Summary
Tools for rapid and efficient transgenesis in “safe harbor” loci in an isogenic context remain important to exploit the possibilities of human pluripotent stem cells (hPSCs). We created hPSC master cell lines suitable for FLPe recombinase-mediated cassette exchange (RMCE) in the AAVS1 locus that allow generation of transgenic lines within 15 days with 100% efficiency and without random integrations. Using RMCE, we successfully incorporated several transgenes useful for lineage identification, cell toxicity studies, and gene overexpression to study the hepatocyte lineage. However, we observed unexpected and variable transgene expression inhibition in vitro, due to DNA methylation and other unknown mechanisms, both in undifferentiated hESC and differentiating hepatocytes. Therefore, the AAVS1 locus cannot be considered a universally safe harbor locus for reliable transgene expression in vitro, and using it for transgenesis in hPSC will require careful assessment of the function of individual transgenes.
Highlights
-
•
RMCE using positive/negative selection allows generation of transgenic hPSC lines in 15 days
-
•
The AAVS1 locus exerts transgene inhibition by inducing de novo DNA methylation
-
•
AAVS1-mediated silencing appears sequence and lineage-specific in vitro, yet is not present in vivo
-
•
AAVS1 does not meet the requirements to be considered a universal safe harbor locus in vitro
Verfaillie, Ordovás, and colleagues report an efficient and fast method to genetically engineer hPSC in the AAVS1 safe harbor locus free of random integration events. Their results demonstrate the suitability of the locus for several applications to study the hepatic lineage, but they uncover a so-far-unknown variable AAVS1-mediated transgene inhibition in undifferentiated and hepatocyte differentiating hESC. Therefore, AAVS1 cannot be considered a safe harbor locus.
Introduction
As it has been the case for studies aiming to understand mouse development, transgenesis is an indispensable tool to fully exploit the potential of human pluripotent stem cells (hPSCs). Recent technological advances using site-specific nucleases (Zinc Finger Nucleases [ZFNs], Transcription Activator-Like Effector Nucleases [TALENs], or clustered regularly interspaced short palindromic repeats [CRISPR]/Cas9 system) have allowed to overcome major hurdles hampering genome editing in hPSCs (Li et al., 2014). Gene targeting constitutes the method of choice for transgenesis in hPSCs as it eliminates the drawbacks of random integration methods linked to possible insertional mutagenesis and epigenetic silencing, which lead to variegated transgene expression in subpopulations of cells (Cherry et al., 2000, Yao et al., 2004).
Despite these advances, gene targeting in hPSCs still remains a laborious process, and the development of tools that allow rapid and versatile genetic modification remains of great interest. Site-specific recombinase-mediated homologous recombination with pre-integrated recombination target sequences in “safe harbor” loci, like the Rosa26 or Hprt1 loci, has been extensively used in mouse transgenesis. Such safe harbor loci are found in ubiquitously expressed genes with transcriptional competent conformation that allows stable transgene expression with no detrimental effect on the biology of the modified cells. In hPSCs, Cre recombinase systems for recombinase-mediated cassette exchange (RMCE) have been developed either in the adeno-associated virus integration site 1 (AAVS1 locus) (Ramachandra et al., 2011, Tay et al., 2013, Zhu et al., 2013) or by random integration (Du et al., 2009), though such methods do not constitute a technical improvement over gene targeting approaches using nucleases.
The AAVS1 locus, located in the first intron of the PPP1R12C gene on chromosome 19 has been described to meet the “safe harbor” requirements in a variety of cell types including hPSCs. Though the function of the PPP1R12C gene has not been fully investigated, hPSCs retain pluripotency after targeting. In addition, transgene expression in the AAVS1 locus appears stable in undifferentiated hPSCs and following differentiation to all three germ layers in vitro and in vivo (DeKelver et al., 2010, Hockemeyer et al., 2009, Lombardo et al., 2011, Qian et al., 2014, Smith et al., 2008).
The goal of this study was to generate an efficient and rapid method of transgenesis in the AAVS1 locus of hPSCs, based on RMCE using positive and negative selection to allow the generation of non-clonal transgenic lines, to enable stable incorporation of lineage-specific promoters, molecular response sensors, or inducible gene overexpression. We focused on validating the applicability of the RMCE in the AAVS1 locus during hepatocyte differentiation as only few studies have used transgenesis to characterize this lineage in human (Davis et al., 2008, Duan et al., 2007, Ishii et al., 2008, Umeda et al., 2013, Wang et al., 2011). Using ZFNs to pre-integrate FRT sequences in the AAVS1, we generated an RMCE system that allows the generation of cell lines with 100% efficiency and without random integrations in ±15 days. We demonstrate the suitability of the locus to support several applications for the study of the hepatocyte lineage, although we found variable transgene inhibition in the pluripotent and differentiated state of hESC in vitro in a potential lineage-dependent manner, which appears to not be present in teratomas in vivo. Our results suggest that the AAVS1 locus is not as safe as generally believed.
Results
Generation of an RMCE-Suitable Master Cell Line and RMCE
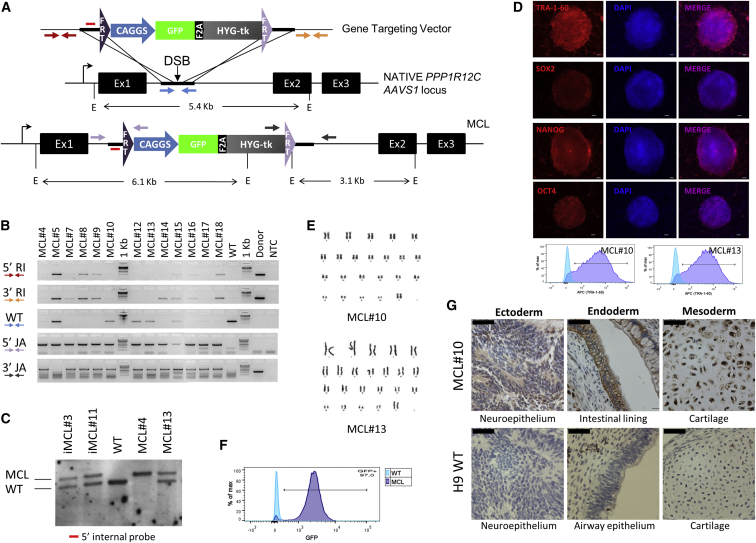
The master cell line (MCL) was generated as explained in the Supplemental Experimental Procedures (Figure 1A). Amplification of the wild-type allele and Southern blotting was performed to determine whether the integration was mono or biallelic and to rule out random integration events (Figures 1B and 1C). Two heterozygously targeted clones were chosen for further characterization of maintenance of pluripotency (teratoma formation assay was carried out using a protocol approved by the Institutional Ethics Committee at KU Leuven) and a normal karyotype (Figures 1D, 1E, and S1A). In agreement with previous studies, GFP was homogeneously expressed in undifferentiated and differentiated cells from the selected clones, which was stable during passaging and differentiation (Figures 1F and 1G).
Figure 1.
Generation and Characterization of FRT-Containing Master Cell Lines in hESC
(A) The AAVS1 gene targeting vector pZ:F3-CAGGS GPHTK-F, containing homology regions to the PPP1R12C locus (thick bars) and flanking FRTs (additional details in the Supplemental Information). The 5′ internal Southern blot probe (red bar) and fragment sizes of DNA digested with EcoRI (E) are indicated.
(B) PCR genotyping of the master cell line (MCL) clones using primer sets depicted in (A) for 5′/3′ random integration (RI), amplification of the wild-type allele (WT), and 5′/3′ junction assays (JA). NTC, negative template control.
(C) Southern blot of the wild-type cells and clones of iPSC (iMCL) and ESC (MCL) master cell lines.
(D) Top: Expression of pluripotency markers of a representative MCL clone by immunocytochemistry (scale bars, 100 μm). Bottom: TRA-1-60 expression in two clones determined by FACS.
(E) Karyogram of two hESC MCL clones.
(F) GFP expression in undifferentiated hESC MCL after more than 20 passages.
(G) Immunohistochemistry of GFP expression in teratoma of one MCL clone and WT cells (scale bars, 50 μm).
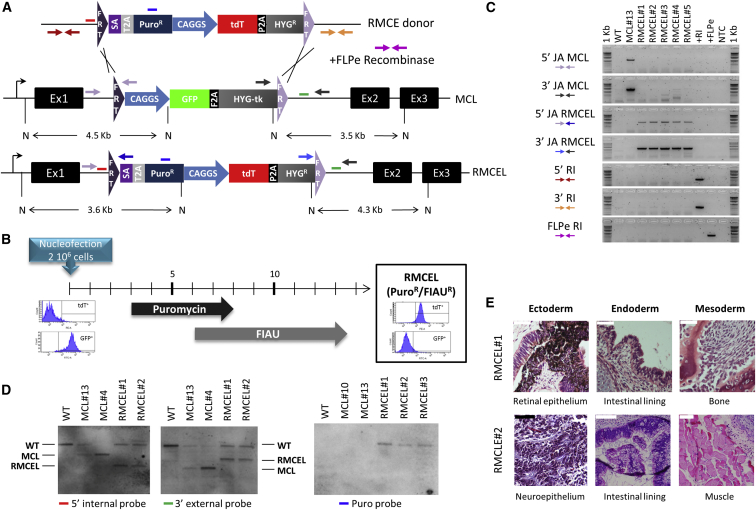
RMCE was then performed by nucleofection of the master cell line with the donor vector pZ:F3-P CAGGS tdTPH-F and the FLPe-expressing vector (Figure 2A). Control conditions without donor or without FLPe were included to evaluate the rate of random integration events and the specificity of the RMCE mediated by FLPe. The cells were selected with progressively increasing concentrations of puromycin, to select for cells integrating the donor plasmid, combined with negative selection with FIAU, to select for exchange events resulting in loss of the HSV-tk suicide gene and to eliminate possible random integrations (Figure 2B). We detected an average of 12.8 ± 6.8 (n = 6) PuroR/FIAUR-resistant RMCE colonies, all homogeneous tdT+/GFP– demonstrating that efficiency of recombination by cassette exchange was 100% (Figure 2B). No resistant colonies were obtained when cells were transfected with the donor alone, indicating that recombination was mediated by FLPe between the FRT sequences and that the negative selection efficiently selects against random integration events. Following selection, the non-clonal newly generated RMCE lines (all PuroR/FIAUR cells) were subjected to PCR and Southern blotting (Figures 2C and 2D) to confirm full RMCE and absence of random integration of either the donor or FLPe plasmids. RMCE lines maintained pluripotency (Figure 2E).
Figure 2.
FLPe-Mediated RMCE Allows Generation of Fully Recombined Lines, Free of Random Integration, with 100% Efficiency
(A) The RMCE donor vector pZ:F3-P CAGGS tdTPH-F (top) flanked by heterotypic FRT sequences (additional details in the Supplemental Information), original master cell line (MCL) and resulting RMCE line (RMCEL) are depicted. Red, blue, and green lines represent the 5′ internal, puromycin, and 3′ external Southern blot probes, respectively. Fragment sizes of DNA digested with NcoI (N) are indicated.
(B) Timeline of RMCEL generation (all PuroR/FIAUR cells) and selection program with FACS histograms representing the cassette exchange.
(C) PCR characterization of independent RMCELs using primer sets depicted in (A) for 5′/3′ JA of the MCL or RMCEL, 5′/3′ RI of the donor or FLPe-expressing vector. Wild-type (WT), MCL#13, and controls for random and FLPe integration (+RI and +FLPe) and no template control (NTC) samples were included.
(D) Southern blotting of hESC wild-type (WT) cells, MCLs and RMCELs.
(E) Teratoma formation assay of two RMCE lines.
Scale bars, 50 μm (black) and 100 μm (white).
A similar master cell line was generated in an iPSC line, and FLPe-mediated RMCE was validated (Figure 1C; Figures S1A–S1E). Therefore, this constitutes a method allowing highly efficient complete RMCE in ±15 days, without random integrations and without the need for single colony characterization after recombination.
Lineage Tracing Using the OCT4 Promoter Reveals Inhibition Exerted by the AAVS1 Locus
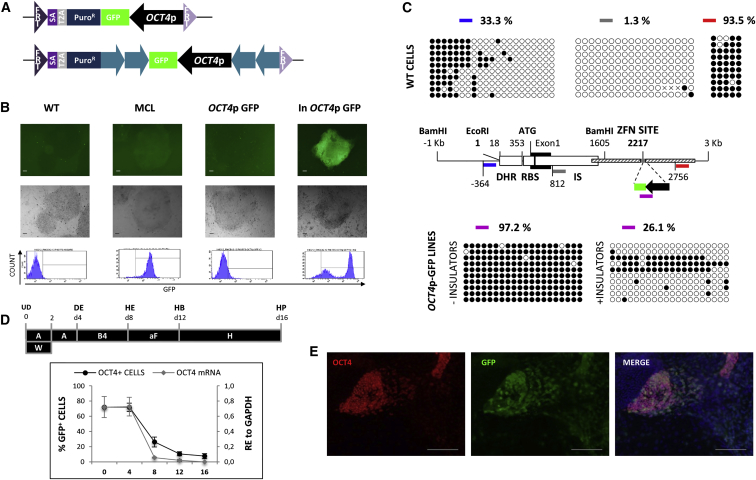
To demonstrate the suitability of the AAVS1 locus for lineage identification and tracing studies, we recombined the OCT4 promoter-GFP RMCE vector pZ:F3-P OCT4p-GFP-F in this locus (Figure 3A, top; Figure S2). Contrary to what was observed when the OCT4p was randomly integrated in hESC (Gerrard et al., 2005), we did not detect OCT4p promoter activity (Figure 3B). Thus, the AAVS1 locus might inhibit or silence the OCT4p despite the insulator activity ascribed to a DNase hypersensitive region (DHR) region present in the AAVS1 (Ogata et al., 2003). The epigenetic status of the AAVS1 was previously described as open chromatin by DNase hypersensitivity and chromatin immunoprecipitation (ChIP) assays, but no study has assessed the DNA methylation in this region. We performed bisulfite sequencing of different fragments of the AAVS1 locus in wild-type, undifferentiated hESCs and observed DNA hypo-methylation upstream of the DHR (33% methylated CpGs), total absence of methylation in the AAVS1 integration site region (1.3% methylated CpGs), but hyper-methylation in the ZFN targeting region (93.5% methylated CpGs) (Figure 3C, top and middle). The integrated OCT4p-GFP DNA sequence was also found nearly 100% methylated, consistent with the notion that de novo DNA methylation could spread from the proximal ZFN target site into the transgene (Figure 3C, bottom). Because insulators are known to have a barrier function and block spreading of adherent methylated domains (Dickson et al., 2010), we flanked the transgene with two inverted tandem repeat copies of the cHS4 insulator (Figure 3A, bottom; Figure S2). As expected, insulators prevented DNA methylation of the transgene (Figure 3C, bottom), and the promoter activity was restored yielding nearly 100% GFP+ cells in undifferentiated cells and loss of GFP during differentiation (Figures 3B, 3D, and 3E). Thus, the AAVS1 locus exerts inhibition in undifferentiated hESC by promoting de novo DNA methylation of the transgene, which can be overcome with insulators.
Figure 3.
The AAVS1 Locus Mediates Transgene Silencing by DNA Methylation in Undifferentiated hESC
(A) The RMCE donor vectors pZ:F3-P OCT4p-GFP-F (top) and pZ:F3-P (cHS4)X4 OCT4p-GFP-F (bottom) contain the OCT4p-GFP-polyA cassette without or with insulators (blue arrows), respectively.
(B) Representative data of GFP expression levels (microscopy and FACS) in undifferentiated wild-type (WT), master cell line (MCL) and OCT4p without (OCT4p GFP) or with insulators (In OCT4p GFP). Scale bar, 100 μm.
(C) Middle: representation of the AAVS1 locus adapted from Ogata et al. (2003). Thin line: 4-kb-long sequence containing the AAVS1 locus. Empty boxes: DNase hypersensitive region (DHR), Rep-binding site (RBS), and integration site (IS). Exon 1 (black box), homology regions used for homologous recombination (striped boxes), and ZFN targeting site (gray line) are depicted. Bisulfite sequenced regions analyzed in wild-type (WT) undifferentiated hESC: blue, gray, and red thick lines. The OCT4p-GFP RMCE lines (+/− insulators) and the fragment analyzed by bisulfite sequencing (purple line) are below the ZFN site. Top and bottom show representative panels and percentages of the results of bisulfite sequencing of the regions highlighted in the middle figure (n = 2 IEs).
(D) Top, scheme of the hepatic differentiation protocol: Activin (A), W (Wnt3a), B4 (BMP4), aF (aFGF), H (HGF), UD (undifferentiated), DE (definitive endoderm), HE (hepatic endoderm), HB (hepatoblasts), and HP (hepatocytes). Bottom, OCT4 expression levels as percentage of GFP+ cells and mRNA expression levels (relative expression [RE], mean ± SEM of n = 3 IEs).
(E) Immunocytochemistry of day 8 differentiated progeny. Scale bar, 100 μm.
Lineage Identification during Hepatocyte Specification
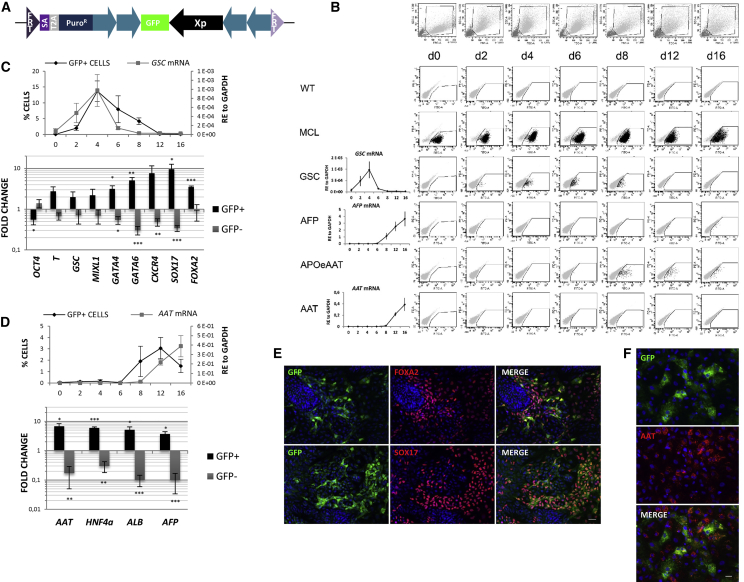
We next tested whether lineage identification during hepatocyte differentiation could be achieved by incorporating cell type specific promoters for a mesoendoderm marker gene, GOOSECOID (GSCp), a hepatocyte endoderm gene, ALFAFETOPROTEIN (AFP) enhancer-promoter (AFPe-p) shown to be active in differentiating hESC (by stable random integration of the transgene) (Ishii et al., 2008) and the more mature hepatocyte marker gene ALPHA-1 ANTITRYPSIN (AAT) with APOLIPOPROTEIN E enhancer and AAT promoter (APOeAATp) or minimal AAT promoter (AATp) shown to be active in human hepatocyte cell lines (by transient transfection) and differentiating hESC (by transduction of the differentiated progeny), respectively (Duan et al., 2007, Lam et al., 2007), all flanked by two inverted tandem repeat copies of the cHS4 insulator (Figure 4A). All these genes were expressed during differentiation at the expected time points (Figure 4B). Stable cell lines for each promoter were generated by RMCE and were subsequently subjected to directed differentiation to the hepatocyte lineage along with the wild-type hESC and the master cell line as negative and positive controls to gate GFP expression, respectively. Following directed differentiation, the master cell line showed continued homogeneous GFP expression (Figure 4B). Approximately 15% of differentiating GSCp cells were GFP+ on day 4, which decreased to ±5% on day 8, and no GFP+ cells were detected by day 12, a pattern that reflected transcript levels (Figure 4C). For the APOeAATp reporter line, we detected 2%–4% GFP+ cells on days 8, 12, and 16 of differentiation (Figure 4B), which underestimated the number of AAT-expressing cells, more significantly in the final maturation stage (day 16) where AAT gene expression increases as compared to day 12 (Figures 4D and 4F). No GFP+ cells were found for AFPe-p and AATp reporter lines, even though when integrated randomly, these promoters were successfully used to isolate AFP and AAT expressing cells from differentiating hESC (Duan et al., 2007, Ishii et al., 2008).
Figure 4.
Activity of Lineage-Specific Promoters during Hepatocyte Commitment
(A) The RMCE donor vector pZ:F3-P (cHS4)X4 X-GFP-F contains lineage-specific promoters (Xp) flanked by insulators (blue arrows) driving GFP.
(B) Timeline of cell morphology (SSC-FSC, top row) and GFP fluorescence (bottom rows) of wild-type (WT), master cell line (MCL), and RMCELs with the indicated promoters during the hepatocyte differentiation. Left graphs: endogenous mRNA expression levels of GSC, AFP, and AAT (relative expression, RE).
(C and D) Top, average percentage of GFP+ cells during differentiation and endogenous mRNA expression levels in GSCp (C) and APOeAATp (D) RMCE lines. Bottom, mRNA expression profile of GFP+ and GFP– sorted cells (relative gene expression to unsorted cells) from GSCp (day 4, C) and APOeAATp (day 12, D) RMCELs. Data as mean ± SEM of n = 3 IEs.
(E and F) Immunocytochemistry of GSCp (day 4, E) and APOeAATp (day 12, F) RMCEL progenies. Areas with concentrated amount of positive cells are shown.
Scale bar, 35 μm. ∗p < 0.05, ∗∗p < 0.001, and ∗∗∗p < 0.0001 by Student’s t test.
On day 4, GFP+ cells were isolated from the differentiating progeny of the GSCp line for further characterization (Figure 4C). Compared with the unsorted population, GSC transcripts were 2-fold higher in the GSC-GFP+ cells and 4-fold lower in the GSC-GFP– cells. GSC-GFP+ cells expressed higher levels of the mesoendodermal markers T and MIXL1 and significantly lower levels of OCT4. Interestingly, transcripts for the definitive endoderm marker genes, GATA4, GATA6, CXCR4, SOX17, and FOXA2, were 3- to 10-fold higher in the GSC-GFP+ cells compared with unsorted cells. The enrichment of chiefly definitive endoderm markers suggests that selection based on GFP transcribed by the GSCp on day 4 of differentiation enriches for cells that are being committed to the endodermal lineage. This was further substantiated by immunocytochemistry, demonstrating co-expression of GFP with FOXA2 and SOX17 (Figure 4E).
We also isolated AAT-GFP+ and AAT-GFP– cells from d12 progeny of the APOeAATp line (Figure 4D). Compared with unsorted cells, transcript levels for AAT were 6-fold higher in the GFP+ cells and 9-fold lower in the GFP– fraction, although AAT transcripts were still abundant in the GFP– cells. Expression of HNF4α, ALB, and AFP was significantly higher in the APOeAATp-GFP+ cells and significantly lower in the GFP– fraction. Immunocytochemistry confirmed that GFP+ cells where AAT+ but also that many AAT+ cells were not identified by the reporter (Figure 4F). As AAT decreased in the negative fraction, the APOeAATp correctly identified a subpopulation of AAT+ cells with highest AAT transcript levels both on day 12 and 16 (Figure S3).
Thus, the GSCp and APOeAATp reporters identify specifically GSC- and AAT-expressing cells, respectively, allowing the identification, isolation, and characterization of these two specific cell populations. However, despite presence of the cHS4 insulators, the APOeAATp reporter only identified a limited fraction of AAT+ cells on day 16, and the AFPp and AATp did not activate in response to differentiation. To investigate whether these limitations were hepatocyte specific, a non-hepatic promoter was tested. The HB9 motor neuron-specific promoter, previously used to identify this lineage by transduction of hESC-derived progeny (Marchetto et al., 2008), yielded similar results as the APOeAATp (Figure S3B) indicating that proper lineage-specific reporter activity in the AAVS1 is most likely promoter sequence dependent.
Use of the AAVS1 Locus as an Isogenic Locus for Molecular Sensors
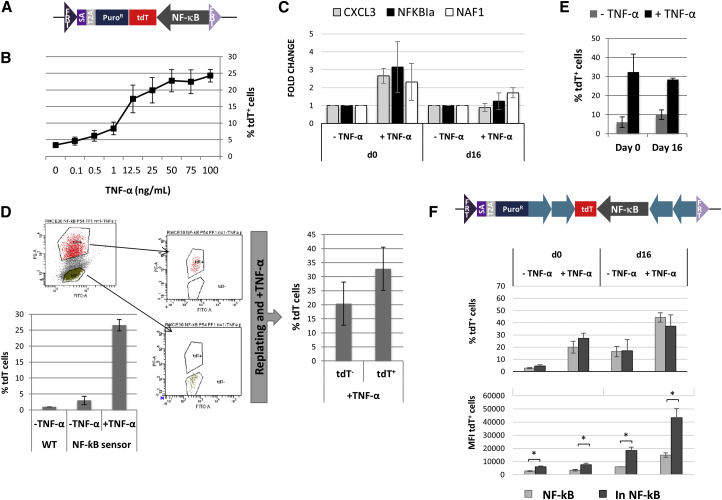
Molecular sensors with reporter cassettes are increasingly used to assess cell function, health, or viability in response to drugs or other stimuli (Lake et al., 2012). As an example of a toxicology cassette suitable for mode of action studies associated with drug toxicity, we recombined a necrosis factor κB (NF-κB) stress sensor into the AAVS1 locus by RMCE (Figure 5A). Following treatment of the cells with tumor necrosis factor alpha (TNF-α) for 48 hr, the fraction of tdT+ cells increased in a dose-dependent manner. A plateau of ±25% tdT+ cells was reached with concentrations of 50 ng/ml and higher (Figure 5B). The relatively low number of cells positive for tdT is likely due to the fact that TNF-α poorly activates NF-κB in hESC, as shown by the low induction of transcript levels of direct NF-κB target genes CXCL3, NAF1, and NFKIα on day 0 (Figure 5C, left). To assess whether the limited response reflects incomplete NF-κB reporter activation, we exposed ESC for 24 hr with 100 ng/ml TNF-α, isolated tdT+ and tdT– cells by fluorescence-activated cell sorting (FACS), replated them, and after 3 days re-challenged with TNF-α. TNF-α induced tdT response to the same extent in the tdT+ and tdT– sorted cells (Figure 5D), demonstrating that TNF-α activates NF-κB heterogeneously in hESC due to poor induction capacity.
Figure 5.
Recombination of a Molecular Response Sensor in the AAVS1 Locus
(A) The RMCE donor vector pZ:F3-P NF-κB-F contains an NF-κB response element followed by tdT.
(B) NF-κB sensor response in undifferentiated cells with increasing doses of TNF-α.
(C) Relative gene expression (to non-induced cells) of NF-κB direct target genes on days 0 or 16.
(D) Sorting of tdT+ and tdT– cells after induction and percentages of tdT+ cells in undifferentiated wild-type (WT) or NF-κB sensor-RMCEL before and after replating.
(E) TNF-α response on day 0 and 16 of hepatocyte differentiation.
(F) RMCE donor vector pZ:F3-P (cHS4)X4 NF-κB tdT-F containing insulators (blue arrows) and response to TNF-α induction (day 0 or 16) without (NF-κB) or with insulators (In NF-κB) determined by FACS and expressed as percentage of tdT+ cells and MFI.
Data: mean ± SEM, n = 3 IEs. ∗p < 0.05 by Student’s t test.
We next tested the function of the NF-κB sensor in ESC differentiated toward the hepatocyte lineage. When day 16 hepatocyte progeny was exposed to 100 ng/ml TNF-α for 24 hr, again almost 30% tdT+ cells could be detected, while non-induced cells showed a basal reporter activity, probably indicating minimal levels of culture-induced stress (Figure 5E). As observed in undifferentiated cells, addition of TNF-α only mildly induced NF-κB direct target genes on day 16 (Figure 5C, right). Nevertheless, this indicated that the NF-κB sensor is active in both undifferentiated and differentiated cells and correctly reports for NF-κB activation.
We also assessed whether the NF-κB stress sensor activity would be increased when the construct was flanked by insulators. However, the percentage of tdT+ cells found following treatment of undifferentiated or day 16 hepatocyte progeny with 100 ng/ml TNF-α was similar to the original construct, while tdT protein expression (as mean fluorescence intensity, MFI) was significantly and robustly improved (Figure 5F). Therefore, in agreement with results seen for OCT4p, the use of insulators improved the reporter activity in the NF-κB sensitive cells.
Inducible Expression from AAVS1 Locus during Hepatocyte Commitment
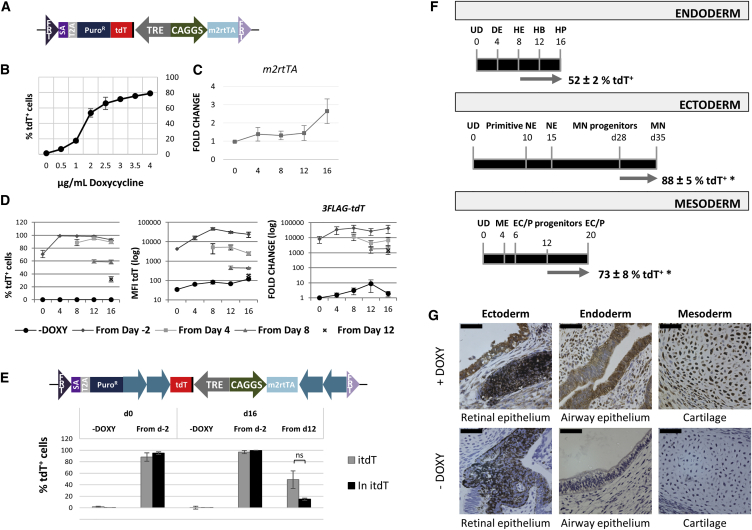
To assess whether it would be possible to generate an efficient inducible expression system in the AAVS1 locus, we generated an “all-in-one” donor vector for inducible expression (Figure 6A). After recombination in the AAVS1 locus, the undifferentiated cells were treated with doxycycline (DOXY) for 48 hr at different doses. No leaky tdT expression was seen in the absence of DOXY, and the percentage of tdT+ cells increased in a dose-dependent manner reaching a plateau of ±75% activation when cells were treated with 3 μg/ml DOXY for 2 days (Figure 6B).
Figure 6.
Inducible Expression from the AAVS1 Locus during Hepatocyte Differentiation
(A) RMCE donor vector pZ:F3-P TetOn 3f-tdT-F for inducible expression (fully described in the Supplemental Information).
(B) Percentage of tdT+ cells in response to increasing doses of DOXY for 48 hr.
(C) Relative gene expression (to day 0) of m2rtTA during hepatocyte differentiation.
(D) Percentage of tdT+ cells, expression levels of tdT per cell (mean fluorescence intensity, MFI) and 3FLAG-tdT mRNA (relative gene expression levels to –DOXY) with 3 μg/ml DOXY from the indicated time points.
(E) RMCE donor vector pZ:F3-P (cHS4)X4 TetOn 3f-tdT-F with insulators (blue arrows). Response to 3 μg/ml DOXY, initiated at the indicated time points (on day 0 and 16), of the RMCELs without (itdT) and with insulators (In itdT) expressed as percentage of tdT+ cells.
(F) Directed differentiation of itdT RMCEL toward endoderm (hepatocytes, HP), ectoderm (motor neurons, MN), and mesoderm (endothelial cells/pericytes, EC/P) with addition of 3 μg/ml DOXY during the last 7–8 days of differentiation (gray arrow). UD, undifferentiated cell; DE, hepatic endoderm; HE, hepatic endoderm; HB, hepatoblast; NE, neuroectoderm; ME, mesoderm.
(G) Immunohistochemistry analysis of 3flag-tdT expression in teratomas of the itdT RMCEL in animals fed with or without DOXY.
Scale bars, 50 μm. Data: mean ± SEM of n ≥ 3 IEs, ∗p < 0.05 by Student’s t test. Statistical significance is not represented in (B) and (D); ns (not significant).
We next determined whether tdT expression could also be induced during differentiation of hPSCs toward the hepatocyte lineage, by adding 3 μg/ml DOXY 2 days prior to the start of differentiation (day 2, undifferentiated cells), or starting on days 4, 8, or 12 during the hepatocyte differentiation process. hPSC progeny expressed stable m2rtTA levels throughout differentiation (Figure 6C). Addition of DOXY (or higher concentrations up to 10 μg/ml) did not affect the differentiation potential of hESC toward hepatocyte-like cells at all studied time points compared to non-treated cells (Figures S4A and S4B). Continued treatment with DOXY from day −2 to day 16 resulted in nearly 100% of differentiating ESC positive for tdT from day 4 onward, with the MFI of tdT+ cells reaching maximal levels on day 8, which was sustained until day 16. When DOXY was started on day 4, nearly 100% of cells were tdT+, but their MFI did not increase over time, and remained lower than when DOXY was started on day −2 (Figure 6D, left and middle panels). More markedly, when DOXY was started from days 8 and 12, only ±60% and ±35% of the ESC progeny was tdT+ on day 16 with a further pronounced decrease in MFI. The percentage of tdT+ cells and protein expression levels correlated well with the levels of 3flag-tdT transcripts (Figure 6D, right panel).
This behavior could be caused by an insufficient amount of DOXY in differentiating cells. Therefore, we tested the effect of 5 and 10 μg/ml DOXY (higher doses showed cytotoxicity). Although we found some increase in the fraction of tdT+ cells and their MFI, this did not reach levels seen when DOXY was added on day −2 in each case (Figure S4C). Thus, although reduced responsiveness could in part be overcome by increasing DOXY concentrations, we hypothesized that the tdT inducible cassette could be specifically inhibited in the AAVS1 locus during hepatocyte differentiation. Therefore, we included insulators flanking the inducible cassette. Interestingly and contrary to what we observed for OCT4p or the NF-κB sensor, insulators did not improve the reporter activity in terms of the fraction of tdT+ cells or reporter expression levels (MFI) in both the undifferentiated state or in day 16 progeny when DOXY was added from d12 (Figure 6E; Figure S4D). Insulators improved reporter activity only when DOXY was maintained for the duration of the differentiation process.
To investigate whether the apparent AAVS1-mediated inhibitory mechanism affected other lineages in vitro, we also tested the inducible reporter expression system in the mesodermal (endothelial cells and pericytes) and ectodermal (motor neurons) lineages. Significantly more tdT+ cells were present in the differentiated progenies of both systems than in the hepatocyte differentiation, indicating that, during in vitro differentiation, inhibition exerted by the AAVS1 locus was hepatocyte specific (Figure 6F).
To further understand the apparent lineage-specific silencing, we assessed the expression of 3flag-tdT in vivo in teratoma formation assays and found homogeneous 3flag expression in tissues representative of all the three germ layers (Figure 6G), even if we could not identify hepatocyte progeny in these teratomas.
In summary, these results suggest that the TRE integrated in the AAVS1 locus may be specifically inhibited during hepatocyte commitment in vitro in the absence of active transcription from the start of differentiation, and that insulators cannot overcome this. However, this mechanism appears to not to affect endodermal lineages in vivo.
Discussion
We describe here the generation and full characterization of hESC and iPSC master cell lines suitable for RMCE in the AAVS1 locus. Previous studies describing RMCE systems in hPSCs use only a positive selectable cassette, which leads to the selection of cells wherein the transgene is recombined in the pre-integrated FRT cassette, but also elsewhere in the genome, requiring screening of individual colonies to exclude random integrations (Du et al., 2009, Ramachandra et al., 2011, Tay et al., 2013, Zhu et al., 2013). We here report a strategy using positive/negative selection that allows for the generation of fully recombined hPSC lines in 15 days with 100% targeting efficiency and free of random integration events, that eliminates the need for colony selection after RMCE and full characterization of pluripotency or genome integrity.
A number of studies have suggested that the AAVS1 is a safe harbor locus in hPSCs, as well as in other cell types. However, our studies demonstrate that the AAVS1 locus is not as “safe” as suggested, as transgene expression was variable in both undifferentiated and hepatocyte committed ESC progeny in vitro. The AAVS1 has been described as an open chromatin locus by DNase hypersensitive assays and ChIP, but these studies did not cover the region targeted by the ZFNs in PSCs (Lamartina et al., 2000, Lombardo et al., 2011, Ogata et al., 2003, van Rensburg et al., 2013). No analysis of the DNA methylation status of the locus was carried out. Here, we demonstrate that the region targeted by the ZFNs used in this study, but also in other studies and by TALENs and CRIPS/Cas9 (Hockemeyer et al., 2009, Hockemeyer et al., 2011, Mali et al., 2013), can exert silencing in vitro via induction of de novo DNA methylation in undifferentiated hESC. The insulator activity and/or range of action described for the DHR region appears therefore not to ensure expression of all transgenes in this site (Ogata et al., 2003). Inhibition was observed for transgenic OCT4p activity, but the universal CAGGS promoter (in the master cell line and RMCE lines) or the NF-κB sensor was stably active without the need of insulators. Hence, inhibition by DNA methylation possibly occurs in a sequence-dependent manner (Feltus et al., 2003).
A number of lineage-specific promoters flanked by insulators showed variable activity during hepatocyte differentiation in vitro: the GSC promoter reported efficiently for GSC-expressing cells, while APOeAATp significantly underestimated the real percentage of AAT-expressing cells, as well as the neural HB9p. Independently of their efficacy when integrated in the AAVS1, all these promoters successfully allowed identification, isolation, and characterization of the respective cell populations. However, activity of the AAT, AFP, and HB9 promoters in the AAVS1 strongly differed from what was previously reported when integrated randomly in the hESC genome to perform identification of AAT+, AFP+, and HB9+ cells, respectively (Duan et al., 2007, Ishii et al., 2008, Wainger et al., 2014). In random transgenesis, cells that robustly activate the transgenic promoters in a specific manner are usually selected. This could be the result of multiple copy insertions or position effects due to proximity to regulatory regions that enhance the specific promoter activity, both improving the promoter activity even if some important regulatory elements are not present in the cloned fragment. The absence of AAVS1-mediated inhibition during neural differentiation (Figure 6F) suggests that the underperformance of the HB9p must be linked to the promoter fragment used in our study and proves that functionally validated promoters do not necessarily reproduce the activity of previous studies when integrated as a single copy in the genome in the AAVS1 locus. Instead, apparent AAVS1-mediated transgene inhibition during hepatic commitment (Figure 6F) may partially explain the malfunctioning of the hepatic promoters independently of the competences of the cloned fragment. Whether additional regulatory elements might be needed to achieve proper activity of the APOeAATp, AATp, AFPp when recombined into the AAVS1 locus is not known, and it is beyond the scope of our work.
Inducible expression during hepatocyte differentiation from the AAVS1 showed that only a fraction of cells responded to DOXY when it was applied later during differentiation, even when using higher concentrations of DOXY or insulators. The observed AAVS1-mediated inhibitory mechanism of the TRE construct is in line with the previously observed notion that reduced gene expression can trigger silencing (Oyer et al., 2009). This may explain our observations that uninsulated transgenes, such as the ones driven by the CAGGS constitutive promoter, the NF-κB sensor, or the continuously active TRE, were not inhibited when introduced in the AAVS1 locus during hepatocyte differentiation. The apparent discrepancy between the inducible TRE construct and the NF-κB sensor activation at later stages of the differentiation might be explained because unlike the un-induced TRE, NF-κB is already activated even in in the absence of TNF-α administration, albeit at low levels, likely due to culture-induced stress (Figure 5E). Interestingly, in contrast with hepatocyte differentiation, the uninsulated inducible tdT cassette could be efficiently activated in hESC-derived motor neurons and mesodermal cells, and ubiquitous reporter expression was seen in cells of the three germ layers in teratomas in vivo, although we cannot definitely state that inhibition does not occur in hepatocytes in vivo. Even if our studies did not show faithful activity of the HB9p, the studies using the inducible system are consistent with other reports demonstrating successful neural and hematopoietic lineage identification by promoter constructs in the AAVS1 locus without the need of insulators (Chang and Bouhassira, 2012, Sullivan et al., 2014, Tiyaboonchai et al., 2014, Wainger et al., 2014) and with previous studies in vivo reporting stable transgene expression (Hockemeyer et al., 2011, Qian et al., 2014). Why cHS4 insulators were not able to block the AAVS1-mediated inhibition of the TRE during hepatocyte differentiation, and potentially the hepatoblast/hepatocyte lineage-specific promoters, but were functional when used with the OCT4p and NF-κB sensor remains unknown. Thus, the lack of proper hepatoblast/hepatocyte-specific promoter activity might be explained by both AAVS1-mediated inhibition during hepatocyte differentiation (not blocked by insulators as for TRE) and lack of important regulatory elements.
One of the parameters that defines a safe harbor locus is its capability to allow stable, faithful, and predictable transgene expression in all different cell types. Independently of the competences of particular lineage-specific promoters, our results demonstrate that complex genetic and/or epigenetic regulatory mechanisms, including DNA methylation and other unknown mechanisms, act in the human AAVS1 locus in vitro in hPSCs in a sequence and lineage-dependent manner. AAVS1 is not able to support faithful and predictable transgene expression in undifferentiated hPSCs (OCT4 promoter without insulators), nor in a lineage-dependent manner (at least in the hepatocyte lineage) in vitro, though transgene silencing is not found in vivo. Mosaicism of transgene expression in safe harbor loci was similarly described for the well-characterized murine Rosa26 or ColA1 loci during in vitro and in vivo differentiation of ESC, even with the use of insulators (Beard et al., 2006, Haenebalcke et al., 2013). Our observations therefore contradict previous reports describing AAVS1 as a universal safe harbor locus for stable transgene expression in hPSCs in vitro (Lombardo et al., 2011, Qian et al., 2014, Smith et al., 2008). However, most of the in vitro studies have used strong constitutive promoters that appear to be refractory to AAVS1-exerted inhibition, as we show here for the CAGGS promoter, or do not assess transgene expression in hepatocytes in vitro. In summary, we demonstrate that the AAVS1 locus in human cells does not allow universal expression of transgenes and hence does not meet the true safe harbor locus definition. Whether targeting in a hypomethylated region within the AAVS1 would allow better transgene expression in hepatocytes in vitro needs to still be assessed.
With the advent of CRISPR-Cas9 technology, creation of knockout or knockin lines has significantly improved. As, for example, knockin (-add-on) strategies constitute the most faithful method to perform lineage identification and tracing, the CRISPR-Cas9 based approach can be considered superior compared with the introduction of lineage tracing and identification cassettes in the AAVS1 locus. In addition, with the generation of the iCRISPR methodology, inducible expression of Cas9 from the AAVS1 (González et al., 2014), creation of transgenic hPSCs has become more robust and efficient than approaches using co-transfection of CRISPR and guide RNAs; one could argue that the utility of master cell lines with docking sites in the AAVS1 locus is no longer scientifically relevant. However, the technology described here still provides a number of advantages over the use of CRISPR-Cas9 or the iCRISPR platform: (1) a transgenic line can be generated in 2 weeks, without the need for identification of off-target effects or characterization of pluripotency and genome integrity of the recombinant lines; and (2) it allows the creation of a series of applications that require the use of a defined isogenic locus, which allows side-by-side comparisons between different transgenes. As a matter of fact, the AAVS1 locus has been proved suitable for most of these applications in hPSCs during the recent past years (Garçon et al., 2013, González et al., 2014, Raitano et al., 2015, Rio et al., 2014, Wang et al., 2012).
In conclusion, we generated an RMCE-based method in the AAVS1 locus of hPSCs (applicable to other locus if desired) that enables fast and efficient generation of transgenic lines, and we demonstrate that the AAVS1 locus is not as safe as believed, though it remains a confirmed valuable tool for transgenesis in hPSCs for several applications in vitro and in vivo. The use of lineage-specific promoters or inducible systems during directed differentiation requires that promoter constructs are tested individually in specific lineages to ensure that they are not impacted by AAVS1-mediated inhibitory effects causing inefficient expression in vitro.
Experimental Procedures
Human ESC/iPSC Differentiation
hESC and hiPSC were harvested using trypsin (GIBCO) or accutase (Sigma), respectively, and plated onto 1.6% matrigel (BD) coated dishes at 7 × 104 cells/cm2 with mTeSR1 medium. 2–3 days after plating, differentiation was initiated as described previously (Roelandt et al., 2012).
Generation of the Master Cell Lines and RMCE Lines
Two million H9-ESC or iPSC were transfected with 10 μg of donor vector and 3 μg of AAVS1 ZFN mRNA using the hESC Nucleofector Solution Kit 2 (Amaxa) and programs A13 or F16 for cells cultured on iMEF or feeder free, respectively. Selection was performed with 25–50 μg/ml of Hygromycin B (Sigma-Aldrich) for 10–15 days. Recombinant colonies were manually picked and expanded for further characterization. RMCE was performed by Nucleofection as described. Further details on constructs generation, description, and molecular characterization of recombinant lines can be found in the Supplemental Information.
NF-κB Reporter Activity Induction
Undifferentiated hESC were incubated with increasing concentrations of TNF-α for 48 hr. For the rest of experiments, day 0 and 16 hepatocyte-progenies were treated with 100 ng/ml TNF-α for 24 hr and were analyzed by FACS.
Flow Cytometry and Cell Sorting
TRA-1-60 staining (BD Biosciences, antibody and isotype control) was performed according to manufacturer’s instructions. FACS analysis and sorting were carried out on FACS Canto and FACS Aria III (BD Biosciences), respectively. Data analysis was done using the FACS Diva (BD) or FlowJo software.
Statistical Analysis
A minimum of three independent experiments (n = 3 independent experiments [IEs]) were carried out. Data values represent average ±SEM and were subject to the two-tailed Student’s t test. Values of ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 were considered statistically significant.
Author Contributions
L.O. conceived the study, generated the data, and wrote the paper. R.B. contributed with the generation and testing of donor vectors and data collection. M.P. carried out the methylation studies and contributed with the generation and testing of donor vectors and data collection. Y.C. generated and characterized the iPSC MCL and contributed with the generation and testing of donor vectors. E.W. and T.S. performed the histological analysis and immunohistochemistry of teratoma. R.S. cloned and tested the GSC promoter. N.H. and J.V. contributed to the cloning and testing of hepatocyte promoters. P.B. developed technical work for the whole article. Q.C. and K.V. contributed with the optimization of gene targeting in hPSCs. K.E. and V.V. carried out the immunocytochemistry. B.Z.S. contributed with the generation and testing of donor vectors. W.G. and L.V.D.B. carried out moneuron differentiations. S.R. performed the teratoma formation assays. Y.N., S.B.-W., and T.C. developed and tested the NF-κB sensor. C.M.V. designed the study and wrote the paper. All the authors have read and edited the manuscript.
Acknowledgments
L.O. was funded by IWT/OZM/090838, IACS BPAMER3/08/04, and Government of Aragon FMI048/08; M.P. by FWO 1288714N; and R.S. by the Dutch Diabetes Foundation. R.B., N.H., J.V., K.C., and Q.C. were supported by IWT fellowships SB-121393, SB-121396, SB-101230, SB-091228, and SB-093228, respectively. B.Z.S. was financed by FP7-PEOPLE-2011-IIF (Proposal No. 298785) and W.G. by the China Scholarship Council. S.B.-W. and T.C. were supported by the FP7-HEMIBIO (266777). Funding to C.M.V. was from FWO G.0667.07 and G.0975.11; KU Leuven (EIW-B4855-EF/05/11, ETH-C1900-PF, EME-C2161-GOA/11/012), IWT-HEPSTEM, BELSPO-IUAP-DEVREPAIR, FP7-HEMIBIO (266777). We thank Prof. P. Collas and Dr. B. Izzi for their discussions related to the DNA methylation studies; Dr. D. Franckaert, Dr. S. Schlenner, and Dr. A. Van Nieuwenhuijze for their availability to operate the sorter as well as M. Welters and R. Van Rossom for technical assistance.
Published: October 8, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, four figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.09.004.
Contributor Information
Laura Ordovás, Email: laura.ordovas@med.kuleuven.be, lau_ordo@hotmail.com.
Catherine M. Verfaillie, Email: catherine.verfaillie@med.kuleuven.be.
Supplemental Information
References
- Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- Chang C.J., Bouhassira E.E. Zinc-finger nuclease-mediated correction of α-thalassemia in iPS cells. Blood. 2012;120:3906–3914. doi: 10.1182/blood-2012-03-420703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S.R., Biniszkiewicz D., van Parijs L., Baltimore D., Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol. Cell. Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.P., Ng E.S., Costa M., Mossman A.K., Sourris K., Elefanty A.G., Stanley E.G. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- DeKelver R.C., Choi V.M., Moehle E.A., Paschon D.E., Hockemeyer D., Meijsing S.H., Sancak Y., Cui X., Steine E.J., Miller J.C. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson J., Gowher H., Strogantsev R., Gaszner M., Hair A., Felsenfeld G., West A.G. VEZF1 elements mediate protection from DNA methylation. PLoS Genet. 2010;6:e1000804. doi: 10.1371/journal.pgen.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z.W., Hu B.Y., Ayala M., Sauer B., Zhang S.C. Cre recombination-mediated cassette exchange for building versatile transgenic human embryonic stem cells lines. Stem Cells. 2009;27:1032–1041. doi: 10.1002/stem.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y., Catana A., Meng Y., Yamamoto N., He S., Gupta S., Gambhir S.S., Zern M.A. Differentiation and enrichment of hepatocyte-like cells from human embryonic stem cells in vitro and in vivo. Stem Cells. 2007;25:3058–3068. doi: 10.1634/stemcells.2007-0291. [DOI] [PubMed] [Google Scholar]
- Feltus F.A., Lee E.K., Costello J.F., Plass C., Vertino P.M. Predicting aberrant CpG island methylation. Proc. Natl. Acad. Sci. USA. 2003;100:12253–12258. doi: 10.1073/pnas.2037852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garçon L., Ge J., Manjunath S.H., Mills J.A., Apicella M., Parikh S., Sullivan L.M., Podsakoff G.M., Gadue P., French D.L. Ribosomal and hematopoietic defects in induced pluripotent stem cells derived from Diamond Blackfan anemia patients. Blood. 2013;122:912–921. doi: 10.1182/blood-2013-01-478321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard L., Zhao D., Clark A.J., Cui W. Stably transfected human embryonic stem cell clones express OCT4-specific green fluorescent protein and maintain self-renewal and pluripotency. Stem Cells. 2005;23:124–133. doi: 10.1634/stemcells.2004-0102. [DOI] [PubMed] [Google Scholar]
- González F., Zhu Z., Shi Z.D., Lelli K., Verma N., Li Q.V., Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenebalcke L., Goossens S., Naessens M., Kruse N., Farhang Ghahremani M., Bartunkova S., Haigh K., Pieters T., Dierickx P., Drogat B. Efficient ROSA26-based conditional and/or inducible transgenesis using RMCE-compatible F1 hybrid mouse embryonic stem cells. Stem Cell Rev. 2013;9:774–785. doi: 10.1007/s12015-013-9458-z. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P., Cost G.J., Zhang L., Santiago Y., Miller J.C. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T., Fukumitsu K., Yasuchika K., Adachi K., Kawase E., Suemori H., Nakatsuji N., Ikai I., Uemoto S. Effects of extracellular matrixes and growth factors on the hepatic differentiation of human embryonic stem cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2008;295:G313–G321. doi: 10.1152/ajpgi.00072.2008. [DOI] [PubMed] [Google Scholar]
- Lake M.C., Nguyen Q.D., Ali S., Aboagye E.O. Development of a novel molecular sensor for imaging estrogen receptor-coactivator protein-protein interactions. PLoS ONE. 2012;7:e44160. doi: 10.1371/journal.pone.0044160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P.Y., Sia K.C., Khong J.H., De Geest B., Lim K.S., Ho I.A., Wang G.Y., Miao L.V., Huynh H., Hui K.M. An efficient and safe herpes simplex virus type 1 amplicon vector for transcriptionally targeted therapy of human hepatocellular carcinomas. Mol. Ther. 2007;15:1129–1136. doi: 10.1038/sj.mt.6300165. [DOI] [PubMed] [Google Scholar]
- Lamartina S., Sporeno E., Fattori E., Toniatti C. Characteristics of the adeno-associated virus preintegration site in human chromosome 19: open chromatin conformation and transcription-competent environment. J. Virol. 2000;74:7671–7677. doi: 10.1128/jvi.74.16.7671-7677.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Suzuki K., Kim N.Y., Liu G.H., Izpisua Belmonte J.C. A cut above the rest: targeted genome editing technologies in human pluripotent stem cells. J. Biol. Chem. 2014;289:4594–4599. doi: 10.1074/jbc.R113.488247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A., Cesana D., Genovese P., Di Stefano B., Provasi E., Colombo D.F., Neri M., Magnani Z., Cantore A., Lo Riso P. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto M.C., Muotri A.R., Mu Y., Smith A.M., Cezar G.G., Gage F.H. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Ogata T., Kozuka T., Kanda T. Identification of an insulator in AAVS1, a preferred region for integration of adeno-associated virus DNA. J. Virol. 2003;77:9000–9007. doi: 10.1128/JVI.77.16.9000-9007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyer J.A., Chu A., Brar S., Turker M.S. Aberrant epigenetic silencing is triggered by a transient reduction in gene expression. PLoS ONE. 2009;4:e4832. doi: 10.1371/journal.pone.0004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian K., Huang C.T., Chen H., Blackbourn L.W., 4th, Chen Y., Cao J., Yao L., Sauvey C., Du Z., Zhang S.C. A simple and efficient system for regulating gene expression in human pluripotent stem cells and derivatives. Stem Cells. 2014;32:1230–1238. doi: 10.1002/stem.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitano S., Ordovàs L., De Muynck L., Guo W., Espuny-Camacho I., Geraerts M., Khurana S., Vanuytsel K., Tóth B.I., Voets T. Restoration of progranulin expression rescues cortical neuron generation in an induced pluripotent stem cell model of frontotemporal dementia. Stem Cell Reports. 2015;4:16–24. doi: 10.1016/j.stemcr.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandra C.J., Shahbazi M., Kwang T.W., Choudhury Y., Bak X.Y., Yang J., Wang S. Efficient recombinase-mediated cassette exchange at the AAVS1 locus in human embryonic stem cells using baculoviral vectors. Nucleic Acids Res. 2011;39:e107. doi: 10.1093/nar/gkr409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio P., Baños R., Lombardo A., Quintana-Bustamante O., Alvarez L., Garate Z., Genovese P., Almarza E., Valeri A., Díez B. Targeted gene therapy and cell reprogramming in Fanconi anemia. EMBO Mol. Med. 2014;6:835–848. doi: 10.15252/emmm.201303374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelandt P., Obeid S., Paeshuyse J., Vanhove J., Van Lommel A., Nahmias Y., Nevens F., Neyts J., Verfaillie C.M. Human pluripotent stem cell-derived hepatocytes support complete replication of hepatitis C virus. J. Hepatol. 2012;57:246–251. doi: 10.1016/j.jhep.2012.03.030. [DOI] [PubMed] [Google Scholar]
- Smith J.R., Maguire S., Davis L.A., Alexander M., Yang F., Chandran S., ffrench-Constant C., Pedersen R.A. Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells. 2008;26:496–504. doi: 10.1634/stemcells.2007-0039. [DOI] [PubMed] [Google Scholar]
- Sullivan S.K., Mills J.A., Koukouritaki S.B., Vo K.K., Lyde R.B., Paluru P., Zhao G., Zhai L., Sullivan L.M., Wang Y. High-level transgene expression in induced pluripotent stem cell-derived megakaryocytes: correction of Glanzmann thrombasthenia. Blood. 2014;123:753–757. doi: 10.1182/blood-2013-10-530725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay F.C., Tan W.K., Goh S.L., Ramachandra C.J., Lau C.H., Zhu H., Chen C., Du S., Phang R.Z., Shahbazi M. Targeted transgene insertion into the AAVS1 locus driven by baculoviral vector-mediated zinc finger nuclease expression in human-induced pluripotent stem cells. J. Gene Med. 2013;15:384–395. doi: 10.1002/jgm.2745. [DOI] [PubMed] [Google Scholar]
- Tiyaboonchai A., Mac H., Shamsedeen R., Mills J.A., Kishore S., French D.L., Gadue P. Utilization of the AAVS1 safe harbor locus for hematopoietic specific transgene expression and gene knockdown in human ES cells. Stem Cell Res. (Amst.) 2014;12:630–637. doi: 10.1016/j.scr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda K., Suzuki K., Yamazoe T., Shiraki N., Higuchi Y., Tokieda K., Kume K., Mitani K., Kume S. Albumin gene targeting in human embryonic stem cells and induced pluripotent stem cells with helper-dependent adenoviral vector to monitor hepatic differentiation. Stem Cell Res. (Amst.) 2013;10:179–194. doi: 10.1016/j.scr.2012.11.003. [DOI] [PubMed] [Google Scholar]
- van Rensburg R., Beyer I., Yao X.Y., Wang H., Denisenko O., Li Z.Y., Russell D.W., Miller D.G., Gregory P., Holmes M. Chromatin structure of two genomic sites for targeted transgene integration in induced pluripotent stem cells and hematopoietic stem cells. Gene Ther. 2013;20:201–214. doi: 10.1038/gt.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger B.J., Kiskinis E., Mellin C., Wiskow O., Han S.S., Sandoe J., Perez N.P., Williams L.A., Lee S., Boulting G. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Rodriguez R.T., Wang J., Ghodasara A., Kim S.K. Targeting SOX17 in human embryonic stem cells creates unique strategies for isolating and analyzing developing endoderm. Cell Stem Cell. 2011;8:335–346. doi: 10.1016/j.stem.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang W.Y., Hu S., Lan F., Lee A.S., Huber B., Lisowski L., Liang P., Huang M., de Almeida P.E. Genome editing of human embryonic stem cells and induced pluripotent stem cells with zinc finger nucleases for cellular imaging. Circ. Res. 2012;111:1494–1503. doi: 10.1161/CIRCRESAHA.112.274969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S., Sukonnik T., Kean T., Bharadwaj R.R., Pasceri P., Ellis J. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol. Ther. 2004;10:27–36. doi: 10.1016/j.ymthe.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Zhu H., Lau C.H., Goh S.L., Liang Q., Chen C., Du S., Phang R.Z., Tay F.C., Tan W.K., Li Z. Baculoviral transduction facilitates TALEN-mediated targeted transgene integration and Cre/LoxP cassette exchange in human-induced pluripotent stem cells. Nucleic Acids Res. 2013;41:e180. doi: 10.1093/nar/gkt721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.