Abstract
We determined the microbicidal activities of antibacterials against nonreplicating Mycobacterium smegmatis grown in a starvation-based Loebel model for persistence. Whereas most drugs lost their activity, fluoroquinolones retained lethal potency. Dose-response characterizations showed a paradoxical more-drug-kills-less Eagle effect. Pretreatment of cultures with chloramphenicol blocked the lethal action of the gyrase inhibitors. These results suggest that fluoroquinolones at low concentrations trigger a protein synthesis-dependent cell death pathway and shut off this suicide pathway at elevated concentrations.
TEXT
The long treatment time required to cure tuberculosis is due to the ability of Mycobacterium tuberculosis to persist in its human host despite extensive chemotherapy. Persister bacilli are thought to be non-drug-susceptible nonreplicating organisms (1, 2). To study persister mycobacteria, a culture model based on nutrient deprivation, known as the Loebel model, was developed (3). When exponentially growing tubercle bacilli are transferred to phosphate-buffered saline (PBS), the bacteria stop replicating but maintain viability for extended periods (4, 5). Drug susceptibility studies revealed that antitubercular drugs show strongly reduced bactericidal activities against these nonreplicating starved bacilli (6–8).
To facilitate the analyses of the molecular mechanisms underlying this nutrient starvation-induced mycobacterial dormancy response, we demonstrated previously that the fast-growing and nonpathogenic relative of the tubercle bacillus Mycobacterium smegmatis mc2155 is also capable of surviving starvation in PBS and thus may be a useful model for the dissection of this process (M.-L. Wu and T. Dick, unpublished data).
Here, we carried out a comparative analysis of the bactericidal activities of antimycobacterials against nongrowing M. smegmatis that were starved for 14 days in PBS versus exponentially growing bacteria to characterize the susceptibility of the nonreplicating form of the organism. To generate starved M. smegmatis culture, the saprophyte was grown in 7H9 liquid medium to an optical density at 600 nm of 0.5, and cells were collected by centrifugation, washed, and resuspended in PBS with 0.025% Tween80 at 107 CFU/ml and incubated in roller bottles at 2 rpm for 14 days. The cultures were then diluted to 106 CFU/ml and treated with 100 μM of various antimycobacterials for 1 day, and the effect on viability was determined via CFU enumeration on 7H10 agar (4). In parallel, exponentially growing cultures in 7H9 medium adjusted to 106 CFU/ml were exposed to the same drugs at the same concentration and for the same time to determine the bactericidal effect on replicating cells. Table 1 shows that most drugs lost their bactericidal activity against nonreplicators, as expected. However, two drug classes, aminoglycosides and fluoroquinolones, retained a significant level of activity against the otherwise nonsusceptible bacilli.
TABLE 1.
Drug susceptibility of 14-day-old starved versus exponentially growing M. smegmatis cultures
| Drug | Fold kill ina: |
|
|---|---|---|
| Growing culture | Nongrowing culture | |
| Isoniazid | 60 | 1 |
| Rifampin | 40 | 1 |
| Ethambutol | 20 | 1 |
| Linezolid | 30 | 1 |
| Tetracycline | 22 | 1 |
| Clarithromycin | 14 | 1 |
| Chloramphenicol | 8 | 1 |
| Erythromycin | 6 | 1 |
| Ciprofloxacinb | >10,000 | 100 |
| Ofloxacin | >10,000 | 30 |
| Moxifloxacinb | >10,000 | 16 |
| Streptomycin | >10,000 | >10,000 |
| Kanamycin | >10,000 | >10,000 |
| Amikacin | >10,000 | >10,000 |
Cultures were exposed to the drugs at 100 μM for 1 day, and survival was determined via CFU enumeration. Fold kill was calculated as the ratio of initial CFU/ml (106)/surviving CFU/ml after 1 day of drug treatment. 1, no kill; >10,000, surviving CFU/ml for the particular drug was below the limit of detection (102 CFU/ml).
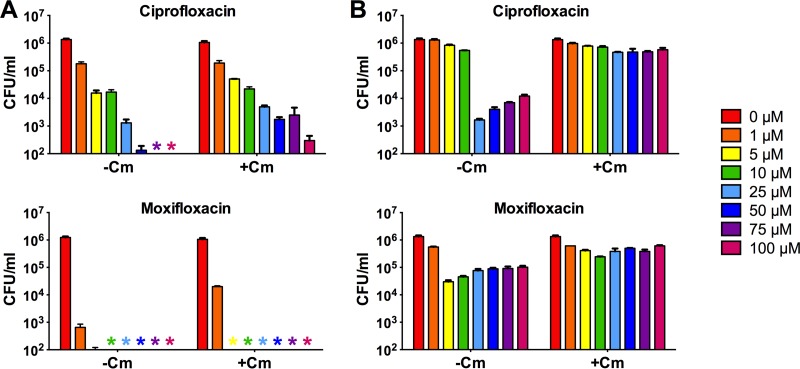
See Fig. 1 for a higher resolution of lethal activities of ciprofloxacin and moxifloxacin.
To characterize the bactericidal activity of these two drug classes against nongrowing bacteria, comparative dose-response experiments were carried out in which exponentially growing and 14-day-old starved cultures were exposed to 1 to 100 μM streptomycin and ciprofloxacin, respectively.
Streptomycin (MIC = 0.6 μM) displayed monophasic kill curves; i.e., higher concentrations of the aminoglycoside drug killed more bacilli in exponentially growing and nonreplicating starved cultures. Exposure to 5 μM streptomycin resulted in a 3-log CFU reduction under both culture conditions. Other translation inhibitors, including linezolid, tetracycline, clarithromycin, and chloramphenicol, did not show this behavior (Table 1), indicating that this aminoglycoside effect might be due to their property of inducing mistranslation or might be protein synthesis independent.
We then characterized the microbicidal activities of ciprofloxacin. Figure 1A (−Cm) shows the concentration-kill curve of the drug (MIC = 0.6 μM) for growing M. smegmatis. Exposure to 5 μM fluoroquinolone resulted in a 2-log CFU reduction compared to the initial inoculum. Higher concentrations of ciprofloxacin (50 to 100 μM) killed the growing culture down to the limit of detection, i.e., >4 logs.
FIG 1.
Concentration-kill curves of ciprofloxacin and moxifloxacin for exponentially growing (A) and nutrient-starved (B) 14-day-old nongrowing M. smegmatis cultures. −Cm, without chloramphenicol pretreatment, cultures were exposed to the fluoroquinolones at 1 to 100 μM for 1 day, and survival was determined via CFU enumeration; +Cm, cultures were pretreated with a sublethal (growth-inhibitory) concentration of chloramphenicol (45 μM) for 1 h to halt growth and then exposed to the fluoroquinolones. Experiments were performed three times independently in triplicate, and representative results are shown as means and standard deviations. Colored asterisks on the x axis indicate that the CFU concentration for the particular drug concentration was below the limit of detection (102 CFU/ml).
Surprisingly, starved nonreplicating mycobacterial cultures showed a triphasic concentration-kill curve (Fig. 1B, −Cm). Ciprofloxacin concentrations of up to 10 μM showed only a mild effect on M. smegmatis viability, while the same concentrations caused a loss of viability of 2 logs in growing cultures. At 25 μM, a 3-log kill was observed. A further increase in ciprofloxacin concentration to 50 μM resulted in less kill. Exposure to 100 μM ciprofloxacin further increased survival by 1 log relative to the 25 μM exposure experiment; i.e., more drug killed less. This paradoxical concentration-kill effect was not observed for exponentially growing cultures (Fig. 1A, −Cm).
Previously, Malik and colleagues (9, 10) studied the effect of fluoroquinolones on nonreplicating mycobacteria employing the protein synthesis inhibitor chloramphenicol at static concentrations to halt growth. To determine whether the paradoxical kill effect of ciprofloxacin observed for starvation-induced nonreplicating M. smegmatis can also be observed for drug-induced nonreplicating bacilli, we exposed exponentially growing cultures to a growth-inhibitory (nonmicrobicidal) concentration of 45 μM chloramphenicol for 1 h to halt growth (10) and then treated the nongrowing culture with 1 to 100 μM ciprofloxacin. Figure 1A (+Cm) shows a monophasic kill curve of the fluoroquinolone for the growth-arrested cultures: higher concentrations of ciprofloxacin killed more; i.e., the paradoxical kill effect observed for starvation-induced nonreplicating bacilli was not seen against chloramphenicol-pretreated log-phase cultures. Consistent with data reported by Malik and colleagues (9, 10), inhibition of protein synthesis had a strong attenuating effect on the lethal activity of ciprofloxacin; i.e., 5 μM of fluoroquinolone, the concentration that killed 2 logs of growing culture, reduced viability of the chloramphenicol-halted culture by merely 1 log (Fig. 1A, +Cm).
To determine whether inhibition of protein synthesis also affects the ciprofloxacin-induced killing of starvation-induced nongrowing culture, we pretreated 14-day-old starved cultures with 45 μM chloramphenicol (nonmicrobicidal) for 1 h and then added 1 to 100 μM ciprofloxacin. Figure 1B (+Cm) shows that inhibition of protein synthesis almost completely abolished the lethal activity of ciprofloxacin against starved cultures, suggesting that the 3-log kill observed with 25 μM ciprofloxacin in starved (chloramphenicol-free) cultures required protein synthesis.
Malik et al. (10) reported intriguing differences between the cell death mechanism of ciprofloxacin (a C-8-H fluoroquinolone) and moxifloxacin (a C-8-methoxy fluoroquinolone). Whereas inhibition of protein synthesis of growing M. smegmatis cultures via pretreatment with chloramphenicol had a strong attenuating effect on the microbicidal activity of ciprofloxacin, the lethal effect of moxifloxacin was almost unaffected by the translation inhibitor. Figure 1A (−Cm/+Cm) shows that this behavior was reproduced under our culture assay conditions. Pretreatment of growing cultures with chloramphenicol had only a minor attenuating effect on the lethal activity of moxifloxacin (MIC = 0.1 μM), thus supporting previous reports that moxifloxacin, in contrast to ciprofloxacin (and gatifloxacin with M. tuberculosis [9]), kills mostly via a protein synthesis-independent pathway (10).
To determine whether the triphasic concentration-kill curve observed for ciprofloxacin in starvation-induced cultures can also be observed for moxifloxacin, we treated starved cultures with the C-8-methoxy fluoroquinolone and measured CFU. Figure 1B (−Cm) shows that moxifloxacin treatment also generated a triphasic kill curve. However, the concentration-kill curve was somewhat compressed along the y axis (CFU/ml) compared to the curve generated by ciprofloxacin. Although the optimum microbicidal concentration for moxifloxacin was lower than that for ciprofloxacin (5 μM versus 25 μM), the drug killed fewer bacilli at this concentration (1 log versus 3 logs). Figure 1B (+Cm) shows that pretreatment of nutrient-starved cultures with chloramphenicol reduced the lethal effect of moxifloxacin. However, the impact of protein synthesis inhibition on the lethal activity of moxifloxacin appeared to be less pronounced than that of ciprofloxacin.
Taken together, our results suggest that the motto “the more, the better” does not apply to fluoroquinolones against starvation-induced nonreplicating M. smegmatis; i.e., increasing the concentration of fluoroquinolones beyond an optimum bactericidal concentration did not increase the kill but actually increased survival. This phenomenon appears to be drug class specific, as it was not observed for aminoglycosides.
The paradoxical less-kills-more bactericidal phenomenon is termed the Eagle effect, according to Harry Eagle, who first described this phenomenon in 1948 (11). Eagle found that penicillin showed an optimum bactericidal concentration beyond which the rate of bacterial death was reduced against many strains of streptococci and staphylococci (11, 12). The observation Eagle made was later expanded to various Gram-negative bacteria, such as Haemophilus influenzae and Proteus species (13). Furthermore, drugs other than β-lactams, including colistin, exhibit this paradoxical effect (14). Interestingly, fluoroquinolones were shown to have an Eagle effect against staphylococci and Escherichia coli (15–18).
It is important to note that Drlica et al. (19) and Dong et al. (20) observed a fluoroquinolone-induced Eagle effect in growing cultures of Mycobacterium bovis BCG. We did not observe this effect in our fluoroquinolone treatments of growing M. smegmatis. Furthermore, it is interesting to note that Malik et al. (17) observed an Eagle effect of quinolones for chloramphenicol-treated E. coli cultures. We did not observe this effect in chloramphenicol-halted M. smegmatis cultures (Fig. 1A, +Cm). The reason for these discrepancies is not clear; differences in incubation time may be involved, since they are known to affect the mechanism of quinolone-mediated killing (21). The fact that we did not see an Eagle effect in growing and chloramphenicol-halted M. smegmatis cultures does not necessarily mean that this phenomenon does not occur under these culture conditions. The extensive killing we observed may have obscured such an effect.
What is known about the cell death mechanisms triggered by the fluoroquinolones in bacteria? The quinolones trap type II topoisomerases on DNA as a complex in which DNA is broken but constrained by protein. A consequence of drug-enzyme-DNA complex formation is a reversible inhibition of DNA replication and growth arrest, not death. Cell death arises from poorly understood subsequent protein synthesis-dependent and -independent events in which bacterial chromosomes are fragmented and toxic reactive oxygen species may be generated (22–29).
The observed killing of starved nonreplicating M. smegmatis reported here confirms previous observations that fluoroquinolones can cause cell death without concurrent DNA replication (23, 25). This appears to be true across bacterial species. Zhao et al. (30) used a temperature-sensitive dnaB mutant of E. coli to show that stopping replication had little effect on the lethal activity of quinolones.
How the fluoroquinolones precisely kill starved nonreplicating mycobacteria and how they block this apparently protein synthesis-dependent cell death pathway at elevated concentrations are under investigation. It appears that induction of efflux pumps as an explanation for the drug-induced nonsusceptibility can be excluded, as the pump inhibitors reserpine and verapamil (31, 32) did not eliminate the observed Eagle effect (data not shown). In contrast to most bacteria, which possess two type II topoisomerases (topoisomerase IV and DNA gyrase, both targets for fluoroquinolones), mycobacteria possess only DNA gyrase (33), which should simplify the molecular dissection of the fluoroquinolone-induced kill-and-rescue phenomenon.
ACKNOWLEDGMENTS
This work was supported by the Singapore Ministry of Health's National Medical Research Council under its TCR flagship grant NMRC/TCR/011-NUHS/2014 to T.D. and is part of the Singapore Programme of Research Investigating New Approaches to Treatment of Tuberculosis (SPRINT-TB; www.sprinttb.org) led by Nick Paton. M.-L.W. received a research scholarship from the Yong Loo Lin School of Medicine.
We thank Jansy Sarathy, Public Health Research Institute, Newark, NJ, for discussion. We also thank the anonymous reviewer for his very insightful and useful comments.
M.-L.W. and T.D. conceived the project, M.-L.W. and J.T. carried out the experiments, and M.-L.W. and T.D. analyzed the data and wrote the manuscript.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Barry CE III, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. 2009. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol 7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lenaerts A, Barry CE III, Dartois V. 2015. Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol Rev 264:288–307. doi: 10.1111/imr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loebel RO, Shorr E, Richardson HB. 1933. The influence of foodstuffs upon the respiratory metabolism and growth of human tubercle bacilli. J Bacteriol 26:139–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gengenbacher M, Rao SP, Pethe K, Dick T. 2010. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology 156:81–87. doi: 10.1099/mic.0.033084-0. [DOI] [PubMed] [Google Scholar]
- 5.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 6.Xie Z, Siddiqi N, Rubin EJ. 2005. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 49:4778–4780. doi: 10.1128/AAC.49.11.4778-4780.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarathy J, Dartois V, Dick T, Gengenbacher M. 2013. Reduced drug uptake in phenotypically resistant nutrient-starved nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:1648–1653. doi: 10.1128/AAC.02202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarathy JP, Lee E, Dartois V. 2013. Polyamines inhibit porin-mediated fluoroquinolone uptake in mycobacteria. PLoS One 8:e65806. doi: 10.1371/journal.pone.0065806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malik M, Drlica K. 2006. Moxifloxacin lethality against Mycobacterium tuberculosis in the presence and absence of chloramphenicol. Antimicrob Agents Chemother 50:2842–2844. doi: 10.1128/AAC.00250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik M, Lu T, Zhao X, Singh A, Hattan CM, Domagala J, Kerns R, Drlica K. 2005. Lethality of quinolones against Mycobacterium smegmatis in the presence or absence of chloramphenicol. Antimicrob Agents Chemother 49:2008–2014. doi: 10.1128/AAC.49.5.2008-2014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eagle H, Musselman AD. 1948. The rate of bactericidal action of penicillin in vitro as a function of its concentration, and its paradoxically reduced activity at high concentrations against certain organisms. J Exp Med 88:99–131. doi: 10.1084/jem.88.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eagle H. 1951. Further observations on the zone phenomenon in the bactericidal action of penicillin. J Bacteriol 62:663–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yourassowsky E, Vander Linden MP, Lismont MJ, Schoutens E. 1978. Qualitative study of paradoxical zone phenomenon of penicillins against 17 bacterial species of clinical importance. Chemotherapy 24:92–96. doi: 10.1159/000237766. [DOI] [PubMed] [Google Scholar]
- 14.Annear DI. 1970. An optimal zone of colistin activity with Serratia marcescens. Med J Aust 2:225–227. [DOI] [PubMed] [Google Scholar]
- 15.Lewin CS, Smith JT. 1989. The bactericidal activity of ofloxacin against staphylococci. J Chemother 1:151–153. [PubMed] [Google Scholar]
- 16.Lewin CS, Morrissey I, Smith JT. 1991. The mode of action of quinolones: the paradox in activity of low and high concentrations and activity in the anaerobic environment. Eur J Clin Microbiol Infect Dis 10:240–248. doi: 10.1007/BF01966996. [DOI] [PubMed] [Google Scholar]
- 17.Malik M, Capecci J, Drlica K. 2009. Lon protease is essential for paradoxical survival of Escherichia coli exposed to high concentrations of quinolone. Antimicrob Agents Chemother 53:3103–3105. doi: 10.1128/AAC.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crumplin GC, Smith JT. 1975. Nalidixic acid: an antibacterial paradox. Antimicrob Agents Chemother 8:251–261. doi: 10.1128/AAC.8.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drlica K, Xu C, Wang JY, Burger RM, Malik M. 1996. Fluoroquinolone action in mycobacteria: similarity with effects in Escherichia coli and detection by cell lysate viscosity. Antimicrob Agents Chemother 40:1594–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong Y, Xu C, Zhao X, Domagala J, Drlica K. 1998. Fluoroquinolone action against mycobacteria: effects of C-8 substituents on growth, survival, and resistance. Antimicrob Agents Chemother 42:2978–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Liu X, Qu Y, Wang X, Li L, Zhao X. 2012. Inhibitors of reactive oxygen species accumulation delay and/or reduce the lethality of several antistaphylococcal agents. Antimicrob Agents Chemother 56:6048–6050. doi: 10.1128/AAC.00754-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drlica K, Malik M, Kerns RJ, Zhao X. 2008. Quinolone-mediated bacterial death. Antimicrob Agents Chemother 52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malik M, Zhao X, Drlica K. 2006. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol Microbiol 61:810–825. doi: 10.1111/j.1365-2958.2006.05275.x. [DOI] [PubMed] [Google Scholar]
- 24.Long Q, Du Q, Fu T, Drlica K, Zhao X, Xie J. 2015. Involvement of Holliday junction resolvase in fluoroquinolone-mediated killing of Mycobacterium smegmatis. Antimicrob Agents Chemother 59:1782–1785. doi: 10.1128/AAC.04434-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao X, Hong Y, Drlica K. 2015. Moving forward with reactive oxygen species involvement in antimicrobial lethality. J Antimicrob Chemother 70:639–642. doi: 10.1093/jac/dku463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Zhao X, Malik M, Drlica K. 2010. Contribution of reactive oxygen species to pathways of quinolone-mediated bacterial cell death. J Antimicrob Chemother 65:520–524. doi: 10.1093/jac/dkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drlica K, Hiasa H, Kerns R, Malik M, Mustaev A, Zhao X. 2009. Quinolones: action and resistance updated. Curr Top Med Chem 9:981–998. doi: 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dwyer DJ, Collins JJ, Walker GC. 2015. Unraveling the physiological complexities of antibiotic lethality. Annu Rev Pharmacol Toxicol 55:313–332. doi: 10.1146/annurev-pharmtox-010814-124712. [DOI] [PubMed] [Google Scholar]
- 30.Zhao X, Malik M, Chan N, Drlica-Wagner A, Wang JY, Li X, Drlica K. 2006. Lethal action of quinolones against a temperature-sensitive dnaB replication mutant of Escherichia coli. Antimicrob Agents Chemother 50:362–364. doi: 10.1128/AAC.50.1.362-364.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasca MR, Guglierame P, Arcesi F, Bellinzoni M, De Rossi E, Riccardi G. 2004. Rv2686c-Rv2687c-Rv2688c, an ABC fluoroquinolone efflux pump in Mycobacterium tuberculosis. Antimicrob Agents Chemother 48:3175–3178. doi: 10.1128/AAC.48.8.3175-3178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasca MR, Guglierame P, De Rossi E, Zara F, Riccardi G. 2005. mmpL7 gene of Mycobacterium tuberculosis is responsible for isoniazid efflux in Mycobacterium smegmatis. Antimicrob Agents Chemother 49:4775–4777. doi: 10.1128/AAC.49.11.4775-4777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouige A, Darmon A, Piton J, Roue M, Petrella S, Capton E, Forterre P, Aubry A, Mayer C. 2013. Mycobacterium tuberculosis DNA gyrase possesses two functional GyrA-boxes. Biochem J 455:285–294. doi: 10.1042/BJ20130430. [DOI] [PubMed] [Google Scholar]