Abstract
Finafloxacin is a novel fluoroquinolone with improved antimicrobial efficacy, especially in an acidic environment. The efficacy of finafloxacin for the inhibition of Helicobacter pylori infection was compared with the efficacies of levofloxacin and moxifloxacin at neutral and acidic pH. The impacts of gyrA point mutation on the efficacy of those three fluoroquinolones were also investigated. A total of 128 clinical H. pylori strains were utilized. MICs of levofloxacin, moxifloxacin, and finafloxacin were determined at pH 5.0 and pH 7.0 by the agar dilution method. The impact of gyrA point mutations that are responsible for fluoroquinolone resistance was analyzed; the results showed 50 strains with an Asn-87 point mutation, 48 strains with an Asp-91 point mutation, and the remaining 30 strains with no gyrA mutations. The use of finafloxacin led to MIC values at pH 5.0 that were lower than the values seen at pH 7.0 for 112 strains (112/128, 87.5%), and this proportion was higher than that seen with moxifloxacin (21/128, 16.4%, P < 0.001). Finafloxacin also demonstrated a rate of susceptibility (MIC, <1 μg/ml) (37.5%, 48/128) at pH 5.0 that was higher than that seen with moxifloxacin (2.3%, 3/128) (P < 0.001). The trends were similar regardless of which of the Asn-87, Asp-91, and A2143 point mutations were present. In conclusion, the superior antimicrobial efficacy of finafloxacin against H. pylori in an acidic environment suggests the possible use of finafloxacin for treatment of H. pylori infection, as has been proposed by its developer, Merlion Pharma.
INTRODUCTION
Helicobacter pylori infection is a cause of recurrent peptic ulcer disease, chronic gastritis, and gastric malignancies (1). It has been proven that the eradication of H. pylori can prevent peptic ulcer recurrence (2). However, drug instability and insufficient diffusion to gastric mucosa and mucus in that highly acidic environment require the combination of antibiotics with a proton pump inhibitor (PPI) for H. pylori eradication (3). PPI-clarithromycin-containing triple therapy was the first-line eradication treatment until recently (4). However, as the failure rate of the 7-day triple therapy has increased progressively, sequential or concomitant therapy has come to be used (3, 5). Unfortunately, the 7-day triple therapy is still regarded as the standard primary therapy in South Korea because there is no proven alternative regimen that can provide more efficient and safer eradication (6, 7). The unsatisfactory response of alternative eradication regimens was mainly caused by antimicrobial resistance and was particularly due to clarithromycin resistance (5, 8). Therefore, the Maastricht IV consensus has recommended that PPI-clarithromycin-containing triple therapy without prior susceptibility testing should be avoided when the clarithromycin resistance rate is higher than 15% to 20% (2). Moreover, the current high prevalence of metronidazole resistance in South Korea (9) indicates the necessity of reestablishment of a new standard first-line eradication therapy.
Introducing new classes of drugs, such as rifabutin or fluoroquinolone, has been tried. Unfortunately, attempts at therapy using rifabutin have been faced with limitations due to insufficient efficacy and considerable side effects. High bioavailability and good compliance were evidence of the considerable superiority of fluoroquinolone to other classes of antibiotics. However, fluoroquinolone resistance in H. pylori has increased and so far has been an unsolved problem. Fluoroquinolone resistance is primarily due to gyrA N87 or D91 point mutations in the quinolone resistance-determining region (QRDR) (10–12). The N87K mutation in gyrA was the most critical mutation among the H. pylori isolates for which the fluoroquinolone-containing eradication treatment was ineffective (10–12).
Finafloxacin is a novel 8-cyano-fluoroquinolone that demonstrates optimal antimicrobial efficacy even in an acidic environment. Usually, the efficacy of common fluoroquinolones is weakened in an acidic environment, but the efficacy of finafloxacin is not weakened under those conditions (13, 14). The spectrum of in vitro activity and the antimicrobial efficacy of finafloxacin in many other microorganisms were demonstrated previously (13–16). In addition, many studies have shown considerable finafloxacin antimicrobial efficacy under slightly acidic conditions, such as pH 5.0 (14, 16). Those studies demonstrated that the use of finafloxacin is advantageous in treating acidic foci of infection. Since the acidic environment of gastric mucosa and mucus is the greatest limitation (17) for treating H. pylori, the use of acid-stable fluoroquinolone might be a good alternative therapy. However, the in vitro activity of finafloxacin compared to that of other, preexisting fluoroquinolones, especially against clinically isolated H. pylori strains, has not been tested.
From this background, the aim of this study was to evaluate the efficacy of finafloxacin compared with those of levofloxacin and moxifloxacin for the inhibition of clinically isolated H. pylori strains at normal pH and acidic pH. In addition, we evaluated the impact of gyrA point mutations on the activity of these three fluoroquinolones, and the factors associated with the susceptibility to finafloxacin at pH 5.0 were analyzed.
MATERIALS AND METHODS
Study subjects.
Between December 2009 and December 2013, patients with H. pylori infection who had never received eradication therapy for H. pylori infection were consecutively enrolled at the Seoul National University Bundang Hospital located in Gyeonggi province near Seoul, South Korea. The patients' demographic and clinical data, including age, sex, history of previous H. pylori eradication treatment, and endoscopic findings, were gathered. H. pylori infection was defined by a positive rapid urease test result (CLO test; Delta West, Bentley, Australia) during a gastric mucosal biopsy procedure performed on the lesser curvature of the midantrum or midbody and/or by a positive result showing histological evidence of H. pylori with modified Giemsa staining in the gastric mucosal biopsy specimen from the lesser curvature and greater curvature of the midantrum and midbody (18).
The isolates obtained from patients without previous H. pylori eradication treatment were defined as primary strains. The isolates from patients with previous H. pylori eradication treatment were classified as secondary strains (10, 19, 20).
PPI-clarithromycin-containing triple therapy consists of clarithromycin administered at 500 mg twice a day (b.i.d.), amoxicillin at 1,000 mg b.i.d., and esomeprazole at 20 mg b.i.d. Moxifloxacin-containing triple therapy consists of moxifloxacin (Avelox; Bayer HealthCare, AG, Wuppertal, Germany) administered at 400 mg once a day (q.d.), amoxicillin at 1,000 mg b.i.d., and esomeprazole at 20 mg b.i.d. (MEA triple therapy). Quadruple therapy consists of tripotassium dicitrate bismuthate administered at 300 mg four times a day (q.i.d.), metronidazole at 500 mg three times a day (t.i.d.), tetracycline at 500 mg q.i.d., and esomeprazole at 20 mg b.i.d. All subjects provided informed consent, and the study protocol was approved by the Ethics Committee at Seoul National University Bundang Hospital.
Culture and antimicrobial susceptibility test.
For H. pylori isolation, mucosal biopsy specimens obtained from a lesser curvature and a greater curvature in the midantrum and the midbody were cultured and drug MICs were determined (1, 17). Between endoscopies, endoscopes (including biopsy channels and forceps) were cleaned thoroughly with detergent and disinfected for 30 min in an Olympus EW-30 unit (Olympus, Tokyo, Japan). The biopsy specimens from the antrum and the body were cultured separately at 37°C on brain heart infusion (Difco Laboratories, Detroit, MI, USA) plates containing 7% horse blood under microaerobic conditions (5% O2, 10% CO2, 85% N2) for 3 to 5 days. The antrum and body biopsy specimens were evaluated separately, and H. pylori bacteria were identified by Gram staining, by colony morphology, and by oxidase, catalase, and urease reactions.
At least one colony of H. pylori was isolated from the antrum and the body, and the drug MIC for each isolate was determined by the agar dilution method (17). The isolates were then subcultured on Mueller-Hinton agar supplemented with 5% defibrinated sheep blood for 48 h. The bacterial suspension, adjusted to 1 × 107 CFU, was directly inoculated onto an antibiotic-containing agar dilution plate. After 72 h of incubation, MICs of the three fluoroquinolones (levofloxacin, moxifloxacin, and finafloxacin) and clarithromycin were determined. Levofloxacin, moxifloxacin, and clarithromycin were from Sigma Chemical Co. (St. Louis, MO, USA), and finafloxacin was from Merlion Pharmaceuticals (Berlin, Germany). The pH of the Mueller-Hinton broth was adjusted by the addition of hydrochloric acid. The range of antibiotic concentrations used to evaluate the resistance of fluoroquinolones was from ≤0.125 to >8 μg/ml. The resistance breakpoint was ≥1 μg/ml for the fluoroquinolones as defined previously (9). The resistance breakpoint for clarithromycin is >1.0 μg/ml, according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.
Determination of mutations of the 23S rRNA gene and gyrA.
To detect H. pylori 23S rRNA mutations related to clarithromycin resistance, H. pylori genomic DNA was extracted from the isolates (18, 21). Specimens were homogenized in proteinase K solution (20 mmol/liter Tris-HCl [pH 8.0], 10 mmol/liter EDTA, 0.5% sodium dodecyl sulfate, and 10 mg/ml proteinase K) by using a sterile micropestle and incubated for 3 h. DNA was isolated by phenol-chloroform extraction and ethanol precipitation. For PCR amplification, oligonucleotide primer sequences were derived from known sequences, which are as follows: for 23S rRNA, 5′-CGTAACTATAACGGTCCTAAG-3′ and 5′-TTAGCTAACAGAAACATCAAG-3′ (GenBank accession no. U27270) (21); and for gyrA, 5′-TTTAGCTTATTCAATGAGCGT-3′ and 5′-GCAGACGGCTTGGTAGAATA-3′ (GenBank accession no. L29481) (10, 22). The amplification was carried out in a thermal cycler (MJ PTC-0200; Bio-Rad Laboratories, Waltham, MA, USA) (10, 23). PCR amplification consisted of 35 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 57°C, and 1 min of extension at 72°C. The amplification conditions for the 23S rRNA and gyrA genes in the present study were the same, but the sizes of the amplified fragments of the 23S rRNA and gyrA genes were 291 and 426 bp, respectively. The amplified products were then purified, and sequencing was performed on the two strands of the nonrestricted amplicons using an ABI Prism 377XL DNA sequencer (Applied Biosystems, Foster City, CA, USA).
Statistical analysis.
SPSS for Windows (version 17.0; SPSS, Chicago, IL, USA) was used to perform statistical analysis. The chi-square test or Fisher's exact test was used to analyze categorical variables. Continuous variables were analyzed using Student's t test. Univariate and multivariate analyses by binary logistic regression were used to determine independent factors associated with finafloxacin susceptibility at pH 5.0. Multiple factors were included in the logistic regression analysis, including age, gender, clinical disease, previous eradication status, and analysis of A2143 from 23S rRNA or gyrA from genotypes of the H. pylori isolates. Null hypotheses of no difference were rejected in cases in which P values were less than 0.05.
RESULTS
Baseline characteristics of 128 patients according to types of gyrA mutation.
A total of 128 patients with H. pylori infection participated. The baseline demographic and clinical characteristics of the 128 patients analyzed on the basis of the types of gyrA mutation are summarized in Table 1. In short, 128 H. pylori strains isolated from the patients were categorized into three groups, which were composed of a group consisting of 50 strains with an Asn-87 mutation, a group consisting of 48 strains with an Asp-91 mutation, and a group consisting of the remaining 30 strains with no gyrA mutations. There was no statistically significant difference among the groups with regard to age, sex, endoscopic diagnosis, and clarithromycin resistance. However, there were statistically significant differences in terms of the history of previous H. pylori eradication regimen and the proportion of isolates containing 23S rRNA mutations (Table 1).
TABLE 1.
Baseline characteristics of 128 patients based on the types of gyrA mutation
| Parametera | Value(s) |
P | ||
|---|---|---|---|---|
| Asn-87 mutation | Asp-91 mutation | No mutation | ||
| No. of patients | 50 | 48 | 30 | |
| Patient age (mean ± SD) | 57.30 ± 10.85 | 58.75 ± 11.61 | 57.41 ± 10.31 | 0.784 |
| No. of males/no. of females | 17/33 | 24/24 | 18/12 | 0.062 |
| No. of patients with primary strains/no. of patients with secondary strainsb | 25/25 | 37/11 | 28/2 | <0.001 |
| No. of patients with indicated previous treatment regimen in cases of secondary strains | ||||
| PPI-clarithromycin-containing triple treatment | 7 | 2 | 1 | 0.129 |
| MEA triple treatment | 9 | 2 | 1 | 0.028 |
| Quadruple treatment | 9 | 7 | 0.047 | |
| No. of patients with clarithromycin-resistant strains/total no. of patients (%) | 34/50 (68.0) | 28/48 (58.3) | 15/30 (50.0) | 0.272 |
| No. of patients with strains with 23S rRNA mutation (A2142 or A2143)/total no. of patients (%) | 27/50 (54.0) | 24/48 (50.0) | 4/30 (13.3) | 0.001 |
| No. of patients with DU/BGU/gastric cancer or dysplasia/gastritis | 5/2/17/26 | 6/4/22/16 | 2/2/20/6 | 0.079 |
PPI, proton-pump inhibitor; MEA, moxifloxacin plus esomeprazole plus amoxicillin; Quadruple, bismuth plus metronidazole plus tetracycline plus esomeprazole; DU, duodenal ulcer; BGU, benign gastric ulcer. Bold characters indicate statistical significance.
H. pylori isolates obtained from the patients without previous H. pylori eradication treatment were defined as primary strains. If the patients had had previous H. pylori eradication treatment then the isolates were defined as primary strains.
Drug MIC distributions for all H. pylori study strains.
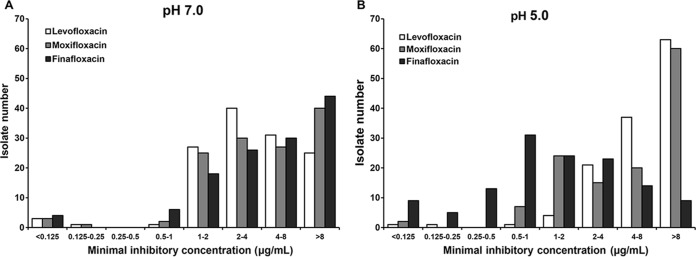
The drug MIC distributions for all 128 H. pylori strains for the three fluoroquinolones (levofloxacin, moxifloxacin, and finafloxacin) are presented in Fig. 1. MIC distributions at pH 7.0 are presented in Fig. 1A; those at pH 5.0 are presented in Fig. 1B. In a pH 7.0 environment, the patterns of MIC distributions among the antibiotics were very similar. In contrast, the MICs of finafloxacin measured at pH 5.0 showed a definite downward shift compared with the MICs measured at pH 7.0 for each isolate. Finafloxacin demonstrated MIC values at pH 5.0 that were lower than those at pH 7.0 for 112 isolates (112/128, 87.5%), and the number of isolates with a finafloxacin MIC reduction was significantly higher than the number with a moxifloxacin MIC reduction (21/128, 16.4%, P < 0.001). At pH 5.0, furthermore, isolates demonstrated a significantly higher rate of susceptibility to finafloxacin (37.5%, 48/128) than to moxifloxacin (2.3%, 3/128, P < 0.001). However, the MICs of levofloxacin and moxifloxacin at pH 5.0 showed a distribution similar to that seen at pH 7.0.
FIG 1.
MIC distribution of fluoroquinolones.
Comparisons of MICs for different pH values, gyrA mutations, and antimicrobial agents.
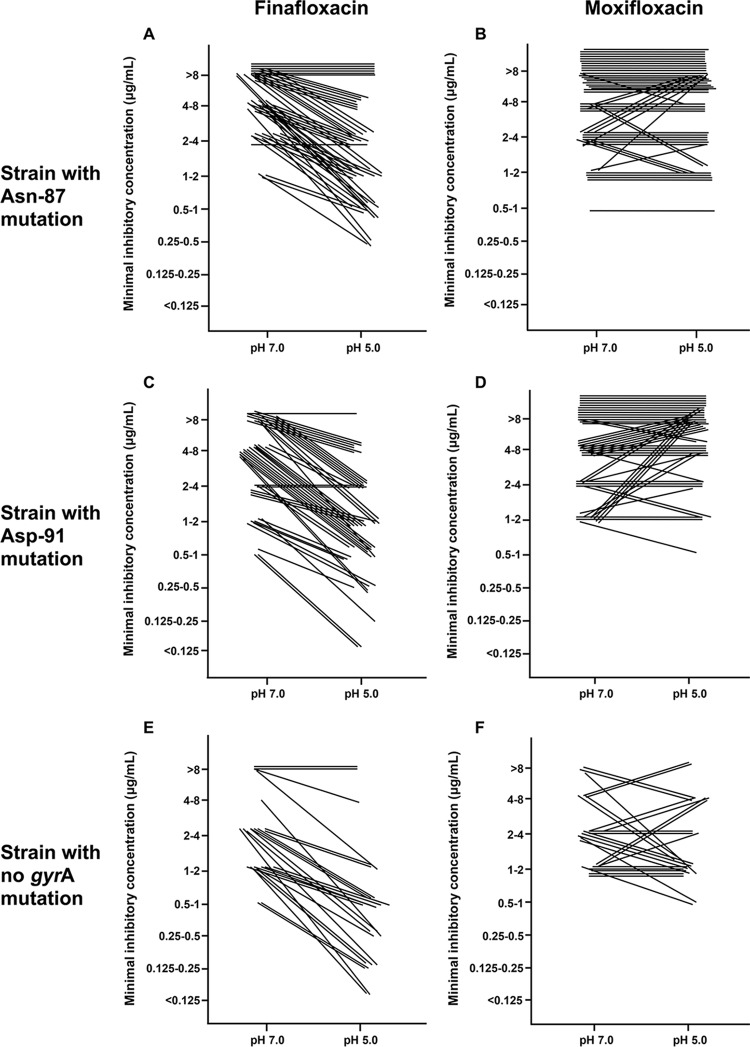
Comparisons between finafloxacin and moxifloxacin MICs for different pH values and gyrA mutations for each isolate are presented in Fig. 2. The results from the strains with gyrA Asn-87 mutations (n = 50) are presented in Fig. 2A and B. As shown in Fig. 2, most of strains showed a greater downward shift of finafloxacin MIC values than of moxifloxacin MIC values for changes of pH from 7.0 to 5.0 (Fig. 2B) (P < 0.001). Similarly, the strains with gyrA Asp-91 mutations (n = 48; Fig. 2C and D) and with no mutation (n = 30; Fig. 2E and F) demonstrated a greater downward shift of finafloxacin MIC values than of moxifloxacin MIC values (P < 0.001 for both comparisons).
FIG 2.
Comparisons among finafloxacin and moxifloxacin MICs regarding different pH and gyrA mutations.
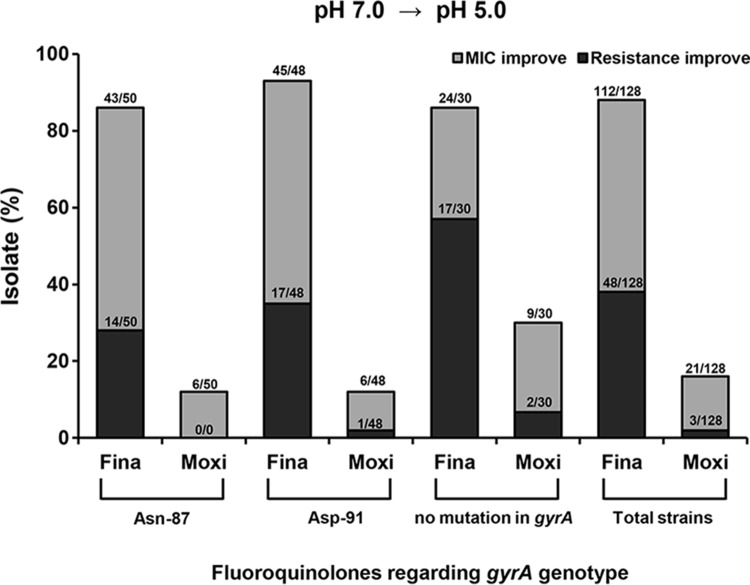
Additionally, the proportion of isolates that demonstrated drug MIC or resistance improvement was calculated based on the pH change from 7.0 to 5.0 (Fig. 3). Improvements of drug MICs and resistance rates were most frequently found in the strains with no gyrA mutations (n = 30) and least frequently in the strains with Asn-87 mutations (n = 50).
FIG 3.
Isolates which undergo changes of fluoroquinolone susceptibility and MICs under conditions of a change from pH 7.0 to pH 5.0. Fina, finafloxacin; Moxi, moxifloxacin.
Finafloxacin susceptibility of primary strains, secondary strains, and strains with A2143 mutations.
A total of 90 strains were classified as primary strains, while an additional 38 strains were secondary strains (Table 2). Among the primary strains, those in the A2143 mutation-positive group (n = 23) demonstrated a higher rate of finafloxacin susceptibility (30.4%, 7/23) than of moxifloxacin susceptibility (0%, 0/23, P = 0.003) at pH 5.0. In the case of the strains in the A2143-negative group (n = 67), the rate of finafloxacin susceptibility (61.2%, 41/67) was higher than the rate of moxifloxacin susceptibility (13.4%, 9/67, P < 0.001). Among the secondary strains (n = 38), moxifloxacin-susceptible strains were not found. However, 25.0% (8/32) of the strains in the A2143-positive group were finafloxacin susceptible and 33.3% (2/6) of the strains in the A2143-negative group were finafloxacin susceptible.
TABLE 2.
Fluoroquinolone susceptibility under pH 5.0 conditions according to primary, secondary, and A2143 mutations
| Strain categorya | A2143 mutation test result | No. (%) of strains with moxifloxacin susceptibility at pH 5.0/total no. of strains | No. (%) of strains with finafloxacin susceptibility at pH 5.0/total no. of strains | P |
|---|---|---|---|---|
| Primary (n = 90) | Positive (n = 23) | 7/23 (30.4) | 0.003 | |
| Negative (n = 67) | 9/67 (13.4) | 41/67 (61.2) | <0.001 | |
| Secondary (n = 38) | Positive (n = 32) | 8/32 (25.0) | 0.002 | |
| Negative (n = 6) | 2/6 (33.3) | 0.003b |
The H. pylori isolates obtained from the patients without previous H. pylori eradication treatment were defined as primary strains. If the patients had had previous H. pylori eradication treatment, then the isolates were defined as secondary strains.
The susceptibility breakpoint for fluoroquinolones by the agar dilution method was <1 μg/ml (Mann-Whitney U test).
Factors associated with finafloxacin susceptibility at pH 5.0.
Results of analysis of predictors associated with finafloxacin susceptibility at pH 5.0 are presented in Table 3. In the univariate analysis, primary strain status, previous quadruple therapy, A2143 mutation, Asn-87 mutation, and absence of a gyrA mutation were factors associated with finafloxacin resistance results with statistical significance. In the multivariate analysis, however, Asn-87 mutation, Asp-91 mutation, and A2143 mutation remained independent factors associated with finafloxacin susceptibility at pH 5.0 (Table 3).
TABLE 3.
Factors associated with finafloxacin susceptibility at pH 5.0
| Parametera | Value(s) for strains with indicated finafloxacin susceptibility at pH 5.0 |
P |
Adjusted OR (95% CI)c | ||
|---|---|---|---|---|---|
| Susceptible (n = 58) | Resistant (n = 70) | Univariate analysis | Multivariate analysis | ||
| Patient age (mean ± SD) | 57.8 ± 12.1 | 57.9 ± 10.0 | 0.951 | ||
| No. of male patients/no. of female patients | 30/28 | 29/41 | 0.287 | ||
| No. of patients with clinical disease | 0.178b | ||||
| BGU | 2 | 5 | |||
| DU | 4 | 5 | |||
| Gastric cancer or dysplasia | 33 | 27 | |||
| Gastritis | 17 | 31 | |||
| No. of patients with primary strains | 48 | 42 | 0.005 | ||
| No. of patients with secondary strains and indicated previous treatment | |||||
| PPI triple | 3 | 7 | 0.510b | ||
| PPI + MEA triple | 3 | 8 | 0.343b | ||
| Quadruple | 3 | 13 | 0.031b | ||
| No. of patients with strains with A2143 mutation | 15 | 40 | 0.001 | 0.028 | 2.532 (1.104–5.809) |
| No. of patients with strains with indicated gyrA mutation | |||||
| Asn-87 | 14 | 36 | 0.002 | 0.001 | 6.901 (2.228–21.378) |
| Asp-91 | 20 | 28 | 0.521 | 0.016 | 3.929 (1.284–12.022) |
| No mutation | 24 | 6 | <0.001 | ||
H. pylori isolates obtained from the patients without previous H. pylori eradication treatment were defined as primary strains. If the patients had had previous H. pylori eradication treatment, then the isolates were defined as secondary strains.
Fisher's exact test.
OD, odds ratio; CI, confidence interval.
DISCUSSION
For the past 2 decades, the first-line regimen established worldwide for the treatment of H. pylori infection has been the 7-day triple therapy that consists of administration of the combination of a proton pump inhibitor, amoxicillin, and clarithromycin (24). However, there has been a progressive decline of the H. pylori eradication rate, and this is related to the increase in clarithromycin resistance (25). Due to this unfavorable outcome, many gastroenterologists have looked for alternative antimicrobial agents, such as fluoroquinolone. Fluoroquinolone-containing eradication regimens have demonstrated favorable results in several countries (26). However, the increase in fluoroquinolone resistance has become a significant limitation for providing effective H. pylori eradication. Newly developed fluoroquinolones, such as gemifloxacin, clinafloxacin, and sitafloxacin, have been studied and demonstrated favorable outcomes for treatment of H. pylori (27). In particular, sitafloxacin has been reported to show outstanding antimicrobial efficacy even for microorganisms with gyrA mutations (28, 29). Unfortunately, sitafloxacin is still not available in most countries, including South Korea.
Meanwhile, considering the instability and insufficient diffusion to the extremely acidic environment of gastric mucosa and mucus, H. pylori eradication therapy requires combinations of potent acid-suppressive drugs (5). Finafloxacin has an advantage over other fluoroquinolones in terms of efficacy, especially for the acidic foci of infection (13, 15), and has shown minimal toxicity issues during preclinical and clinical trials (15, 16). Given that background, the present investigators assumed that finafloxacin might have better activity than conventional fluoroquinolones for treatment of H. pylori infections. This might be the first study to compare the in vitro activity of finafloxacin with that of other fluoroquinolone agents against clinical H. pylori isolates. Also, the present investigators have an additional hypothesis; namely, that finafloxacin could potentially overcome the resistance mechanism represented by a gyrA mutation. Moreover, the applicability of finafloxacin for patients suffering from failure of first- or second-line eradication therapy could be addressed.
Finafloxacin demonstrated activity against H. pylori that was superior to that of the conventional fluoroquinolones with respect to MIC values and resistance rates in the present study. It is a very meaningful discovery because moxifloxacin was previously seen by gastroenterologists as the fluoroquinolone whose results have been the most favorable for H. pylori eradication so far (30). A new antimicrobial agent with better efficacy such as finafloxacin might well be welcomed, because recent studies of H. pylori eradication strategies have presented a dilemma of increased efficacy resulting from adding more drugs to the regimen versus the consequent side effects. Moreover, finafloxacin demonstrated better activity in the secondary strains, which derived from the patients with a history of eradication failure. The results imply that finafloxacin might have better efficacy than other fluoroquinolones, especially among patients who have experienced previous eradication treatment failure.
In addition, the main antibacterial mechanism of fluoroquinolone is the interruption of DNA replication by interfering with DNA gyrase and topoisomerase activities. Finafloxacin is a potent inhibitor of both DNA gyrase and DNA topoisomerase IV. However, H. pylori bacteria do not have the parC or parE genes that encode topoisomerase IV (31). Therefore, mutation of DNA gyrase genes, especially gyrA, is known to play an important role in strain susceptibility. This may be one reason that H. pylori strains with single-point mutations of gyrA appear to be more difficult for finafloxacin to overcome than other microorganisms, such as Escherichia coli or Acinetobacter baumannii, where inhibition of topoisomerase IV still negatively affects the growth of pathogens (13, 16).
Even though finafloxacin demonstrates superior efficacy against some H. pylori isolates, finafloxacin efficacy improvement was limited to the strains with gyrA Asn-87 and Asp-91 mutations. Regarding a previous report on gyrA mutation, about 75% of the fluoroquinolone-resistant H. pylori isolates had either a gyrA Asn-87 mutation or a gyrA Asp-91 mutation (10). Since a number of fluoroquinolone-resistant H. pylori isolates have gyrA Asn-87 and Asp-91 mutations, efforts aimed at overcoming the gyrA mutation by the use of finafloxacin therapy encounter difficulty. From the present results, it is inferred that the expected superior efficacy of finafloxacin-containing treatment compared to treatments containing other fluoroquinolones might be weakened for patients infected with H. pylori strains with a gyrA Asn-87 or Asp-91 mutation. Moreover, the better efficacy of finafloxacin was limited to the strains with A2143 mutations. Although there was no direct association between two different resistance mechanisms, the presence of an A2143 point mutation in the 23S rRNA gene and the presence of a gyrA mutation in the quinolone resistance-determining region with respect to clarithromycin resistance, isolates with an A2143 mutation could have another resistance mechanism, such as a multidrug efflux pump mechanism, that could interfere with fluoroquinolone activity more frequently (32, 33). For this reason, careful patient selection and an efficacy comparison study performed with conventional fluoroquinolones are required for determining proper indications of finafloxacin efficacy.
Even though the superiority of finafloxacin treatment was proven for H. pylori in in vitro infections, it should also be proven by clinical trial for humans. However, pivotal clinical studies of H. pylori eradication with finafloxacin have not yet commenced. Ongoing observation of clinical results will be needed.
In conclusion, in an acidic environment, finafloxacin demonstrates antimicrobial activity against H. pylori that is superior to that seen with other, preexisting fluoroquinolones. Although gyrA mutations might reduce the efficacy of finafloxacin, finafloxacin appears to be better for treating H. pylori than other preexisting fluoroquinolones. Further investigation with clinical trials should be performed, and it is necessary to confirm anti-H. pylori efficacy in patients as well as the appropriate clinical finafloxacin application.
ACKNOWLEDGMENTS
This work was supported by grant no 06-2012-221 from the Seoul National University Bundang Hospital Research fund.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Mentis A, Lehours P, Mégraud F. 2015. Epidemiology and diagnosis of Helicobacter pylori infection. Helicobacter 20(Suppl 1):1–7. doi: 10.1111/hel.12250. [DOI] [PubMed] [Google Scholar]
- 2.Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T, El-Omar EM, Kuipers EJ. 2012. Management of Helicobacter pylori infection—the Maastricht IV/Florence consensus report. Gut 61:646–664. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 3.Vakil N. 2009. H. pylori treatment: new wine in old bottles? Am J Gastroenterol 104:26–30. doi: 10.1038/ajg.2008.91. [DOI] [PubMed] [Google Scholar]
- 4.Kim SG, Jung HK, Lee HL, Jang JY, Lee H, Kim CG, Shin WG, Shin ES, Lee YC; Korean College of Helicobacter and Upper Gastrointestinal Research. 2014. Guidelines for the diagnosis and treatment of Helicobacter pylori infection in Korea, 2013 revised edition. J Gastroenterol Hepatol 29:1371–1386. doi: 10.1111/jgh.12607. [DOI] [PubMed] [Google Scholar]
- 5.Graham DY, Fischbach L. 2010. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut 59:1143–1153. doi: 10.1136/gut.2009.192757. [DOI] [PubMed] [Google Scholar]
- 6.Choi HS, Chun HJ, Park SH, Keum B, Seo YS, Kim YS, Jeen YT, Um SH, Lee HS, Kim CD, Ryu HS. 2012. Comparison of sequential and 7-, 10-, 14-d triple therapy for Helicobacter pylori infection. World J Gastroenterol 18:2377–2382. doi: 10.3748/wjg.v18.i19.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon K, Kim N, Nam RH, Suh JH, Lee S, Kim JM, Lee JY, Kwon YH, Choi YJ, Yoon H, Shin CM, Park YS, Lee DH. 2015. The ultimate eradication rate of H. pylori after 1st, 2nd or 3rd line therapy in Korea. J Gastroenterol Hepatol 30:490–495. doi: 10.1111/jgh.12839. [DOI] [PubMed] [Google Scholar]
- 8.Fischbach L, Evans EL. 2007. Meta-analysis: the effect of antibiotic resistance status on the efficacy of triple and quadruple first-line therapies for Helicobacter pylori. Aliment Pharmacol Ther 26:343–357. doi: 10.1111/j.1365-2036.2007.03386.x. [DOI] [PubMed] [Google Scholar]
- 9.Lee JW, Kim N, Kim JM, Nam RH, Chang H, Kim JY, Shin CM, Park YS, Lee DH, Jung HC. 2013. Prevalence of primary and secondary antimicrobial resistance of Helicobacter pylori in Korea from 2003 through 2012. Helicobacter 18:206–214. doi: 10.1111/hel.12031. [DOI] [PubMed] [Google Scholar]
- 10.Lee JW, Kim N, Nam RH, Park JH, Kim JM, Jung HC, Song IS. 2011. Mutations of Helicobacter pylori associated with fluoroquinolone resistance in Korea. Helicobacter 16:301–310. doi: 10.1111/j.1523-5378.2011.00840.x. [DOI] [PubMed] [Google Scholar]
- 11.Miyachi H, Miki I, Aoyama N, Shirasaka D, Matsumoto Y, Toyoda M, Mitani T, Morita Y, Tamura T, Kinoshita S, Okano Y, Kumagai S, Kasuga M. 2006. Primary levofloxacin resistance and gyrA/B mutations among Helicobacter pylori in Japan. Helicobacter 11:243–249. doi: 10.1111/j.1523-5378.2006.00415.x. [DOI] [PubMed] [Google Scholar]
- 12.Rimbara E, Noguchi N, Kawai T, Sasatsu M. 2012. Fluoroquinolone resistance in Helicobacter pylori: role of mutations at position 87 and 91 of GyrA on the level of resistance and identification of a resistance conferring mutation in GyrB. Helicobacter 17:36–42. doi: 10.1111/j.1523-5378.2011.00912.x. [DOI] [PubMed] [Google Scholar]
- 13.Higgins PG, Stubbings W, Wisplinghoff H, Seifert H. 2010. Activity of the investigational fluoroquinolone finafloxacin against ciprofloxacin-sensitive and -resistant Acinetobacter baumannii isolates. Antimicrob Agents Chemother 54:1613–1615. doi: 10.1128/AAC.01637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Idelevich EA, Kriegeskorte A, Stubbings W, Kahl BC, Peters G, Becker K. 2011. Comparative in vitro activity of finafloxacin against staphylococci displaying normal and small colony variant phenotypes. J Antimicrob Chemother 66:2809–2813. doi: 10.1093/jac/dkr393. [DOI] [PubMed] [Google Scholar]
- 15.Stubbings W, Leow P, Yong GC, Goh F, Korber-Irrgang B, Kresken M, Endermann R, Labischinski H. 2011. In vitro spectrum of activity of finafloxacin, a novel, pH-activated fluoroquinolone, under standard and acidic conditions. Antimicrob Agents Chemother 55:4394–4397. doi: 10.1128/AAC.00833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emrich NC, Heisig A, Stubbings W, Labischinski H, Heisig P. 2010. Antibacterial activity of finafloxacin under different pH conditions against isogenic strains of Escherichia coli expressing combinations of defined mechanisms of fluoroquinolone resistance. J Antimicrob Chemother 65:2530–2533. doi: 10.1093/jac/dkq375. [DOI] [PubMed] [Google Scholar]
- 17.Lee JY, Kim N, Kim MS, Choi YJ, Lee JW, Yoon H, Shin CM, Park YS, Lee DH, Jung HC. 2014. Factors affecting first-line triple therapy of Helicobacter pylori including CYP2C19 genotype and antibiotic resistance. Dig Dis Sci 59:1235–1243. doi: 10.1007/s10620-014-3093-7. [DOI] [PubMed] [Google Scholar]
- 18.Kim SE, Park YS, Kim N, Kim MS, Jo HJ, Shin CM, Lee SH, Hwang JH, Kim JW, Jeong SH, Lee DH, Jung HC. 2013. Effect of Helicobacter pylori eradication on functional dyspepsia. J Neurogastroenterol Motil 19:233–243. doi: 10.5056/jnm.2013.19.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JM, Kim JS, Kim N, Kim SG, Jung HC, Song IS. 2006. Comparison of primary and secondary antimicrobial minimum inhibitory concentrations for Helicobacter pylori isolated from Korean patients. Int J Antimicrob Agents 28:6–13. doi: 10.1016/j.ijantimicag.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 20.Chang WL, Sheu BS, Cheng HC, Yang YJ, Yang HB, Wu JJ. 2009. Resistance to metronidazole, clarithromycin and levofloxacin of Helicobacter pylori before and after clarithromycin-based therapy in Taiwan. J Gastroenterol Hepatol 24:1230–1235. doi: 10.1111/j.1440-1746.2009.05829.x. [DOI] [PubMed] [Google Scholar]
- 21.Hwang TJ, Kim N, Kim HB, Lee BH, Nam RH, Park JH, Lee MK, Park YS, Lee DH, Jung HC, Song IS. 2010. Change in antibiotic resistance of Helicobacter pylori strains and the effect of A2143G point mutation of 23S rRNA on the eradication of H. pylori in a single center of Korea. J Clin Gastroenterol 44:536–543. [DOI] [PubMed] [Google Scholar]
- 22.Gerrits MM, van Vliet AH, Kuipers EJ, Kusters JG. 2006. Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis 6:699–709. doi: 10.1016/S1473-3099(06)70627-2. [DOI] [PubMed] [Google Scholar]
- 23.Tankovic J, Lascols C, Sculo Q, Petit JC, Soussy CJ. 2003. Single and double mutations in gyrA but not in gyrB are associated with low- and high-level fluoroquinolone resistance in Helicobacter pylori. Antimicrob Agents Chemother 47:3942–3944. doi: 10.1128/AAC.47.12.3942-3944.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham DY, Lee YC, Wu MS. 2014. Rational Helicobacter pylori therapy: evidence-based medicine rather than medicine-based evidence. Clin Gastroenterol Hepatol 12:177–186. e173; discussion e112–e173. doi: 10.1016/j.cgh.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JW, Kim N, Kim JM, Nam RH, Kim JY, Lee JY, Lee DH, Jung HC. 2014. A comparison between 15-day sequential, 10-day sequential and proton pump inhibitor-based triple therapy for Helicobacter pylori infection in Korea. Scand J Gastroenterol 49:917–924. doi: 10.3109/00365521.2014.896409. [DOI] [PubMed] [Google Scholar]
- 26.Federico A, Nardone G, Gravina AG, Iovene MR, Miranda A, Compare D, Pilloni PA, Rocco A, Ricciardiello L, Marmo R, Loguercio C, Romano M. 2012. Efficacy of 5-day levofloxacin-containing concomitant therapy in eradication of Helicobacter pylori infection. Gastroenterology 143:55–61. e51; quiz e13–e54. doi: 10.1053/j.gastro.2012.03.043. [DOI] [PubMed] [Google Scholar]
- 27.Chang WL, Kao CY, Wu CT, Huang AH, Wu JJ, Yang HB, Cheng HC, Sheu BS. 2012. Gemifloxacin can partially overcome quinolone resistance of H. pylori with gyrA mutation in Taiwan. Helicobacter 17:210–215. doi: 10.1111/j.1523-5378.2012.00935.x. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki H, Nishizawa T, Muraoka H, Hibi T. 2009. Sitafloxacin and garenoxacin may overcome the antibiotic resistance of Helicobacter pylori with gyrA mutation. Antimicrob Agents Chemother 53:1720–1721. doi: 10.1128/AAC.00049-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furuta T, Sugimoto M, Kodaira C, Nishino M, Yamade M, Uotani T, Sahara S, Ichikawa H, Yamada T, Osawa S, Sugimoto K, Watanabe H, Umemura K. 2014. Sitafloxacin-based third-line rescue regimens for Helicobacter pylori infection in Japan. J Gastroenterol Hepatol 29:487–493. doi: 10.1111/jgh.12442. [DOI] [PubMed] [Google Scholar]
- 30.Yoon H, Kim N, Lee BH, Hwang TJ, Lee DH, Park YS, Nam RH, Jung HC, Song IS. 2009. Moxifloxacin-containing triple therapy as second-line treatment for Helicobacter pylori infection: effect of treatment duration and antibiotic resistance on the eradication rate. Helicobacter 14:77–85. [DOI] [PubMed] [Google Scholar]
- 31.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 32.Hirata K, Suzuki H, Nishizawa T, Tsugawa H, Muraoka H, Saito Y, Matsuzaki J, Hibi T. 2010. Contribution of efflux pumps to clarithromycin resistance in Helicobacter pylori. J Gastroenterol Hepatol 25(Suppl 1):S75–S79. doi: 10.1111/j.1440-1746.2009.06220.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Liu ZQ, Zheng PY, Tang FA, Yang PC. 2010. Influence of efflux pump inhibitors on the multidrug resistance of Helicobacter pylori. World J Gastroenterol 16:1279–1284. doi: 10.3748/wjg.v16.i10.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]