethylating agent 5-aza-deoxycytidine (5-aza-CdR). The results showed increased expression of all of the studied miRNAs in 4C3− cells, which might suggest that these miRNAs are epigenetically regulated and are more susceptible to destabilization during cell transformation. Taken together, these data suggest that epigenetic alterations that occur early during malignant transformation might be a result from the modulation of miR-26, miR-29, miR-203, and the consequent effects on key genes involved in the epigenetic machinery.
2. Materials and Methods
2.1. Cell Culture, Treatment with 5-aza-CdR, and Transfection with miR-203
Nonmetastatic (4C3−) and metastatic (4C3+) murine melanoma cells were obtained from melan-a nontumorigenic melanocytes [34] through repetitive anchorage blockade cycles, as described by Oba-Shinjo et al. [26]. Melan-a, 4C3−, and 4C3+ cells were grown in RPMI medium, pH 6.9 (Gibco, Carlsbad, CA), supplemented with 5% fetal bovine serum (Vitrocell, Campinas, SP, Brazil) and 1% penicillin-streptomycin solution (Life Technologies, Carlsbad, CA) at 37°C under 5% CO2. In addition, phorbol myristate acetate (PMA, Sigma, St. Louis, MO) was added at a final concentration of 200 nM to the complete medium for melan-a cells. The cells were treated with 10 μM 5-aza-deoxycytidine (5-aza-CdR, Sigma, St. Louis, MO) for 48 hours, followed by RNA or total protein extraction.
An aliquot of 3,5 μg of PSilencer 4.1-CMV-Puro plasmid containing the sequence of mmu-miR-203 or scrambled was kindly provided by Dr. Candi, Faculty of Medicine, Department of Experimental Medicine and Surgery, Rome [35]. They were transfected into cells with Fugene 6 (Promega), and culture was maintained in medium with 4 ug/mL puromycin through 14 days in order to select transfected resistant cells, followed by RNA and total protein extraction.
2.2. Methylation-Sensitive Single-Nucleotide Primer Extension Assay (Ms-SNuPE)
Global methylation status was inferred by methylation-sensitive single nucleotide primer extension (Ms-SNuPE) method. As previously described [27], the average of methylated CpG sites located at two different repetitive elements (retrovirus-C long terminal repeats, RC-LTR, and intracisternal A-particle elements, IAPy) was performed.
2.3. Retrotranscription and Quantitative Real-Time PCR
Total RNA extracted using TRIzol (Life Technologies, Carlsbad, CA) was treated with DNase I (Promega, Fitchburg, Wisconsin) and retrotranscribed using the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Carlsbad, CA), and the corresponding loop-primer of the miRNA Assay Kit (Life Technologies, Carlsbad, CA), according to the manufacturer's instructions. The reverse transcription reaction was followed by quantitative real-time PCR (qRT-PCR) using specific primers included in miRNA Assay kit (Life Technologies, Carlsbad, CA) with TaqMan PCR Master Mix (Life Technologies, Carlsbad, CA).
2.4. Western Blotting
Total protein was extracted using RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) and protease inhibitors (1 mM PMSF, 10 μg/mL aprotinin, 10 μg/mL leupeptin, and 200 μM sodium orthovanadate). The obtained protein concentration was evaluated in a spectrophotometer. Denatured protein was loaded onto an SDS-PAGE gel and subsequently transferred to a PVDF membrane (Bio-Rad, Hercules, CA). The membrane was incubated with anti-Dnmt3a (1 : 5000, Abcam, Cambridge, UK, #ab13887); anti-Dnmt3b (1 : 800, Santa Cruz Biotechnology, Santa Cruz, CA, #sc-70984); anti-Mecp2 (1 : 1,000, Calbiochem-Merck, Darmstadt, Germany, #472520); or anti-Ezh2 (1 : 2000, Abcam, Cambridge, UK, #ab3748) antibodies. For normalization, the membrane was also incubated with an anti-β-actin antibody (1 : 5,000, Santa Cruz Biotechnology, Santa Cruz, CA, #sc-101017). Subsequently, the membrane was incubated with an anti-mouse or anti-rabbit secondary antibody (Life Technologies, Carlsbad, CA) and visualized through chemiluminescence using the Supersignal West Pico Chemiluminescent Substrate (Pierce, Thermo Scientific, Rockford).
2.5. Statistical Analysis
Statistical analyses of the gene expression and methylation assays were performed by nonparametric one-way ANOVA, succeeded by Tukey's post hoc test, using the GraphPad Prism 5.0 program. At least two independent replicates were performed for each analysis. In all figures, the error bars in the histograms represent the mean standard error of the obtained results.
3. Results and Discussion
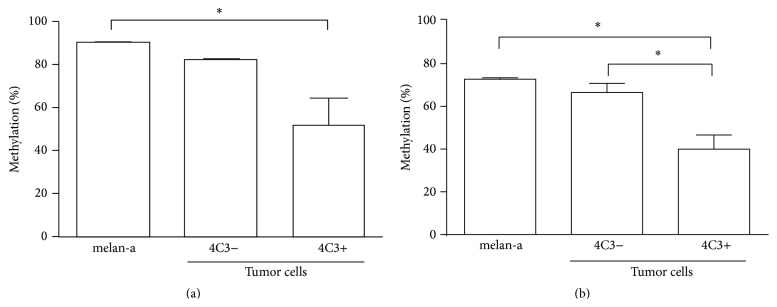
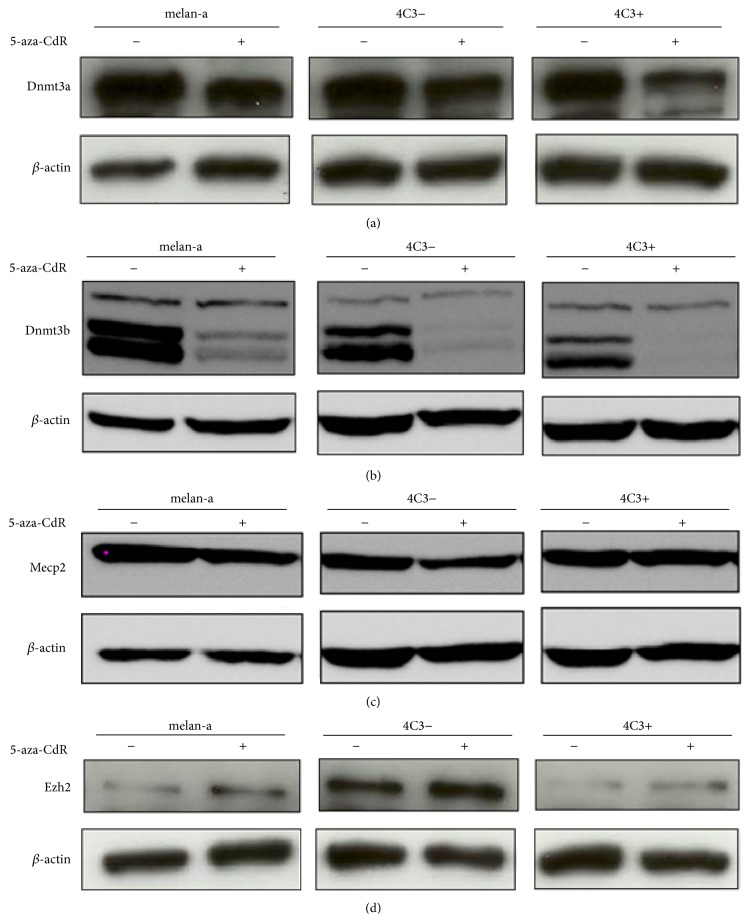
In a previous study, using a murine melanoma model obtained through sequential anchorage blockade cycles in melan-a melanocytes, Molognoni and coworkers [27] demonstrated alterations in the expression of genes involved in the epigenetic machinery and changes in DNA methylation levels in premalignant melanocytes, suggesting that epigenetic reprogramming occurs prior to malignant transformation in these cells. To verify the contribution of miRNAs to this phenomenon, the expression of key components of the epigenetic machinery was evaluated in parental, melan-a, and melan-a-derived melanoma cells (the nonmetastatic 4C3− cell line and its metastatic derivative, 4C3+). These 4C3 cells have been characterized elsewhere [26, 36]. To verify whether the 4C3− and 4C3+ melanoma cell lines exhibit an altered pattern of DNA methylation, methylation of repetitive DNA sequences (retrovirus-C and intracisternal A particle elements) was assessed using the Ms-SNuPE method. The metastatic 4C3+ cell line showed a significant reduction of global methylation compared with melan-a and nonmetastatic 4C3− cells (Figure 1). This result is consistent with the global demethylation observed in most tumor types [2, 37] and in other melanoma cells in the same murine melanoma model [27]. Hence, the expression of Dnmt3a, Dnmt3b, Mecp2, and Ezh2 (Figure 2) of the parental melan-a and melanoma 4C3− and 4C3+ cell lines was assessed at the protein level. Dnmt3b expression was reduced in 4C3− cells compared to normal melanocytes melan-a and even more reduced in 4C3+ metastatic cells (Figure 2(b)). The same pattern was seen in Dnmt3a and Mecp2, however, in an attenuated manner (Figures 2(a) and 2(c)). Taken together, these results show that the global DNA hypomethylation observed during the progression of melanoma is accompanied by downregulation of Dnmt3a, Dnmt3b, and Mecp2 expression. Whether DNMT3a and DNMT3b are downregulated during cancer development is a debated issue. Indeed, most tumor types overexpress DNMTs [38]. However, there is evidence that DNMTs are mutated or weakly expressed in cancer cells [39–41]. Our findings are consistent with those of Molognoni et al. [27], who also observed Mecp2 downregulation during malignant transformation. However, these authors found that the expression of Dnmt3b was not statistically significantly different between melan-a, premalignant, nonmetastatic, and metastatic melanoma cells and Dnmt3a was upregulated in the metastatic cell line, possibly reflecting clonal differences. Additionally, we have assessed the expression of Ezh2 at the protein level (Figure 2(d)) in all 3 studied cell lines. In melan-a and in 4C3+, Ezh2 was found poorly expressed but overexpressed in 4C3−. Accordingly, this expression fluctuation was also seen in Molognoni et al. [27], showing that Ezh2 might be involved in the primary events of melanocytes transformation.
Figure 1.
Metastatic melanoma cells are hypomethylated compared with nontumorigenic melan-a cells. The methylation status of three specific CpGs associated with A-repeats (a) and two CpGs in the retrovirus-C repetitive sequence (b) were evaluated using Ms-SNuPE in the melan-a, 4C3−, and 4C3+ cell lines. The percentage of methylation was determined by calculating the average at the different CpG sites analyzed. Ma: nontumorigenic melan-a melanocyte line; 4C3−: nonmetastatic melanoma cell line; 4C3+: metastatic melanoma cell line. ∗ p < 0.05.
Figure 2.
Dnmt3a (a), Dnmt3b (b), Mecp2 (c), and Ezh2 (d) expression profile in melan-a, 4C3−, and 4C3+ before and after treatment with 5-aza-CdR. The protein expression was evaluated by Western blotting in melan-a, 4C3−, and 4C3+ cells that were untreated or treated with 10 μM 5-aza-CdR for 48 hours. The expression of β-actin was used for normalization. melan-a: nontumorigenic melan-a melanocyte lineage; 4C3−: nonmetastatic melanoma cell line; and 4C3+: metastatic melanoma cell line.
Recently, it has been reported that numerous miRNAs are involved in the silencing of components of the epigenetic machinery (reviewed in [42]). Moreover, it is been predicted that some miRNAs are involved in the regulation of genes related to the same pathway [43]. Indeed, the miR-29 family is particularly interesting, as overexpression of its members in lung tumor cells significantly reduces Dnmt3a and Dnmt3b expression [30]. More specifically, miR-29b reduces DNMT1, DNMT3A, and DNMT3B expression in acute myeloid leukemia (AML) [44] and germ cells [45]. miR-29b and miR-29c also target YY1, a chromatin remodeling protein that recruits PRC2 and a histone deacetylase, HDCA, to suppress the expression of specific loci [46]. miR-26 is also associated with the downregulation of DNMT3B, since transfection of miR-26b in human breast cancer cells showed diminished expression of DNMT3B. On the other hand, transfection of miR-26b antagomiR showed an overexpression of DNMT3B [32]. miR-26a also targets Ezh2 mRNA [31], and both members of miR-26 family (miR-26a and miR-26b) could target Mecp2/MECP2 mRNA, as predicted by several miRNA target prediction programs (Microrna.org; TargetScan 4.2; Pictar; and DIANA/miRGen). Moreover, miR-203 has been validated as a regulator of Bmi-1, which is a member of the Polycomb repressive complex 1 (PRC1) family of proteins and is also involved in epigenetic modifications [47]. Additionally, the expression of miR-203 has been inversed correlated to DNMT3B in breast cancer cells [32, 33]. This miRNA might also target Mecp2/MECP2, as predicted using the same four prediction programs. Thus, these miRNAs are good candidates for the regulation of genes involved in the epigenetic machinery during melanocyte transformation.
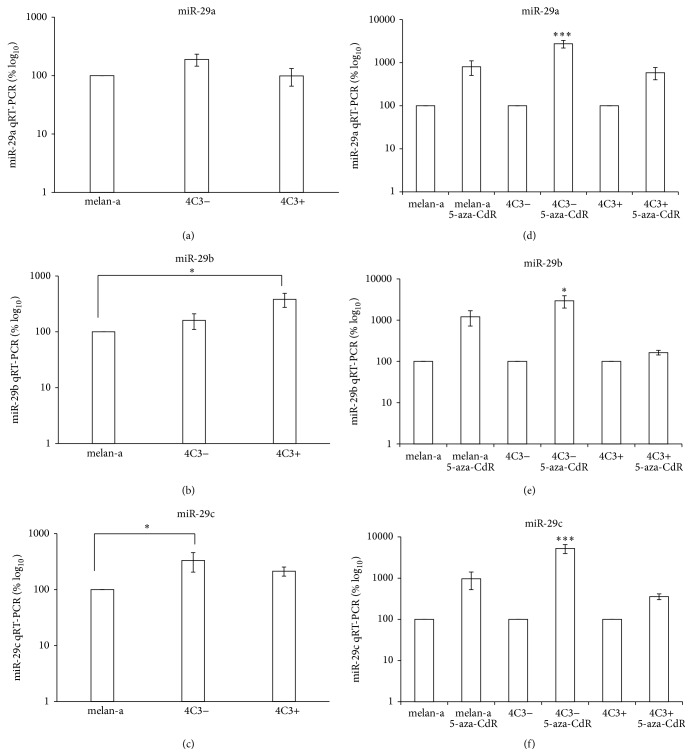
Since miR-29 family was validated to target Dnmt3a and Dnmt3b [30], the expression profile of the miR-29 family was evaluated and showed that miR-29a did not contribute to the phenotype of the malignant cells, as there were no statistically significant differences observed between the 3 studied cell lines (Figure 3(a)). Nevertheless, miR-29b and miR-29c were significantly upregulated in 4C3+ and 4C3− cells, respectively, compared with melan-a cells (Figures 3(b) and 3(c), resp.). Since Dnmt3a and Dnmt3b are downregulated in 4C3− and in 4C3+ cells (Figures 2(a) and 2(b)), it is conceivable that these miRNAs regulate the de novo Dnmts expression during melanocyte transformation.
Figure 3.
miR-29 expression during melanocyte transformation and 5-aza-CdR treatment. miR-29a (a), miR-29b (b), and miR-29c (c) expression was evaluated in the three cell lines via qRT-PCR. The cells were treated with 10 μM 5-aza-CdR for 48 hours, and the expression of miR-29a (d), miR-29b (e), and miR-29c (f) was subsequently evaluated. U6 was used for normalization. Ma: nontumorigenic melan-a melanocyte lineage; 4C3−: nonmetastatic melanoma cell line; and 4C3+: metastatic melanoma cell line. ∗ p < 0.05; ∗∗∗ p < 0.001.
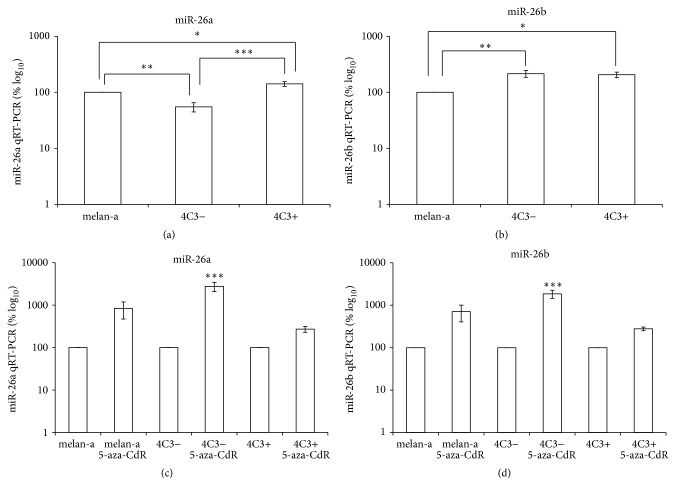
The expression of miR-26a was also assessed, showing a significant reduction in its expression in 4C3− cells compared with parental cells. However, upregulation was observed in metastatic melanoma cells compared with melan-a and 4C3− cells (Figure 4(a)). The observed pattern of miR-26a expression is opposed to its validated target Ezh2 (Figure 2(d)). Since the latter is upregulated in nonmetastatic cells compared with melanocytes and metastatic melanoma cells, it is suggestive that miR-26a might regulate Ezh2 expression during the melanocytes malignant transformation. This histone methyltransferase is a part of the Polycomb Complex Repressor 2 (PCR2) system, which represses gene expression through interactions with unmethylated CpG islands in normal cells (reviewed in [48]). Ezh2 has been associated with the aberrant silencing of CpG islands near the promoters of tumor suppressor genes [49]. This site-specific hypermethylation could result from the interaction of Ezh2 with Dnmts to position these proteins near CpG islands, promoting de novo methylation (reviewed in [48]). The overexpression of Ezh2 during cell transformation [50] might promote the assembly of Dnmts to the CpG islands providing a more stable silencing mark. Since overexpression of Dnmts is not obligatory to promote aberrant hypermethylation in CpG island in cancer cells [41], low levels of these proteins might be sufficient to silence CpG islands near suppressor gene promoters that are more susceptible to DNA methyltransferases in the presence of abundant Ezh2 [51]. Conversely, downregulation of Dnmt3a, Dnmt3b, and Mecp2 could favor the derepression of repetitive DNA and the global demethylation observed in tumor cells, promoting genomic instability. Thus, overexpression of the epigenetic machinery might not be necessary to maintain the transformed state.
Figure 4.
miR-26 expression during melanocyte transformation and 5-aza-CdR treatment. miR-26a (a) and miR-26b (b) expression was examined in the three cell lines using qRT-PCR. The cells were treated with 10 μM 5-aza-CdR for 48 hours, and the expression of miR-26a (c) and miR-26b (d) was subsequently evaluated. U6 was used for normalization. Ma: nontumorigenic melan-a melanocyte lineage; 4C3−: nonmetastatic melanoma cell line; and 4C3+: metastatic melanoma cell line. ∗ p < 0.05; ∗∗ p < 0.01; ∗∗∗ p < 0.001.
Further, the upregulation of miR-26a seen in 4C3+ cells and miR-26b seen in 4C3− and 4C3+ cells (Figures 4(a) and 4(b)) is opposed to Mecp2 expression in these cells compared with melan-a cells (Figure 2(c)). Because the miR-26 family has been predicted to target Mecp2/MECP2 mRNAs, these miRNAs might be relevant to Mecp2 downregulation and are good candidates to further validation as its regulators. Finally, miR-26b and DNMT3b showed an inverse correlation expression, whereas DNMT3b is downregulated and miR-26b is upregulated in melanoma cells compared to melanocytes, which corroborates to previous evidence that DNMT3b is regulated by miR-26b [32].
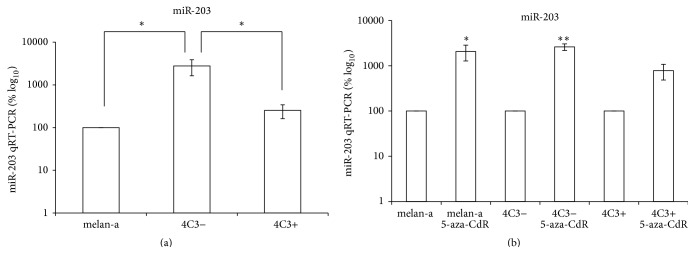
The analysis of the expression of other miRNA that potentially targets Mecp2 and Dnmt3b, miR-203, revealed that it was significantly upregulated in 4C3− cells compared with the melan-a and 4C3+ cell lines (Figure 5(a)). This finding supports the hypothesis that miR-203 regulates both genes and might be involved in the induction of the transformed state as only the nonmetastatic cell line showed an upregulation of miR-203 expression. Alternatively, miR-203 could be overexpressed in 4C3− cells as a feedback mechanism to avoid transformation, as miR-203 has been previously described as an antimetastatic agent in human prostate cancer [52].
Figure 5.
miR-203 expression during melanocyte transformation and 5-aza-CdR treatment. miR-203 (a) expression was evaluated in the three cell lines using qRT-PCR. The cells were treated with 10 μM 5-aza-CdR for 48 hours, and the expression of miR-203 (b) was then evaluated. U6 was used for normalization. Ma: nontumorigenic melan-a melanocyte lineage; 4C3−: nonmetastatic melanoma cell line; and 4C3+: metastatic melanoma cell line. ∗ p < 0.05; ∗∗ p < 0.01.
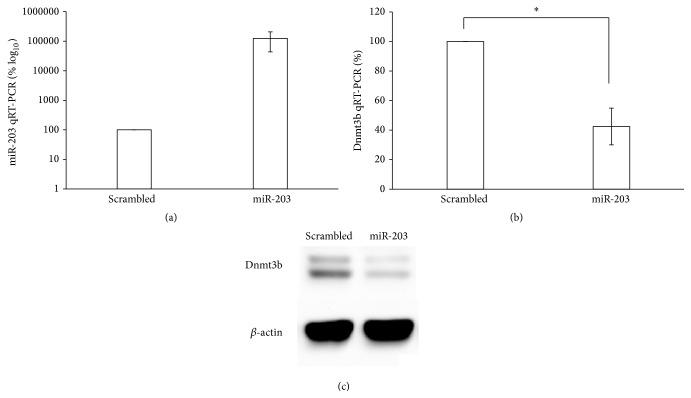
To test the hypothesis that miR-203 targets Dnmt3b, we overexpress the miR-203 in melan-a cells. A significant downregulation of Dnmt3b at RNA and protein levels was observed (Figure 6), being the first evidence of miR-203 regulating Dnmt3b.
Figure 6.
Dnmt3b expression after exogenous overexpression of miR-203 in melan-a cells. (a) miR-203 and (b) Dnmt3b expression was evaluated by qRT-PCR in melan-a cells transfected with pSilencer-mmu-miR-203 or pSilencer-Scrambled. The expression of Dnmt3b at protein level (c) was also evaluated after the same treatment by Western blotting. ∗ p < 0.05.
To verify whether these miRNAs could be epigenetically regulated, each cell line was treated with 5-aza-CdR. After treatment, increased miR-203 expression was observed in melan-a and 4C3− cells (Figure 5(b)). These results are consistent with the literature reporting that miR-203 is epigenetically silenced in human tumor cells [53, 54]. miR-26 and miR-29 family members were also overexpressed in 4C3− cells compared with untreated cells (Figures 4(c), 4(d), 3(d), 3(e), and 3(f), resp.). Given that miR-26 and miR-29 exhibit expression patterns similar to that of miR-203 in 4C3− cells treated with 5-aza-CdR, it is conceivable that these miRNAs are also epigenetically regulated in these cells. The expression of these miRNAs was not statistically significantly different in treated metastatic 4C3+ cells compared with untreated 4C3+ cells, possibly because the untreated counterpart cells are already demethylated. An in silico analysis of CpG islands near the miRNA genes of Mus musculus and their promoters was performed using Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway) and the Transcriptional Regulatory Element Database (http://rulai.cshl.edu/cgi-bin/TRED/tred.cgi?process=home). The analysis showed that miR-26a-1 is hosted within a gene, whose promoter overlaps with a CpG island of 1230 bp. The intragenic miRNAs, miR-26a-2 (miR-26a-2 and miR-546) cluster and miR-26b, are both associated with CpG islands of 1752 and 1098 bp near the transcription start site (TSS) in the host gene or overlapping with the predicted host gene promoter, respectively. The miR-29a/29b-1 cluster is intergenic and exhibits 4 small CpG islands near the TSS, ranging from 206 to 836 bp. It has been predicted that this cluster is imprinted, as it is embedded into the imprinted Copg 2 cluster [55]. The only exception to this pattern is the intragenic miR-29b-2/29c cluster, which presents a 558 bp CpG island distant from its TSS. miR-203, by its turn, is intergenic and seems to be embedded in a CpG island of 804 bp. These analyses showed that all miRNAs studied here are near of regions commonly regulated epigenetically.
To our knowledge, this study provides the first evidence that miR-26a, miR-26b, miR-29b, and miR-29c might be epigenetically regulated in mouse and is in agreement with study of Desjobert and colleagues that showed miR-29a methylated in human lymphoma cells [56]. An estimated 60% of human coding genes have been associated with CpG islands, while approximately half of known miRNA genes are located near or embedded within a CpG island, indicating that methylation is an important mechanism for the regulation of miRNA transcription [57]. However, upregulation of miRNAs following 5-aza-CdR treatment was observed only in the nonmetastatic melanoma cell line, potentially reflecting the unstable epigenetic marks in these cells. The expression of Dnmt3b was assessed from total protein extracts, demonstrating the efficiency of 5-aza-CdR treatment in inducing Dnmt3b degradation (Figure 2(b)). In contrast, no differences in Mecp2 expression were observed at the protein (Figure 2(c)) following 5-aza-CdR treatment compared with untreated cells. Taken together, these findings support the idea that these miRNAs are epigenetically regulated. Nevertheless, they must be further characterized.
In conclusion, this study demonstrated that miR-203 overexpression promotes Dnmt3b downregulation. Additionally, it highlights the importance of miR-26, miR-29, and miR-203 in promoting the imbalanced expression of genes involved in the epigenetic machinery during the malignant transformation of melanocytes. These results indicate that the miRNA methylation status plays an important role in the establishment of the altered epigenetic state observed in tumors.
Acknowledgments
The authors would like to thank Dr. Ana Cristina Krepischi (Department of Genetics and Evolutionary Biology, University of Sao Paulo), Dr. Helena Bonciani Nader (Department of Biochemistry, Federal University of Sao Paulo), Dr. Peter Jones and Dr. Gangning Liang (Norris Comprehensive Cancer Center, University of Southern California), and Fernanda Molognoni and Adriana Taveira da Cruz (Department of Pharmacology, Federal University of Sao Paulo) for helpful advice. The authors also thank Dr. Eleanora Candy, Faculty of Medicine, Department of Experimental Medicine and Surgery, Rome, who kindly provided pSilencer plasmid constructs. This work was supported through funding from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Walsh C. P., Chaillet J. R., Bestor T. H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genetics. 1998;20(2):116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 2.Gaudet F., Hodgson J. G., Eden A., et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300(5618):489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M., Almouzni G. How epigenetics integrates nuclear functions. Workshop on epigenetics and chromatin: transcriptional regulation and beyond. EMBO Reports. 2005;6(7):624–628. doi: 10.1038/sj.embor.7400456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 5.Jones P. A., Baylin S. B. The fundamental role of epigenetic events in cancer. Nature Reviews Genetics. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 6.Straussman R., Nejman D., Roberts D., et al. Developmental programming of CpG island methylation profiles in the human genome. Nature Structural & Molecular Biology. 2009;16(5):564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe D., Suetake I., Tada T., Tajima S. Stage- and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mechanisms of Development. 2002;118(1-2):187–190. doi: 10.1016/s0925-4773(02)00242-3. [DOI] [PubMed] [Google Scholar]
- 8.Fahrner J. A., Eguchi S., Herman J. G., Baylin S. B. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Research. 2002;62(24):7213–7218. [PubMed] [Google Scholar]
- 9.Jones P. A., Baylin S. B. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohm J. E., McGarvey K. M., Yu X., et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nature Genetics. 2007;39(2):237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tate P., Skarnes W., Bird A. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nature Genetics. 1996;12(2):205–208. doi: 10.1038/ng0296-205. [DOI] [PubMed] [Google Scholar]
- 12.Nan X., Bird A. The biological functions of the methyl-CpG-binding protein MeCP2 and its implication in Rett syndrome. Brain and Development. 2001;23(supplement 1):S32–S37. doi: 10.1016/s0387-7604(01)00333-3. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen C. T., Gonzales F. A., Jones P. A. Altered chromatin structure associated with methylation-induced gene silencing in cancer cells: correlation of accessibility, methylation, MeCP2 binding and acetylation. Nucleic Acids Research. 2001;29(22):4598–4606. doi: 10.1093/nar/29.22.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobosy J. R., Selker E. U. Emerging connections between DNA methylation and histone acetylation. Cellular and Molecular Life Sciences. 2001;58(5-6):721–727. doi: 10.1007/pl00000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monteggia L. M., Kavalali E. T. Rett syndrome and the impact of MeCP2 associated transcriptional mechanisms on neurotransmission. Biological Psychiatry. 2009;65(3):204–210. doi: 10.1016/j.biopsych.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoder J. A., Walsh C. P., Bestor T. H. Cytosine methylation and the ecology of intragenomic parasites. Trends in Genetics. 1997;13(8):335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 17.Widschwendter M., Jiang G., Woods C., et al. DNA hypomethylation and ovarian cancer biology. Cancer Research. 2004;64(13):4472–4480. doi: 10.1158/0008-5472.CAN-04-0238. [DOI] [PubMed] [Google Scholar]
- 18.Deng G., Nguyen A., Tanaka H., et al. Regional hypermethylation and global hypomethylation are associated with altered chromatin conformation and histone acetylation in colorectal cancer. International Journal of Cancer. 2006;118(12):2999–3005. doi: 10.1002/ijc.21740. [DOI] [PubMed] [Google Scholar]
- 19.Jun H. J., Woolfenden S., Coven S., et al. Epigenetic regulation of c-ROS receptor tyrosine kinase expression in malignant gliomas. Cancer Research. 2009;69(6):2180–2184. doi: 10.1158/0008-5472.can-08-3351. [DOI] [PubMed] [Google Scholar]
- 20.Esteller M., Herman J. G. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. The Journal of Pathology. 2002;196(1):1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 21.Weber M., Davies J. J., Wittig D., et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature Genetics. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 22.Costello J. F., Frühwald M. C., Smiraglia D. J., et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nature Genetics. 2000;24(2):132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M., Corn P. G., Baylin S. B., Herman J. G. A gene hypermethylation profile of human cancer. Cancer Research. 2001;61(8):3225–3229. [PubMed] [Google Scholar]
- 24.Sjöblom T., Jones S., Wood L. D., et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 25.Feinberg A. P., Ohlsson R., Henikoff S. The epigenetic progenitor origin of human cancer. Nature Reviews Genetics. 2006;7(1):21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 26.Oba-Shinjo S. M., Correa M., Ricca T. I., et al. Melanocyte transformation associated with substrate adhesion impediment. Neoplasia. 2006;8(3):231–241. doi: 10.1593/neo.05781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molognoni F., Cruz A. T., Meliso F. M., et al. Epigenetic reprogramming as a key contributor to melanocyte malignant transformation. Epigenetics. 2011;6(4):450–464. doi: 10.4161/epi.6.4.14917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman R. C., Farh K. K.-H., Burge C. B., Bartel D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peschansky V. J., Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9(1):3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabbri M., Garzon R., Cimmino A., et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung F. W., Tellam R. L. MicroRNA-26a targets the histone methyltransferase Enhancer of zeste homolog 2 during myogenesis. The Journal of Biological Chemistry. 2008;283(15):9836–9843. doi: 10.1074/jbc.m709614200. [DOI] [PubMed] [Google Scholar]
- 32.Sandhu R., Rivenbark A. G., Coleman W. B. Loss of post-transcriptional regulation of DNMT3b by microRNAs: a possible molecular mechanism for the hypermethylation defect observed in a subset of breast cancer cell lines. International Journal of Oncology. 2012;41(2):721–732. doi: 10.3892/ijo.2012.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rupninder S., Rivenbark A. G., Mackler R. M., Livasy C. A., Coleman W. B. Dysregulation of microRNA expression drives aberrant DNA hypermethylation in basal-like breast cancer. International Journal of Oncology. 2014;44(2):563–572. doi: 10.3892/ijo.2013.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett D. C., Cooper P. J., Hart I. R. A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. International Journal of Cancer. 1987;39(3):414–418. doi: 10.1002/ijc.2910390324. [DOI] [PubMed] [Google Scholar]
- 35.Lena A. M., Shalom-Feuerstein R., di Val Cervo P. R., et al. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death and Differentiation. 2008;15(7):1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 36.Silva A. G., Graves H. A., Guffei A., et al. Telomere-centromere-driven genomic instability contributes to karyotype evolution in a mouse model of melanoma. Neoplasia. 2010;12(1):11–19. doi: 10.1593/neo.91004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narayan A., Ji W., Zhang X.-Y., et al. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. International Journal of Cancer. 1998;77(6):833–838. doi: 10.1002/(sici)1097-0215(19980911)77:660;833::aid-ijc662;3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 38.Rhee I., Bachman K. E., Park B. H., et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416(6880):552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 39.Fanelli M., Caprodossi S., Ricci-Vitiani L., et al. Loss of pericentromeric DNA methylation pattern in human glioblastoma is associated with altered DNA methyltransferases expression and involves the stem cell compartment. Oncogene. 2008;27(3):358–365. doi: 10.1038/sj.onc.1210642. [DOI] [PubMed] [Google Scholar]
- 40.Ley T. J., Ding L., Walter M. J., et al. DNMT3A mutations in acute myeloid leukemia. The New England Journal of Medicine. 2010;363(25):2424–2433. doi: 10.1056/nejmoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eads C. A., Danenberg K. D., Kawakami K., Saltz L. B., Danenberg P. V., Laird P. W. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Research. 1999;59(10):2302–2306. [PubMed] [Google Scholar]
- 42.Sato F., Tsuchiya S., Meltzer S. J., Shimizu K. MicroRNAs and epigenetics. The FEBS Journal. 2011;278(10):1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 43.Shirdel E. A., Xie W., Mak T. W., Jurisica I. NAViGaTing the micronome—using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS ONE. 2011;6(2) doi: 10.1371/journal.pone.0017429.e17429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garzon R., Liu S., Fabbri M., et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113(25):6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takada S., Berezikov E., Choi Y. L., Yamashita Y., Mano H. Potential role of miR-29b in modulation of Dnmt3a and Dnmt3b expression in primordial germ cells of female mouse embryos. RNA. 2009;15(8):1507–1514. doi: 10.1261/rna.1418309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H., Garzon R., Sun H., et al. NF-κB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14(5):369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wellner U., Schubert J., Burk U. C., et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature Cell Biology. 2009;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 48.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature Reviews Genetics. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 49.Velichutina I., Shaknovich R., Geng H., et al. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116(24):5247–5255. doi: 10.1182/blood-2010-04-280149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varambally S., Dhanasekaran S. M., Zhou M., et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 51.Hoffmann M. J., Engers R., Florl A. R., Otte A. P., Müller M., Schulz W. A. Expression changes in EZH2, but not in BMI-1, SIRT1, DNMT1 or DNMT3B, are associated with DNA methylation changes in prostate cancer. Cancer Biology and Therapy. 2007;6(9):1403–1412. doi: 10.4161/cbt.6.9.4542. [DOI] [PubMed] [Google Scholar]
- 52.Saini S., Majid S., Yamamura S., et al. Regulatory role of mir-203 in prostate cancer progression and metastasis. Clinical Cancer Research. 2011;17(16):5287–5298. doi: 10.1158/1078-0432.CCR-10-2619. [DOI] [PubMed] [Google Scholar]
- 53.Kozaki K.-I., Imoto I., Mogi S., Omura K., Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Research. 2008;68(7):2094–2105. doi: 10.1158/0008-5472.can-07-5194. [DOI] [PubMed] [Google Scholar]
- 54.Furuta M., Kozaki K.-I., Tanaka S., Arii S., Imoto I., Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2009;31(5):766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 55.Peters J., Robson J. E. Imprinted noncoding RNAs. Mammalian Genome. 2008;19(7-8):493–502. doi: 10.1007/s00335-008-9139-4. [DOI] [PubMed] [Google Scholar]
- 56.Desjobert C., Renalier M.-H., Bergalet J., et al. MiR-29a down-regulation in ALK-positive anaplastic large cell lymphomas contributes to apoptosis blockade through MCL-1 overexpression. Blood. 2011;117(24):6627–6637. doi: 10.1182/blood-2010-09-301994. [DOI] [PubMed] [Google Scholar]
- 57.Weber B., Stresemann C., Brueckner B., Lyko F. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6(9):1001–1005. doi: 10.4161/cc.6.9.4209. [DOI] [PubMed] [Google Scholar]