Abstract
The cephalosporinase CMY-107, a Tyr199Cys mutant form of CMY-2 encoded by an IncI self-transferable plasmid carried by an Escherichia coli clinical strain, was characterized. The enzyme hydrolyzed oximino-cephalosporins and aztreonam more efficiently than CMY-2 did.
TEXT
Of the five groups of plasmid-borne AmpC β-lactamases (cephalosporinases), the Citrobacter freundii-derived CMY-type enzymes are the most widespread. CMY-2 is considered the progenitor of numerous variants (www.lahey.org/Studies/other.asp#table1) (1–3), most of which efficiently hydrolyze penicillins and older cephalosporins, while their activity against expanded-spectrum cephalosporins and aztreonam is relatively low. There have, however, been variants classified as extended-spectrum AmpCs (ESACs) (4), such as CMY-30, -32, -42, -94, and -95, exhibiting enhanced hydrolysis against oximino-β-lactams mostly because of replacements in conserved Ω-loop residues (5–8). An additional group of naturally occurring CMY-2-derived ESACs, including CMY-33 and CMY-44, are capable of efficiently hydrolyzing cefepime, a property that has been attributed to deletions in the H-10 helix (4, 9, 10).
We describe here CMY-107, a Tyr199Cys mutant form of CMY-2 with extended-spectrum (ES) properties that is produced by an Escherichia coli clinical strain of sequence type 2013 (ST2013), as determined by an established multilocus sequence typing scheme (11).
E. coli EL-495 was isolated in 2012 in an Athens hospital from the urine of an outpatient with vesicoureteral reflux who had been repeatedly treated with cefuroxime. The β-lactam resistance profile of EL-495 indicated cephalosporinase production (Table 1). The strain was susceptible to aminoglycosides, fluoroquinolones, and co-trimoxazole. The resistance pattern of EL-495 was transferred by conjugation to E. coli 1R716 (lac Strr) (Table 1) (12). Whole-cell DNA preparations from EL-495 and a transconjugant clone (Trc-495) were used as templates in PCR assays for various bla genes (13, 14). Sequencing of the PCR products showed that both strains carried a blaCMY-2-like gene (see below) and blaTEM-1. Isoelectric focusing of cell extracts obtained by sonication showed the production of two β-lactamases with isoelectric points of 5.4 (TEM-1 penicillinase) and 9.0 corresponding to the CMY-2-type enzyme. It was therefore concluded that resistance to oximino-β-lactams was due primarily to the latter cephalosporinase.
TABLE 1.
Etest MICs of β-lactams tested against E. coli strains carrying wild-type plasmid pEL495 and CMY-encoding recombinant plasmids
| β-Lactam | MIC (μg/ml) for E. coli strain: |
||||||
|---|---|---|---|---|---|---|---|
| EL-495(pEL495)c | Trc-495(pEL495)c | 1R716 | DH5α(pAC-cmy107)c | DH5α(pAC-cmy2)d | DH5α(pACYC184) | DH5α | |
| Ticarcillin | >256 | >256 | 1.5 | >256 | >256 | 2 | 2 |
| Ticarcillin + CLa | >256 | >256 | 1.5 | >256 | >256 | 2 | 2 |
| Piperacillin | 64 | 48 | 1.5 | >256 | >256 | 2 | 1.5 |
| Piperacillin + TZb | 8 | 8 | 0.5 | 12 | 16 | 1 | 1 |
| Cefoxitin | 128 | 128 | 2 | >256 | >256 | 2 | 2 |
| Cefuroxime | >256 | >256 | 2 | >256 | >256 | 2 | 2 |
| Ceftazidime | 128 | 128 | 0.12 | >256 | 128 | 0.12 | 0.19 |
| Cefotaxime | 32 | 32 | ≤0.06 | 256 | 32 | ≤0.06 | ≤0.06 |
| Aztreonam | 16 | 12 | ≤0.06 | 64 | 24 | ≤0.06 | ≤0.06 |
| Cefepime | 0.5 | 0.5 | ≤0.06 | 1 | 1 | ≤0.06 | ≤0.06 |
| Imipenem | 0.5 | 0.5 | 0.19 | 0.5 | 0.5 | 0.12 | 0.12 |
CL, clavulanic acid (at a fixed concentration of 2 μg/ml).
TZ, tazobactam (at a fixed concentration of 4 μg/ml).
Produces CMY-107.
Produces CMY-2.
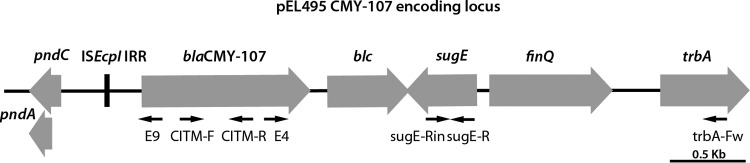
Electrophoresis of plasmid DNA derived from EL-495 and trc-EL-495 by alkaline lysis (15) demonstrated the transfer of a 70-kb plasmid (pEL495) that hybridized with a blaCMY-2 probe (16) and was classified into the IncI family by PCR-based replicon typing (17). The blaCMY locus was characterized by PCR mapping and sequencing of the amplicons, as well as primer walking directly on pEL495 (Fig. 1; see Table S1 in the supplemental material) (14, 18–20). The blaCMY gene differed from blaCMY-2 (GenBank accession no. X91840) by an A-to-G transition at position 656 of the coding sequence, resulting in a Cys-for-Tyr199 substitution in the mature protein. The novel variant was designated CMY-107. blaCMY-107 was part of a 2,590-bp segment similar to those commonly found in plasmid-borne loci that encode CMY-2-like enzymes (Fig. 1). Upstream of blaCMY-107, a 159-bp fragment of ISEcp1 (that included the right inverted repeat, as well as the promoter region provided by ISEcp1) and the pndC gene (belonging to the IncI scaffold) were found. An identical structure has been described in pSTHV23035, a CMY-2-encoding IncIγ plasmid from Salmonella enterica serovar Typhimurium (GenBank accession no. GQ398239) (21). Upstream of sugE, a sequence comprising the open reading frames encoding the FinQ (fertility inhibition system) and TrbA (plasmid transfer function) proteins was also identified, as in pSTHV23035 (Fig. 1).
FIG 1.
Structure of the CMY-107-encoding locus carried by pEL495. The sequencing primers used in this study are indicated by the black arrows. For the sequences of the oligonucleotide used for PCR mapping and sequencing, see Table S1 in the supplemental material. The gene arrangement is identical to that of the respective locus of pSTHV23035.
The fact that the CMY-107-encoding locus of pEL495 was identical to that of pSTHV23035—and rather rare among blaCMY-carrying IncI replicons—indicated a common ancestry. To document this notion, an E. coli K802N transconjugant harboring pSTHV23035 (21) was used to purify the plasmid as it was done for pEL495, and the two molecules were compared by restriction fragment length polymorphism analysis after EcoRI digestion. Their electrophoretic profiles were similar, with the sole difference being an additional band (∼3,000 bp) in the digests of pSTHV23035. Moreover, hybridization with a blaCMY-2-specific probe showed that blaCMY was located in a 5,500-bp fragment in both plasmids (see Fig. S1 in the supplemental material).
Previously described plasmids pBC-cmy2 and pAC-cmy2, derived from pBC-SK(+/−) and pACYC184, respectively (5), were used to construct pBC-cmy107 and pAC-cmy107 by site-directed mutagenesis with the QuikChange mutagenesis kit (Stratagene) and two mutagenic primers, Y199C-F (5′-CAG AAC GAA CAA AAA GAT TGTGCC TGG GGC TAT CGC GAA-3′, where boldface and underlining indicate the altered nucleotide and codon 199 in blaCMY-107, respectively) and Y199C-R (5′-TTC GCG ATA GCC CCA GGC ACA ATC TTT TTG TTC GTT CTG-3′). The recombinant plasmids were used to transform E. coli DH5α. The resulting clones produced CMY-2 and CMY-107 and were used for cephalosporinase purification [pBC-SK(+/−) derivatives] and comparative determination of β-lactam MICs under isogenic conditions (pACYC184 derivatives). CMY-107 and CMY-2 were purified to near homogeneity (>95%) after two ion-exchange chromatography steps with Q- and S-Sepharose columns (22). The activities of the enzymes against various β-lactam substrates were assessed by UV spectrophotometry (23, 24). The hydrolysis constants of penicillin G, cephalothin, and cefoxitin were similar for CMY-107 and CMY-2. CMY-107 was more active against cefuroxime and cefotaxime than CMY-2 was because of its much higher kcat, which was partially compensated for by higher Km values. The turnover rate (kcat) of CMY-107 for ceftazidime was at least 1 order of magnitude higher than that of CMY-2. Also, measurable hydrolysis of aztreonam was observed with CMY-107, whereas CMY-2 was inactive against this compound (Table 2). The rates of cefepime and imipenem hydrolysis by both enzymes were too low to be measured accurately. The differences between the kinetic parameters of CMY-107 and CMY-2 were compatible with the MICs of β-lactams (Table 1), confirming the ES properties of the former cephalosporinase. The inhibition profile of CMY-107 was typical for an AmpC (the Ki values for cloxacillin, aztreonam, RO-48-1220, and tazobactam were 0.8, 7.0, 110, and 820 nM, respectively), without remarkable differences from that of CMY-2.
TABLE 2.
Kinetic parameters of CMY-107 and CMY-2 cephalosporinases on various β-lactam substratesa
| Substrate | CMY-107 (Cys199) |
CMY-2 (Tyr199) |
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM)b | kcat/Km (μM−1 s−1) | kcat (s−1) | Km (μM)b | kcat/Km (μM−1 s−1) | |
| Penicillin G | 20 ± 3 | 0.8 ± 0.1 | 25 | 24 ± 3 | 0.6 ± 0.08 | 40 |
| Cephalothin | 202 ± 14 | 13 ± 3.0 | 16 | 267 ± 10 | 17 ± 1.0 | 16 |
| Cefoxitin | 0.30 ± 0.02 | 0.12 ± 0.02 | 2.5 | 0.35 ± 0.04 | 0.15 ± 0.02 | 2.3 |
| Cefuroxime | 0.56 ± 0.07 | 0.04 ± 0.01 | 14 | 0.014 ± 0.005 | 0.005 ± 0.001 | 2.8 |
| Cefotaxime | 0.80 ± 0.1 | 0.075 ± 0.02 | 10.7 | <0.01 ± 0.002 | 0.005 ± 0.001 | <2 |
| Ceftazidime | 0.14 ± 0.05 | 0.15 ± 0.02 | 0.9 | 0.01 ± 0.002 | 0.02 ± 0.003 | 0.5 |
| Aztreonam | 0.018 | 0.007 | 2.6 | NHc | ||
Values are the means of three independent measurements.
When the Km was <10 μM, the respective values were determined as Kis with cephalothin as the reporter substrate.
NH, no hydrolysis observed after 1 h of incubation of 250 μΜ aztreonam with 0.3 μΜ enzyme.
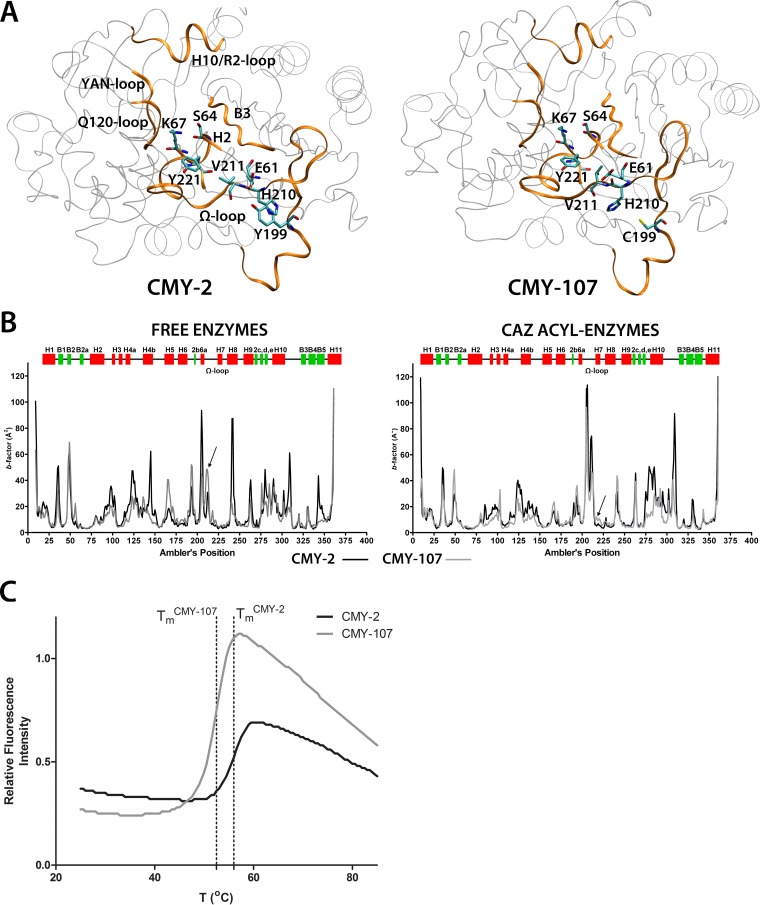
The enzymatic activity of CMY-107 resembles that of the Val211Gly (CMY-30) and Val211Ser (CMY-42) ES variants of CMY-2 (5, 7). Analysis of the crystal structure of CMY-2 (Protein Data Bank entry 1ZC2), as well as molecular dynamics (MD) simulations (25), showed that tyrosine at position 199 in CMY-2 (located at the amino end of the Ω loop) is “bridged” with position 211 as its side chain interacts with (i) His210 through stacking and quadrupole interactions and (ii) the carboxylate of Glu61, which also interacts with the main-chain nitrogen of Val211 (Fig. 2A, left). Replacement of Tyr199 with cysteine in CMY-107 would lead to the abolishment of these interactions, with the side chain of His210 being free to move and, at the same time, Glu61 interacting less frequently with the amide group of Val211 (Fig. 2A, right), as indicated by 5-ns MD simulations of a model of the enzyme performed as described previously (25).
FIG 2.
(A) Three-dimensional structures of CMY-2 (left) and CMY-107 (right) corresponding to the average minimized structures of the two enzymes obtained after 5-ns MD simulations. Structures forming the active site are highlighted (orange ribbons), and the side chains of residues of interest are shown as sticks. Tyr199 in CMY-2 was interacting with His210, located in the middle of the Ω loop. In CMY-107, the side chain of Cys199 could not participate in any interaction with His210, while the side chain of Clu61 interacted less frequently with the amide of Val211 (an interaction that was strong in CMY-2 simulations). (B) Thermal-factor profiles of the two enzymes in their free forms and in ceftazidime (CAZ) acyl enzymes. In CMY-107, the regions around position 211 in free enzymes and Tyr221 in CAZ acyl enzymes (indicated by arrows) were more flexible than in the respective species of CMY-2. (C) Thermal denaturation of CMY-2 and CMY-107 in the presence of SYPRO orange performed in a Bio-Rad MiniOpticon instrument. The ES enzyme was less stable than CMY-2 (melting temperature [Tm] of CMY-2, 56.3 ± 0.3°C; Tm of CMY-107, 52.8 ± 0.3°C [values obtained in three independent experiments]).
Disruption of the His210-Tyr199-Glu61-Val211(-NH) system in CMY-107 by the Tyr199Cys substitution seemed to affect the MD of the enzyme in ways similar to those in which Val211Gly and Val211Ser did (22). The thermal factors of the mutant enzyme's backbone in its free form were significantly elevated in Ω-loop region at positions 210 to 213 compared to those of CMY-2 (Fig. 2B, left). Moreover, in ceftazidime acyl-enzyme species, CMY-107 exhibited increased flexibility in the Tyr221 region (Fig. 2B, right). Interpretation of atomic fluctuations through principal-component analysis revealed some additional traits common to CMY-107 and the other Ω-loop ES CMY-2 variants regarding the main inner protein movements across the MD trajectories. In free CMY-107, the Ω loop exhibited an intense concerted movement that was correlated with the Q120 loop and H10 helix/R2 loop fluctuations (see Fig. S2, left panel, in the supplemental material), while in ceftazidime acyl enzymes, the bound substrate exhibited increased vibrations that were correlated with either the H10/R2 loop (through the dihydrothiazine ring of ceftazidime) or the movements of the Ω loop and especially those of His210, Val211, and Tyr221 (see Fig. S2, right panels).
Molecular simulations of CMY-107 indicated that the enzyme possesses elements that are more flexible than those of CMY-2. Thermal denaturation experiments (22) showed that this flexibility is translated into decreased thermal stability, with CMY-107 exhibiting a melting temperature 3.5°C lower than that of CMY-2 (Fig. 2C). As the same was observed for CMY-30 and CMY-42, there are clear indications of a correlation between reduced thermal stability and increased hydrolysis of oximino-β-lactams by CMY-2-derived ESACs. A plausible hypothesis is that the increased flexibility/“destabilization” of the active site in oximino-β-lactam/acyl-enzyme species of CMY-107 would lead to faster hydrolysis of the ester through more frequent adaptation of conformations resembling a transition state.
The clinical significance of CMY-2-type ESACs is largely unknown. Their frequency may be higher than suggested by their sporadic occurrence. The transcription of blaCMY-2-type genes is controlled by the strong promoter provided by ISEcp1, resulting in increased cephalosporinase production and, consequently, increased resistance to newer β-lactams. This, in turn, could mask the effects of ESAC production. Nevertheless, the occurrence of CMY-107, as well as the CMY-94 (Leu293Ser), CMY-95 (Leu293Ser/Val211Ala), and CMY-33 (Leu293-Ala294 deletion) ESACs, mutant forms of CMY-2 that emerged during ceftazidime, aztreonam, and cefepime chemotherapy (8, 10), suggests that prolonged treatment of patients colonized with CMY-2-expressing members of the family Enterobacteriaceae with oximino-β-lactams may lead to the selection of ES mutants. Spread of the respective genes is also evident, as the Tyr199Cys mutant form of CMY-2 was first identified in a surveillance study in France during 2004 to 2008 (26). The CMY-30 and CMY-42 ESACs have also been isolated more than once (27–30). The E. coli EL495 isolate described here belongs to a rare ST (ST2013) (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli), a single-locus variant of ST73 that, to our knowledge, has not previously been associated with blaCMY-2. Findings presented here underscore the mobility of the blaCMY-2-type genes, as well as the possibility of selection of diverse ESACs under the appropriate pressure. Apart from their potential clinical importance, studies of ESACs provide insights regarding the structure-function relationships of AmpCs.
Nucleotide sequence accession number.
The nucleotide sequences described in this study have been assigned accession no. KR061886 in the GenBank database.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Luisa Peixe for kindly providing E. coli strain K802N carrying the pSTHV23035 plasmid.
This work was supported by the InfeNeuTra project (MIS 450598), GSRT's KRIPIS action, funded by Greece and the European Regional Development Fund of the European Union under the O. P. Competitiveness and Entrepreneurship, NSRF 2007-2013.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01793-15.
REFERENCES
- 1.Philippon A, Arlet G, Jacoby GA. 2002. Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother 46:1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlow M, Hall BG. 2002. Origin and evolution of the AmpC beta-lactamases of Citrobacter freundii. Antimicrob Agents Chemother 46:1190–1198. doi: 10.1128/AAC.46.5.1190-1198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacoby GA. 2009. AmpC β-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Mammeri H. 2007. Extended-spectrum cephalosporinases: structure, detection and epidemiology. Future Microbiol 2:297–307. doi: 10.2217/17460913.2.3.297. [DOI] [PubMed] [Google Scholar]
- 5.Kotsakis SD, Papagiannitsis CC, Tzelepi E, Tzouvelekis LS, Miriagou V. 2009. Extended-spectrum properties of CMY-30, a Val211Gly mutant of CMY-2 cephalosporinase. Antimicrob Agents Chemother 53:3520–3523. doi: 10.1128/AAC.00219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endimiani A, Doi Y, Bethel CR, Taracila M, Adams-Haduch JM, O'Keefe A, Hujer AM, Paterson DL, Skalweit MJ, Page MG, Drawz SM, Bonomo RA. 2010. Enhancing resistance to cephalosporins in class C beta-lactamases: impact of Gly214Glu in CMY-2. Biochemistry 49:1014–1023. doi: 10.1021/bi9015549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hentschke M, Kotsakis SD, Wolters M, Heisig P, Miriagou V, Aepfelbacher M. 2011. CMY-42, a novel plasmid-mediated CMY-2 variant AmpC beta-lactamase. Microb Drug Resist 17:165–169. doi: 10.1089/mdr.2010.0137. [DOI] [PubMed] [Google Scholar]
- 8.Crémet L, Caroff N, Giraudeau C, Reynaud A, Caillon J, Corvec S. 2013. Detection of clonally related Escherichia coli isolates producing different CMY β-lactamases from a cystic fibrosis patient. J Antimicrob Chemother 68:1032–1035. doi: 10.1093/jac/dks520. [DOI] [PubMed] [Google Scholar]
- 9.Doi Y, Paterson DL, Adams-Haduch JM, Sidjabat HE, O'Keefe A, Endimiani A, Bonomo RA. 2009. Reduced susceptibility to cefepime among clinical isolates producing novel variants of CMY-2 β-lactamase. Antimicrob Agents Chemother 53:3159–3161. doi: 10.1128/AAC.00133-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pires J, Taracila M, Bethel CR, Doi Y, Kasraian S, Tinguely R, Sendi P, Bonomo RA, Endimiani A. 21 September 2015. In vivo evolution of CMY-2 to CMY-33 β-lactamase in Escherichia coli ST131: characterization of an acquired extended-spectrum AmpC (ESAC) conferring resistance to cefepime. Antimicrob Agents Chemother doi: 10.1128/AAC.01804-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vatopoulos AC, Philippon A, Tzouvelekis LS, Komninou ZZ, Legakis NJ. 1990. Prevalence of a transferable SHV-5 type β-lactamase in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Greece. J Antimicrob Chemother 26:635–648. doi: 10.1093/jac/26.5.635. [DOI] [PubMed] [Google Scholar]
- 13.Mavroidi A, Miriagou V, Malli E, Stefos A, Dalekos GN, Tzouvelekis LS, Petinaki E. 2012. Emergence of Escherichia coli sequence type 410 (ST410) with KPC-2 β-lactamase. Int J Antimicrob Agents 39:247–250. doi: 10.1016/j.ijantimicag.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Pérez-Pérez FJ, Hanson ND. 2002. Detection of plasmid-mediated ampC β-lactamase genes in clinical isolates by using multiplex PCR. J Antimicrob Chemother 40:2153–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Russel DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 16.Zioga A, Whichard JM, Joyce KJ, Tzelepi E, Tzouvelekis LS, Miriagou V. 2008. Evidence for chromosomal and plasmid location of CMY-2 cephalosporinase gene in Salmonella serotype Typhimurium. J Antimicrob Chemother 61:1389–1390. doi: 10.1093/jac/dkn116. [DOI] [PubMed] [Google Scholar]
- 17.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 18.Kotsakis SD, Tzouvelekis LS, Lebessi E, Doudoulakakis A, Bouli T, Tzelepi E, Miriagou V. 2015. Characterization of a mobilizable IncQ plasmid encoding cephalosporinase CMY-4 in Escherichia coli. Antimicrob Agents Chemother 59:2964–2966. doi: 10.1128/AAC.05017-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Andrea MM, Literacka E, Zioga A, Giani T, Baraniak A, Fiett J, Sadowy E, Tassios PT, Rossolini GM, Gniadkowski M, Miriagou V. 2011. Evolution and spread of a multidrug-resistant Proteus mirabilis clone with chromosomal AmpC-type cephalosporinases in Europe. Antimicrob Agents Chemother 55:2735–2742. doi: 10.1128/AAC.01736-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Fernández A, Chiaretto G, Bertini A, Villa L, Fortini D, Ricci A, Carattoli A. 2008. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother 61:1229–1233. doi: 10.1093/jac/dkn131. [DOI] [PubMed] [Google Scholar]
- 21.Antunes P, Coque TM, Peixe L. 2010. Emergence of an IncIγ plasmid encoding CMY-2 β-lactamase associated with the international ST19 OXA-30-producing β-lactamase Salmonella Typhimurium multidrug-resistant clone. J Antimicrob Chemother 65:2097–2100. doi: 10.1093/jac/dkq293. [DOI] [PubMed] [Google Scholar]
- 22.Kotsakis SD, Caselli E, Tzouvelekis LS, Petinaki E, Prati F, Miriagou V. 2013. Interactions of oximino-substituted boronic acids and beta-lactams with the CMY-2-derived extended-spectrum cephalosporinases CMY-30 and CMY-42. Antimicrob Agents Chemother 57:968–976. doi: 10.1128/AAC.01620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauvois C, Shimizu Ibuka A, Celso A, Alba J, Ishii Y, Frere J-M, Galleni M. 2005. Kinetic properties of four plasmid-mediated AmpC β-lactamases. Antimicrob Agents Chemother 49:4240–4246. doi: 10.1128/AAC.49.10.4240-4246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Power P, Galleni M, Ayala JA, Gutkind G. 2006. Biochemical and molecular characterization of three new variants of AmpC β-lactamases from Morganella morganii. Antimicrob Agents Chemother 50:962–967. doi: 10.1128/AAC.50.3.962-967.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotsakis SD, Tzouvelekis LS, Petinaki E, Tzelepi E, Miriagou V. 2011. Effects of the Val211Gly substitution on molecular dynamics of CMY-2 cephalosporinase: implications on hydrolysis of expanded-spectrum cephalosporins. Proteins 79:3180–3192. doi: 10.1002/prot.23150. [DOI] [PubMed] [Google Scholar]
- 26.Corvec S, Cremet L, Leprince C, Dauvergne S, Reynaud A, Lepelletier D, Caroff N. 2010. Epidemiology of Escherichia coli clinical isolates producing AmpC plasmidic beta-lactamase during a 5-year period in a French teaching hospital. Diagn Microbiol Infect Dis 67:277–281. doi: 10.1016/j.diagmicrobio.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Helmy MM, Wasfi R. 2014. Phenotypic and molecular characterization of plasmid mediated AmpC beta-lactamases among Escherichia coli, Klebsiella spp., and Proteus mirabilis isolated from urinary tract infections in Egyptian hospitals. Biomed Res Int 2014:171548. doi: 10.1155/2014/171548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pannaraj PS, Bard JD, Cerini C, Weissman SJ. 2015. Pediatric carbapenem-resistant Enterobacteriaceae in Los Angeles, California, a high-prevalence region in the United States. Pediatr Infect Dis J 34:11–16. doi: 10.1097/INF.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel L, Schrenzel J, Cherkaoui A, Bernabeu S, Renzi G, Nordmann P. 2011. Molecular analysis of NDM-1-producing enterobacterial isolates from Geneva, Switzerland. J Antimicrob Chemother 66:1730–1733. doi: 10.1093/jac/dkr174. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Lou D, Xu Y, Shang Y, Li D, Huang X, Li Y, Hu L, Wang L, Yu F. 2013. First identification of coexistence of blaNDM-1 and blaCMY-42 among Escherichia coli ST167 clinical isolates. BMC Microbiol 13:282. doi: 10.1186/1471-2180-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.