Abstract
Chemical modification of 16S rRNA can confer exceptionally high-level resistance to a diverse set of aminoglycoside antibiotics. Here, we show that the pathogen-derived enzyme NpmA possesses dual m1A1408/m1G1408 activity, an unexpected property apparently unique among the known aminoglycoside resistance 16S rRNA (m1A1408) methyltransferases. Although the biological significance of this activity remains to be determined, such mechanistic variation in enzymes acquired by pathogens has significant implications for development of inhibitors of these emerging resistance determinants.
TEXT
Methylation of 16S rRNA has emerged as a significant new threat to the efficacy of aminoglycoside antibiotics, particularly against Gram-negative pathogens (1), with two families of resistance methyltransferase defined by the modifications they produce, m7G1405 and m1A1408 (Fig. 1A). Structural and functional studies of enzymes from each family, and of both aminoglycoside producer and pathogenic origin, have firmly established them as class I methyltransferases (2) and have begun to reveal the molecular details of their interactions with the cosubstrate S-adenosyl-l-methionine (SAM) and the 30S subunit substrate (3–7).
FIG 1.
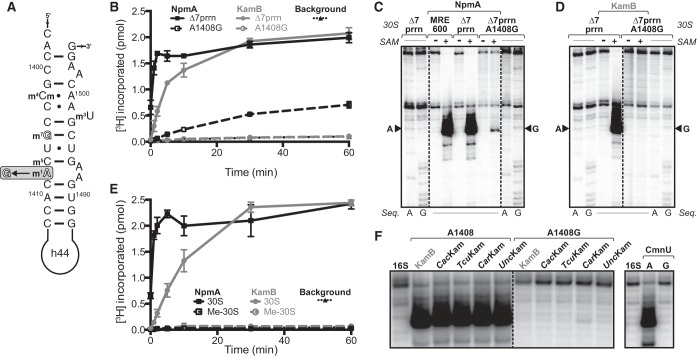
NpmA methylates 30S subunits with G1408. (A) Sequence and secondary structure of the h44 region containing the target nucleotides (outline font) of the m7G1405 and m1A1408 aminoglycoside-resistance 16S rRNA methyltransferases. The mutation in the 30S-A1408G subunit (shaded box) used in this study and the locations of intrinsic 16S rRNA modifications in this region are also indicated. (B) In vitro methylation in the presence of [3H]SAM of Δ7prrn wild-type and 30S-A1408G subunits by NpmA (black) and KamB (gray). NpmA, but not KamB, can methylate 30S-A1408G subunits (dashed black line). (C and D) RT analysis of 16S rRNA methylation by NpmA (C) and KamB (D) using 30S subunits from E. coli MRE600 (A1408), Δ7prrn (A1408), and Δ7prrn-A1408G. Sequencing lanes (Seq.) contain the complementary dideoxynucleotide in the RT reaction. The position of modified 1408 is marked by arrowheads. (E) In vitro methylation in the presence of [3H]SAM of wild-type unmethylated (30S) and m1A1408 premethylated (Me-30S) subunits by NpmA (black) and KamB (gray). (F) RT analysis of modification of wild-type or 30S-A1408G subunits by five other 16S rRNA (m1A1408) methyltransferases (13, 14); each enzyme is capable of incorporating only the m1A1408 modification. The origins of the enzymes not described in the text are Catenulispora acidiphila (CacKam; UniProt accession C7Q5P8), Thermomonospora curvata (TcuKam; D1A6K4), Candidatus Arthromitus sp. SFB-mouse-NYU (CarKam; F9VLU6), an uncultured bacterium (UncKam; K2DC64), and Saccharothrix mutabilis subsp. capreolus (CmnU; A6YEH1).
Despite these advances, important questions remain about the molecular mechanisms of action of these enzymes. For example, studies of the pathogen-derived 16S rRNA (m1A1408) methyltransferase NpmA (8) and the orthologous enzyme Kmr from Sorangium cellulosum So ce56 (9) revealed a critical but undefined role for the 30S in promoting SAM binding and/or catalysis. We therefore sought to establish a system to dissect the mechanism by which the 30S subunit regulates m1A1408 methyltransferase activity. To this end, we obtained the wild-type (Δ7prrn) Escherichia coli strain SQZ10 and its 16S rRNA A1408G variant (Δ7prrn-A1408G), in which all chromosomal rRNA operons are replaced by a single plasmid-borne copy (10, 11). Our expectation was that subunits isolated from the A1408G strain would allow analysis of 30S enzyme-SAM interactions in the absence of the modification reaction.
We first used an established in vitro methylation assay with [3H]SAM (12), to verify that neither NpmA nor KamB, a 16S rRNA (m1A1408) methyltransferase from the tobramycin producer Streptoalloteichus tenebrarius, had activity against the 30S-A1408G subunits. To our surprise, however, we found that NpmA, but not KamB, did in fact appear to methylate the variant 30S subunits (Fig. 1B), and we decided to examine this unexpected enzymatic activity.
To identify the site of modification in 30S-A1408G, we performed two additional assays. First, reverse transcription (RT) of 16S rRNA extracted from untreated and NpmA- or KamB-treated wild-type and 30S-A1408G subunits was used to visualize the modified nucleotide (Fig. 1C and D). Correlating with the [3H]SAM assays, a strong stop corresponding to m1A1408 modification was observed for both enzymes with wild-type 30S, whereas only NpmA treatment resulted in a band at the equivalent position for 30S-A1408G subunits. Additionally, we observed no difference in NpmA modification of wild-type 30S isolated from the Δ7prrn strain and E. coli MRE600, typically used in these assays (12, 13). Second, we isolated wild-type 30S subunits from E. coli BL21(DE3) cells grown without and with KamB overexpression, and tested the activity of NpmA and KamB against these unmodified and m1A1408 premodified substrates using the [3H]SAM assay (Fig. 1E). Neither NpmA nor KamB could incorporate 3H into subunits previously modified with m1A1408, demonstrating that NpmA does not possess activity at an alternative 16S rRNA nucleotide outside the region examined in our RT assay. From these experiments, we conclude that NpmA methylates 30S-A1408G subunits at nucleotide 1408 and thus possesses dual A1408/G1408 modification activity.
To address the question of whether this novel activity is unique to NpmA, we tested the ability of Kmr and five other 16S rRNA (m1A1408) family members (13, 14) to modify the 30S-A1408G subunits. Although all enzymes robustly modified wild-type subunits in vitro, none possessed catalytic activity with the 30S-A1408G substrate (Fig. 1F). Thus, among the collection of enzymes tested, dual A1408/G1408 activity is unique to the sole pathogen-derived 16S rRNA (m1A1408) methyltransferase, NpmA.
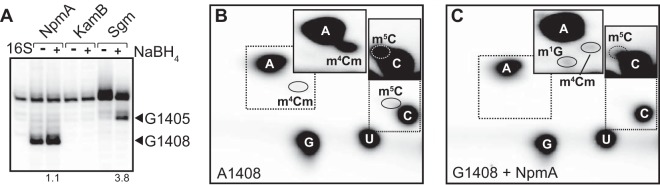
The m1A1408 methyltransferases are structurally most similar to the tRNA methyltransferase TrmB (3, 5), which catalyzes an m7G modification (at tRNA position 46). NpmA might therefore feasibly catalyze either m1G or m7G modification, which would require either altered catalysis (but with similar target base orientation) or substantially altered target positioning (with similar chemistry), respectively. Modification at the guanine 7 position produces a weak RT stop but can be enhanced by sodium borohydride (NaBH4) reduction of the modification and subsequent cleavage of the RNA chain by β-elimination at the abasic site generated (15, 16). We measured the effect of NaBH4 treatment on the intensity of RT stop generated by NpmA modification of G1408 and the adjacent m7G1405 modification catalyzed by Sgm (17, 18) as a positive control. Whereas the band corresponding to m7G1405 was enhanced 3.8-fold, no significant change was observed for G1408 (∼1.1-fold increase) (Fig. 2A). Next, to directly examine the modified nucleotide, we isolated the rRNA fragment corresponding to nucleotides C1378-G1432 using a complementary deoxyribonucleotide (18) and examined the rRNA nucleotide composition using an established thin-layer chromatography (TLC) system (solvents A and B in reference 19). Comparison of unmodified wild-type and NpmA-modified 30S-A1408G rRNA revealed a new spot at the known position of m1G, confirming this as the modification incorporated by NpmA (Fig. 2B and C).
FIG 2.
NpmA methylates G and A at nucleotide 1408 at the same position (N1). (A) RT analysis of (left to right) unmodified 16S rRNA (16S), NpmA or KamB in vitro-modified 16S rRNA extracted from Δ7prrn-A1408G 30S subunits with and without subsequent sodium borohydride treatment, and equivalent analysis of in vivo-modified 16S rRNA from wild-type E. coli BL21(DE3) cells expressing the m7G1405 methyltransferase Sgm. The values below the gel are fold enhancement with NaBH4 treatment (+). (B and C) TLC analysis of the nucleoside composition of the 16S rRNA fragment corresponding to nucleotides 1378 to 1432 from unmodified wild-type (B) and NpmA-modified Δ7prrn-A1408G (C) 30S subunits. Regions boxed with a dotted line are duplicated in the insets (solid lines) with 20×-increased contrast to allow visualization of the low-abundance modified nucleotides.
The TLC analyses also identified the previously unmapped position of the m4Cm modification in this solvent system (Fig. 2B and C). The reduction in intensity of the spot derived from m4Cm1402 and that from m5C1407 (Fig. 1) further suggests that A1408G mutation and/or N1 methylation directly impacts the activity of the enzymes (RsmH/RsmI and RsmF, respectively) which otherwise stoichiometrically modify these rRNA residues (20).
Several instances have been documented of RNA methyltransferases that catalyze the same modification on more than one site, either in the same or on two different substrates. However, dual nucleotide specificity may be much rarer, with only a single example of a class IV (SPOUT family) methyltransferase from the euryarchaeon Thermococcus kodakaraensis being reported to catalyze tRNA m1A9/m1G9 modification (21). Our finding that the class I NpmA possesses such dual activity is particularly remarkable given that NpmA makes only a single direct base interaction with the 16S rRNA (to the A1408 N6 amine), which should uniquely select for adenine (7). How NpmA adjusts its active site to accommodate the guanine base and accomplish methylation of the protonated N1 atom remains to be determined. We speculate that NpmA may have retained ancestral m1G activity, but given that A1408G itself confers exceptionally high-level aminoglycoside resistance, it is unclear why NpmA would have retained this activity. G1408 is found in a small fraction of bacteria, and we speculated that N1 methylation might overcome a growth deficit caused by guanine at this position in the ribosomal decoding center. However, we could not detect a significant difference in growth between the wild-type and A1408G Δ7prrn strains.
In conclusion, we have found that the pathogen-derived 16S rRNA (m1A1408) methyltransferase NpmA possesses dual nucleotide specificity and is capable of catalyzing both m1A1408 and m1G1408 modifications. While the biological significance of NpmA's dual specificity is presently unclear, the finding that a pathogen-derived enzyme might possess a catalytic capacity significantly different from that of orthologous enzymes from other bacteria has the potential to confound development of specific inhibitors of such resistance determinants.
ACKNOWLEDGMENTS
The E. coli Δ7prrn and Δ7prrn(A1408G) strains were a generous gift from Kurt Fredrick. We are also grateful to Christine Dunham for useful discussions during the preparation of the manuscript.
This work was supported by grant R01-AI088025 from the National Institute of Allergy and Infectious Diseases.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
REFERENCES
- 1.Wachino J, Arakawa Y. 2012. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updat 15:133–148. doi: 10.1016/j.drup.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Schubert HL, Blumenthal RM, Cheng XD. 2003. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci 28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Husain N, Obranic S, Koscinski L, Seetharaman J, Babic F, Bujnicki JM, Maravic-Vlahovicek G, Sivaraman J. 2011. Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit. Nucleic Acids Res 39:1903–1918. doi: 10.1093/nar/gkq1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Husain N, Tkaczuk KL, Tulsidas SR, Kaminska KH, Cubrilo S, Maravic-Vlahovicek G, Bujnicki JM, Sivaraman J. 2010. Structural basis for the methylation of G1405 in 16S rRNA by aminoglycoside resistance methyltransferase Sgm from an antibiotic producer: a diversity of active sites in m7G methyltransferases. Nucleic Acids Res 38:4120–4132. doi: 10.1093/nar/gkq122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macmaster R, Zelinskaya N, Savic M, Rankin CR, Conn GL. 2010. Structural insights into the function of aminoglycoside-resistance A1408 16S rRNA methyltransferases from antibiotic-producing and human pathogenic bacteria. Nucleic Acids Res 38:7791–7799. doi: 10.1093/nar/gkq627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitt E, Galimand M, Panvert M, Courvalin P, Mechulam Y. 2009. Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases. J Mol Biol 388:570–582. doi: 10.1016/j.jmb.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 7.Dunkle JA, Vinal K, Desai PM, Zelinskaya N, Savic M, West DM, Conn GL, Dunham CM. 2014. Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA. Proc Natl Acad Sci U S A 111:6275–6280. doi: 10.1073/pnas.1402789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wachino J, Shibayama K, Kurokawa H, Kimura K, Yamane K, Suzuki S, Shibata N, Ike Y, Arakawa Y. 2007. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother 51:4401–4409. doi: 10.1128/AAC.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savic M, Sunita S, Zelinskaya N, Desai PM, Macmaster R, Vinal K, Conn GL. 2015. 30S Subunit-dependent activation of the Sorangium cellulosum So ce56 aminoglycoside resistance-conferring 16S rRNA methyltransferase Kmr. Antimicrob Agents Chemother 59:2807–2816. doi: 10.1128/AAC.00056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asai T, Zaporojets D, Squires C, Squires CL. 1999. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc Natl Acad Sci U S A 96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin D, Fredrick K. 2009. Control of translation initiation involves a factor-induced rearrangement of helix 44 of 16S ribosomal RNA. Mol Microbiol 71:1239–1249. doi: 10.1111/j.1365-2958.2009.06598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zelinskaya N, Rankin CR, Macmaster R, Savic M, Conn GL. 2011. Expression, purification and crystallization of adenosine 1408 aminoglycoside-resistance rRNA methyltransferases for structural studies. Protein Expr Purif 75:89–94. doi: 10.1016/j.pep.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witek MA, Conn GL. 2014. Expansion of the aminoglycoside-resistance 16S rRNA (m(1)A1408) methyltransferase family: expression and functional characterization of four hypothetical enzymes of diverse bacterial origin. Biochim Biophys Acta 1844:1648–1655. doi: 10.1016/j.bbapap.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felnagle EA, Rondon MR, Berti AD, Crosby HA, Thomas MG. 2007. Identification of the biosynthetic gene cluster and an additional gene for resistance to the antituberculosis drug capreomycin. Appl Environ Microbiol 73:4162–4170. doi: 10.1128/AEM.00485-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zueva VS, Mankin AS, Bogdanov AA, Baratova LA. 1985. Specific fragmentation of tRNA and rRNA at a 7-methylguanine residue in the presence of methylated carrier RNA. Eur J Biochem 146:679–687. doi: 10.1111/j.1432-1033.1985.tb08704.x. [DOI] [PubMed] [Google Scholar]
- 16.Wintermeyer W, Zachau HG. 1975. Tertiary structure interactions of 7-methylguanosine in yeast tRNA Phe as studied by borohydride reduction. FEBS Lett 58:306–309. doi: 10.1016/0014-5793(75)80285-7. [DOI] [PubMed] [Google Scholar]
- 17.Kojic M, Topisirovic L, Vasiljevic B. 1992. Cloning and characterization of an aminoglycoside resistance determinant from Micromonospora zionensis. J Bacteriol 174:7868–7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savic M, Lovric J, Tomic TI, Vasiljevic B, Conn GL. 2009. Determination of the target nucleosides for members of two families of 16S rRNA methyltransferases that confer resistance to partially overlapping groups of aminoglycoside antibiotics. Nucleic Acids Res 37:5420–5431. doi: 10.1093/nar/gkp575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grosjean H, Keith G, Droogmans L. 2004. Detection and quantification of modified nucleotides in RNA using thin-layer chromatography. Methods Mol Biol 265:357–391. [DOI] [PubMed] [Google Scholar]
- 20.Cubrilo S, Babic F, Douthwaite S, Maravic Vlahovicek G. 2009. The aminoglycoside resistance methyltransferase Sgm impedes RsmF methylation at an adjacent rRNA nucleotide in the ribosomal A site. RNA 15:1492–1497. doi: 10.1261/rna.1618809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kempenaers M, Roovers M, Oudjama Y, Tkaczuk KL, Bujnicki JM, Droogmans L. 2010. New archaeal methyltransferases forming 1-methyladenosine or 1-methyladenosine and 1-methylguanosine at position 9 of tRNA. Nucleic Acids Res 38:6533–6543. doi: 10.1093/nar/gkq451. [DOI] [PMC free article] [PubMed] [Google Scholar]