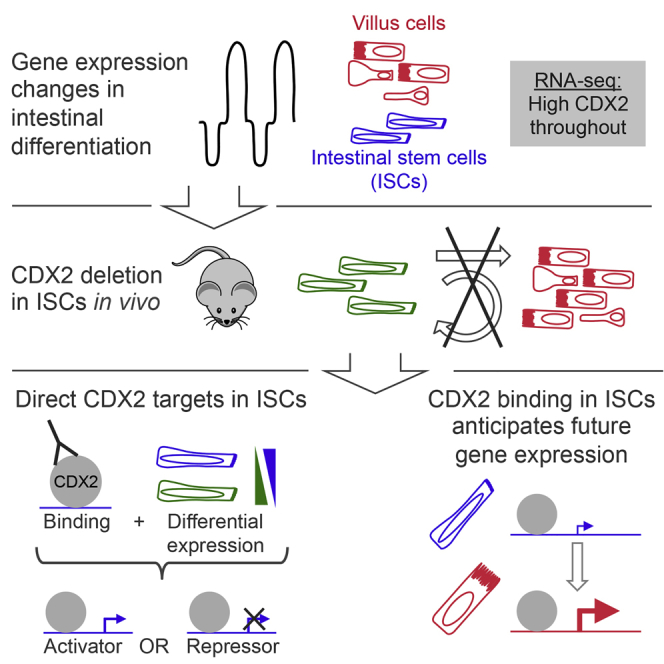
Summary
Lgr5-expressing intestinal stem cells (ISCs) renew the adult gut epithelium by producing mature villus cells (VCs); the transcriptional basis for ISC functions remains unclear. RNA sequencing analysis identified transcripts modulated during differentiation of Lgr5+ ISCs into VCs, with high expression of the intestine-restricted transcription factor (TF) gene Cdx2 in both populations. Cdx2-deleted mouse ISCs showed impaired proliferation and long-term inability to produce mature lineages, revealing essential ISC functions. Chromatin immunoprecipitation sequencing analysis of CDX2 in Lgr5+ ISCs, coupled with mRNA profiling of control and Cdx2−/− ISCs, identified features of CDX2 regulation distinct from VCs. Most CDX2 binding in ISCs occurs in anticipation of future gene expression, but whereas CDX2 primarily activates VC genes, direct ISC targets are activated and repressed. Diverse CDX2 requirements in stem and differentiated cells may reflect the versatility of TFs that specify a tissue in development and control the same tissue in adults.
Graphical Abstract
Highlights
-
•
The TF Cdx2 is highly expressed in intestinal stem cells and villus cells
-
•
CDX2 is required for proliferation and differentiation of intestinal stem cells
-
•
CDX2 binding in stem cells anticipates future villus cell gene expression
-
•
Direct CDX2 targets in ISCs may be activated or repressed
Shivdasani and colleagues demonstrate an essential role for transcription factor CDX2 in replication and differentiation of intestinal stem cells (ISCs). Considering CDX2 binding together with transcriptional changes during epithelial differentiation, they show that binding in ISCs anticipates future gene expression. Through analysis of Cdx2-null ISCs, they identify direct target genes, which are activated or repressed and may underlie the defects.
Introduction
Intestinal stem cells (ISCs) produce diverse mature cell types. The best characterized and most abundant ISC pool consists of Lgr5+ cells at the crypt base (Barker et al., 2007). These cells replicate daily, giving rise to highly proliferative transit-amplifying (TA) progenitors, which occupy higher crypt tiers and eventually produce terminally differentiated villus cells (VCs), including absorptive enterocytes and secretory (goblet and enteroendocrine) cells. Lgr5+ ISCs contribute to all mature VC lineages for months (Barker et al., 2007) and generate “organoids” that self-renew in culture (Sato et al., 2009). Other ISC populations at different crypt positions replicate less frequently, respond to tissue injury, and express markers such as Bmi1, Hopx, Lrig1, and Tert (Clevers, 2013); Lgr5+ ISCs may co-express the same marker genes (Itzkovitz et al., 2012, Muñoz et al., 2012). Indefinite self-renewal and the potential to yield distinct daughter lineages require particular transcriptional programs, but the transcription factors (TFs) that drive them and the associated chromatin states are insufficiently understood. Although knockout mice have implicated several TFs, including ASCL2, KLF5, and YY1, in control of various ISC functions (Nandan et al., 2015, Perekatt et al., 2014, van der Flier et al., 2009), their target genes and mechanisms remain unclear, in part because it is difficult to isolate enough ISCs for biochemical studies.
Using RNA sequencing (RNA-seq) to profile transcripts in intestinal VCs and Lgr5+ ISCs, we observed that Cdx2, a homeobox TF gene implicated in gut epithelial development (Gao et al., 2009, Grainger et al., 2010) and adult enterocyte maturation (Verzi et al., 2010b), was highly and almost equally expressed in both cell populations. Conditional null mice revealed an autonomous proliferation deficit in Cdx2−/− ISCs, and instead of producing mature intestinal cell types, these cells formed undifferentiated cystic structures in vivo and failed to generate proper organoids in culture. Analysis of CDX2 occupancy in wild-type ISCs, integrated with transcriptional changes in purified Cdx2−/− ISCs, identified candidate direct target genes and revealed a likely dual role in gene activation and repression. Most CDX2 binding in ISCs, however, occurs in anticipation of gene activity in future cell generations.
Results
Differential Gene Expression in ISCs and VCs
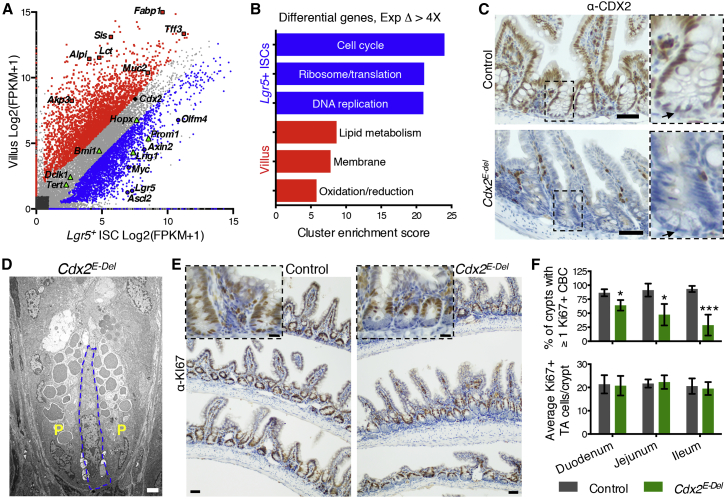
To identify gene expression changes during intestinal epithelial differentiation, we isolated Lgr5+ ISCs from Lgr5EGFP-CreERT2 knockin mice (Lgr5KI) (Barker et al., 2007) and mature VCs, comprised mainly of enterocytes. As RNA-seq revealed modest 3′ bias in ISC samples, we applied a 3′ tag counting method to restrict FPKM (fragments per kilobase of exon per million fragments mapped) calculations to reads mapping to the most 3′ 1,000 bp of each gene (Sigurgeirsson et al., 2014). We observed significant differences in 3,317 VC-enriched and 2,889 ISC-enriched transcripts (Q < 0.05, fold change > 2; Figure 1A). VC-enriched genes included enterocyte (Sis, Alpi/Akp3, Lct, Fabp1) and goblet-cell (Muc2, Tff3) markers, while genes such as Lgr5, Ascl2, Axin2, Myc, and Olfm4 were enriched in ISCs (Figure 1A). Genes enriched >4-fold in ISCs belonged in Gene Ontology terms related to cell-cycle control, protein translation, and DNA replication, while VC-enriched terms included lipid metabolism, membrane transport, and oxidation-reduction (Figure 1B), indicating that the data are reliable. Genes previously ascribed to alternative ISC populations—Bmi1, Hopx, Lrig1, and Prom1—were highly expressed in both Lgr5+ ISCs and VCs (Figure 1A), supporting recent reports that these markers are not exclusive (Itzkovitz et al., 2012, Muñoz et al., 2012). Levels of Cdx2, an intestine-restricted TF gene necessary for enterocyte maturation (Verzi et al., 2010b), were almost as high in purified ISCs as in VCs (Figure 1A); immunostaining confirmed CDX2 expression in ISCs (Figure 1C).
Figure 1.
Loss of Cdx2 mRNA Selectively Impairs ISC Proliferation
(A) RNA-seq data from three biological replicates of Lgr5+ ISCs and VCs identify modulated genes. Grey, 6,752 genes with comparable expression in both cell types; red, 3,317 VC-enriched genes; blue, 2,889 ISC-enriched genes (>2-fold; Q < 0.05). Markers of enterocytes or goblet cells (red), Lgr5+ ISCs (blue), and other ISC populations (green) are highlighted. Cdx2 is highly expressed in both cell types.
(B) Gene Ontology clusters of genes with >4-fold change in VCs and ISCs.
(C) CDX2 immunohistochemistry (IHC) in control and Cdx2E-Del mouse ileum 3 days after final TAM treatment, confirming complete epithelial loss, including ISCs (magnified to right; arrows).
(D) Electron microscopy of Cdx2E-Del ileum confirms the presence of ISCs (blue) among Paneth (P) cells.
(E) KI67 IHC reveals loss of proliferation in ISCs (magnified in Inset), but not in TA cells.
(F) Fraction of crypts with at least one KI67+ ISC in different regions of control and Cdx2E-Del intestines (∗p < 0.01; ∗∗∗p < 0.0001; t test). The average number of KI67+ TA cells is not significantly altered. At least 30–50 crypts were counted in each of at least four biological replicates.
Scale bars represent 50 μm (C and E), 20 μm (inset in E), and 2 μm (D).
Loss of CDX2 Markedly and Selectively Impairs ISC Proliferation and Differentiation
To investigate a possibly separate CDX2 role in ISCs, we examined Cdx2Fl/Fl;Villin-CreERT2 mice, where tamoxifen (TAM) deletes Cdx2 in all small intestine epithelial cells (Cdx2E-Del for epithelium deleted; Figure 1C). Although ISCs with characteristic ultrastructural features were present at the crypt base (Figure 1D), the number of crypts with at least one KI67+ ISC was reduced throughout the Cdx2E-Del small intestine, most significantly in the ileum (Figures 1E and 1F), where CDX2 levels are normally highest (James and Kazenwadel, 1991). This defect was confined to crypt base ISCs; TA cells in higher tiers showed a normal mitotic fraction (Figures 1E and 1F).
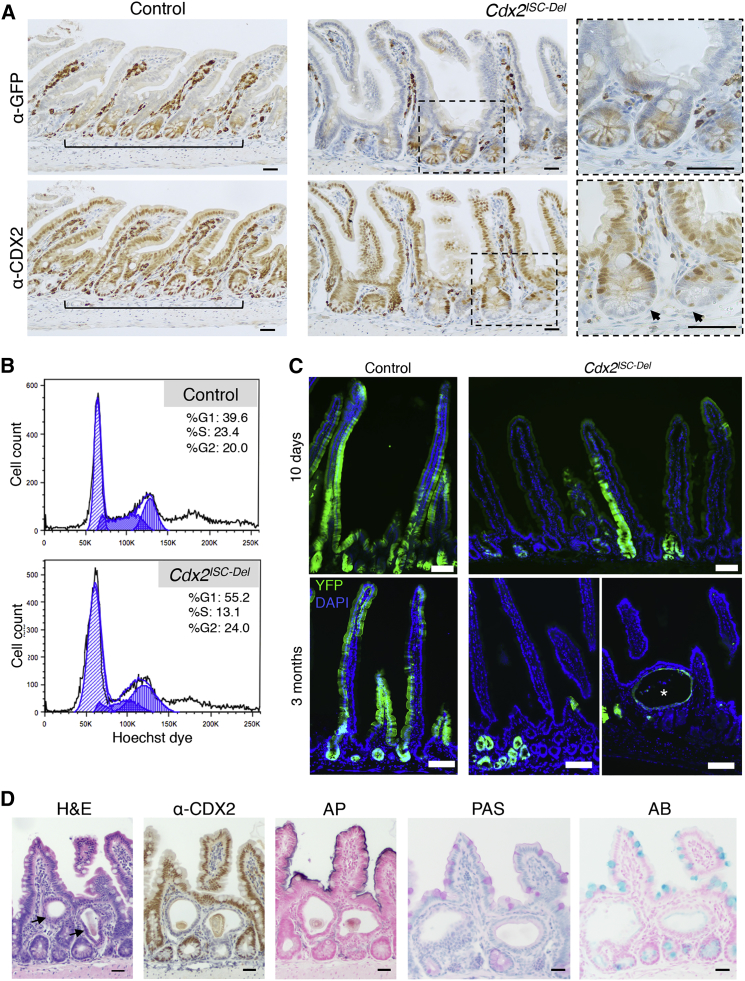
Cdx2Fl/Fl;Lgr5KI mice (Cdx2ISC-Del for ISC deleted) showed CDX2 loss from all GFP+ crypts after TAM treatment (Figure 2A). As Lgr5KI mice are mosaic (Barker et al., 2007), many crypts retained CDX2 in all cells, and because Paneth cells do not express Lgr5 (Barker et al., 2007) and turn over slowly (Ireland et al., 2005), they retained CDX2 (Figure 2A). In the proximal small intestine, where mosaic Lgr5KI expression is highest (Barker et al., 2007), we observed fewer LGR5-GFPHI cells in S-phase and more cells in G1 (Figure 2B). Thus, independent measures reveal a significant defect in replication of Cdx2 null ISCs. Because CDX2 is expressed only in epithelial cells and not in the lamina propria, this defect must be cell autonomous.
Figure 2.
CDX2 Is Required for Lgr5+ ISC Proliferation and Differentiation
(A) Serial tissue sections from control and Cdx2ISC-Del intestines 4 days after TAM, showing CDX2 loss in patches of LGR5-GFP-Cre+ crypts (dashed areas magnified to right). CDX2 is retained in some Paneth cells in these crypts (arrows).
(B) Representative cell-cycle analysis of Lgr5+ ISCs from control and Cdx2ISC-Del proximal small bowel. Flow cytometry of Cdx2Del ISCs stained with Hoechst dye reveals accumulation in G1, with a reduced S-phase fraction.
(C) Cdx2Fl/Fl;Lgr5KI;R26RYFP and control mice were injected with TAM for 5 days and examined at the indicated times. YFP fluorescence at 10 days indicates that traced Cdx2Del crypts retain some ability to contribute to villi. At 90 days, tracing in Cdx2ISC-Del intestines is confined to cysts (asterisk) and unproductive crypts.
(D) H&E staining of the cysts shows luminal debris. IHC confirms that traced cysts lack CDX2 and are surrounded by CDX2+ crypts that did not express the Lgr5KI allele. Alkaline phosphatase (AP), periodic acid-Schiff (PAS), and Alcian blue (AB) staining show absence of mature enterocytes and goblet cells in the cystic structures.
Scale bars represent 30 μm (A and D) and 50 μm (C). See also Figure S1.
Intact TA cell replication suggested that early events following ISC differentiation may not require CDX2. To test this possibility, we crossed a Rosa26lox-STOP-lox-YFP allele (R26RYFP [Srinivas et al., 2001]) onto the Cdx2Fl/Fl;Lgr5KI background to enable lineage tracing. After treating mice with TAM, we followed YFP expression serially (Figure 2C), noting again that mosaic Lgr5KI expression would delete Cdx2 and activate Yfp in a minority of crypts. As TAM induction of the Lgr5-CreERT2 irreversibly marks cells with YFP, continuous Lgr5 expression is not required to monitor the lineage tracing. Ten days later, YFP was present in crypts and villi in control (Cdx2+/+;Lgr5KI;R26RYFP) and, at slightly lower frequency, in Cdx2ISC-Del mice (Figure 2C). However, starting at 1 month and continuing for at least 6 months after Cre activation, YFP expression in Cdx2ISC-Del intestines was confined to crypts and cystic inclusions, and never detected in villi (Figures 2C, S1A, and S1B). These cysts, similar to those previously noted in Cdx2−/− crypts (Stringer et al., 2012), showed total absence of CDX2 and of mature VC features such as alkaline phosphatase, which marks enterocytes, or avidity for periodic acid-Schiff or Alcian blue, which marks goblet cells (Figure 2D). Cells lining the cysts had a cuboidal morphology, with round nuclei (Figure S1C); EZR and NHERF localization indicated intact apico-basal polarity (Figure S1D). LGR5-GFP expression was typically confined to one side of the cysts, near proliferating TA-like cells, which is reminiscent of normal crypt organization, and non-replicating cells commonly were sloughed from the upper cyst wall, collecting as luminal debris (Figure S1E). Thus, Cdx2Del ISCs proliferate poorly, and although they spawn replication-competent TA cells, maturation of their progeny is severely impaired. Of note, intestinal crypt cells also express CDX1 (James and Kazenwadel, 1991), a homolog with redundant activity in TA cells (Verzi et al., 2011). The specific defects in Cdx2−/− crypts reveal a crucial, non-redundant requirement in the stem cell compartment.
Lgr5+ ISCs generate organoid bodies when cultured in defined media (Sato et al., 2009). To test whether Cdx2 loss affects this property, we treated Cdx2Fl/Fl;Villin-CreERT2 intestinal crypts with TAM in culture. Eight days later, organoids from control Cre− crypts were large and showed crypt-like evaginations, whereas Cdx2-deficient crypts formed few organoids; when present, these were small and spherical, with few protrusions, and totally lacked CDX2, but showed signs of multilineage differentiation in the form of mature enterocyte and goblet cell markers (Figures S1F–S1H). Thus, significant deficits in cell replication, lineage tracing, organoid formation, and generation of mature intestinal cells reveal a CDX2 requirement in ISC functions, ostensibly distinct from CDX2’s activity in mature enterocytes or its redundant functions with CDX1 in TA cells.
CDX2 Loss in ISCs Affects Genes that Regulate Cell Proliferation
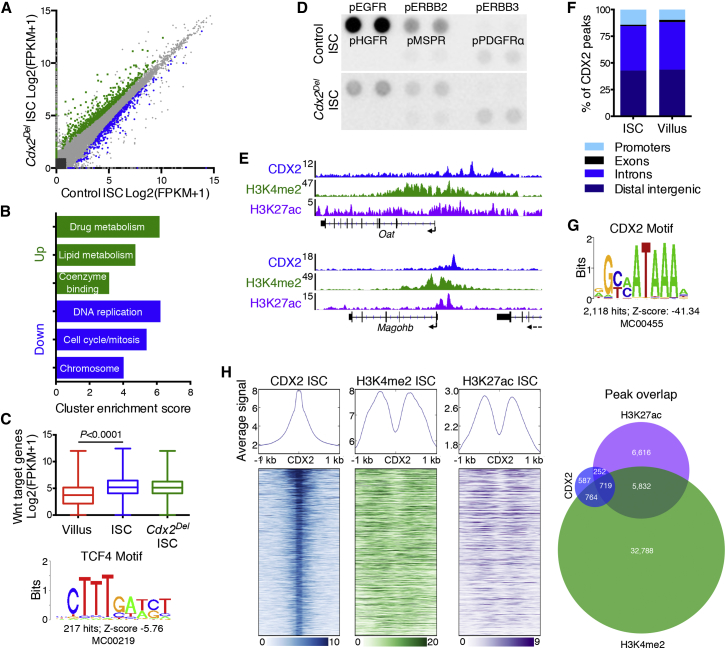
To identify genes dysregulated in CDX2 null ISCs, we isolated GFP+ cells from Cdx2ISC-Del and control (Cdx2+/+;Lgr5KI) intestinal crypts after 4 days of TAM exposure. RNA-seq confirmed loss of most transcripts containing loxP-flanked exon 2 in Cdx2Del ISCs (Figure S2A). In addition, 412 transcripts were decreased and 639 were increased at least 1.5-fold (Q < 0.05; Figure 3A). Genes with reduced expression were enriched for terms related to mitosis, consistent with reduced ISC replication, whereas genes that increased in expression were enriched for terms related to drug and lipid metabolism (Figure 3B). Because CDX2 may mediate intestine-specific Wnt signaling (Verzi et al., 2010a), we assessed the consequences of CDX2 loss on a curated list of 203 intestinal Wnt target genes (van de Wetering et al., 2002). These transcripts showed higher average expression in control ISCs than in VCs, but average expression was unperturbed in Cdx2Del ISCs (Figure 3C); only 24 individual Wnt target genes were significantly affected, with no obvious pattern (Figure S2B). Although 217 CDX2 binding sites contain a TCF/LEF motif (Figure 3C), the data suggest that CDX2 activity in ISCs is largely Wnt independent, possibly compensated by the partially redundant factor CDX1. We also probed arrays containing 39 phosphorylated receptor tyrosine kinases with protein lysates from purified control and Cdx2Del ISCs. Strong phosphorylation signals for the epidermal growth factor receptor (EGFR) and ERBB2 were reproducibly lower in Cdx2Del ISCs (Figure 3D). Together, these findings reveal that CDX2 controls select ISC genes, some of which mediate cell surface events in EGFR signaling. A single dominant target gene is, however, unlikely to explain CDX2 requirements in vivo. We therefore sought to determine features of CDX2 regulation by mapping its binding sites in ISCs.
Figure 3.
Genes Perturbed by CDX2 Loss and Delineation of CDX2 Binding in ISCs
(A) RNA-seq data from isolated control and Cdx2Del ISCs, using three biological replicates. 639 transcripts are elevated in Cdx2−/− ISCs (green), and 412 transcripts are reduced (blue). Grey, genes with <1.5-fold change or Q > 0.05.
(B) Gene Ontology cluster enrichment among genes reduced or increased in Cdx2Del ISCs.
(C) Average RNA-seq FPKM values of 203 intestinal Wnt target genes in control VCs and ISCs and in Cdx2Del ISCs. Significance was assessed by a t test. Bottom: the TCF4 motif found in 217 CDX2 binding sites.
(D) Phospho-receptor tyrosine kinase arrays probed with lysates from purified control or Cdx2Del ISCs. Signals for pEGFR and pERBB2 are reduced in Cdx2Del ISCs.
(E) ChIP-seq signals for CDX2, H3K4me2, and H3K27ac near the Oat and Magohb genes; the y axis represents aligned ChIP-seq tag counts.
(F) Distribution of CDX2 binding sites among genomic regions is similar in ISCs and VCs.
(G) The consensus CDX2 motif is the most enriched (Z score: −41.34) in CDX2-bound sites.
(H) Heatmaps of ChIP signals for CDX2, H3K4me2, and H3K27ac around CDX2 binding sites in purified ISCs. Right: overlap of CDX2 binding sites with called H3K4me2 and H3K27ac peaks, which underestimate the overlap evident in heatmaps.
Gene Targets of CDX2 Regulation in ISCs
In contrast with >12,000 sites previously detected in VCs (Verzi et al., 2013), chromatin immunoprecipitation sequencing (ChIP-seq) on purified wild-type Lgr5+ ISCs identified 2,371 regions enriched for CDX2 binding (e.g., Figure 3E). The distribution of CDX2 binding sites in promoters and distant sites in Lgr5+ ISCs resembled that in VCs (Figure 3F); these areas showed evolutionary conservation (Figure S2C), and the CDX2 consensus motif was the most recurring (Figure 3G). Moreover, binding sites in ISCs coincided with presence of H3K4me2 (Kim et al., 2014) and H3K27ac (Sheaffer et al., 2014) histone marks, indicating that they represent bona fide active cis-regulatory elements (Figures 3E and 3H).
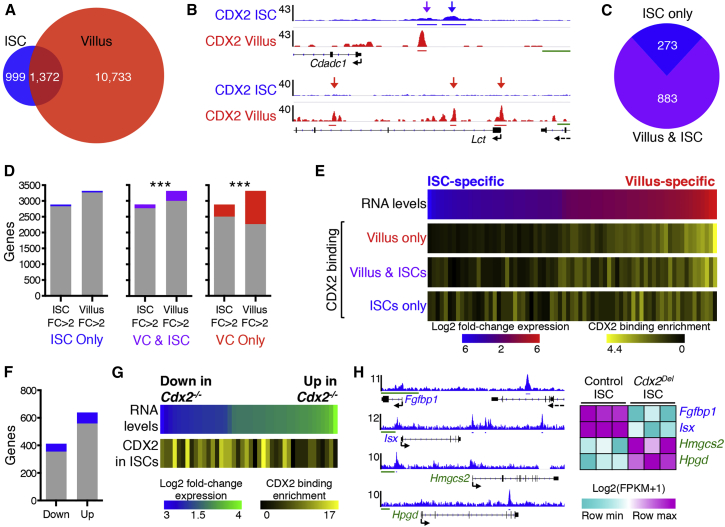
Most CDX2 binding sites in VCs (Verzi et al., 2013) gave weak or no signals in Lgr5+ ISCs (Figure 4A), reflecting either truly absent binding or the challenges inherent in detecting low-level occupancy. Conversely, at least 60% of CDX2 binding sites in purified ISCs were also evident in mature VCs (Figures 4A and 4B), likely representing occupancy at genes expressed in both cell types or ISC binding in anticipation of VC activity. The remaining ∼1,000 binding sites in Lgr5+ ISCs were considerably weaker or absent in VCs (Figures 4A and 4B) and may represent sites of ISC-specific activity. To distinguish among these possibilities, we first assigned CDX2 binding sites to the nearest gene within 30 kb. Of the 1,156 genes showing at least 1 binding site in ISCs, 883 genes were also occupied in VCs (Figure 4C). We first approached these findings from the perspective of genes with nearby CDX2 binding (Figure 4D). The small proportion of genes bound only in ISCs was similar among ISC-enriched and VC-expressed genes, whereas genes bound only in VCs or in both ISCs and VCs were more highly expressed in VCs. Approaching the data next from the perspective of genes differentially expressed in ISCs and VCs (Figure 4E), we see that villus-expressed genes are enriched for CDX2 binding in both ISCs and VCs. In contrast, genes with unique CDX2 binding in ISCs are distributed across the expression spectrum (Figure 4E). Notably, genes bound in both ISCs and VCs are highly enriched for “transcriptional activity” (Gene Ontology cluster enrichment score: 3.38) and include Cdx1/2, Gata4/6, Hnf4a/g, Hnf1b, Klf5, Vdr, and Yy1 (Figure S2D). Thus, much of the CDX2 binding in ISCs persists in differentiated cells, near genes that are highly expressed in the latter. This binding thus precedes future VC activity rather than serving functions unique to ISCs.
Figure 4.
Contrasting Features of CDX2 Binding in ISCs and VCs
(A) Number of CDX2 binding sites identified exclusively in ISCs (blue), only in VCs (red), or in both cell types with ≥1-bp overlap (purple).
(B) Illustrative data traces for CDX2 binding sites of each class (arrows). y axis, ChIP-seq tag counts; x axis: genome coordinates.
(C) Among the 1,156 genes that bind CDX2 within 30 kb in ISC, 273 are bound only in ISCs and 883 are also bound in VCs.
(D) Genes bound only in VCs (red) or in both VCs and ISCs (purple) are mostly expressed in VCs (∗∗∗p < 0.0001; χ2 test); genes bound only in ISCs (blue) are not enriched for expression in either compartment.
(E) Genes ordered by the degree of differential expression in ISCs and VCs (>2-fold, Q < 0.05, from data in Figure 1A) were binned into groups of 100 (blue–red heatmap). Yellow heatmaps represent average number of CDX2 binding sites in VCs only, in ISCs only, or in both populations within 30 kb of genes in each bin, normalized for average CDX2 binding across all expressed genes.
(F) CDX2 binds near similar proportions of genes (p = 0.57, Fisher’s exact test) that show reduced or increased transcript levels in Cdx2−/− ISCs.
(G) Genes ordered by the degree of differential expression in Cdx2Del versus control ISCs (from data in Figure 3A) were binned into groups of 25 (blue–green heatmap). Below: average number of CDX2 binding sites in ISCs within 30 kb of the genes in each bin, divided by the average CDX2 binding across all expressed genes.
(H) CDX2 binding data at representative upregulated (Hmgcs2 and Hpgd) and downregulated (Fgfpb1 and Vdr) genes. Right: heatmap of corresponding RNA-seq data.
Scale bars represent 1 kb (green line in B) and 5 kb (H). See also Figures S2 and S3.
Because direct transcriptional targets in ISCs should show both CDX2 binding and perturbed mRNA levels in its absence, we considered ISC binding in the light of differential gene expression in Cdx2ISC-Del ISCs. Most CDX2-bound genes were unaffected in transcript levels, implying that CDX2 is dispensable for their regulation. Among the 137 genes with CDX2 binding and altered mRNA levels in Cdx2−/− ISCs, 80 genes were increased and 57 were decreased (Figures 4F–4H and S3). In VCs, by contrast, CDX2 binds almost exclusively at loci that are downregulated upon Cdx2 loss (Verzi et al., 2013). Thus, whereas CDX2 predominantly activates VC genes, in ISCs it may activate some genes and repress others.
Discussion
We demonstrate an early, cell-autonomous role for CDX2 in promoting ISC replication in vivo and Cdx2−/− cells’ steadily diminished ability to produce differentiated VCs. Previous studies of Cdx2Del cysts and organoid cultures have suggested that Cdx2 loss activates stomach epithelial marker genes and hence compromises intestinal identity (Hryniuk et al., 2012, Simmini et al., 2014, Stringer et al., 2012). However, our RNA-seq analysis of Cdx2Del ISCs revealed elevated levels of only 2 of 11 common stomach-specific transcripts (Table S1), and we did not detect CDX2 binding near these genes in ISCs. Thus, gastric transdifferentiation does not seem to be a prominent early feature of Cdx2−/− ISCs, though other target genes, including transcriptional and epigenetic regulators, may perturb stomach-specific genes over time.
Feedback mechanisms for ISC homeostasis are poorly understood. By revealing different CDX2 requirements in various crypt cell populations, our findings provide useful insights. First, combined loss of CDX1 and CDX2 markedly affects TA cell replication (Verzi et al., 2011), but these cells withstand loss of CDX2 alone. Second, the outcome when Lgr5+ ISCs are incapacitated by CDX2 loss—but remain viable—differs from that when Lgr5+ ISCs are killed using Diphtheria toxin. Whereas reserve Bmi1+ ISCs are rapidly recruited into an active role in the latter case (Tian et al., 2011), Cdx2−/− cysts persist for months, avoiding replacement by CDX2+ cells, such as the progeny of quiescent ISCs in Cdx2ISC-Del mice. One possibility is that leaky Lgr5-Cre expression in quiescent ISCs triggered deletion of Cdx2, which is also required in this ISC pool, but Lgr5 expression is notably absent in quiescent ISCs in Lgr5KI mice (Barker et al., 2007, Tian et al., 2011). We therefore favor the alternative explanation that persistent ISCs and TA cells in Cdx2ISC-Del intestines are sufficient to avoid the feedback signals that elicit compensatory activity from quiescent ISCs.
This study advances understanding of CDX2 mechanisms in ISC maintenance and differentiation. CDX2 occupies hundreds of genomic sites exclusively in ISCs and thousands of additional sites in VCs. Unlike in VCs, however, CDX2 binding in ISCs correlates poorly with ISC-expressed genes. Instead, much of it overlaps with binding sites in VCs, occurring at genes activated in subsequent cell generations. Moreover, while CDX2 behaves almost exclusively as a transactivator in VCs, it may directly activate or repress ISC genes. This could occur through interactions with different co-factors, alternative post-translational modifications, or differential chromatin access. Studies of other TFs and chromatin in purified ISCs are necessary to resolve these possibilities.
Experimental Procedures
Mice
Cdx2Fl/Fl, Villin-CreERT2, Lgr5-EGFP-IRES-CreERT2, and Rosa26-lox-STOP-lox-YFP mice were described previously (Barker et al., 2007, el Marjou et al., 2004, Srinivas et al., 2001, Verzi et al., 2010b). For recombination of conditional alleles, mice were injected intraperitoneally for 4–5 days with 1–2 mg tamoxifen (Sigma). All procedures followed protocols approved by an Institutional Animal Care and Use Committee.
Epithelial Cell Isolation
Crypts from the proximal small intestine were isolated by scraping off villi and incubating in 5 mM EDTA. Crypts were trypsinized into a single cell suspension and passed through a FACSAria III instrument (BD Biosciences) to sort single live LGR5-GFPHI cells. Alternatively, intestines were incubated in EDTA and VCs were captured using 70-μm filters.
RNA-Seq
More than 105 control ISCs, Cdx2Del ISCs, or VCs were collected from single mice, and isolated RNA was sequenced (see the Supplemental Experimental Procedures for RNA isolation, library preparation, sequencing, and analysis protocols). Three biological replicates for each condition were compared, and significantly dysregulated genes were analyzed for Gene Ontology term enrichment.
Histology
Intestine segments were fixed overnight in 4% paraformaldehyde at 4°C, and they were dehydrated for paraffin embedding or incubated overnight in 30% sucrose and frozen in optimum cutting temperature (OCT) compound (Tissue-Tek). 5-μm paraffin sections were stained with H&E, alkaline phosphatase, alcian blue, periodic acid-Schiff, or specific antibodies (Ab) to CDX2, KI67, GFP, EZR, or NHERF (more information in the Supplemental Experimental Procedures). 10-μm cryosections were mounted with DAPI to image native YFP.
ChIP-Seq
5 × 106 LGR5-GFPHI ISCs were pooled from several mice and cross-linked in 1% formaldehyde at room temperature for 25 min, followed by lysis, sonication, and immunoprecipitation with 3 μg CDX2 Ab (Bethyl Laboratories). See the Supplemental Experimental Procedures for ChIP, sequencing, and analysis protocols. CDX2 binding sites were identified from aligned sequencing reads and assigned to the single nearest gene within 30 kb. Published ChIP-seq data were analyzed in parallel.
Additional details of protocols can be found in the Supplemental Experimental Procedures.
Acknowledgments
This work was supported by NIH awards R01DK082889 (R.A.S.), R01DK084056 (D.T.B.), and F31CA180784 (A.K.S.R.) and a NSF graduate research fellowship (A.K.S.R.). We thank S. Robine for Villin-CreERT2 mice; P. Cejas and T.-H. Kim for sharing expertise and reagents; and M. Verzi and N. O’Neill for valuable discussions.
Published: October 15, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, three figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.09.006.
Accession Numbers
The accession number for all sequencing data reported in this paper is GEO: GSE70766.
Supplemental Information
References
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- el Marjou F., Janssen K.P., Chang B.H., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D., Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- Gao N., White P., Kaestner K.H. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger S., Savory J.G.A., Lohnes D. Cdx2 regulates patterning of the intestinal epithelium. Dev. Biol. 2010;339:155–165. doi: 10.1016/j.ydbio.2009.12.025. [DOI] [PubMed] [Google Scholar]
- Hryniuk A., Grainger S., Savory J.G.A., Lohnes D. Cdx function is required for maintenance of intestinal identity in the adult. Dev. Biol. 2012;363:426–437. doi: 10.1016/j.ydbio.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Ireland H., Houghton C., Howard L., Winton D.J. Cellular inheritance of a Cre-activated reporter gene to determine Paneth cell longevity in the murine small intestine. Dev. Dyn. 2005;233:1332–1336. doi: 10.1002/dvdy.20446. [DOI] [PubMed] [Google Scholar]
- Itzkovitz S., Lyubimova A., Blat I.C., Maynard M., van Es J., Lees J., Jacks T., Clevers H., van Oudenaarden A. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat. Cell Biol. 2012;14:106–114. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James R., Kazenwadel J. Homeobox gene expression in the intestinal epithelium of adult mice. J. Biol. Chem. 1991;266:3246–3251. [PubMed] [Google Scholar]
- Kim T.H., Li F., Ferreiro-Neira I., Ho L.L., Luyten A., Nalapareddy K., Long H., Verzi M., Shivdasani R.A. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz J., Stange D.E., Schepers A.G., van de Wetering M., Koo B.K., Itzkovitz S., Volckmann R., Kung K.S., Koster J., Radulescu S. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandan M.O., Ghaleb A.M., Bialkowska A.B., Yang V.W. Krüppel-like factor 5 is essential for proliferation and survival of mouse intestinal epithelial stem cells. Stem Cell Res. (Amst.) 2015;14:10–19. doi: 10.1016/j.scr.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perekatt A.O., Valdez M.J., Davila M., Hoffman A., Bonder E.M., Gao N., Verzi M.P. YY1 is indispensable for Lgr5+ intestinal stem cell renewal. Proc. Natl. Acad. Sci. USA. 2014;111:7695–7700. doi: 10.1073/pnas.1400128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Sheaffer K.L., Kim R., Aoki R., Elliott E.N., Schug J., Burger L., Schübeler D., Kaestner K.H. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 2014;28:652–664. doi: 10.1101/gad.230318.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurgeirsson B., Emanuelsson O., Lundeberg J. Sequencing degraded RNA addressed by 3′ tag counting. PLoS ONE. 2014;9:e91851. doi: 10.1371/journal.pone.0091851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmini S., Bialecka M., Huch M., Kester L., van de Wetering M., Sato T., Beck F., van Oudenaarden A., Clevers H., Deschamps J. Transformation of intestinal stem cells into gastric stem cells on loss of transcription factor Cdx2. Nat. Commun. 2014;5:5728. doi: 10.1038/ncomms6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer E.J., Duluc I., Saandi T., Davidson I., Bialecka M., Sato T., Barker N., Clevers H., Pritchard C.A., Winton D.J. Cdx2 determines the fate of postnatal intestinal endoderm. Development. 2012;139:465–474. doi: 10.1242/dev.070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., de Sauvage F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A.P. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- van der Flier L.G., van Gijn M.E., Hatzis P., Kujala P., Haegebarth A., Stange D.E., Begthel H., van den Born M., Guryev V., Oving I. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- Verzi M.P., Hatzis P., Sulahian R., Philips J., Schuijers J., Shin H., Freed E., Lynch J.P., Dang D.T., Brown M. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. Proc. Natl. Acad. Sci. USA. 2010;107:15157–15162. doi: 10.1073/pnas.1003822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M.P., Shin H., He H.H., Sulahian R., Meyer C.A., Montgomery R.K., Fleet J.C., Brown M., Liu X.S., Shivdasani R.A. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell. 2010;19:713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M.P., Shin H., Ho L.-L., Liu X.S., Shivdasani R.A. Essential and redundant functions of caudal family proteins in activating adult intestinal genes. Mol. Cell. Biol. 2011;31:2026–2039. doi: 10.1128/MCB.01250-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M.P., Shin H., San Roman A.K., Liu X.S., Shivdasani R.A. Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding. Mol. Cell. Biol. 2013;33:281–292. doi: 10.1128/MCB.01185-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.