Abstract
The purpose of this study was to determine the prevalence and molecular characterization of plasmid-mediated quinolone resistance (PMQR) genes (qnrA, qnrB, qnrS, aac(6′)-Ib-cr, and qepA) among ESBL-producing Klebsiella pneumoniae isolates in Kashan, Iran. A total of 185 K. pneumoniae isolates were tested for quinolone resistance and ESBL-producing using the disk diffusion method and double disk synergy (DDST) confirmatory test. ESBL-producing strains were further evaluated for the bla CTX-M genes. The PCR method was used to show presence of plasmid-mediated quinolone resistance genes and the purified PCR products were sequenced. Eighty-seven ESBL-producing strains were identified by DDST confirmatory test and majority (70, 80.5%) of which carried bla CTX-M genes including CTX-M-1 (60%), CTX-M-2 (42.9%), and CTX-M-9 (34.3%). Seventy-seven ESBL-producing K. pneumoniae isolates harbored PMQR genes, which mostly consisted of aac(6′)-Ib-cr (70.1%) and qnrB (46.0%), followed by qnrS (5.7%). Among the 77 PMQR-positive isolates, 27 (35.1%) and 1 (1.3%) carried 2 and 3 different PMQR genes, respectively. However, qnrA and qepA were not found in any isolate. Our results highlight high ESBL occurrence with CTX-M type and high frequency of plasmid-mediated quinolone resistance genes among ESBL-producing K. pneumoniae isolates in Kashan.
1. Introduction
Klebsiella pneumoniae (K. pneumoniae) is one of the most common causes of nosocomial infections in adults and children and the mortality rate because it is high [1, 2]. Prolonged hospitalization, protracted admission in Intensive Care Unit (ICU), long term exposure to antibiotics especially to third-generation cephalosporins, and residing invasive devices are among the risk factors that have been reported for acquisition of infections due to extended-spectrum β-lactamases- (ESBL-) producing K. pneumoniae in adults [2]. In a study, female sex, corticosteroid therapy, infection with Klebsiella species, and recent exposure to third-generation cephalosporins have been reported as the relevant risk factors for acquisition of bloodstream infections with ESBL-producing organisms in children [3]. ESBL-positive K. pneumoniae is often resistant to non-β-lactam antibiotics (especially fluoroquinolones), so treatment of infections due to these multidrug-resistant strains appears to be very difficult [4, 5]. CTX-M (“CTX” implies activity against cefotaxime, “M” refers to Munich) producing K. pneumoniae is becoming increasingly prevalent in many countries of the world [5]. Fluoroquinolone resistance among Gram-negative pathogens developed shortly after these antimicrobials were introduced [6, 7].
Although the preliminary mechanism of fluoroquinolone resistance in members of the family Enterobacteriaceae is traditionally mediated by the mutation of chromosomal genes encoding DNA gyrase, topoisomerase IV, regulatory efflux pumps, and/or porins, recent reports indicate that quinolone resistance may also be related to plasmid-mediated quinolone resistance genes (qnrA, qnrB, qnrS, aac(6′)-Ib-cr, and qepA) [8, 9]. The quinolones inhibit DNA gyrase as their target site of action and cause bacterial cell death. Quinolone resistance genes including qnrA, qnrB, and qnrS code for proteins belonging to the pentapeptide repeat family interacting with DNA gyrase and topoisomerase IV enzymes to prevent quinolone inhibition [6]. The second PMQR mechanism captures a variant of aminoglycoside acetyltransferase (AAC(6′)-Ib-cr), which can diminish a fluoroquinolone activity by adding an acetyl group to this antimicrobial agent [10]. The last mechanism of PMQR is the quinolone efflux pump (QepA), a proton-dependent transporter, which causes hydrophilic quinolone resistance, especially to norfloxacin, ciprofloxacin, and enrofloxacin [9]. Plasmid-mediated quinolone resistance (PMQR) determinants have been recognized worldwide with a thoroughly high prevalence among extended-spectrum β-lactamases- (ESBLs-) producing K. pneumoniae [1, 6, 7, 11–13]. Little is known about the frequency of PMQR genes in K. pneumoniae isolates recovered from clinical specimens from humans in Iran. Therefore, the aim of this study was to determine the prevalence of PMQR genes in ESBLs-positive K. pneumoniae isolated from clinical specimens of patients referred to Shahid Beheshti hospital in Kashan, Iran.
2. Material and Methods
2.1. Sample Collection
A cross-sectional study was conducted between April 1, 2013, and April 17, 2014. One hundred eighty-five nonrepetitive clinical isolates (urine, blood, wound, sputum, etc.) suspected to be K. pneumoniae and isolated from patients referred to Shahid Beheshti Hospital in Kashan were tested in the study. Approval of the study protocol was received from the Ethical Review Board of Kashan University of Medical Sciences. Written informed consent was obtained from all study participants or their parents/guardians. Of the patients that participated in the study, 114 (61.6%) were females and 71 (38.4%) were males.
2.2. Bacterial Isolates
One clinical specimen was obtained for culture and microbial characterization from each patient and was used for bacterial culture. Each specimen was immediately inserted into Tryptic Soy Broth (TSB) medium (Merck, catalogue number: 105459) and sent to laboratory and, after incubation at 37°C for 6–12 hours, cultured on MacConkey agar (Merck, catalogue number: 105465) for the isolation of bacteria.
The cultured plates were incubated at 37°C and were examined after overnight incubation. Characteristic colonies of K. pneumoniae were confirmed by gram staining, oxidase, and biochemical tests including Indole, Methyl Red, Voges Proskauer, and Citrate (IMVIC) tests [14]. The confirmed K. pneumoniae isolates were used for study of resistance pattern and DNA extraction.
2.3. Antimicrobial Susceptibility Testing and Determination of ESBL Production
Antimicrobial susceptibility and detection of quinolone resistance were determined by disk diffusion method using Mueller Hinton agar according to the Clinical and Laboratory Standards Institute (CLSI) recommendations [15]. The following disks were used: imipenem (S: ≥16 mm, I: 14-15 mm, and R: ≤13 mm), aztreonam (S: ≥21 mm, I: 18–20 mm, and R: ≤17 mm), ampicillin (S: ≥17 mm, I: 14–16 mm, and R: ≤13 mm), ciprofloxacin (S: ≥31 mm, I: 21–30 mm, and R: ≤20 mm), cephalothin (S: ≥18 mm, I: 15–17 mm, and R: ≤14 mm), cefotaxime (S: ≥26 mm, I: 23–25 mm, and R: ≤22 mm), cefoxitin (S: ≥18 mm, I: 15–17 mm, and R: ≤14 mm), ceftriaxone (S: ≥23 mm, I: 20–22 mm, and R: ≤19 mm), gentamicin (S: ≥15 mm, I: 13-14 mm, and R: ≤12 mm), nalidixic acid (S: ≥19 mm, I: 14–18 mm, and R: ≤13 mm), ceftazidime (S: ≥21 mm, I: 18–20 mm, and R: ≤17 mm), and amoxicillin-clavulanic acid (S: ≥18 mm, I: 14–17 mm, and R: ≤13 mm). The reference strain E. coli ATCC 25922 was used as a control. Results were interpreted as susceptible, intermediate, or resistant according to the criteria recommended by the CLSI and the manufacturer protocols (Mast, UK). Double disk synergy test was accomplished in strains which were resistant to ceftriaxone, ceftazidime, cefotaxime, and aztreonam by disk diffusion method to confirm production of extended-spectrum β-lactamases (ESBLs) using ceftazidime discs with and without clavulanic acid, which were placed on the center of a plate containing Mueller Hinton agar medium at a distance of 25 mm from each other [16].
2.4. DNA Extraction
Crude DNA was extracted from each K. pneumoniae isolate extracted from the bacterial suspension by boiling method. The template DNA stored at −20°C until polymerase chain reaction (PCR) amplification was performed.
2.5. Detection of ESBL CTX-M Type by PCR
K. pneumoniae isolates that were ESBL-positive by DDST confirmatory test were subjected to amplification by PCR method using bla CTX-M specific primers (Table 1). Final reaction mixtures volume of 25 μL was prepared with 10 pmol of each primer, 200 mM of dNTP, 1 unit of Taq polymerase, 2.5 μL of 10x reaction buffer, 1.5 mM MgCl2 in final concentration, and 100 ng DNA template. Amplification reactions were carried out in a thermocycler (Eppendorf master cycler, MA) under the following conditions: 94°C for 5 min, followed by 30 cycles of 94°C for 25 sec, 52°C for 40 sec, 72°C for 50 sec, and 72°C for 6 min for the final elongation step [17]. The amplified products were electrophoresed on 2% agarose gels. The gels were stained in ethidium bromide (0.5 mg/mL) visualized in gel document system (Biorad, UK).
Table 1.
Primers used for polymerase chain reaction and sequencing.
| Primer | Target gene | Sequence (5′ → 3′) | Amplification product (bp) | Reference |
|---|---|---|---|---|
| CTX-MA CTX-MB |
bla
CTX-M
bla CTX-M |
CGCTTTGCGATGTGCAG ACCGCGATATCGTTGGT |
590 | [17] |
| M-1F M-1R |
bla
CTX-M-1
bla CTX-M-1 |
AAAAATCACTGCGCCAGTTC AGCTTATTCATCGCCACGTT |
415 | [17] |
| M-2 M-2R |
bla
CTX-M-2
bla CTX-M-2 |
CGACGCTACCCCTGCTAT CCAGCGTCAGATTTTTCAGG |
552 | [17] |
| M-8F M-8R |
bla
CTX-M-8
bla CTX-M-8 |
ACGCTCAACACCGCGATC CGTGGGTTCTCGGGGATAA |
490 | [17] |
| M-9F M-9R |
bla
CTX-M-9
bla CTX-M-9 |
CAAAGAGAGTGCAACGGATG ATTGGAAAGCGTTCATCACC |
205 | [17] |
| qnrA F qnrA R |
qnrA
qnrA |
ATTTCTCACGCCAGGATTTG GATCGGCAAAGGTTAGGTCA |
516 | [13] |
| qnrB F qnrB R |
qnrB
qnrB |
GATCGTGAAAGCCAGAAAGG ACGATGCCTG¬GTAGTTGTCC |
469 | [13] |
| qnrS F qnrS R |
qnrS
qnrS |
ACGACATTCGTCAACTGCAA TAAATTGGCACCCTGTAGGC |
417 | [13] |
| aac(6′)-Ib-cr F aac(6′)-Ib-cr R |
aac(6′)-Ib-cr
aac(6′)-Ib-cr |
TGACCAACAGCAACGATTCC TTAGGCATCACTGCGTGTTC |
554 | [14] |
| qepA F qepA R |
qepA
qepA |
GGACATCTACGGCTTCTTCG AGCTGCAGGTACTGCGTCAT |
720 | [15] |
2.6. Characterization of PMQR-Harboring Strains
PMQR genes, including qnrA, qnrB, qnrS, aac(6′)-Ib-cr, and qepA, were identified by PCR and Sanger sequencing. Pairs of primers used for PCR and sequencing are shown in Table 1. Amplification for qnrA, qnrB, and qnrS was carried out under the following conditions: initial denaturation at 94°C for 5 min, followed by 32 cycles of amplification at 94°C for 45 sec, 53°C for 45 sec, and 72°C for 60 sec with final extension at 72°C for 5 min [10]. PCR conditions for aac(6′)-Ib-cr were as follows: initial denaturation at 95°C for 5 min, followed by 32 cycles of amplification at 95°C for 20 sec, 59°C for 40 sec, and 70°C for 30 sec, with final extension at 72°C for 5 min [18].
The amplification conditions used for detection of qepA were as follows: initial denaturation at 94°C for 5 min, followed by 30 cycles of amplification at 94°C for 1 min, annealing at 59°C for 30 sec, and extension at 72°C for 1 min, with final extension at 72°C for 5 min [19]. The PCR was performed on total volume of 50 μL containing 100 ng genomic DNA from K. pneumoniae culture, 200 mM each of dNTP, 1x PCR buffer (20 mM Tris-HCl, pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.5 mM of each primer, and 1.5 U of Taq polymerase. Amplified samples (10 μL) were electrophoresed on 1.0% agarose gel in TBE buffer. The gel was stained with ethidium bromide 0.5 mg/mL. The amplified bands were visualized under ultraviolet light and photographed. Reaction mixtures without a DNA template served as negative controls. All positive amplicons were sequenced to confirm the PMQR genes.
2.7. DNA Sequencing and Sequence Analysis
PCR products that contain the positive desired genes (qnrB, qnrS, and aac(6′)-Ib-cr) were sequenced using the ABI Capillary System (Macrogen Research, Seoul, Republic of Korea). The sequences were analyzed using Chromas Pro version 1.7.5 Technelysium (http://technelysium.com.au/) and carried out online using the BLAST program of the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/).
2.8. Statistical Analysis
All statistical analyses were carried out using SPSS for windows version 15.0. Differences by the χ 2 test were considered significant, if P < 0.05. Prevalence data is presented with 95% confidence intervals (CI).
3. Results
Of the 185 K. pneumoniae species included in this study, 127 (68.6%) were nosocomial and were isolated from patients who had been hospitalized for two days or more in the Shahid Beheshti Hospital of Kashan and had no signs of infection with K. pneumoniae at the beginning of hospitalization. K. pneumoniae species were isolated from urine, 119 (64.3%), wound, 9 (4.8%), blood, 6 (3.2%), respiratory tract samples, 46 (24.9%) (including 5 of sputum, 3 of bronchoalveolar lavage, 37 of respiratory chip, and 1 of nasal discharge), cerebrospinal fluid, 2 (1.1%), and catheter, 3 (1.6%).
The mean age of the patients was between 48 and 52 years (50.36 ± 3.80). The prevalence rate of ESBL-producing K. pneumoniae infection was 47.0% (87/185). Of these ESBL-positive K. pneumoniae isolates, 82 (94.3%) were nosocomial. The majority of ESBL-positive patients were female, 48 (55.2%), versus 39 male (44.8%) and the most ESBL-producing strains (% 64.4) were recovered from elderly patients (<50 years of age) whereas the rates of ESBL-producing K. pneumoniae isolated from younger patients (≤50 years) were lower (35.6%). Among 87 ESBL-positive K. pneumoniae isolates, 78 (89.7%) were fluoroquinolone resistant, of which 100% demonstrated multidrug-resistant (MDR) pattern and were resistant to at least one agent in three or more antimicrobial categories. Among ESBL-positive K. pneumoniae isolates, the most prevalence of resistance was seen against ceftriaxone, cephalothin, cefotaxime, and ampicillin (Figure 1). Of isolates that were deemed ESBL-producing bacteria by DDST test, 80.5% (70/87) contained bla CTX-M genes. PCR assays and sequencing of bla CTX-M genes revealed that 60% (n = 42), 42.9% (n = 30%), and 34.3% (n = 24) of these isolates were identified as CTX-M-1, CTX-M-2, and CTX-M-9, respectively; also none of the isolates were positive for CTX-M-8. PMQR determinants were using PCR in 77 (88.5%) ESBL-positive isolates, which mostly consisted of aac(6′)-Ib-cr and qnrB, followed by qnrS genes. Among the PMQR-positive isolates, 30 (39%) and 1 (1.3%) of which coharbored 2 and 3 different PMQR genes, respectively. However, qnrA and qepA were not found in any isolate (Table 2). The nucleotide sequences of the PCR products of PMQR genes were all identical to those qnrB1, qnrS8, and aac(6′)-Ib-cr in the GenBank nucleotide database (http://www.ncbi.nlm.nih.gov/blast/) and accession numbers obtained for them in current study are KP340790, KP340791, and KP340792, respectively.
Figure 1.
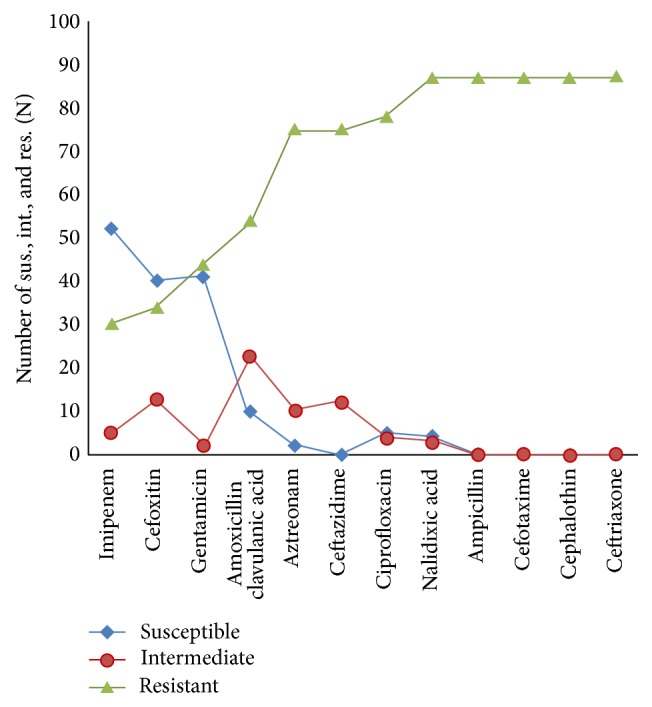
Antimicrobial resistance of ESBL-producing Klebsiella pneumoniae isolates was measured by disk diffusion method.
Table 2.
Prevalence of the plasmid-mediated quinolone resistance genes among extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates.
| PMQR genes | Positive isolates n (%) |
Negative isolates n (%) |
Total n (%) |
|---|---|---|---|
| qnrA | 0 (0%) | 87 (100%) | 87 (100%) |
| qnrB | 40 (46.0%) | 47 (54.0%) | 87 (100%) |
| qnrS | 5 (5.7%) | 82 (94.3%) | 87 (100%) |
| aac(6′)-Ib-cr | 61 (70.1%) | 26 (29.9%) | 87 (100%) |
| qepA | 0 (0%) | 87 (100%) | 87 (100%) |
The statistical analysis confirmed that ICU admission, more than fifty years of age and urinary infection, and infection with fluoroquinolones-resistant and multidrug-resistant strains (P < 0.001) were clinical factors that significantly associated with the presence of K. pneumoniae isolates that would yield positive results for ESBL molecular testing (Table 3).
Table 3.
Association between patient characteristics and acquisition of extended-spectrum β-lactamase-producing Klebsiella pneumoniae infection.
| Patient characteristic | ESBL-negative n = 98 (%) |
ESBL-positive n = 87 (%) |
P value |
|---|---|---|---|
| Age | |||
| >50 years | 36 (36.7) | 56 (64.4) | ≤0.001 |
| ≤50 years | 62 (63.3) | 31 (35.6) | |
| Sex | |||
| Male | 27 (27.6) | 39 (44.8) | 0.05 |
| Female | 71 (72.4) | 48 (55.2) | |
| Ward | |||
| ICU | 15 (33.3) | 23 (28) | ≤0.001 |
| Internal medicine | 7 (15.6) | 10 (12.2) | |
| Infectious | 5 (11.1) | 14 (17.1) | |
| Surgery | 6 (13.3) | 12 (14.6) | |
| Gynecology | 4 (8.9) | 7 (8.5) | |
| Pediatric | 5 (11.1) | 7 (8.5) | |
| Emergency | 1 (2.2) | 6 (7.3) | |
| CCU | 2 (4.4) | 3 (3.7) | |
| Sample type | 0.001 | ||
| Urine | 74 (75.5) | 45 (51.7) | |
| CSF | 1 (1) | 1 (1.1) | |
| Catheters | 1 (1) | 2 (2.3) | |
| Blood | 5 (5.1) | 1 (1.1) | |
| Wound | 3 (3.1) | 6 (6.9) | |
| Respiratory | 14 (14.3) | 32 (36.8) | |
| Drug resistance | |||
| Fluoroquinolones resistance | 5 (5.1) | 78 (89.7) | ≤0.001 |
| Multidrug resistance | 62 (63.3) | 87 (100.0) |
4. Discussion
Plasmid-mediated quinolone resistance mechanisms play an important role in the expansion of quinolone and fluoroquinolone resistance among ESBL-producing K. pneumoniae [20]. Infections caused by such resistant isolates can be difficult to treat [21]. Over the past 10 years, PMQR determinants have appeared as an important issue [22].
In this study, the resistance rates to nalidixic acid and ciprofloxacin were very high. Since the development of nalidixic acid in 1962, the drug has been used for the treatment of infections caused by K. pneumoniae [23]. The resistance rate for nalidixic acid, therefore, is expected to be higher than that for fluoroquinolones. Ciprofloxacin is frequently used for treatment of K. pneumoniae infections in Iran [24, 25]. Other studies in most parts of the world have indicated that fluoroquinolone resistance in ESBL-producing K. pneumoniae isolates is increasing [26]. Although the exact causes of the high ciprofloxacin resistance rate found in the present study compared to previous reports [26–29] are not known, the differences in the culture methodology between studies may explain the differences in the prevalence.
Our results showed the prevalence rates of ESBL-producing K. pneumoniae isolates were comparable to those recently conducted in Tehran, Iran [25]. A higher prevalence was noted in Mangalore (48.53%); however, lower prevalence was noted in China (33.3%), Saudi Arabia (12.2%), the United States (8%), and Canada (5%) [22, 26, 30–32].
In terms of association between patient characteristics and acquisition of ESBL-producing Klebsiella infection, we found that ESBL production was significantly higher among elderly patients. Similar results have been recorded in studies from Ghana and Saudi Arabia [33, 34]. The increased ESBL prevalence in elderly patients could be due to higher rates of prior antimicrobial therapy in this group of ages that led to greater antibiotic pressure among them. To date, qnr genes have been widely discovered in southern and eastern Asia, North and South America, and Europe [10]. Our study demonstrated the high prevalence (51.7%) of qnr genes among 87 ESBL-producing isolates of K. pneumoniae that were comparable with those noted in Morocco (50.0%) and China (65.5%) [20, 35]. However, low frequencies were found in Tunisia (15.0%), Mexico (13.7%), and USA (11.1%) [6, 13, 32]. In current study, the highest prevalence of qnr genes was related to qnrB. In Brazil, qnrB has been reported as the most widespread qnr determinant [36, 37]. No K. pneumoniae isolate harbored qnrA gene, although the prevalence of this gene was high among E. coli isolates in our previous study [24]. The low prevalence of qnrS and the absence of qnrA observed in our study were in agreement with studies from France and Tunisia [6, 38]. AAC(6′)-Ib-cr, a novel PMQR protein, was first reported in 2003 but is now recognized to be widely distributed. Overall, aac(6′)-Ib-cr genes were the most frequent PMQR determinant in our study. The high prevalence of aac(6′)-Ib-cr among K. pneumoniae isolates in our region may be related to clonal spread of a single clone, although further studies using molecular typing methods such as pulsed field gel electrophoresis (PFGE) are needed to confirm this statement. The absence of qepA observed in this study was comparable to the most recent studies conducted in Thailand, Korea, and Mexico [8, 9, 13]. The qepA gene has been documented only from Japan, France, and China and mostly detected among E. coli strains [39]. Our finding showed some PMQR-positive K. pneumoniae isolates coharbored different PMQR genes. This finding could indicate that these PMQR genes were located on the same plasmid, although plasmid analysis is required to confirm this. The majority (60%) of CTX-M-type ESBL producer isolates were CTX-M-1. In a study in Russian hospitals, in agreement with our findings, 92.9% of CTX-M β-lactamases were found to be CTX-M-1 type [17]. Association of qnr genes with extended-spectrum beta-lactamases (ESBLs) has been described [40]. Our qnr-positive ESBL producer K. pneumoniae isolate showed high positive rates of beta-lactamases such as CTX-M-1 and CTX-M-2. This was primarily attributable to the qnrB1 subtype. We suspect that qnr genes possibly contribute to the widespread distribution of CTX-M-1 and CTX-M-2 in our region where fluoroquinolones are widely used. Our results also revealed that some bla CTX-M negative isolates were ESBL-producing showing that ESBL genes of other families may be involved. Another interesting finding was that fluoroquinolone-resistant ESBL-producing K. pneumoniae strains which carried qnrB, qnrS, and aac(6′)-Ib-cr genes were specifically disseminated in ICU ward.
This result is alarming because hospitalized ICU patients are vulnerable and tend to have more risk factors than the other patients; also selective pressure exerted by extensive antibiotic use in ICU ward favors selection and transmission of such multidrug-resistant strains.
5. Conclusion
The results showed high ESBL occurrence with CTX-M type and high frequency of plasmid-mediated quinolone resistance genes among ESBL-producing K. pneumoniae isolates in our region. The aac(6′)-Ib-cr variant and qnrB gene were widely distributed among K. pneumoniae isolates. The screening of ESBL-producing K. pneumoniae for PMQR carriage could be helpful in both treatment and prevention of spread of resistant strains.
Acknowledgments
The technical assistance of Microbiology Department personnel's and staff of Shahid Beheshti Hospital is acknowledged with pleasure.
Disclosure
The results described in this paper were a part of MSc student thesis of Ms. Ehsaneh Shams.
Conflict of Interests
The authors report no conflict of interests in this work.
References
- 1.Rakotonirina H. C., Garin B., Randrianirina F., Richard V., Talarmin A., Arlet G. Molecular characterization of multidrug-resistant extended-spectrum β-lactamase-producing Enterobacteriaceae isolated in Antananarivo, Madagascar. BMC Microbiology. 2013;13(85):1–10. doi: 10.1186/1471-2180-13-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benner K. W., Prabhakaran P., Lowros A. S. Epidemiology of infections due to extended-spectrum beta-lactamase–producing bacteria in a pediatric intensive care unit. The Journal of Pediatric Pharmacology and Therapeutics. 2014;19(2):83–90. doi: 10.5863/1551-6776-19.2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaoutis T. E., Goyal M., Chu J. H., et al. Risk factors for and outcomes of bloodstream infection caused by extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella species in children. Pediatrics. 2005;115(4):942–949. doi: 10.1542/peds.2004-1289. [DOI] [PubMed] [Google Scholar]
- 4.Drawz S. M., Bonomo R. A. Three decades of beta-lactamase inhibitors. Clinical Microbiology Reviews. 2010;23(1):160–201. doi: 10.1128/cmr.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padilla E., Llobet E., Doménech-Sánchez A., Martínez-Martínez L., Bengoechea J. A., Albertí S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrobial Agents and Chemotherapy. 2010;54(1):177–183. doi: 10.1128/aac.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferjani S., Saidani M., Amine F. S., Boubaker I. B. B. Prevalence and characterization of plasmid-mediated quinolone resistance genes in extended-spectrum β-lactamase-producing Enterobacteriaceae in a Tunisian hospital. Microbial Drug Resistance. 2015;21(2):158–166. doi: 10.1089/mdr.2014.0053. [DOI] [PubMed] [Google Scholar]
- 7.Kim H. B., Park C. H., Kim C. J., Kim E. C., Jacoby G. A., Hooper D. C. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob Agents Chemother. 2009;53(2):639–645. doi: 10.1128/aac.01051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park K. S., Kim M. H., Park T. S., Nam Y. S., Lee H. J., Suh J. T. Prevalence of the plasmid-mediated quinolone resistance genes, aac(6′)-Ib-cr, qepA, and oqxAB in clinical isolates of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae in Korea. Annals of Clinical and Laboratory Science. 2012;42(2):191–197. [PubMed] [Google Scholar]
- 9.Pasom W., Chanawong A., Lulitanond A., Wilailuckana C., Kenprom S., Puang-Ngern P. Plasmid-mediated quinolone resistance genes, aac(6′)-Ib-cr, qnrS, qnrB, and qnrA, in urinary isolates of Escherichia coli and Klebsiella pneumoniae at a teaching hospital, Thailand. Japanese Journal of Infectious Diseases. 2013;66(5):428–432. doi: 10.7883/yoken.66.428. [DOI] [PubMed] [Google Scholar]
- 10.Jiang Y., Zhou Z., Qian Y., et al. Plasmid-mediated quinolone resistance determinants qnr and aac(6′)-Ib-cr in extended-spectrum α-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. Journal of Antimicrobial Chemotherapy. 2008;61(5):1003–1006. doi: 10.1093/jac/dkn063. [DOI] [PubMed] [Google Scholar]
- 11.Okade H., Nakagawa S., Sakagami T., et al. Characterization of plasmid-mediated quinolone resistance determinants in Klebsiella pneumoniae and Escherichia coli from Tokai, Japan. Journal of Infection and Chemotherapy. 2014;20(12):778–783. doi: 10.1016/j.jiac.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Robicsek A., Jacoby G. A., Hooper D. C. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infectious Diseases. 2006;6(10):629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 13.Silva-Sánchez J., Cruz-Trujillo E., Barrios H., et al. Characterization of plasmid-mediated quinolone resistance (PMQR) genes in extended-spectrum β-lactamase-producing Enterobacteriaceae pediatric clinical isolates in Mexico. PLoS ONE. 2013;8(10) doi: 10.1371/journal.pone.0077968.e77968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahon C. R., Lehman D. C., Manuselis G. Textbook of Diagnostic Microbiology. 4th. Saunders Company; 2011. [Google Scholar]
- 15.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; 22nd Informational Supplement M100-S22. Wayne, Pa, USA: CLSI; 2013. [Google Scholar]
- 16.Singh R. M., Singh H. L. Comparative evaluation of six phenotypic methods for detecting extended-spectrum beta-lactamase-producing Enterobacteriaceae. Journal of Infection in Developing Countries. 2014;8(4):408–415. doi: 10.3855/jidc.4052. [DOI] [PubMed] [Google Scholar]
- 17.Edelstein M., Pimkin M., Palagin I., Edelstein I., Stratchounski L. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrobial Agents and Chemotherapy. 2003;47(12):3724–3732. doi: 10.1128/aac.47.12.3724-3732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin S. Y., Kwon K. C., Park J. W., et al. Characteristics of aac(6′)-IB-cr gene in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from Chungnam area. Korean Journal of Laboratory Medicine. 2009;29(6):541–550. doi: 10.3343/kjlm.2009.29.6.541. [DOI] [PubMed] [Google Scholar]
- 19.Kang H. Y., Tamang M. D., Seol S. Y., Kim J. Dissemination of plasmid-mediated qnr, aac(6′)-Ib-cr, and qepA genes among 16S rRNA methylase producing enterobacteriaceae in Korea. Journal of Bacteriology and Virology. 2009;39(3):173–182. doi: 10.4167/jbv.2009.39.3.173. [DOI] [Google Scholar]
- 20.Bouchakour M., Zerouali K., Claude J. D. P. G., et al. Plasmid-mediated quinolone resistance in expanded-spectrum beta lactamase producing enterobacteriaceae in Morocco. The Journal of Infection in Developing Countries. 2010;4(12):799–803. doi: 10.3855/jidc.796. [DOI] [PubMed] [Google Scholar]
- 21.Hooper D. C. Emerging mechanisms of fluoroquinolone resistance. Emerging Infectious Diseases. 2010;7(2):337–341. doi: 10.3201/eid0702.010239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du J., Li P., Liu H., Lü D., Liang H., Dou Y. Phenotypic and molecular characterization of multidrug resistant Klebsiella pneumoniae isolated from a university teaching hospital, China. PLoS ONE. 2014;9(4) doi: 10.1371/journal.pone.0095181.e95181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emmerson A. M., Jones A. M. The quinolones: decades of development and use. Journal of Antimicrobial Chemotherapy. 2003;51(1):13–20. doi: 10.1093/jac/dkg208. [DOI] [PubMed] [Google Scholar]
- 24.Firoozeh F., Zibaei M., Soleimani-Asl Y. Detection of plasmid-mediated qnr genes among the quinolone-resistant Escherichia coli isolates in Iran. The Journal of Infection in Developing Countries. 2014;8(7):818–822. doi: 10.3855/jidc.3746. [DOI] [PubMed] [Google Scholar]
- 25.Raei F., Eftekhar F., Feizabadi M. M. Prevalence of quinolone resistance among extended-spectrum β-lactamase producing uropathogenic Klebsiella pneumoniae . Jundishapur Journal of Microbiology. 2014;7(6) doi: 10.5812/jjm.10887.e10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pai H., Seo M.-R., Choi T. Y. Association of QnrB determinants and production of extended-spectrum β-lactamases or plasmid-mediated AmpC β-lactamases in clinical isolates of Klebsiella pneumoniae . Antimicrobial Agents and Chemotherapy. 2007;51(1):366–368. doi: 10.1128/aac.00841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhary N. K., Mahadeva Murthy S. Prevalence of multidrug resistance in uropathogenic Klebsiella species with reference to extended spectrum β-lactamases production. Research Journal of Pharmaceutical, Biological and Chemical Sciences. 2013;4(3):728–735. [Google Scholar]
- 28.Fu Y., Zhang W., Wang H., et al. Specific patterns of gyrA mutations determine the resistance difference to ciprofloxacin and levofloxacin in Klebsiella pneumoniae and Escherichia coli . BMC Infectious Diseases. 2013;13(1, article 8):6. doi: 10.1186/1471-2334-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aitta A. A., El Said M., Gamal D., et al. Biotyping and molecular characterization of Klebsiella pneumoniae producing extended-spectrum beta-lactamase in Cairo, Egypt: a multicenter study. Researcher. 2013;5(9):1–11. [Google Scholar]
- 30.Shiju M. P., Yashavanth R., Narendra N. Detection of extended spectrum beta-lactamase production and multidrug resistance in clinical isolates of E. coli and K. pneumoniae in Mangalore. Journal of Clinical and Diagnostic Research. 2010;4(3):2442–2445. [Google Scholar]
- 31.Kader A. A., Kumar A. Prevalence and antimicrobial susceptibility of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a general hospital. Annals of Saudi Medicine. 2005;25(2):239–242. doi: 10.5144/0256-4947.2005.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M., Sahm D. F., Jacoby G. A., Hooper D. C. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrobial Agents and Chemotherapy. 2004;48(4):1295–1299. doi: 10.1128/aac.48.4.1295-1299.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obeng-Nkrumah N., Twum-Danso K., Krogfelt K. A., Newman M. J. High levels of extended-spectrum beta-lactamases in a major teaching hospital in Ghana: the need for regular monitoring and evaluation of antibiotic resistance. American Journal of Tropical Medicine and Hygiene. 2013;89(5):960–964. doi: 10.4269/ajtmh.12-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panhotra B. R., Saxena A. K., Al-Ghamdi A. M. Extended-spectrum beta-lactamases producing Klebsiella pneumoniae hospital acquired bacteremia. Risk factors and clinical outcome. Saudi Medical Journal. 2004;25(12):1871–1876. [PubMed] [Google Scholar]
- 35.Yang H., Chen H., Yang Q., Chen M., Wang H. High prevalence of plasmid-mediated quinolone resistance genes qnr and aac(6′)-Ib-cr in clinical isolates of Enterobacteriaceae from nine teaching hospitals in China. Antimicrobial Agents and Chemotherapy. 2008;52(12):4268–4273. doi: 10.1128/aac.00830-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marra A. R., Wey S. B., Castelo A., et al. Nosocomial bloodstream infections caused by Klebsiella pneumoniae: impact of extended-spectrum β-lactamase (ESBL) production on clinical outcome in a hospital with high ESBL prevalence. BMC Infectious Diseases. 2006;6(24):1–8. doi: 10.1186/1471-2334-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minarini L. A. R., Poirel L., Cattoir V., Darini A. L. C., Nordmann P. Plasmid-mediated quinolone resistance determinants among enterobacterial isolates from outpatients in Brazil. Journal of Antimicrobial Chemotherapy. 2008;62(3):474–478. doi: 10.1093/jac/dkn237. [DOI] [PubMed] [Google Scholar]
- 38.Crémet L., Caroff N., Dauvergne S., Reynaud A., Lepelletier D., Corvec S. Prevalence of plasmid-mediated quinolone resistance determinants in ESBL Enterobacteriaceae clinical isolates over a 1-year period in a French hospital. Pathologie Biologie. 2011;59(3):151–156. doi: 10.1016/j.patbio.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Wong M. H., Chan E. W., Liu L. Z., Chen S. PMQR genes oqxAB and aac(6′)Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella typhimurium . Frontiers in Microbiology. 2014;5(521):1–7. doi: 10.3389/fmicb.2014.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong H. S., Bae I. K., Shin J. H., et al. Prevalence of plasmid-mediated quinolone resistance and its association with extended-spectrum beta-lactamase and AmpC beta-lactamase in Enterobacteriaceae. Korean Journal of Laboratory Medicine. 2011;31(4):257–264. doi: 10.3343/kjlm.2011.31.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]


